Abstract
Background
Thermophilic filamentous fungus Myceliophthora thermophila has great capacity for biomass degradation and is an attractive system for direct production of enzymes and chemicals from plant biomass. Its industrial importance inspired us to develop genome editing tools to speed up the genetic engineering of this fungus. First-generation CRISPR–Cas9 technology was developed in 2017 and, since then, some progress has been made in thermophilic fungi genetic engineering, but a number of limitations remain. They include the need for complex independent expression cassettes for targeting multiplex genomic loci and the limited number of available selectable marker genes.
Results
In this study, we developed an Acidaminococcus sp. Cas12a-based CRISPR system for efficient multiplex genome editing, using a single-array approach in M. thermophila. These CRISPR–Cas12a cassettes worked well for simultaneous multiple gene deletions/insertions. We also developed a new simple approach for marker recycling that relied on the novel cleavage activity of the CRISPR–Cas12a system to make DNA breaks in selected markers. We demonstrated its performance by targeting nine genes involved in the cellulase production pathway in M. thermophila via three transformation rounds, using two selectable markers neo and bar. We obtained the nonuple mutant M9 in which protein productivity and lignocellulase activity were 9.0- and 18.5-fold higher than in the wild type. We conducted a parallel investigation using our transient CRISPR–Cas9 system and found the two technologies were complementary. Together we called them CRISPR–Cas-assisted marker recycling technology (Camr technology).
Conclusions
Our study described new approaches (Camr technology) that allow easy and efficient marker recycling and iterative stacking of traits in the same thermophilic fungus strain either, using the newly established CRISPR–Cas12a system or the established CRISPR–Cas9 system. This Camr technology will be a versatile and efficient tool for engineering, theoretically, an unlimited number of genes in fungi. We expect this advance to accelerate biotechnology-oriented engineering processes in fungi.
Keywords: CRISPR–Cas12a, CRISPR–Cas9, Genome editing, Myceliophthora thermophila, Marker recycling, Cellulase
Background
Filamentous fungi are important decomposers that contribute plant biomass to the biological carbon cycle [1, 2]. The natural ability of these microorganisms to secrete enzymes, organic acids, and secondary metabolites has been harnessed for high-level protein production in biotechnology, food, textile, and pharmaceutical industries [3–5]. Genetic engineering can be a powerful approach for filamentous fungi not only to gain deep elucidation of gene function, but to also improve production levels and minimize unwanted by-product formation [6, 7]. However, the typical efficiency of homologous integration is very low using classical genetic approaches. CRISPR–Cas systems have recently enabled a wide range of applications for genome editing in many organisms [8–12]. Remarkably, and in just the past few years, the CRISPR–Cas9 system has emerged as a significantly efficient strategy to solve the problem of low gene editing frequency in filamentous fungi [13–24]. Most CRISPR–Cas9 systems use plasmids or autonomous replicating vectors to introduce Cas9 and single-guide RNA (sgRNA) into fungal genomes [13–23], although some researchers have successfully used purified RNA/Cas9 protein complexes [24, 25]. The Cas9–sgRNA complex binds to the corresponding target site of the protospacer in a genome and specifically induces double-strand breaks. These breaks can be used as a basis for site-specific mutagenesis mediated by non-homologous end-joining or for the introduction of precise mutation or integration via homology-directed repair.
The other restrictive factor for genetic and metabolic engineering in filamentous fungi arises from the limited number of dominant selectable markers, including the low number of antibiotic resistance and auxotrophic genes. Therefore, it is necessary to identify markers for recycling systems for the introduction of multiple expression constructs or sequential gene deletions. Marker recycling systems have been developed in some filamentous fungi by excision of the marker pyrG/amdS by counter selection with 5-fluoroorotic acid/5-fluoroacetamide [26–28] or site-specific recombination systems, such as Cre-loxP [29–33] and FLP/FRT [34, 35], in which the expression of Cre recombinase or flippase (FLP) eliminated the marker cassette flanked by loxP or FRT sites. The counter-selection method requires auxotrophic strains that can be time-consuming and laborious to build, and therefore the Cre-loxP recombination system has been more widely adapted for marker rescue in fungi, such as Aspergillus nidulans, Neurospora crassa, Neotyphodium sp., Aspergillus oryzae, Cryphonectria parasitica, Metarhizium robertsii, Penicillium oxalicum, and Fusarium graminearum [29–33]. Recently, Katayama et al. [36] established an efficient multiple genetic engineering technique in A. oryzae that was based on the CRISPR–Cas9 system and recycling of an AMA1-based plasmid harboring the drug-resistance marker ptrA, allowing for repeatable genetic manipulation. More recently, Leynaud-Kieffer et al. [37] reported a simple Cas9-based gene targeting method that provided selectable, iterative, and marker-free generation of genomic editing using the auxotrophic marker pyrG.
We previously developed a CRISPR–Cas9 system that efficiently edited the thermophilic filamentous fungi Myceliophthora thermophila (Synonym: Thermothelomyces thermophilus) and Myceliophthora heterothallica, enabling simultaneous multigene disruptions of up to four loci using selectable markers bar and neo [20]. M. thermophila is a potential reservoir of novel industrial thermostable enzymes and has the exceptional potential to produce proteins, chemicals, and biofuels directly from renewable biomass [38–42]. Multiple mutants of M. thermophila with double-, triple- and quadruple-deletions and significantly increased cellulase production were generated using this CRISPR–Cas9 genome-engineering tool. However, stacking additional traits to improve lignocellulase production in such mutants was limited, because only one selectable marker (hph, conferring hygromycin resistance) remained after the markers bar and neo were used to generate the primary multiplex-locus deletions. Further, using hph might produce a dead-end strain in which no further selective markers are available for additional genetic manipulation. This limitation was ameliorated in this study by implementing an approach that allowed for marker recycling.
Recently, Cas12a (also known as Cpf1), a member of the class 2 type V-A CRISPR system family, uses a single RuvC catalytic domain for guide RNA double-stranded DNA cleavage and has been harnessed for genome editing [43–45]. Distinct from Cas9, Cas12a possesses several unique and attractive features [9–11] and provides for substantial expansion of the genomic editing toolbox in some eukaryotic organisms [43–51] and bacteria [52–55]. For instance, Cas12a is a single-RNA-guided nuclease that does not need a trans-activating CRISPR RNA (crRNA). Cas12a enzymes mature the CRISPR–RNA array itself without additional RNase [43], recognize a T-rich protospacer adjacent motif (PAM) [44], and generate staggered ends in its PAM-distal target site [44]. Compared to Cas9, one major advantage of Cas12a is its ability to encode two or more crRNAs in a multiplex single transcript by using customized CRISPR arrays [45]. These features generated interest in Cas12a as an expanded minimalistic CRISPR–Cas system for convenient multiplex genes editing and regulation by incorporating multiple crRNAs insulated by short direct repeats [45, 50, 51, 54, 55]. Among the Cas12a (Cpf1) orthologs, three (Francisella novicida U112 FnCpf1, Acidaminococcus sp. BV3L6 AsCpf1, and Lachnospiraceae bacterium ND2006 LbCpf1) have been studied the most both in vivo [45–55] and in vitro [43, 44, 56]. The crRNA used with Cas12a orthologs typically composed of a 23–25 nt guide sequence and a 19 nt direct repeat [43–50]. Thus far, the CRISPR–Cas12a system has been developed only in A. nidulans and A. niger [57], and T. thermophilus [58] as far as we know in filamentous fungi. To test whether the Cas12a effector can be used as an attractive alternative genome editing tool in thermophilic filamentous fungi, in this study, we firstly developed a new efficient CRISPR–Cas12a (AsCpf1) system in M. thermophila. Secondly, we established a marker recycling approach based on transient introduction of the new CRISPR–Cas12a system or our previous CRISPR–Cas9 system in M. thermophila (Fig. 1). Thirdly, we generated 11 gene modifications in the wild-type strain through three rounds of manipulations as proof-of-concept, which resulted in hyper-cellulase production. We showed that the CRISPR–Cas (Cas12a or Cas9) system is a versatile technology that can rapidly and conveniently generate multi-trait strains, in which iterative stacking of industrially relevant traits is not limited by the selectable marker availability.
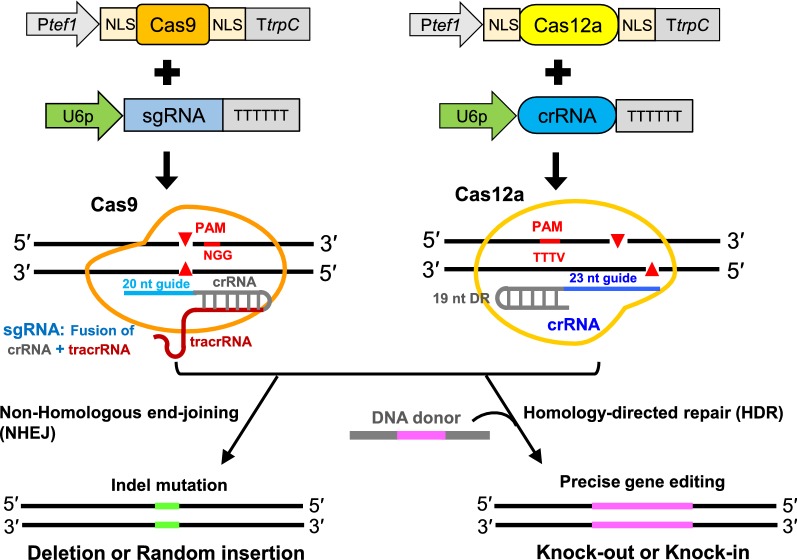
Fig. 1.
Schematic overview of CRISPR–Cas12a- and CRISPR–Cas9-mediated genome editing in the thermophilic fungus M. thermophila. To edit the M. thermophila genome, either the Cas9 and sgRNA-expressing cassettes or Cas12a and crRNA expression cassettes were introduced into the recipient protoplasts. For the CRISPR–Cas9 system, the single-guide RNA (sgRNA) contains crRNA and tracrRNA. As compared to Cas9, Cas12a is a single-RNA-guided nuclease that does not require a tracrRNA and the crRNA consists of 19-nt direct repeat and 23-nt guide. Cas12a has a T-rich PAM sequence located at the 5′ end of the protospacer and cleaves DNA distal from the PAM and generates staggered ends. Once Cas9 or Cas12a nuclease generates the sequence-specific double-stranded DNA break (DSB), non-homologous end-joining (NHEJ) or homology-directed repair (HDR) processes mediate DNA modification at the cleavage locus. NHEJ produced small random insertion or deletion (indels) cleaved site, whereas HDR uses DNA donor template for the precise recombination. DR, 19 nt direct repeat; U6p, MtU6 promoter; Ptef1, Tef1 promoter; NLS, nuclear localization signal; TtrpC, trpC Terminator. TTTTTT, poly-T sequence used as terminator
Results
Construction of CRISPR–Cas12a system in M. thermophila
Two Cas12a (Cpf1) orthologs (AsCpf1 and LbCpf1) were reported to have robust DNA cleavage activity in higher eukaryotes with a 5′-TTTV-3′ PAM, where V can be A, C, or G [43]. To investigate its potential editing activity in filamentous fungi, we designed and developed a Cas12a-mediated genome editing tool in M. thermophila. Previously, we successfully harnessed the tef1 promoter Ptef1 and U6 snRNA promoter U6p to drive the expression of Cas9 and sgRNAs, respectively [20]. Thus, in this study, the codon-optimized Cas12a (AsCpf1) gene and the corresponding crRNA were expressed under the control of Ptef1 and U6p, respectively (Additional file 1: Fig. S1). The CRISPR–Cas12a system consisted of separate Cas12a and crRNA expression cassettes (Fig. 1). Briefly, the PCR products of Ptef1-Cas12a-TtrpC and U6p-crRNA were mixed with or without the homology template and co-transformed into M. thermophila protoplasts.
CRISPR–Cas12a-mediated genetic mutation by non-homologous end-joining
The site-specific DNA cleavage created by Cas12a allowed the creation of frameshift insertion and deletion (indel) mutations by non-homologous end-joining repair, which led to loss-of-function of the target genes. To easily determine whether Cas12a was functional in M. thermophila, we designed a crRNA expression cassette to target amdS, a gene that is essential for growth on acetamide as the only nitrogen source, deletion of which resulted in resistance to fluoroacetamide (FAA). This allowed us to deliver the PCR products of the Ptef1-Cas12a-TtrpC and U6p-amdS-crRNA cassettes into protoplasts of the recipient M. thermophila strain M1 [20], which contained amdS and was sensitive to FAA (Fig. 2a). FAA-resistant transformants were obtained and mutations in amdS were verified by DNA sequencing. We obtained six Cas12a-induced indel mutations at sites distal from the PAM and these frameshift mutations were the result of deletions of several nucleotides (Fig. 2b). In a control experiment in which transformation was performed using the Cas12a expressing cassette only, no colonies were obtained on FAA. Together these results demonstrate that delivery of transient PCR products of Cas12a and crRNA cassettes can efficiently mediate mutation of the target gene via non-homologous end-joining repair in M. thermophila.
Fig. 2.
CRISPR–Cas12a system as a new robust genome editing tool in M. thermophila. a Schematic illustration of the mutagenesis of amdS for measuring Cas12a-mediated DNA cleavage in M. thermophila. Positive transformants were selected on plates containing 2 mg mL−1 FAA. b Indel patterns at the amdS target locus of FAA-resistant transformants. The number on the right of each sequence is the indel length (−, deletion). Blue, crRNA base-pairing site; red, PAM sequences; WT, wild-type sequence. c Schematic illustration of CRISPR–Cas12a-donor DNA-mediated deletion of the target gene cre-1 based on homology-directed repair (HDR). Twenty transformants were selected and verified by PCR analysis. The expected length of the deletion mutant was 1.9 kb, whereas that of the wild-type strain (WT), which was the negative control, was 1.0 kb (rightmost lane). Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant
CRISPR–Cas12a-mediated single gene editing by homology-directed repair (HDR)
Gene editing by HDR is an efficient strategy for precise deletion and insertion in a desired locus. To further evaluate the potential of Cas12a-assisted homology-mediated mutagenesis, we used Cas12a to facilitate loss-of-function of cre-1 (carbon catabolite repression transcription factor) using a DNA repair template. The PCR products of Ptef1-Cas12a-TtrpC, U6p-cre1-crRNA, and donor-cre1-neo were introduced simultaneously into protoplasts of wild-type M. thermophila. Then, 20 transformants were picked randomly by G418 selection to determine the gene editing efficiency by PCR analysis using specific primer sets (Fig. 2c). The homologous recombination (HR) efficiencies of gene replacement (including both homokaryons and heterokaryons) for all tested transformants are summarized in Table 1. Compared with transformation with donor DNA alone (15%), the HR frequency of gene replacement (12 homokaryons + 6 heterokaryons) was as high as 90% after co-transformation using the CRISPR–Cas12a system and HR donor DNA (Table 1; Fig. 2c; and Additional file 2: Fig. S1). The HR rates were also low (15% or 10%) when Cas12a or crRNA alone with donor DNA was used for the transformation (Table 1; Additional file 2: Fig. S1). As shown in Fig. 2c, 12 out of 20 (60% deletion efficiency) transformants were confirmed as positive mutant with cre-1 successfully deleted, whereas the control transformation with donor DNA alone or with Cas12a or crRNA alone led to no final correct deletion (0 out 20, 0% deletion efficiency) but only 2 or 3 heterokaryotic mutants of cre-1 gene (Table 1; Additional file 2: Fig. S1). Six out of 20 transformants displayed the heterokaryotic phenotype in our PCR analysis, which showed two bands of correct integration band and wild-type band (Fig. 2c). Based on the multinucleate nature of filamentous fungi, fungal protoplasts contained an uncertain number of nuclei, there are difficulties producing homokaryotic transformants from multinucleate tissue and most of transformants were often heterokaryons through classic method. Streaking conidia may serve to achieve the separation, using sorbose and selective media, and confirming purity by repeated streaking [59]. Cumulatively, these results indicate that similar to the previous M. thermophila CRISPR–Cas9 system, the Acidaminococcus sp. CRISPR–Cas12a system was very efficient in promoting homologous recombination-mediated gene editing and generating homokaryotic deleted mutants.
Table 1.
Summary of the genomic editing in M. thermophila using the CRISPR–Cas system
| Host straina | Target locus | Elements in co-transformation | No. of analyzed transformants | No. of HR transformantsb | HR efficiency (%)c | Each gene disruption efficiency (%) |
|---|---|---|---|---|---|---|
| WT | cre-1 | Donor-cre1-neo | 20 | 3 | 15 | 0 |
| WT | cre-1 | Cas12a +donor-cre1-neo | 20 | 3 | 15 | 0 |
| WT | cre-1 | crRNA +donor-cre1-neo | 20 | 2 | 10 | 0 |
| WT | cre-1 | Cas12a + crRNA + donor-cre1-neo | 20 | 18 | 90 | 60 |
| WT | cre-1, res-1, gh1-1 | Donor DNA of cre-1, res-1 and gh1-1 | 23 | 0 | 0 |
0 0 0 |
| WT | cre-1, res-1, gh1-1 | Cas12a + pooled three sets of crRNA + donor DNA of cre-1, res-1 and gh1-1 | 20 | 8 | 40 |
35 25 25 |
| WT | cre-1, res-1, gh1-1 | Cas12a + array1 + donor DNA of cre-1, res-1 and gh1-1 | 22 | 7 | 32 |
41 32 27 |
| WT | cre-1, res-1, gh1-1 | Cas9 + three sets of sgRNA + donor DNA of cre-1, res-1 and gh1-1 | 23 | 9 | 39 |
39 35 39 |
| M3 | neo,alp-1, rca-1, hcr-1 | Donor DNA of neo, alp-1, rca-1 and hcr1 | 23 | 0 | 0 |
0 0 0 0 |
| M3 | neo,alp-1, rca-1, hcr -1 | Cas12a + array2 + donor DNA of neo, alp-1, rca-1 and hcr-1 | 23 | 5 | 22 |
26 30 13 22 |
| M3 | neo, alp-1, rca-1, hcr-1 | Cas9 + four sets of sgRNA + donor DNA of neo, alp-1, rca-1 and hcr-1 | 23 | 5 | 22 |
35 30 13 30 |
| M7 | bar, ap-3, prk-6 | Donor DNA of bar, ap-3 and prk-6 | 22 | 0 | 0 |
0 0 0 |
| M7 | bar, ap-3, prk-6 | Cas12a + array3 + donor DNA of bar, ap-3 and prk-6 | 22 | 9 | 41 |
32 27 23 |
| M7 | Bar, ap-3, prk-6 | Cas9 + three sets of sgRNA + donor DNA of bar, ap-3 and prk-6 | 21 | 8 | 38 |
33 29 19 |
aWT, wild-type strain; M3, triple-mutant Δcre1Δres1Δgh1-1; M7, septuple mutant ∆cre1Δres1Δgh1-1ΔneoΔalp1Δrca1::xyr1Δhcr1
bHR, homologous recombination
cHR efficiency, HR frequency of gene replacement (including both homokaryons and heterokaryon)
CRISPR–Cas12a-mediated multiplex genome editing using both the pooled crRNA cassettes and a single crRNA array
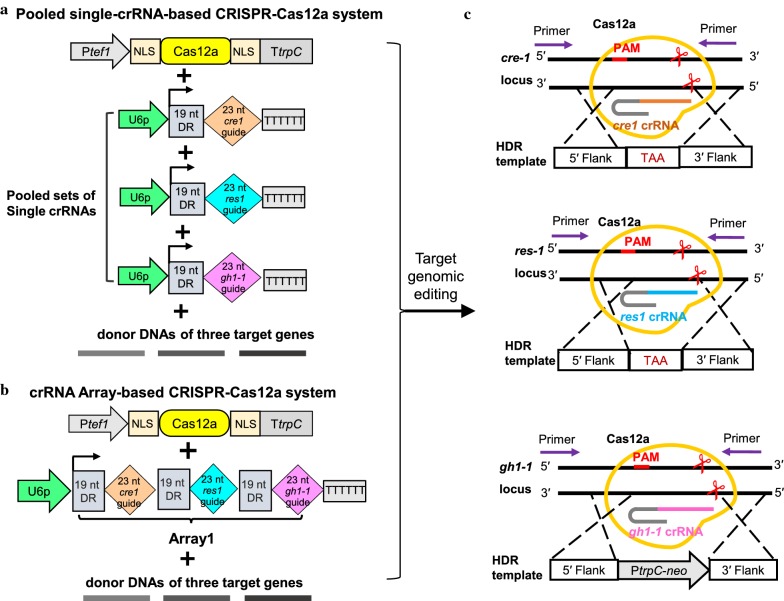
To test whether this CRISPR–Cas12a system could efficiently target multiplex genes in M. thermophila, we explored its ability to target three different loci simultaneously. Besides cre-1, two other genes (res-1 and gh1-1) involved in cellulase production were chosen as targets. Genome editing using HDR is well suited to modifying genes without introducing unwanted changes. Two markerless donor DNA sequences (donor-cre1-TAA and donor-res-1-TAA) were designed as repair templates for precise seamless gene deletion by introducing stop codon (TAA) centrally in the flank sequence, as well as the donor-gh1-1-neo construct to target gh1-1 to allow G418 selection of transformants (Fig. 3a). Unlike Cas9, Cas12a required only one short 42 nt crRNA composed of 19 nt direct repeat and 23 nt guide, so no trans-activating crRNA was needed. Distinct from Cas9, one major advantage of Cpf1 possesses the ability to process a customized crRNA array both in vitro and in vivo [45]. In order to test whether the our CRISPR–Cas12a system maintains precursor crRNA array processing and mediate efficient multiplex gene editing by only using a single customized CRISPR array in thermophilic fungus M. thermophila, we built a crRNA array expressing pre-crRNAs (array 1) in the order cre1–res1–gh1-1 (Fig. 3b) as well as three single crRNA cassettes (Fig. 3a). We delivered the Cas12, crRNA (pooled crRNAs or array 1), and donor DNA in a one-step transformation (Fig. 3a, b). Then 20 or 22 transformants were picked randomly for PCR analysis (Additional file 3: Fig. S2). Some transformants showed two PCR bands (both of wild-type and knockout) were heterokaryon and subsequent separate of pure homokaryotic mutants from the transformants might be frequently necessary. Compared to the control transformation with donor DNAs alone (Additional file 4: Fig. S3), using both of Pooled single-crRNA-based and crRNA Array-based CRISPR–Cas12a systems, 40% (8 heterokaryons) and 32% (2 homokaryon + 6 heterokaryons) of the tested transformants displayed simultaneous triple homologous recombination efficiency in the three loci, cre-1, res-1, and gh1-1. In contrast to control experiment (0 out 23, 0% efficiency), pool- and array-based editing system showed the deletion efficiencies of cre-1, res-1 and gh1-1 were 35% (7 out 20) and 41% (9 out 22), 25% (5 out 20) and 32% (7 out 22), and 25% (5 out 20) and 27% (6 out 22), respectively (Table 1; Additional file 3: Fig. S2; Additional file 4: Fig. S3). This gene set has been tested using our CRISPR–Cas9 system (Additional file 5: Fig. S4), making it easy to compare the two systems (Table 1). By using our CRISPR–Cas9 system, 9 (including 3 homokaryons and 6 heterokaryons) out of 23 (39%) transformants displayed the triple-gene homologous recombination efficiency, in which 39%, 35% and 39% colonies showed deletions of cre-1, res-1, and gh1-1, respectively. The resulting triple-mutant Δcre1Δres1Δgh1-1 was named M3 and used as a host strain for subsequent engineering. Remarkably, all three target genes were equally well edited using either the crRNA array or pooled single-crRNA cassettes. Overall, these results demonstrate that the CRISPR–Cas12a system can efficiently mediate multiplex gene deletions in M. thermophila through a simple single crRNA array.
Fig. 3.
CRISPR–Cas12a-mediated multiplex genome editing in M. thermophila. a Schematic illustration of the simultaneous deletion of three target genes, cre-1, res-1 and gh1-1, using the CRISPR–Cas12a system with pooled single-crRNA cassettes. b Schematic illustration of the simultaneous deletion of three target genes, cre-1, res-1 and gh1-1, using the CRISPR–Cas12a system with crRNA array1 expressing cassette in the order cre1–res1–gh1-1. c Once Cas12a nuclease introduces the sequence-specific double-stranded DNA break, homology-directed repair (HDR) mediate precise gene deletion using donor DNA template at the cleavage locus of cre-1, res-1 and gh1-1, respectively. DR, 19 nt direct repeat; U6p, MtU6 promoter; Ptef1, Tef1 promoter; NLS, nuclear localization signal; TtrpC, trpC Terminator
CRISPR–Cas-assisted marker recycling (Camr) technology for iterative multiplex genome editing in consecutive steps
Cas nuclease-mediated double-strand breaks can efficiently improve the HR frequency when they occur in sites of interest and induce alterations. Hence, we reasoned that this recombination process could potentially rescue the selectable marker neo. The new crRNA target on neo could replace the existing neo cassette with any desired DNA sequence through CRISPR–Cas-assisted HDR. Thus, successive genome editing can be performed using only two selectable markers in a “ping-pong” style. We used this methodology to create a CRISPR–Cas-assisted marker recycling system (Fig. 4).
Fig. 4.
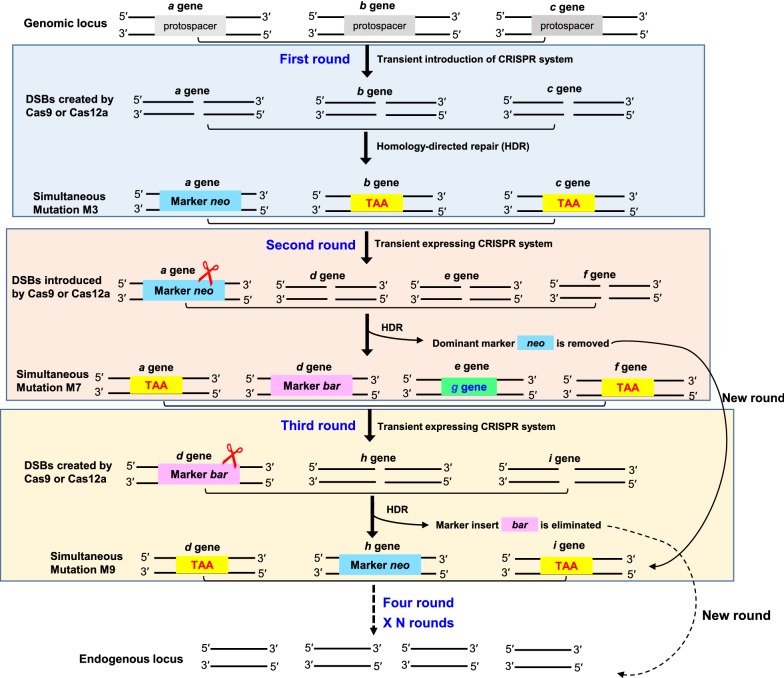
Schematic strategy of CRISPR–Cas-mediated marker recycling approach for iterative multiplex genome editing in M. thermophila. The hendecuple mutants were constructed in three successive transformations by alternatively using two selectable markers neo and bar. In the first transformation, a triple mutant was created by transient CRISPR–Cas12a/Cas9-mediated homology-directed repair (HDR). The neo cassette replaces an endogenous locus, and two target genes are both markerless deletions. In the second transformation, the insert of neo is removed and bar replaces a new endogenous locus in the triple-mutant host strain via HDR by the CRISPR–Cas12a/Cas9 system. In addition, two other gene loci undergo seamless gene replacement and markerless gene disruption. For the third transformation, the octuple mutant is selected for genetic manipulation using the CRISPR system. The selectable marker neo is inserted into a new endogenous locus, and the marker bar is scarlessly removed and the other new gene undergoes markerless deletion. This allows the marker bar to be used again in a new transformation round. DSBs, double-strand breaks; M, mutant
To test this idea, we selected nine key genes in the cellulase production pathway, including cre-1 (MYCTH_2310085), res-1 (MYCTH_2302052), gh1-1 (MYCTH_115968), alp-1 (MYCTH_2303011), Mtxyr-1 (Mycth_2310145), rca-1 (Mycth_2300719), hcr-1 (Mycth_2309600), ap-3 (Mycth_2307451), and prk-6 (Mycth_2303559) [61‒65], and generated marker-recycling multiple knockouts or knockins using only two markers, neo and bar. For each round of transformations, either the single marker gene neo or bar was used for selection of antibiotic-resistant fungal transformation on medium plates supplemented with G418 (for neo gene resistance) or phosphinothricin (for bar gene resistance). In order to efficiently excise the marker gene in the next round transformation, we used only one marker and integrated this marker gene into a single random locus of target genes per each round of edits. In the first transformation, we targeted three genes including cre-1, res-1, and gh1-1 and only locus of gh1-1 was randomly selected for marker gene neo integration, which is convenient for neo knockout in successive transformation. In the second transformation, we targeted four genes (neo, alp-1, rca-1 and hcr-1) and only locus of alp-1 was selected for second marker gene bar integration, which is convenient for bar deletion in the third transformation. In successive transformation, we engineered three genes including bar, ap-3 and prk-6 and only locus of ap-3 was selected for first marker gene neo integration, which is convenient for neo deletion in the next iteration.
Three successive round of experiments were designed to test a proof-of-concept in generating nine genes mutants by using our “ping-pong” style of marker recycling strategy, in which the first maker gene neo can be eliminated in the second transformation after its initial use and then this neo gene can be used again in the third transformation, just as it was used in the first transformation. Based on this design, we can generate the nonuple mutant through three round of edits (Fig. 5). First, three genes, cre-1, res-1, and gh1-1, were targeted, resulting in the triple-mutant M3 (Figs. 3, 5). The second transformation was carried out with the host strain M3. We built a crRNA array expressing pre-crRNAs in the order neo–alp1–rca1–hcr1 (array 2). After co-delivering Cas12a, array 2, and donor templates into M3, 23 putative transformants were picked randomly and analyzed for indels by PCR using primers flanking the targeted loci (Additional file 6: Fig. S5). We found that alp-1 was replaced with the selectable marker bar, which conferred phosphinothricin resistance, whereas the marker neo and hcr-1 were seamlessly deleted. In addition, the essential (hemi-)cellulase regulator xyr-1 was inserted into the rca-1 locus. Five colonies (1 homokaryon + 4 heterokaryon, 22%) were identified with these four loci modified, while none of colonies (0 out of 23, 0%) were shown simultaneous quadruple recombination in the control transformation with donor DNA alone (Table 1; Additional file 7: Fig. S6). In the second round transformation by using our CRISPR–Cas12a system, the gene disruption frequency of neo, alp-1, rca1-1 and hcr-1 were 26%, 30%, 13%, and 22%, respectively. We obtained four combinations of homokaryon mutation genotypes (Additional file 6: Fig. S5), namely one quadruple mutant Δcre1Δres1Δgh1-1Δhcr1 (M4), one quintuple mutants Δcre1Δres1Δgh1-1ΔneoΔalp1Δhcr1 (M5 with disrupted neo) and one sextuple mutant Δcre1Δres1Δgh1-1Δalp1Δrca1::xyr1 (M6), and one septuple mutant Δcre1Δres1Δgh1-1ΔneoΔalp1Δrca1::xyr1Δhcr1 (M7 with disrupted neo). The M5 and M7 mutants were very sensitive to G418, indicating that the marker neo was successfully removed and can be used again in the next genetic engineering operation.
Fig. 5.
CRISPR–Cas12a-assisted marker recycling editing of eleven target genes, including nine genes involved in the cellulase production pathway, via three successive round transformations. The 11 genes were cre-1, res-1, gh1-1, alp-1, xyr-1, rca-1, hcr-1, ap-3, prk-6, and two selectable marker genes neo and bar. HDR, homology-directed repair; WT, wild type; DR, 19 nt direct repeat; U6p, U6 promoter; Ptef1, tef1 promoter; TtrpC, trpC terminator. M, mutant strain. PtrpC, trpC promoter. TAA, stop codon. TTTTTT, poly-T sequence
Strain M7, which lacked the neo marker, was used for the third transformation. We constructed another crRNA array (array 3) to target three loci, the selectable marker bar and two other genes (ap-3 and prk-6) involved in the cellulase production pathway. In the third round of HDR, the marker bar was excised, and ap-3 was replaced by the marker neo, which was rescued in last round; prk-6 was disrupted (Fig. 5). Nine transformants (1 homokaryon + 8 heterokaryons) showed HR in three gene loci and the efficiency of triple recombination was about 41% (Table 1; Additional file 8: Fig. S7), whereas none of transformants (0 out of 23) were displayed all triple-gene homologous recombination modified in control experiment with donor DNA alone (Table 1; Additional file 9: Fig. S8). We obtained three combinations of homokaryotic deletion strains, namely two octuple mutants Δcre1Δres1Δgh1-1ΔneoΔalp1Δrca1::xyr1Δhcr1ΔbarΔap3 (M8-1, eight cellulase production-related genes edited with disrupted copies of neo and bar) and Δcre1Δres1Δgh1-1ΔneoΔalp1Δrca1::xyr1Δhcr1ΔbarΔprk6 (M8-2), and one nonuple mutant Δcre1Δres1Δgh1-1ΔneoΔalp1Δrca1::xyr1Δhcr1ΔbarΔap3Δprk-6 (M9, nine cellulase production pathway genes edited with disrupted copies of neo and bar; the functional neo was added into the ap3 loci, whereas the selectable marker bar was rescued in this transformation round).
On the basis of these results for Cas12a, we tested and optimized our previously reported Cas9 system using the marker recycling approach to obtain more flexible Cas9-based mutagenesis in M. thermophila (Fig. 4). The genetic manipulation was performed by co-delivery of the PCR products of Cas9, sgRNAs, and donor DNA in three sequential rounds of transformation (Additional file 13: Fig. S5; Additional file 10: Fig. S9; Additional file 11: Fig. S10). The HR efficiency of each round in generating Cas9-mediated mutants is summarized in Table 1. The Cas9-based efficiency of simultaneously homologous recombination of three and four genes was approximately 38–39% and 22%, respectively, which were similarly observed in the transformation experiments by using array-based CRISPR–Cas12a system (Table 1), suggesting that crRNA array-based CRISPR–Cas12a system might be more cheaper and convenient for multiplex genome editing. Three successive rounds of transformations and selections resulted in one homokaryon nonuple mutant (M9, nine cellulase production pathway genes edited with disrupted copies of neo and bar), in which gain-of-function of neo but loss-of function of bar were confirmed.
Together, these results indicate that the CRISPR–Cas12a and CRISPR–Cas9 systems are both efficient tools for mediating marker recycling in M. thermophila. We named this CRISPR–Cas12a/Cas9-assisted marker recycling system as “Camr” technology.
Evaluation of cellulolytic enzyme production in M. thermophila mutants obtained via our Camr technology
The quadruple mutant Δcre1Δres1Δgh1-1Δalp1 from our previous study exhibited a pronounced hyper-cellulase secretion phenotype [20], therefore, we used it to assess whether the dose effects of other cellulolytic factors such as xyr-1, rca-1, hcr-1, ap-3 and prk-6 (Fig. 6a) would further enhance cellulase production in this quadruple mutant. We examined the cellulolytic phenotypes on cellulose medium of the eight mutants obtained through the three sequential rounds of transformation described above. As expected, all eight mutants displayed significantly enhanced secreted protein production (~ 602.2 to 1201.9 mg L−1) compared with the wild type WT strain (~ 133.1 mg L−1); in particularly, the secretome of M9 (~ 1201.9 mg L−1) was approximately 9.0-fold higher than that of the WT (Fig. 6b; Additional file 12: Fig. S11). Consistent with the increased secreted protein levels, cellulolytic enzyme activity in the eight mutants was remarkably higher than in WT (Fig. 6c, d). Notably, the endoglucanase and xylanase activity in mutants M8-1, M8-2, and M9 was 9.5- to 10.1-fold and 17.2- to 18.5-fold higher, respectively, than in the WT. Together, these results indicate that the dose-controlled pathway of cellulase expression and secretion is a promising strategy for cellulolytic fungi to develop enzyme hyper-producers via our Camr technology.
Fig. 6.
Cellulolytic phenotype analysis of eight deletion mutants and wild-type strain (WT). a Schematic model illustrating the coordinated action of cellulolytic factors under cellulose induction. Under cellulose conditions, the positive regulators CLR-1, CLR-2, and XYR-1 have been identified as key transcription factors regulating cellulose degradation. Preferred carbon sources such as glucose activate the essential transcription factor CRE-1, which represses the expression of cellulase genes, a phenomenon called carbon catabolite repression (CCR). Lack of the major intracellular β-glucosidase GH1-1 facilitates the accumulation of intracellular cellobiose, which can trigger signaling cascades that include expression of cellulase genes repressed by CRE-1 and activated by CLR-1/2 and XYR-1. The newly identified regulators RES-1, HCR-1, RCA-1, AP-3, the major secreted protease ALP-1, and protein kinase PRK-6 were shown to negatively affect cellulase production. b Secreted protein production of all strains after 6-days cultivation in 2% Avicel medium supplemented with 0.75% yeast extract. Genotype of the eight deletion mutants as compared to the WT strain. The 11 target genes edited by our CRISPR–Cas12a system included cre-1, res-1, gh1-1, neo, alp-1, rca-1, xyr-1, hcr-1, bar, ap-3 and prk-6. The symbol of +, or − or OE indicated the target gene is present, or deleted or overexpressed. c, d Assays for CMCase and xylanase activity in the eight deletion mutants and WT strains in 2% Avicel plus 0.75% yeast extract after 6 days culture. Bars marked by asterisks in each group differ significantly from the unmarked bars (Tukey’s HSD, p < 0.001). Error bars represent SD from three replicates
Discussion
A first-generation genome editing tool using the CRISPR–Cas9 system was developed in thermophilic fungi M. thermophila and M. heterothallica [20]. This CRISPR–Cas9 system was difficult to use when there were more than five target genes because all the sgRNA-expressing cassettes had to be constructed and delivered at the same time. This issue was addressed by the CRISPR–Cas12a system. Although the CRISPR–Cas12a system has been intensively and successfully utilized in higher eukaryotes for genome editing [44–49], this system has been comparatively underexplored in microbes. Until now, Cas12a-based genomic editing tools have been demonstrated only in yeast, a few fungi species [50, 51, 57, 58], and several bacteria [52–55], and have not yet been widely established in thermophilic filamentous fungi. While this manuscript was in revision, a functional CRISPR-Cpf1 system for gene editing was published in T. thermophilus (synonym of M. thermophila) [58] demonstrating that this system may work widely in filamentous fungi.
In this study, we developed a Cas12a-based system from Acidaminococcus sp. BV3L6 as an efficient genomic editing tool for the thermophilic fungus M. thermophila (Fig. 1). We found that the CRISPR components of the Cas12a and crRNA expression cassettes were able to function when introduced transiently and without direct selection. PCR analysis indicated that the CRISPR–Cas12a cassettes were not integrated into the genome (data not shown). This transiently expressed CRISPR–Cas12a system efficiently enabled the generation of indel mutations in the M. thermophila genome (Figs. 2, 5). The transient CRISPR–Cas9 system produced similar results in M. thermophila (Additional file 13: Fig. S5; Additional file 10: Fig. S9; Additional file 11: Fig. S10). These data are consistent with previous reports in Magnaporthe oryzae and Candida albicans, which showed the transient introduction of the CRISPR–Cas9 system efficiently mediated gene editing [24, 60]. The efficient and successful application of transient CRISPR–Cas technology has useful implications. First, this system may minimize the problems associated with Cas toxicity (such as Cas9 and FnCpf1) in some fungi species [24, 50] and off-target cleavage activity. Second, gene edited mutants can be generated rapidly using PCR-amplified cassettes, which obviates the need to construct Cas12a or Cas9 expressing strains, thereby dramatically reducing the time, cost, and use of selectable marker. Although multiplex editing of homologous recombination genes occurred a little more frequently in colonies transformed with pooled crRNAs than in those transformed with array1, the Cas12a-mediated system produced frequencies similar to those of the Cas9 system in colonies transformed with array 1 (Table 1). In addition, the three target genes, cre-1, res-1, and gh1-1, were equally edited and the editing efficiency of each gene was not affected by using either a single-array approach or pooled cassettes. Previous reports in mammalian and yeast cells have found that the position of the crRNA on the array and the number of target genes (up to four) were not crucial for editing efficiency [45, 50]. Thus, we used a single-array approach to perform the Cas12a-mediated multiplex gene editing in the further manipulations. Our results expand the genome editing toolbox to efficiently generate mutations in filamentous fungi.
Previously, we obtained multiple deletion mutants with G418-resistance marker neo, which was integrated simultaneously into each target loci via the CRISPR–Cas9 system, and the copy number of the integrated marker was in good accordance with the number of target genes [20]. Hence, we reasoned that triple or quadruple mutants could be generated by integrating a selectable marker in only one target locus via the CRISPR–Cas12a/Cas9 system, thereby minimizing the copy number of the marker gene, and creating seamless gene deletion or insertion in other target loci. Our results confirm that the acquisition of triple-mutant M3 was achieved by integrating the selectable marker gene neo at the gh1-1 locus and that markerless deletion of cre-1 and res-1 was achieved via one-step transformation using either the CRISPR–Cas12a or CRISPR–Cas9 systems. Additionally, our data indicate that the recombination efficiencies of the target genes did not rely on the integration of a selective marker into the locus, allowing for efficient markerless genomic modification as desired. A similar strategy could be used to exchange or modify target promoters to alter gene expression for industrial applications.
New approaches for marker recycling based on the powerful CRISPR–Cas9 system have been demonstrated recently in fungi, namely S. cerevisiae, C. albicans, A. oryzae and A. niger [36, 37, 61, 62]. Most of these approaches combined Cas9 editing with the auxotrophic marker pryG/ura3 or recombinase-promoted excision. These methods require additional steps for either constructing auxotrophic strains or autonomous replicating plasmids carrying the corresponding recombinase [36, 37, 62], which is complicated and time-consuming. Our results for generating the nonuple mutant M9 (Fig. 5) by marker recycling and iterative stacking of traits in the same strain through editing both the previously used marker and other different loci via the CRISPR/Cas system is a feasible new approach. We employed this strategy to design our Camr technology through alternative use of two selectable markers in a “ping-pong” style where the first marker is rescued when the second marker is used (Fig. 4). As shown in Table 1, the Cas9-based efficiency of simultaneously homologous recombination of three and four genes was approximately 38%, 39% and 22%, respectively, which were similarly observed in the transformation experiments by using array-based CRISPR–Cas12a system, suggesting that crRNA array-based CRISPR–Cas12a system might be more cheap and convenient for multiplex genome editing. Recent work by Zhang and co-workers performed a computational analysis of the targeting range of the CRISPR–Cas12a system and CRISPR–Cas9 system in the human genome [63]. The authors demonstrated that the targeting range of wild-type Cas12a to one target site was per ~ 33 bp in human coding sequences, while the targeting range of Cas9 to one cleavage site was per ~ 7 bp in human coding sequences, suggesting the target frequency of Cas12a is more rare than Cas9 [63]. However, Zhang and co-workers engineered Cas12a variants to exist with altered PAM specificities including 5′ TYCV and 5′ TATV, 5′ MCCC and 5′ RATR, which have expanded the targeting range of Cas12a to one target site per ~ 7 bp in human coding sequences [63]. Moreover, the T-rich PAMs of the Cas12a allow for applications in genome editing in organisms with particularly AT-rich genomes or sequence areas of interest with AT enrichment, so the T-rich-dependent PAMs of Cas12a proteins expand the targeting range of genome editing nucleases and become a useful complement to CRISPR–Cas9 system for genetic engineering.
The traditional methods in filamentous fungi for obtaining multiple gene mutants from continuous single deletion engineering (usually per ~ 4‒5 weeks) to multiple genes are very complicated, laborious and time-consuming, and also limited in the ability to knock out multiple genes at the same time. Compared with the utilization of conventional methods to edit multiple gene, our Camr technology can significantly reduce the time and labor to engineer multiple genes (more than 10) via two or three successive transformations. The manipulation time for our procedure was reduced to 6 days for each round of modification, and up to three or four gene targets can be modified simultaneously. For instance, the triple-gene disruptions M3 mutant (Δcre1Δres1Δgh1-1) was obtained by our Camr technology within 6 days and an additional 7 days were required for fungal conidia growth. We can also obtain multiple mutant genotypes with different combinations of disrupted four genes, such as M4, M5, M6, and M7, in second round transformation (time spending another 6 + 7 days from the host strain M3), which provides additional opportunities to obtain desired mutant strains of industrial interest and thus greatly improves the efficiency and time of genome editing. Taken together, we successfully targeted nine genes in the cellulase production pathway and generated the mutant M9 within ~ 5 weeks from the wild-type strain, in which all nine selected genes were edited correctly, via three successive transformations using markers neo and bar in our Cas12a and Cas9 systems (Fig. 5).
In saprophytic fungi, lignocellulolytic enzyme production is mainly regulated at both the transcriptional and post-translational levels which involves combinatorial action of several transcriptional activators and repressors [1, 2, 64–67]. Genetically engineering the regulatory network at both levels, such as overexpression of transcriptional activators and deletion of repressors, represents an efficient and promising strategy for significantly improving cellulases production in cellulolytic fungi including T. reesei, Penicillium oxalicum, N. crassa and M. thermophila [1, 7, 20, 68]. Generally, the cellulase production is induced by cellulose-derived oligosaccharides (e.g., cellobiose) and repressed by preferentially utilized saccharides (e.g., glucose), a phenomenon called carbon catabolite repression (CCR). Therefore, the carbon catabolite repressor CreA/Cre1 is a well-known highly conserved cellulase repressor throughout the fungal kingdom and elimination of the function of CreA/Cre1 resulted in significantly improved lignocellulolytic enzyme production in Aspergillus spp., T. reesei, N. crassa, P. oxalicum and M. thermophila [1, 2, 7, 20, 68]. Additionally, elimination of β-glucosidases which hydrolyze cellobiose to glucose is able to improve the expression and production of cellulolytic enzymes. For example, disruption of the major intracellular β-glucosidase enhanced cellulase production on cellulose in N. crassa [1], P. oxalicum [7, 69] and M. thermophila [20]. In addition, our recent studies in N. crassa demonstrated that single deletion of the transcriptional regulator res-1 [66], hcr-1 [70] or rca-1 [64], adaptor protein ap-3 [67] or protein kinase prk-6 [67] improved cellulase production and activities under cellulolytic condition.
Our previous report in M. thermophila [20] showed that the single deletion of cre-1, gh1-1, res-1, or alp-1 resulted in significant improvement of cellulase production by about 3.3-, 2.3-, 2.6- or 2.1- fold higher amounts (~ 381.8, ~ 266.2, ~ 306.6 or ~ 245.2 mg L−1) compared with WT strain under 5 days 2% Avicel cultivation, implying that CRE-1 play the largest phenotypic effect on cellulase production in M. thermophila. Based on these above obvious improvement, we hypothesized that there might be a significant synergistic and additive reinforcement effect in cellulase production by genetic integrating modification of these cellulolytic factors. Therefore, in this study, we generated the mutants M3 (Δcre1Δres1Δgh1-1), M7 (M3 + ΔneoΔalp1Δrca1::xyr1Δhcr1), and M9 (M7 + ΔbarΔap3Δprk-6) through using our Camr technology. As expected, the cellulase secretion and activity phenotypes of M3 (~ 602.2 mg L−1, ~ 4.5-fold), M7 (~ 813.5 mg L−1, ~ 6.1-fold) and M9 (~ 1201.9 mg L−1, ~ 9.0-fold) were significantly enhanced compared with those of the WT strain (~ 133.1 mg L−1) (Fig. 6), suggesting that the key regulator CRE-1 play a master role in improving cellulase production and other factors also have positive and synergistic effects on increase of cellulase production. As depicted above, since one key regulator CRE-1 has greatly profound effects on cellulase expression and secretion in M. thermophila and the resulting strain M3 led to significantly higher cellulase production. Thus, the latter engineered strains M7 and M9 produced moderately enhanced cellulase production due to this very high background level of host strain M3.
These enhanced cellulase production levels by the engineered strains developed here are comparable to those of strains engineered in the other well-known cellulase production species P. oxalicum [69], in which the quadruple mutants RE-29 (deleting bgl2 and creA, along with over-expressing the gene clrB and xlnR/xyr1) showed more cellulolytic enzyme activities and secretion abilities than the triple-mutant RE-10 (deleting bgl2 and creA, along with over-expressing the gene clrB). This phenomenon suggests that engineering the key factors involved cellulolytic regulatory networks perform the synergistic improvement of cellulase production. In summary, this result indicates that the regulatory network of cellulase expression and secretion can be genetically engineered using the Camr technology as a simple and efficient strategy to improve cellulase production in cellulolytic fungi within a short time. Therefore, the Camr approach can be used for iterative genomic manipulations in both basic research and industrial strain engineering for biotechnological applications.
Conclusions
In this study, we report an Acidaminococcus sp. Cas12a-based CRISPR system for multiplex genome editing, using a single-array approach in thermophilic filamentous M. thermophila. These CRISPR–Cas12a cassettes worked well for simultaneous multiple gene deletions/insertions. We also developed new approaches that allow easy and efficient marker recycling and iterative stacking of traits in the same thermophilic fungus strain either, using the newly established transiently expressed CRISPR–Cas12a system or the established CRISPR–Cas9 system to make DNA breaks in selected markers. Together we called them CRISPR–Cas-assisted marker recycling technology (Camr technology). We demonstrated its performance by targeting nine genes involved in the cellulase production pathway in M. thermophila via three transformation rounds, using two selectable markers neo and bar. The nonuple mutant M9 were then obtained, in which protein productivity and lignocellulase activity were 9.0- and 18.5-fold higher than in the wild type. We expect this advance to accelerate biotechnology-oriented engineering processes in fungi.
Methods
Strains and growth conditions
Myceliophthora thermophila ATCC 42464 was obtained from the American Type Culture Collection (ATCC). M. thermophila strains were cultured on Vogel’s MM supplemented with 2% sucrose at 45 °C for 7 days to obtain conidia. Antibiotics were added when needed to screen for transformants. For flask culture, M. thermophila conidia at 106 mL−1 were inoculated in 100 mL medium (containing 1 × Vogel’s salt, 2% Avicel, and 0.75% yeast extract) at 45 °C with shaking at 150 rpm. For vector manipulation and propagation, Escherichia coli DH5α (Invitrogen, Shanghai, China) was cultured at 37 °C in Luria–Bertani broth with kanamycin or ampicillin (100 μg mL−1) for plasmid selection.
Plasmid construction for genetic engineering
All the primer sequences used in this study are listed in Additional file 1: Table S1. All the PCR products were amplified using Phusion high-fidelity DNA polymerase (Thermo Fisher, Waltham, MA, USA).
To generate the Cas12a expression plasmid, a codon-optimized Cas12a gene (AsCpf1, GenBank: U2UMQ6) with attached HAC-1 (GenPept: MYCTH_2310995) and SV40 nuclear localization signal (NLS-Cas12a-NLS) was synthesized artificially by the Life Science Research Services Company (Genewiz, Suzhou, China). The strong constitutive tef1 (translation elongation factor EF-1, MYCTH_2298136) promoter Ptef1 of M. thermophila was used to express Cas12a. The TtrpC terminator was cloned from the vector pNA52-1N (GenBank number: Z32697). The synthetic NLS-Cas12a-NLS, Ptef1 promoter [20], and TtrpC terminator were amplified and assembled into a p0380-bar plasmid [41] to form a Ptef1-Cas12a-TtrpC cassette by using a NEB Gibson assembly kit. The sequence of Ptef1-Cas12a-TtrpC expressing cassette is provided in Additional file 13.
The crRNA expression cassette comprised the M. thermophila U6 promoter [20], the target sequence of 23 nt, a short direct repeat of 19 nt (5′-AATTTCTACTCTTGTAGAT-3′) [43] and ploy T sequence used as terminator. The target genes were amdS (GenBank: M16371.1), cre-1 (MYCTH_2310085), res-1 (MYCTH_2302052), gh1-1 (MYCTH_115968), alp-1 (MYCTH_2303011), rca-1 (Mycth_2300719), hcr-1 (Mycth_2309600), ap-3 (Mycth_2307451), prk-6 (Mycth_2303559), and the selectable markers neo (GenBank: HQ416708) and bar (GenBank: X17220). Specific crRNA target sites were designed by using the CRISPR-offinder tool [71], which is available at the BiooTools website (http://www.biootools.com). The target sequence with low off-target probability were selected. The 23-nt protospacer sequences of guide crRNAs are provided in Additional file 14: Table S2. Briefly, the single crRNA expression cassettes, U6p-amdS-crRNA, U6p-cre1-crRNA, U6p-res1-crRNA, and U6p-gh1-1-crRNA, were generated by PCR amplification with primer pairs in Additional file 1: Table S1 and cloned into vector pJET1.2 for sequencing (CloneJET PCR Cloning Kit; Thermo Fisher Scientific, Waltham, MA USA). Three crRNA arrays expressed by a U6 promoter (U6p-array1-cre1-res1-gh1-1, U6p-array2-neo-alp1-rca1-hcr1 and U6p-array3-bar-ap3-prk6) were synthesized by the Genewiz gene synthesis service. Sequence for the all the single crRNAs and crRNA arrays are provided in Additional file 15.
To construct plasmids expressing sgRNA, specific sgRNA target sites in cre-1, res-1, gh1-1, alp-1, rca-1, hcr-1, ap-3, prk-6 were identified using the sgRNACas9 tool [72] with the M. thermophila genome sequence and the target gene as the inputs. The target sequence with low off-target probability were selected. The sgRNA expression cassettes, U6p-rca1-sgRNA, U6p-hcr1-sgRNA, U6p-ap3-sgRNA, U6p-prk6-sgRNA, U6p-neo-sgRNA, and U6p-bar-sgRNA, were constructed as described previously [20]. The target sequences for all the sgRNAs are provided in Additional file 2: Table S2.
To construct the donor DNAs, the 5′ and 3′ flanking fragments of cre-1 (600 bp/539 bp), res-1 (600 bp/600 bp), gh1-1 (600 bp/600 bp), alp-1 (600 bp/600 bp), rca-1 (522 bp/608 bp), hcr-1 (600 bp/600 bp), neo (563 bp/600 bp), ap-3 (600 bp/580 bp), prk-6 (547 bp/600 bp), and bar (599 bp/560 bp) were amplified separately. The selectable marker cassettes PtrpC-neo and PtrpC-bar were amplified from the p0380-neo and p0380-bar plasmids [41]. To construct the markerless templates, the 5′ and 3′ fragments were assembled and ligated into pUC118 using a NEB Gibson kit to generate donor-cre1-TAA, donor-res1-TAA, donor-neo-TAA, donor-hcr1-TAA, donor-prk6-TAA, and donor-bar-TAA. The “TAA” in the donor DNA indicated introducing a stop codon in the target genes. The 5′ and 3′ fragments and PtrpC-neo or PtrpC-bar were assembled and inserted into pUC118 to generate donor-cre1-neo, donor-gh1-1-neo, donor-alp1-bar, and donor-ap3-neo. To overexpress Mtxyr-1 (Mycth_2310145) in the rca-1 locus, the 1200-bp promoter of hsp70 (heat shock protein 70, Mytcth_112686) and the full-length sequence of Mtxyr-1 were amplified separately. These two fragments and the 5′ and 3′ fragments of rca-1 were assembled and ligated into the pUC118 using Gibson kit to generate donor-rca1-Mtxyr-1.
Transformation of M. thermophila protoplasts
The PEG-mediated transformation of M. thermophila protoplasts was performed as described previously [41]. For amdS mutagenesis, the amdS expression strain M1 [20] was used as the host strain. Briefly, 10 μg of the PCR products of Ptef1-Cas12a-TtrpC (9.2 μg) and U6p-amdS-crRNA (0.8 μg) at the same molar concentration ratio were co-transformed into M1 protoplasts. The amdS mutants were inoculated onto MM plates that included 2 mg mL−1 fluoroacetamide (FAA). FAA-resistant mutants were isolated and tested for growth on acetamide medium, followed by PCR sequencing. For cre-1 deletion, total 10 μg PCR cassettes of Ptef1-Cas12a-TtrpC (6.8 μg), U6p-cre1-crRNA (0.6 μg), and donor-cre1-neo (2.6 μg) were mixed at the same molar concentration ratio and added to the protoplasts of WT. Transformants were screened for neo resistance with 80 μg mL−1 G418 after 3 days of culture, followed by PCR identification.
For multiplex gene editing by Cas12a using pooled single or array crRNAs, the 11.5‒12.4 μg PCR products of three pooled crRNAs cassettes (0.5 μg for each crRNA) or array 1 (0.6 μg) and donor DNAs of cre-1 (1.2 μg), res-1 (1.2 μg), and gh1-1 (2.5 μg) were mixed with Ptef1-Cas12a-TtrpC (6.0 μg) at the same molar concentration ratio and co-transformed into WT protoplasts. For multiple gene editing with the transient CRISPR–Cas9 system, the 12.7 μg PCR products of sgRNAs (0.6 μg for each sgRNA), donor DNAs of cre-1 (1.2 μg), res-1 (1.2 μg), and gh1-1 (2.5 μg), and Ptef1-Cas9-TtrpC (6.0 μg) were co-transformed into WT protoplasts. The putative transformants were selected with 80 μg mL−1 G418 and confirmed by PCR, generating the triple deletion strain Δcre1Δres1Δgh1-1 (M3).
To remove marker cassette neo, second round manipulation was performed with the transient Cas12a or Cas9 system. Briefly, total ~ 17.0‒19.0 μg PCR cassettes of array 2 (0.65 μg) or sgRNAs (0.6 μg for each sgRNA) and Ptef1-Cas12a-TtrpC (6.0 μg) or Ptef1-Cas9-TtrpC (6.0 μg) were mixed with donor DNA cassettes of neo (1.2 μg), alp-1 (2.5 μg), rca-1 (5.5 μg), and hcr-1 (1.2 μg) and co-transformed into protoplasts of the M3 strain. Putative transformants were selected on phosphinothricin (100 μg mL−1) and confirmed by PCR, creating the septuple deletion strain Δcre1Δres1Δgh1-1ΔneoΔalp1Δrca1::xyr1Δhcr1 (M7). For third round manipulation, the 11.5‒12.7 μg PCR products of array 3 (0.6 μg) or sgRNAs (0.6 μg for each sgRNA) and Ptef1-Cas12a-TtrpC (6.0 μg) or Ptef1-Cas9-TtrpC (6.0 μg) were mixed with donor DNAs of bar (1.2 μg), ap-3 (2.5 μg) and prk-6 (1.2 μg) and co-transformed into M7 protoplasts. Putative transformants were selected on G418 for 3 days and confirmed by PCR.
For single gene editing, control experiments were performed by adding 2.6 μg of donor-cre1-neo alone, or only the Cas12a cassette (6.8 μg) and donor-cre1-neo (2.6 μg), or only U6p-cre1-crRNA (0.6 μg) and donor-cre1-neo (2.6 μg) to the fungal protoplasts. For multiplex gene editing, control experiments were performed by adding 4.9‒10.4 μg of donor DNAs alone without CRISPR–Cas9/12a expression cassettes to the fungal protoplasts. Transformants were screened for bar resistance with phosphinothricin (100 μg mL−1) or neo resistance with G418 (80 μg mL−1), followed by PCR identification with paired primers (Additional file 1: Table S1).
Protein and enzyme assays
The protein concentration in the supernatants was determined using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). Absorbance was measured at 595 nm and bovine serum albumin was used as the standard. For protein gel electrophoresis, 20-μL unconcentrated culture supernatant was loaded onto a polyacrylamide gel (Novex® NuPAGE® Pre-cast Protein Gels, Thermo Fisher Scientific) for sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Endoglucanase and endo-1,4-β-xylanase activity in the culture supernatants was determined using an Azo-cm-cellulose assay kit (Megazyme) and an Azo-xylan kit (Megazyme) in accordance with the manufacturer’s instructions. All estimates were performed in three repeated assays. The statistical significance of differences among WT and mutant strains was assessed by one-way analysis of variance.
Supplementary information
Additional file 1: Table S1. List of PCR primers used in this study.
Additional file 2: Figure S1. Verification of cre-1 gene deletions in selected transformants with co-transformation of only Cas12a and donor DNA, only crRNA and donor DNA, or only donor DNA. (A) Schematic of homologous recombination (HR) of target gene cre-1. (B–D) PCR analysis of cre-1 deletion with one primer (cre1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other (cre1-in-R) located in the 3′ flanking region of genomic DNA. The expected length of disrupted transformants was 1.9 kb, while that of the WT host strain, used as a negative control, was 1.0 kb (rightmost lane). Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). HDR, homology-directed repair; WT, wild type.
Additional file 3: Figure S2. Verification of triple-gene deletions of cre-1, res-1 and gh1-1 in selected transformants by using Pooled single-crRNA-based CRISPR–Cas12a system (A) or crRNA Array-based CRISPR–Cas12a system (B). PCR analysis of triple-gene deletion of cre-1, res-1 and gh1-1 in selected transformants using one primer (cre1/res1/gh1-1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (cre1/res1/gh1-1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of cre-1, res-1 and gh1-1 were 0.8, 0.7 and 1.9 kb, respectively, while those of WT strain (rightmost lane) was 1.2, 0.9 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair; WT, wild type. U6p, U6 promoter; Ptef1, tef1 promoter; TtrpC, trpC Terminator.
Additional file 4: Figure S3. Verification of triple-gene deletions of gh1-1, cre-1 and res-1 in selected transformants with co-transformation of only three donor-DNAs without CRISPR expressing cassettes. (A) Schematic of homologous recombination (HR) of target genes mediated by each donor DNA. (B) PCR analysis of triple-gene deletion of gh1-1, cre-1 and res-1 in selected 23 transformants using one primer (cre1/gh1-1/res1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (cre1/gh1-1/res1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of gh1-1, cre-1 and res-1 were 1.9, 0.8 and 0.7 kb, respectively, while those of the host strain (rightmost lane) was 1.0, 1.2 and 0.9 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout).
Additional file 5: Figure S4. First round of target genomic editing by CRISPR–Cas9 system. (A) Schematic of homologous recombination (HR) of cre-1, res-1 and gh1-1 mediated by Cas9, sgRNAs and donor DNA. (B) PCR analysis of triple-gene deletion of cre-1, res-1 and gh1-1 in selected transformants using one primer (cre1/res1/gh1-1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (cre1/res1/gh1-1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of cre-1, res-1 and gh1-1 were 0.8, 0.7 and 1.9 kb, respectively, while those of WT strain (rightmost lane) was 1.2, 0.9 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair; WT, wild type.
Additional file 6: Figure S5. Second round of target genomic editing by CRISPR–Cas12a system. (A) Schematic of homologous recombination (HR) of neo, alp-1, rca-1 and hcr-1 mediated by Cas12a, array2 and donor DNA. (B) PCR analysis of quadruple-gene deletion of neo, alp-1, rca-1 and hcr-1 in selected transformants using one primer (gh1-1-out-F2, alp1/rca1/hcr1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (gh1-1-in-R2, alp1/rca1/hcr1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of neo, alp1, rca-1 and hcr-1 were 0.8, 1.6, 5.0 and 0.7 kb, respectively, while those of the host strain (rightmost lane) was 1.9, 1.0, 0.6 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair; symbol star indicated deletion mutant.
Additional file 7: Figure S6. Verification of quadruple-gene deletions of neo, alp-1, rca-1 and hcr-1 in selected transformants with co-transformation of only four donor-DNAs without CRISPR expressing cassettes. (A) Schematic of homologous recombination (HR) of target genes mediated by each donor DNA. (B) PCR analysis of quadruple-gene deletion of neo, alp-1, rca-1 and hcr-1 in selected transformants using one primer (gh1-1-out-F2, alp1/rca1/hcr1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (gh1-1-in-R2, alp1/rca1/hcr1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of neo, alp1, rca-1 and hcr-1 were 0.8, 1.6, 5.0 and 0.7 kb, respectively, while those of the host strain (rightmost lane) was 1.9, 1.0, 0.6 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout).
Additional file 8: Figure S7. Third round of target genomic editing by CRISPR–Cas12a system. (A) Schematic of homologous recombination (HR) of bar, ap-3 and prk-6 mediated by Cas12a, array2 and donor DNA. (B) PCR analysis of triple-gene deletion of bar, ap3 and prk6 in selected 22 transformants using one primer (alp1-out-F2, ap3/prk6-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (alp1-in-R2, gh1-1/res1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of bar, ap-3 and prk-6 were 0.8, 2.0 and 0.8 kb, respectively, while those of the host strain (rightmost lane) was 2.0, 1.2 and 1.2 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair.
Additional file 9: Figure S8. Verification of triple-gene deletions of bar, ap-3 and prk-6 in selected 22 transformants with co-transformation of three donor-DNAs without CRISPR expressing cassettes. (A) Schematic of homologous recombination (HR) of target genes mediated by donor DNA. (B) PCR analysis of triple-gene deletion of bar, ap-3 and prk-6 in selected 22 transformants using one primer (alp1-out-F2, ap3/prk6-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (alp1-in-R2, gh1-1/res1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of bar, ap-3 and prk-6 were 0.8, 2.0 and 0.8 kb, respectively, while those of the host strain (rightmost lane) was 2.0, 1.2 and 1.2 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). HDR, homology-directed repair.
Additional file 10: Figure S9. Second round of target genomic editing by CRISPR–Cas9 system. (A) Schematic of homologous recombination (HR) of neo, alp-1, rca-1 and hcr-1 mediated by Cas12a, array2 and donor DNA. (B) PCR analysis of quadruple-gene deletion of neo, alp-1, rca-1 and hcr-1 in selected transformants using one primer (gh1-1-out-F2, alp1/rca1/hcr1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (gh1-1-in-R2, alp1/rca1/hcr1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of neo, alp1, rca-1 and hcr-1 were 0.8, 1.6, 5.0 and 0.7 kb, respectively, while those of the host strain (rightmost lane) was 1.9, 1.0, 0.6 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). Symbol star indicated deletion mutant. HDR, homology-directed repair.
Additional file 11: Figure S10. Third round of target genomic editing by CRISPR–Cas9 system. (A) Schematic of homologous recombination (HR) of bar, ap-3 and prk-6 mediated by Cas12a, array2 and donor DNA. (B) PCR analysis of triple-gene deletion of bar, ap-3 and prk-6 in selected 22 transformants using one primer (alp1-out-F2, ap3/prk6-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (alp1-in-R2, gh1-1/res1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of bar, ap3 and prk6 were 0.8, 2.0 and 0.8 kb, respectively, while those of the host strain (rightmost lane) was 2.0, 1.2 and 1.2 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair.
Additional file 12: Figure S11. SDS-PAGE of secreted protein from the eight mutant strains and wild type strain (WT) cultured for 6 days in Avicel inducing medium supplemented with 0.75% yeast extract.
Additional file 13: Table S2. List of Cas12a or Cas9 guide and PAM sequences in this study.
Additional file 14: Nucleotide sequence of the Cas12a expression cassette. Purple letters indicate the tef1 (translation elongation factor EF-1, MYCTH_2298136) promoter Ptef1. Blue letters indicate the nuclear localization signal. Red letters indicate the Cas12a gene. Gray letters indicate the TtrpC terminator from A. nidulans trpC gene.
Additional file 15. Nucleotide sequence of the crRNA expression cassettes. Blue letters indicate the RNA polymerase III U6 snRNA promoter. Red letters indicate the 19 nt direct repeat. Green letters indicate the target sequence of amdS, cre-1, res-1, gh1-1, alp-1, neo, rca-1, hcr-1, bar, ap-3, or prk-6, respectively.
Acknowledgements
This study was supported financially by the National Key Research & Developmental Program of China (2018YFA0900500 and 2018YFA0901400), National Natural Science Foundation of China (31771386, 31670042, 31761133018, and 31972878), Chinese Academy of Sciences (XDA21060900) and Youth Innovation Promotion Association of the Chinese Academy of Sciences (Grant No. 2019180).
Abbreviations
- CRISPR
clustered regularly interspaced short palindromic repeats
- Camr technology
CRISPR–Cas-assisted marker recycling technology
- HR
homologous recombination
- sgRNA
single chimeric guide RNA
- crRNA
CRISPR RNA
- tracrRNA
a trans-activating CRISPR RNA
- PAM
protospacer adjacent motif
- DSB
double-strand break
- NHEJ
nonhomologous end joining
- HDR
homology-directed repair
- FAA
fluoroacetamide
- CCR
carbon catabolite repression
- WT
wild type
Authors’ contributions
QL, LZ, and CT designed the project. QL, LZ, and YL carried out the experiments. QL, LZ, JL, and WS analyzed the data. QL and CT wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported financially by the National Key Research & Developmental Program of China (2018YFA0900500 and 2018YFA0901400), National Natural Science Foundation of China (31771386, 31670042, 31761133018, and 31972878), Chinese Academy of Sciences (XDA21060900) and Youth Innovation Promotion Association of the Chinese Academy of Sciences (Grant No. 2019180).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information file.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
A patent involved in genome editing in filamentous fungi was applied by Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qian Liu and Yongli Zhang contributed equally to this work
Contributor Information
Qian Liu, Email: liu_q1@tib.cas.cn.
Yongli Zhang, Email: zhangyl@tib.cas.cn.
Fangya Li, Email: lify@tib.cas.cn.
Jingen Li, Email: li_jg@tib.cas.cn.
Wenliang Sun, Email: sun_wl@tib.cas.cn.
Chaoguang Tian, Email: tian_cg@tib.cas.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13068-019-1637-y.
References
- 1.Glass NL, Schmoll M, Cate JH, Coradetti S. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 2.Kubicek CP, Starr TL, Glass NL. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol. 2014;52:427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- 3.Ward OP. Production of recombinant proteins by filamentous fungi. Biotechnol Adv. 2012;30:1119–1139. doi: 10.1016/j.biotechadv.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Kubicek CP, Kubicek EM. Enzymatic deconstruction of plant biomass by fungal enzymes. Curr Opin Chem Biol. 2016;35:51–57. doi: 10.1016/j.cbpa.2016.08.028. [DOI] [PubMed] [Google Scholar]
- 5.He Y, Wang B, Chen W, Cox RJ, He J, Chen F. Recent advances in reconstructing microbial secondary metabolites biosynthesis in Aspergillus spp. Biotechnol Adv. 2018;36:739–783. doi: 10.1016/j.biotechadv.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Meyer V. Genetic engineering of filamentous fungi-progress, obstacles and future trends. Biotechnol Adv. 2008;26:177–185. doi: 10.1016/j.biotechadv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Liu G, Qu Y. Engineering of filamentous fungi for efficient conversion of lignocellulose: tools, recent advances and prospects. Biotechnol Adv. 2019;37:519–529. doi: 10.1016/j.biotechadv.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR–Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright AV, Nunez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakociunas T, Jensen MK, Keasling JD. CRISPR/Cas9 advances engineering of microbial cell factories. Metab Eng. 2016;34:44–59. doi: 10.1016/j.ymben.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Chen L, Jiang YP, Zhou ZH, Zou G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015;1:15007–15017. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller KK, Chen S, Loros JJ, Dunlap JC. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot Cell. 2015;14:1073–1080. doi: 10.1128/EC.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arazoe T, Miyoshi K, Yamato T, Ogawa T, Ohsato S, Arie T, Kuwata S. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng. 2015;112:2543–2549. doi: 10.1002/bit.25662. [DOI] [PubMed] [Google Scholar]
- 16.Nodvig CS, Nielsen JB, Kogle ME, Mortensen UH. A CRISPR–Cas9 system for genetic engineering of filamentous fungi. PLoS ONE. 2015;10:e0133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuster M, Schweizer G, Reissmann S, Kahmann R. Genome editing in Ustilago maydis using the CRISPR–Cas system. Fungal Genet Biol. 2016;89:3–9. doi: 10.1016/j.fgb.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Katayama T, Tanaka Y, Okabe T, Nakamura H, Fujii W, Kitamoto K, Maruyama J. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotech Lett. 2016;38:637–642. doi: 10.1007/s10529-015-2015-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Meng X, Wei X, Lu L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet Biol. 2016;86:47–57. doi: 10.1016/j.fgb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q, Gao R, Li J, Lin L, Zhao J, Sun W, Tian C. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol Biofuels. 2017;10:1. doi: 10.1186/s13068-016-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nodvig CS, Hoof JB, Kogle ME, Jarczynska ZD, Lehmbeck J, Klitgaard DK, Mortensen UH. Efficient oligo nucleotide mediated CRISPR–Cas9 gene editing in Aspergilli. Fungal Genet Biol. 2018;115:78–89. doi: 10.1016/j.fgb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Zheng X, Zheng P, Zhang K, Cairns TC, Meyer V, Sun J, Ma Y. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger. ACS Synth Biol. 2018 doi: 10.1021/acssynbio.7b00456. [DOI] [PubMed] [Google Scholar]
- 23.Pohl C, Kiel JA, Driessen AJ, Bovenberg RA, Nygard Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth Biol. 2016;5:754–764. doi: 10.1021/acssynbio.6b00082. [DOI] [PubMed] [Google Scholar]
- 24.Foster AJ, Martin-Urdiroz M, Yan X, Wright HS, Soanes DM, Talbot NJ. CRISPR–Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci Rep. 2018;8:14355. doi: 10.1038/s41598-018-32702-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Tao W, Wen S, Li Z, Yang A, Deng Z, Sun Y. In Vitro CRISPR/Cas9 system for efficient targeted DNA editing. MBio. 2015;6:e01714–e01715. doi: 10.1128/mBio.01714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho ND, Arentshorst M, Jin Kwon M, Meyer V, Ram AF. Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl Microbiol Biotechnol. 2010;87:1463–1473. doi: 10.1007/s00253-010-2588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon J, Maruyama J, Kitamoto K. Disruption of ten protease genes in the filamentous fungus Aspergillus oryzae highly improves production of heterologous proteins. Appl Microbiol Biotechnol. 2011;89:747–759. doi: 10.1007/s00253-010-2937-0. [DOI] [PubMed] [Google Scholar]
- 28.Steiger MG, Vitikainen M, Uskonen P, Brunner K, Adam G, Pakula T, Penttila M, Saloheimo M, Mach RL, Mach-Aigner AR. Transformation system for Hypocrea jecorina (Trichoderma reesei) that favors homologous integration and employs reusable bidirectionally selectable markers. Appl Environ Microbiol. 2011;77:114–121. doi: 10.1128/AEM.02100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forment JV, Ramon D, MacCabe AP. Consecutive gene deletions in Aspergillus nidulans: application of the Cre/loxP system. Curr Genet. 2006;50:217–224. doi: 10.1007/s00294-006-0081-2. [DOI] [PubMed] [Google Scholar]
- 30.Florea S, Andreeva K, Machado C, Mirabito PM, Schardl CL. Elimination of marker genes from transformed filamentous fungi by unselected transient transfection with a Cre-expressing plasmid. Fungal Genet Biol. 2009;46:721–730. doi: 10.1016/j.fgb.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Mizutani O, Masaki K, Gomi K, Iefuji H. Modified Cre-loxP recombination in Aspergillus oryzae by direct introduction of Cre recombinase for marker gene rescue. Appl Environ Microbiol. 2012;78:4126–4133. doi: 10.1128/AEM.00080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang DX, Lu HL, Liao X, St Leger RJ, Nuss DL. Simple and efficient recycling of fungal selectable marker genes with the Cre-loxP recombination system via anastomosis. Fungal Genet Biol. 2013;61:1–8. doi: 10.1016/j.fgb.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Twaruschek K, Sporhase P, Michlmayr H, Wiesenberger G, Adam G. New plasmids for Fusarium transformation allowing positive-negative selection and efficient Cre-loxP mediated marker recycling. Front Microbiol. 2018;9:1954. doi: 10.3389/fmicb.2018.01954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khrunyk Y, Munch K, Schipper K, Lupas AN, Kahmann R. The use of FLP-mediated recombination for the functional analysis of an effector gene family in the biotrophic smut fungus Ustilago maydis. New Phytol. 2010;187:957–968. doi: 10.1111/j.1469-8137.2010.03413.x. [DOI] [PubMed] [Google Scholar]
- 35.Bloemendal S, Loper D, Terfehr D, Kopke K, Kluge J, Teichert I, Kuck U. Tools for advanced and targeted genetic manipulation of the beta-lactam antibiotic producer Acremonium chrysogenum. J Biotechnol. 2014;169:51–62. doi: 10.1016/j.jbiotec.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 36.Katayama T, Nakamura H, Zhang Y, Pascal A, Fujii W, Maruyama JI. Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus Aspergillus oryzae. Appl Environ Microbiol. 2019;85:e01896. doi: 10.1128/AEM.01896-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leynaud-Kieffer LMC, Curran SC, Kim I, Magnuson JK, Gladden JM, Baker SE, Simmons BA. A new approach to Cas9-based genome editing in Aspergillus niger that is precise, efficient and selectable. PLoS ONE. 2019;14:e0210243. doi: 10.1371/journal.pone.0210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visser H, Joosten V, Punt PJ, Gusakov AV, Olson PT, Joosten R, Bartels J, Visser J, Sinitsyn AP, Emalfarb MA, et al. Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium lucknowense C1. Ind Biotechnol. 2011;7:10. doi: 10.1089/ind.2011.7.214. [DOI] [Google Scholar]
- 39.Berka RM, Grigoriev IV, Otillar R, Salamov A, Grimwood J, Reid I, Ishmael N, John T, Darmond C, Moisan MC, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 2011;29:922–927. doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- 40.Singh B. Myceliophthora thermophila syn. Sporotrichum thermophile: a thermophilic mould of biotechnological potential. Crit Rev Biotechnol. 2016;36:59–69. doi: 10.3109/07388551.2014.923985. [DOI] [PubMed] [Google Scholar]
- 41.Gu S, Li J, Chen B, Sun T, Liu Q, Xiao D, Tian C. Metabolic engineering of the thermophilic filamentous fungus Myceliophthora thermophila to produce fumaric acid. Biotechnol Biofuels. 2018;11:323. doi: 10.1186/s13068-018-1319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Lin L, Sun T, Xu J, Ji J, Liu Q, Tian C. Direct production of commodity chemicals from lignocellulose using Myceliophthora thermophila. Metab Eng. 2019 doi: 10.1016/j.ymben.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 44.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu M, Lin L, Cheng Y, He X, Sun H, Xie H, Fu J, Liu C, Li J, Chen D, et al. A ‘new lease of life’: FnCpf1 possesses DNA cleavage activity for genome editing in human cells. Nucleic Acids Res. 2017;45:11295–11304. doi: 10.1093/nar/gkx783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun. 2017;8:14406. doi: 10.1038/ncomms14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- 49.Moreno-Mateos MA, Fernandez JP, Rouet R, Vejnar CE, Lane MA, Mis E, Khokha MK, Doudna JA, Giraldez AJ. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun. 2017;8:2024. doi: 10.1038/s41467-017-01836-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swiat MA, Dashko S, den Ridder M, Wijsman M, van der Oost J, Daran JM, Daran-Lapujade P. FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res. 2017;45:12585–12598. doi: 10.1093/nar/gkx1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang JL, Peng YZ, Liu D, Liu H, Cao YX, Li BZ, Li C, Yuan YJ. Gene repression via multiplex gRNA strategy in Y. lipolytica. Microb Cell Fact. 2018;17:62. doi: 10.1186/s12934-018-0909-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang Y, Qian F, Yang J, Liu Y, Dong F, Xu C, Sun B, Chen B, Xu X, Li Y, et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat Commun. 2017;8:15179. doi: 10.1038/ncomms15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun B, Yang J, Yang S, Ye RD, Chen D, Jiang Y. A CRISPR-Cpf1-assisted non-homologous end joining genome editing system of Mycobacterium smegmatis. Biotechnol J. 2018;13:e1700588. doi: 10.1002/biot.201700588. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Wei K, Zheng G, Liu X, Chen S, Jiang W, Lu Y. CRISPR-Cpf1-assisted multiplex genome editing and transcriptional repression in Streptomyces. Appl Environ Microbiol. 2018;84:e00827. doi: 10.1128/AEM.00827-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tak YE, Kleinstiver BP, Nunez JK, Hsu JY, Horng JE, Gong J, Weissman JS, Joung JK. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat Methods. 2017;14:1163–1166. doi: 10.1038/nmeth.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lei C, Li SY, Liu JK, Zheng X, Zhao GP, Wang J. The CCTL (Cpf1-assisted cutting and Taq DNA ligase-assisted Ligation) method for efficient editing of large DNA constructs in vitro. Nucleic Acids Res. 2017;45:e74. doi: 10.1093/nar/gkw1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanegas KG, Jarczynska ZD, Strucko T, Mortensen UH. Cpf1 enables fast and efficient genome editing in Aspergilli. Fungal Biol Biotechnol. 2019;6:6. doi: 10.1186/s40694-019-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwon MJ, Schutze T, Spohner S, Haefner S, Meyer V. Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi. Fungal Biol Biotechnol. 2019;6:15. doi: 10.1186/s40694-019-0079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ebbole D, Sachs MS. A rapid and simple method of isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet Newslett. 1990;37:17–18. [Google Scholar]
- 60.Min K, Ichikawa Y, Woolford CA, Mitchell AP. Candida albicans gene deletion with a transient CRISPR–Cas9 system. mSphere. 2016;1:e00130. doi: 10.1128/mSphere.00130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang MY, Mitchell AP. Marker recycling in Candida albicans through CRISPR–Cas9-induced marker excision. mSphere. 2017;2:e00050. doi: 10.1128/mSphere.00050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vyas VK, Bushkin GG, Bernstein DA, Getz MA, Sewastianik M, Barrasa MI, Bartel DP, Fink GR. New CRISPR Mutagenesis Strategies Reveal Variation in Repair Mechanisms among Fungi. mSphere. 2018;3:e00154. doi: 10.1128/mSphere.00154-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35:789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang B, Cai PG, Sun WL, Li JG, Tian CG, Ma YH. A transcriptomic analysis of Neurospora crassa using five major crop residues and the novel role of the sporulation regulator rca-1 in lignocellulase production. Biotechnol Biofuels. 2015;8:21. doi: 10.1186/s13068-015-0208-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Craig JP, Coradetti ST, Starr TL, Glass NL. Direct target network of the Neurospora crassa plant cell wall deconstruction regulators CLR-1, CLR-2, and XLR-1. MBio. 2015;6:e01452. doi: 10.1128/mBio.01452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan FY, Ma GL, Li JG, Liu Q, Benz JP, Tian CG, Ma YH. Genome-wide analysis of the endoplasmic reticulum stress response during lignocellulase production in Neurospora crassa. Biotechnol Biofuels. 2015;8:66. doi: 10.1186/s13068-015-0248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pei X, Fan F, Lin L, Chen Y, Sun W, Zhang S, Tian C. Involvement of the adaptor protein 3 complex in lignocellulase secretion in Neurospora crassa revealed by comparative genomic screening. Biotechnol Biofuels. 2015;8:124. doi: 10.1186/s13068-015-0302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Druzhinina IS, Kubicek CP. Genetic engineering of Trichoderma reesei cellulases and their production. Microb Biotechnol. 2017;10:1485–1499. doi: 10.1111/1751-7915.12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z, Yao G, Wu R, Gao L, Kan Q, Liu M, Yang P, Liu G, Qin Y, Song X, et al. Synergistic and dose-controlled regulation of cellulase gene expression in Penicillium oxalicum. PLoS Genet. 2015;11:e1005509. doi: 10.1371/journal.pgen.1005509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li JG, Lin LC, Li HY, Tian CG, Ma YH. Transcriptional comparison of the filamentous fungus Neurospora crassa growing on three major monosaccharides d-glucose, d-xylose and l-arabinose. Biotechnol Biofuels. 2014;7:31. doi: 10.1186/1754-6834-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao CZ, Zheng XG, Qu WB, Li GL, Li XY, Miao YL, Han XS, Liu XD, Li ZH, Ma YL, et al. CRISPR-offinder: a CRISPR guide RNA design and off-target searching tool for user-defined protospacer adjacent motif. Int J Biol Sci. 2017;13:1470–1478. doi: 10.7150/ijbs.21312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE. 2014;9(6):e100448. doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. List of PCR primers used in this study.
Additional file 2: Figure S1. Verification of cre-1 gene deletions in selected transformants with co-transformation of only Cas12a and donor DNA, only crRNA and donor DNA, or only donor DNA. (A) Schematic of homologous recombination (HR) of target gene cre-1. (B–D) PCR analysis of cre-1 deletion with one primer (cre1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other (cre1-in-R) located in the 3′ flanking region of genomic DNA. The expected length of disrupted transformants was 1.9 kb, while that of the WT host strain, used as a negative control, was 1.0 kb (rightmost lane). Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). HDR, homology-directed repair; WT, wild type.
Additional file 3: Figure S2. Verification of triple-gene deletions of cre-1, res-1 and gh1-1 in selected transformants by using Pooled single-crRNA-based CRISPR–Cas12a system (A) or crRNA Array-based CRISPR–Cas12a system (B). PCR analysis of triple-gene deletion of cre-1, res-1 and gh1-1 in selected transformants using one primer (cre1/res1/gh1-1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (cre1/res1/gh1-1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of cre-1, res-1 and gh1-1 were 0.8, 0.7 and 1.9 kb, respectively, while those of WT strain (rightmost lane) was 1.2, 0.9 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair; WT, wild type. U6p, U6 promoter; Ptef1, tef1 promoter; TtrpC, trpC Terminator.
Additional file 4: Figure S3. Verification of triple-gene deletions of gh1-1, cre-1 and res-1 in selected transformants with co-transformation of only three donor-DNAs without CRISPR expressing cassettes. (A) Schematic of homologous recombination (HR) of target genes mediated by each donor DNA. (B) PCR analysis of triple-gene deletion of gh1-1, cre-1 and res-1 in selected 23 transformants using one primer (cre1/gh1-1/res1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (cre1/gh1-1/res1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of gh1-1, cre-1 and res-1 were 1.9, 0.8 and 0.7 kb, respectively, while those of the host strain (rightmost lane) was 1.0, 1.2 and 0.9 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout).
Additional file 5: Figure S4. First round of target genomic editing by CRISPR–Cas9 system. (A) Schematic of homologous recombination (HR) of cre-1, res-1 and gh1-1 mediated by Cas9, sgRNAs and donor DNA. (B) PCR analysis of triple-gene deletion of cre-1, res-1 and gh1-1 in selected transformants using one primer (cre1/res1/gh1-1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (cre1/res1/gh1-1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of cre-1, res-1 and gh1-1 were 0.8, 0.7 and 1.9 kb, respectively, while those of WT strain (rightmost lane) was 1.2, 0.9 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair; WT, wild type.
Additional file 6: Figure S5. Second round of target genomic editing by CRISPR–Cas12a system. (A) Schematic of homologous recombination (HR) of neo, alp-1, rca-1 and hcr-1 mediated by Cas12a, array2 and donor DNA. (B) PCR analysis of quadruple-gene deletion of neo, alp-1, rca-1 and hcr-1 in selected transformants using one primer (gh1-1-out-F2, alp1/rca1/hcr1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (gh1-1-in-R2, alp1/rca1/hcr1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of neo, alp1, rca-1 and hcr-1 were 0.8, 1.6, 5.0 and 0.7 kb, respectively, while those of the host strain (rightmost lane) was 1.9, 1.0, 0.6 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair; symbol star indicated deletion mutant.
Additional file 7: Figure S6. Verification of quadruple-gene deletions of neo, alp-1, rca-1 and hcr-1 in selected transformants with co-transformation of only four donor-DNAs without CRISPR expressing cassettes. (A) Schematic of homologous recombination (HR) of target genes mediated by each donor DNA. (B) PCR analysis of quadruple-gene deletion of neo, alp-1, rca-1 and hcr-1 in selected transformants using one primer (gh1-1-out-F2, alp1/rca1/hcr1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (gh1-1-in-R2, alp1/rca1/hcr1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of neo, alp1, rca-1 and hcr-1 were 0.8, 1.6, 5.0 and 0.7 kb, respectively, while those of the host strain (rightmost lane) was 1.9, 1.0, 0.6 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout).
Additional file 8: Figure S7. Third round of target genomic editing by CRISPR–Cas12a system. (A) Schematic of homologous recombination (HR) of bar, ap-3 and prk-6 mediated by Cas12a, array2 and donor DNA. (B) PCR analysis of triple-gene deletion of bar, ap3 and prk6 in selected 22 transformants using one primer (alp1-out-F2, ap3/prk6-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (alp1-in-R2, gh1-1/res1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of bar, ap-3 and prk-6 were 0.8, 2.0 and 0.8 kb, respectively, while those of the host strain (rightmost lane) was 2.0, 1.2 and 1.2 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair.
Additional file 9: Figure S8. Verification of triple-gene deletions of bar, ap-3 and prk-6 in selected 22 transformants with co-transformation of three donor-DNAs without CRISPR expressing cassettes. (A) Schematic of homologous recombination (HR) of target genes mediated by donor DNA. (B) PCR analysis of triple-gene deletion of bar, ap-3 and prk-6 in selected 22 transformants using one primer (alp1-out-F2, ap3/prk6-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (alp1-in-R2, gh1-1/res1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of bar, ap-3 and prk-6 were 0.8, 2.0 and 0.8 kb, respectively, while those of the host strain (rightmost lane) was 2.0, 1.2 and 1.2 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). HDR, homology-directed repair.
Additional file 10: Figure S9. Second round of target genomic editing by CRISPR–Cas9 system. (A) Schematic of homologous recombination (HR) of neo, alp-1, rca-1 and hcr-1 mediated by Cas12a, array2 and donor DNA. (B) PCR analysis of quadruple-gene deletion of neo, alp-1, rca-1 and hcr-1 in selected transformants using one primer (gh1-1-out-F2, alp1/rca1/hcr1-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (gh1-1-in-R2, alp1/rca1/hcr1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of neo, alp1, rca-1 and hcr-1 were 0.8, 1.6, 5.0 and 0.7 kb, respectively, while those of the host strain (rightmost lane) was 1.9, 1.0, 0.6 and 1.0 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). Symbol star indicated deletion mutant. HDR, homology-directed repair.
Additional file 11: Figure S10. Third round of target genomic editing by CRISPR–Cas9 system. (A) Schematic of homologous recombination (HR) of bar, ap-3 and prk-6 mediated by Cas12a, array2 and donor DNA. (B) PCR analysis of triple-gene deletion of bar, ap-3 and prk-6 in selected 22 transformants using one primer (alp1-out-F2, ap3/prk6-out-F) located upstream of the 5′ flanking region of genomic DNA and the other primer (alp1-in-R2, gh1-1/res1-in-R) located in the 3′ flanking region of genomic DNA. The expected lengths of disrupted transformants of bar, ap3 and prk6 were 0.8, 2.0 and 0.8 kb, respectively, while those of the host strain (rightmost lane) was 2.0, 1.2 and 1.2 kb, respectively. Heterokaryotic transformants showed two PCR bands (both of wild-type and knockout). The symbol of star indicated deletion mutant. HDR, homology-directed repair.
Additional file 12: Figure S11. SDS-PAGE of secreted protein from the eight mutant strains and wild type strain (WT) cultured for 6 days in Avicel inducing medium supplemented with 0.75% yeast extract.
Additional file 13: Table S2. List of Cas12a or Cas9 guide and PAM sequences in this study.
Additional file 14: Nucleotide sequence of the Cas12a expression cassette. Purple letters indicate the tef1 (translation elongation factor EF-1, MYCTH_2298136) promoter Ptef1. Blue letters indicate the nuclear localization signal. Red letters indicate the Cas12a gene. Gray letters indicate the TtrpC terminator from A. nidulans trpC gene.
Additional file 15. Nucleotide sequence of the crRNA expression cassettes. Blue letters indicate the RNA polymerase III U6 snRNA promoter. Red letters indicate the 19 nt direct repeat. Green letters indicate the target sequence of amdS, cre-1, res-1, gh1-1, alp-1, neo, rca-1, hcr-1, bar, ap-3, or prk-6, respectively.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information file.