Abstract
Bjarnsholt T, Buhlin K, Dufrenê YF, Gomelsky M, Moroni A, Ramstedt M, Rumbaugh KP, Schulte T, Sun L, Åkerlund B, Römling U (University of Copenhagen; Copenhagen University Hospital, Copenhagen, Denmark; Karolinska Institutet, Huddinge, Sweden; Université catholique de Louvain, Louvain-la-Neuve, Belgium; University of Wyoming, Laramie, Wyoming, USA; Universitá degli Studi di Milano, Milano, Italy; Umeå University, Umeå, Sweden; Texas Tech University Health Sciences Center, Lubbock, Texas, USA; Karolinska Institutet, Karolinska University Hospital, Stockholm, Sweden). Biofilm formation – what we can learn from recent developments.
Although biofilms have been observed early in the history of microbial research, their impact has only recently been fully recognized. Biofilm infections, which contribute to up to 80% of human microbial infections, are associated with common human disorders, such as diabetes mellitus and poor dental hygiene, but also with medical implants. The associated chronic infections such as wound infections, dental caries and periodontitis significantly enhance morbidity, affect quality of life and can aid development of follow-up diseases such as cancer. Biofilm infections remain challenging to treat and antibiotic monotherapy is often insufficient, although some rediscovered traditional compounds have shown surprising efficiency. Innovative anti-biofilm strategies include application of anti-biofilm small molecules, intrinsic or external stimulation of production of reactive molecules, utilization of materials with antimicrobial properties and dispersion of biofilms by digestion of the extracellular matrix, also in combination with physical biofilm breakdown. Although basic principles of biofilm formation have been deciphered, the molecular understanding of the formation and structural organization of various types of biofilms has just begun to emerge. Basic studies of biofilm physiology have also resulted in an unexpected discovery of cyclic dinucleotide second messengers that are involved in interkingdom crosstalk via specific mammalian receptors. These findings even open up new venues for exploring novel anti-biofilm strategies.
Keywords: antimicrobial strategies, biofilm formation, cyclic di-nucleotide second messengers, extracellular matrix, underlying diseases
Introduction
The 60th Nobel Conference on Biofilm formation – its clinical impact and potential treatment, was held at the Nobel Forum, Karolinska Institutet in August, 2013 [1]. In May 2015, a national follow-up conference covering different aspects of biofilm research was organized in the Birke aula at Karolinska Institutet, Campus Flemingsberg. The topics of the meeting included the impact of biofilm-associated diseases in clinical settings, the structures of biofilms, biofilm regulation, development of new tools for biofilm research and novel anti-biofilm strategies. While biofilm formation is often described as the natural mode of microbial growth, it is clear that we still have a lot to learn about this unique microbial sessile life style. Much of the microbiological research during the last century has focused on the investigation of the planktonic lifestyle of microorganisms. This has been without doubt a tremendously successful endeavour and unravelled basic principles of metabolism and physiology. The challenge now is to decipher the specific features of the underresearched biofilm lifestyle, because its ubiquitous nature and its importance in the clinic. Biofilm communities appear to be more complex than planktonic cultures. Not only is the cell physiology different, but any biofilm is composed of cells with diverse physiology one extreme being represented by dormant, metabolically silent, and persister cells. Therefore, new experimental tools that have a much higher resolution than previously anticipated and new physiologically relevant models are necessary to fully understand the high complexity of biofilms.
Diagnosis of biofilm infections
Observations of microbial biofilm formation by van Leeuwenhook, Pasteur and other pioneers date back to the roots of microbiology and infectious disease research [2]. The impact of a foreign implant on the infection process was already recognized in 1956 as the presence of a foreign material required a 7.5×104 lower dose of Staphylococcus aureus to cause a subcutaneous infection in humans which even then did not resolve [3]. This early study demonstrated major hallmarks of many biofilm infections: the presence of a foreign body, which promotes a persistent infection with a low level of inflammation that subsequently leads to tissue destruction. Such were hard to resolve necrotizing soft tissue infections caused by group A Streptococcus pyogenes, usually a rapidly progressing acute infection, recently observed to be associated with in situ biofilm formation, accompanied by a higher bacterial load and an elevated immune response [4].
The 1956 study also already pointed out biofilm-associated infections to be characterized by spatially restricted microbial persistence, which causes a localized immune response. The basic concept of deleterious biofilms as microbial colonization of the ‘wrong type of bacteria at the wrong place’, can be even more dramatically exemplified in the arising association between biofilms and the development of certain cancers. Recently gathered evidence indicates the unusual tumour anaplastic large cell lymphoma, associated with breast implants, to be connected with a Ralstonia dominated microbiome on the implants [5]. Head and neck cancer is associated with hygiene-dependent alterations in oral biofilm formation, exaggerated by risk factors such as smoking and alcohol consumption, which negatively affect health-associated biofilm properties and thus contribute to the etiology of tumours [6]. Furtheron, co-colonization by two toxin-producing bacteria promotes early onset of polyp formation in individuals with familial predisposition for adenomatous polyposis [7].
Thus, challenges in the diagnosis of ‘under radar’ infections caused by biofilms include not only the choice of sample and the sampling procedure, identification of the organism, and, eventually, visualization of the usually small biofilm foci [8], but are to be extended to the microbial composition in biofilms [5–7]. The European Society for Clinical Microbiology and Infectious Diseases has compiled guidelines to reliably diagnose some of the major biofilm infections as well as has identified research needs to improve diagnosis, treatment and prevention of biofilm infections [8].
Treatment strategies for biofilm infections
Although some antibiotics such as rifampicin for Gram-positive bacteria and fluoroquinolones for Gram-negative bacteria, show a superior ability to counteract biofilms [6], a total eradication of a biofilm infection is still a treatment challenge. For example, an established Pseudomonas aeruginosa lung infection in cystic fibrosis patients cannot be eradicated, even following intensive year-long local and systemic antibiotic therapy. Thus, during a 20 year long infection, daily inhalation of tobramycin/colistin and regular 2-week antimicrobial therapy results in the consumption of 1 kg tobramycin, 1 kg colistin and 10 kg of beta-lactam antibiotic [9], without effective eradication of the biofilm-forming pathogens. A consensus agreement exists that only a combination therapy can potentially eliminate biofilm infections [9, 10]. Thereby, treatment with the last resort polymyxin antibiotic colistin is more effective compared to many other antibiotics under anaerobic conditions [11]. In vitro treatment of biofilms, though, induces the formation of a subpopulation of colistin-tolerant cells that can subsequently be killed combining colistin with erythromycin, ciprofloxacin or tetracycline [12–14].
On the other hand, when revisited, widely used natural antimicrobial compounds demonstrated a broad efficacy against biofilm-forming microorganisms ([14–17]; Table 1). For example, the established antimicrobial preservative acetic acid is highly effective in the treatment of chronic wounds infected with biofilm-forming P. aeruginosa [18]. The application of acetic acid might be extended to combination treatment of other biofilm-associated diseases including prosthetic joint infection [18]. Another strategy exploits the antibiofilm and immunomodulatory properties of naturally occurring antimicrobial peptides to develop effective anti-biofilm agents with or without antiinflammatory properties in combination with established antibiotic treatment [19, 20]. The mechanical dispersion of a mature biofilm can already significantly enhance antimicrobial susceptibility. Therefore, the efficiency of antibiotic treatment can be increased by a combination therapy with biofilm matrix degrading enzymes such as DNases, proteases and glycoside hydrolases ([20, 21]; Table 1). The applicability of antimatrix and sequestration molecules has been recently reviewed [20].
Table 1.
Anti-biofilm approaches discussed in the text
| Anti-biofilm approach | Optimization | Anti-biofilm effect | Potential application | References |
|---|---|---|---|---|
| Antimicrobial surface material | State-of-the-art machine learning | Avoidance of cell attachment and biofilm formation on artificial surfaces | Artificial devices such as urinary catheters, pacemakers, cochlear implants | [68] |
| DNases, proteases, glycosidase | Enzyme cocktail adapted to the extracellular matrix components specific to the biofilm | Dispersion of the biofilm by destruction of the extracellular matrix in combination with antimicrobials | Skin and wound biofilm, cystic fibrosis lung infection | [20] |
| Application of antimicrobial peptides | Enhancement of the antibiofilm effect while diminishing the pro-inflammatory effect | Dispersion of established biofilms and avoidance of biofilm formation on artificial surfaces | Artificial devices such as urinary catheters | [19, 20] |
| Photodynamic therapy | Use of high energy blue antimicrobial light | Targeting of photoreactive compounds to create reactive oxygen and nitrogen species | Skin biofilm, wound infection | [27] |
| Atmospheric cold plasma | Production of reactive oxygen and nitrogen species | Reactive oxygen and nitrogen species target lipids, proteins and other macromolecules | Wound infection | [28] |
| Low frequency ultrasonic therapy | To optimize amplitude, frequency, contact time, instrument configuration | Physical intervention to disperse the biofilm with mechanical energy to reach an anti-microbial effect in combination with chemical therapy | Wound infection, periprosthetic joint infection | [29–31] |
| Acetic acid | Combination treatment | Traditional effective antimicrobial, in use to treat infections like Swimmer’s ear and chronic wounds | Other types of biofilm infections like prosthetic joint infection | [18] |
| Nitric oxide, NO | Dose-dependent antimicrobial and antibiofilm effect | Innate immune and signalling molecule that is sensed by sensory protein domains to subsequently induce biofilm dispersion in a broad range of bacteria | Inhalation to treat cystic fibrosis lung infection caused by P. aeruginosa | [16, 100] |
| Hypochlorous acid, HOCl | Innate antimicrobial agent in combination with physical interventions | Peroxidase-generated anti-bacterial innate immune molecule | Wound infection | [17, 101] |
A major hurdle for a rationalized strategy for dispersal by biofilm matrix hydrolysis is the lack of a detailed understanding of the composition of the extracellular matrix (ECM) and the arrangement of macromolecules in clinically relevant biofilms. Small molecules can aid in the characterization of the composition and arrangement of the ECM. Those molecules can either interact selectively with common components of the ECM of biofilms or bind to different unrelated matrix molecules to cause specific spectral shifts. Established compounds that have been used to characterize the ECM of biofilms are, for example, thioflavin T, which binds to amyloid fibers and Calcofluor white (fluorescence brightener 28), which binds to 1→3 and 1→4 β-glucans such as the exopolysaccharide cellulose. On the other hand, Congo red can bind to both, exopolysaccharides and amyloid fibers [22–25]. In the search for alternative compounds, luminescent conjugated oligothiophene derivatives were recently identified through compound library screening approaches to selectively interact with curli amyloid fibres and cellulose, respectively [26]. If successfully broad-ened, this approach could aid the in-depth characterization of biofilm formation in vitro and in vivo.
Several biophysical approaches for the treatment of biofilm infections have been implemented. One of them involves photodynamic therapy, the application of light, in particular blue light, to activate photosensitive compounds such as (proto)porphyrins, ubiquitously present in bacterial organisms (Table 1). The FDA approved therapeutic blue light has a high energy and is effectively absorbed by photosensitive molecules. As such, blue light causes a substantial decrease in cell viability in clinically significant biofilms in vitro and in vivo due to the powerful cytotoxic effect based on electron transfer leading to the creation of reactive oxygen species [27]. Chemically reactive species are also effective as anti-biofilm agents in the external application of atmospheric cold plasma against biofilms ([28]; Table 1). Low frequency ultrasonic therapy, clinically applied to improve chronic wounds, has been shown effective against biofilm-forming microorganisms in experimental infections, probably through the generation of mechanical forces [29–31]. It is worth noting, though, that any successful antimicrobial therapy needs to be supported by a functional immune system to eradicate the foci of infection. Indeed, the future challenge is not only to develop successful strategies to eradicate biofilm infections by antimicrobial therapy, but also to aid a self-healing immune response that is compromised in many patients (see below; [32]).
Prevalent biofilm infections
Skin and subcutaneous infections, including burn wounds, surgical-site infections and nonhealing pressure, venous, arterial and diabetic ulcers are common health problems in industrialized and developing countries that pose a significant economical burden for the society. Chronically infected diabetic foot ulcer, often associated with polymicrobial biofilm formation, is considered the most significant wound care problem in the world affecting up to 25% of diabetic individuals at least once in their lifetime [33]. As diabetes is increasing especially in developing countries [34], the problem of diabetic foot infection with a potential risk for amputation is rapidly growing, with challenges in diagnosis and treatment [35]. Investigation of the microbial wound ecology has revealed that host factors substantially modulate the infection progress promoting a unique biofilm ecology. Under laboratory conditions, P. aeruginosa can kill S. aureus in mixed culture. In the wound, however, albumin at physiological levels promotes co-colonization of these two most prevalent wound pathogens preventing killing of S. aureus by P. aeruginosa (Fig. 1a; [36, 37]). Further on, P. aeruginosa and S. aureus co-colonization enhances tolerance to some antibiotics [37]. On the other hand, insulin treatment, which has known immunomodulatory effects, enhances P. aeruginosa biofilm formation and antibiotic tolerance [38]. In addition to chronic wounds, diabetes patients have been observed to harbour bacterial biofilms in arterial artherosclerotic plaques, which are considered to be a major factor compromising circulation and negatively affecting disease progression [38]. Initial characterization of the physiology of the major wound pathogen P. aeruginosa by transcriptome profiling and genome-wide mutant fitness profiling by Tn-seq revealed in vivo metabolic requirements such as long chain fatty acids as a major carbon source. In these studies, chemotactic flagellar motility was identified as a major contributor to virulence in acute, but not in chronic wound infections [39].
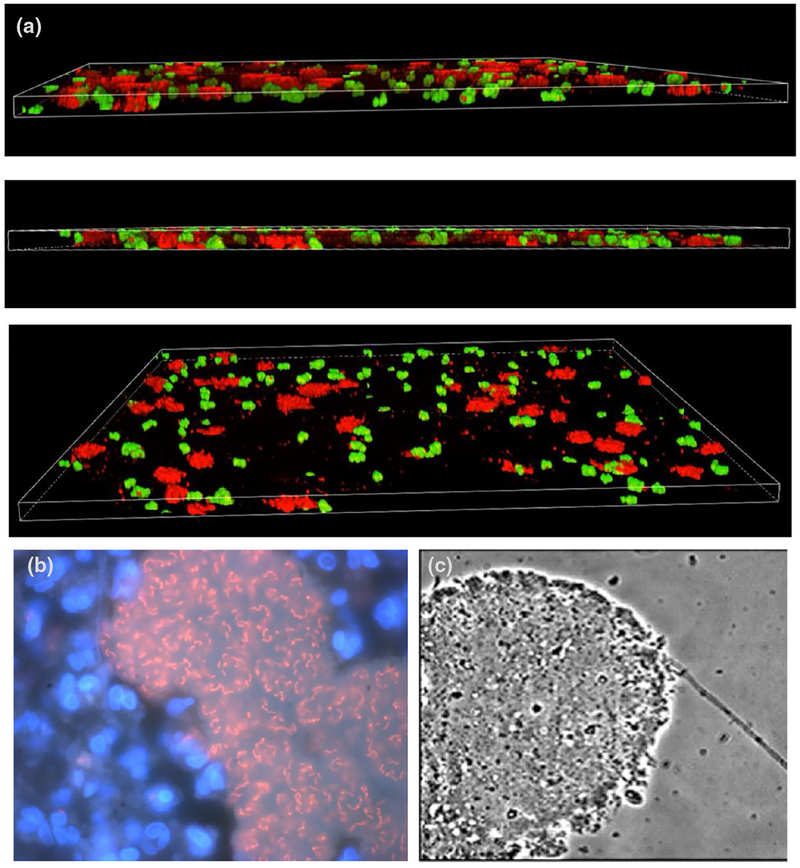
Fig. 1.
(a) P. aeruginosa (red) and S. aureus (green) grown together in a ‘wound-like’ media that supports their co-culture as patchy distinct microcolonies. A three-dimension segment of the medium is shown from different angles. Image by Cody Fell (Rumbaugh laboratory), unpublished. (b) Visualization of P. aeruginosa biofilms in a CF lung of a CF male, 41 years of age, chronic P. aeruginosa mucoid and nonmucoid infection for 28 years, 46 precipitating antibodies, 114 2-week anti-P. aeruginosa treatment courses. Intraluminal P. aeruginosa biofilms surrounded by polymorphonuclear leukocytes (PMNs) visualized using peptide nucleic acid – fluorescence in situ hybridization (PNA-FISH) and DNA staining with 4′, 6-Diamidin-2-phenylindol, (DAPI). Adapted from [9]. (c) P. aeruginosa grown in artificial sputum medium. Adapted from [95] with permission.
The oral biofilm is one of the most complex microbial community in the human body. Over 700 species have been estimated to contribute to dental plaque biofilm formation, which have been classified into ‘so-called’ colour-coded complexes on the basis of sequential colonization in combination with their impact on oral health [40]. Oral diseases such as dental caries, gingivitis, periodontitis and periimplantitis are among the most common biofilm diseases [41, 42]. Periodontitis, a chronic, tissue-destructive inflammation, which degrades the attachment apparatus of the teeth, can cause tooth loss and, in its most severe form, edentulousness, the complete loss of all natural teeth. Microbial plaque (biofilm) forms on hard and soft tissues, first supragingivally then subgingivally with mainly the bacterial pathogens of the orange and red complex that strongly associate with periodontitis such as Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Treponema denticola as etiological agents [43]. This subgingival biofilm formation, which brings a massive bacterial load close to the blood vessels, in combination with inflammation and inflammatory mediators in the periodontium, is associated with a range of chronic diseases [44, 45]. As such, oral health does not only have an impact on the oral cavity, but can be involved in the etiology of various diseases such as cardiovascular diseases e.g. endocarditis and stroke and systemic inflammatory diseases such as rheumatoid arthritis [46, 47].
Elucidation of the molecular basis of biofilm formation
Major regulatory circuits controlling biofilm formation have been unravelled and a range of biofilm-associated extracellular matrix (ECM) components discovered in well-established in vitro and in vivo models (Fig. 1b, c; [48, 49]). ECM components such as exopolysaccharides, proteinaceous pili/fimbriae, other protein components, nucleic acids and lipids are most often stabilized through inter-molecular networks involving specific binding proteins or networking between ECM components [22, 50, 51].
The impact of extracellular DNA (eDNA) for the integrity of biofilm structures was discovered as DNase treatment dissolved early and established biofilms [52, 53]. Thereby, choline-binding proteins can act as positively charged bridges between eDNA and the cell surface and so can cytoplasmic proteins, released and surface exposed in a ‘moon-lighting’, second independent, function [54, 55].
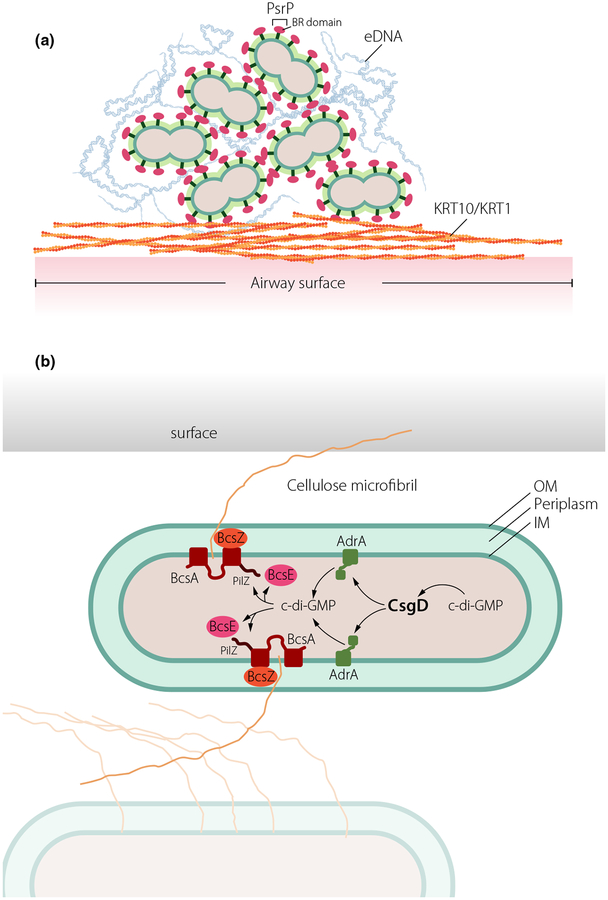
The surface-associated Pneumococcal Serine Rich repeat Protein PsrP, a member of the serine rich repeat protein (SRRP) family, exemplifies one eDNA-receptor that promotes cell clumping and biofilm formation (Fig. 2a). While the C-terminal serine rich-repeat (SRR) region is anchored in the capsular surface, the unique functional binding region (BR) domain mediates adherence to lung associated keratin 10 and promotes biofilm formation [56, 57]. The crystal structure of the BR domain of PsrP revealed a novel variant of the DEv-IgG fold, typical for microbial surface components recognizing adhesive matrix molecule (MSCRAMM) adhesins [58]. The BR domain weakly associates into a β-sheet dimer resembling a molecular saddle with a highly basic concave under-surface that snuggly fits the acidic helical structures of eDNA or keratin 10 [58]. However, molecular structures of BR in complex with its ligands are required to confirm the suggested binding models.
Fig. 2.
Novel molecular mechanisms involved in regulation of biofilm formation. (a) The giant protein PsrP of Streptococcus pneumoniae promotes colonization in the airways through multiple binding events. PsrP, via its BR domain located outside of the capsule, binds to biofilm-associated eDNA and adheres to surface accessible keratin (KRT) 10/KRT 1 as well as self-associates via BR domains. (b) The cellulase BcsZ located in the periplasm regulates cellulose production independently of cyclic di-GMP in S. typhimurium. Cyclic di-GMP (c-di-GMP) regulates expression of csgD encoding the major biofilm regulator. Subsequently, csgD activates transcription of the diguanylate cyclase AdrA to synthesize the cyclic di-GMP involved in production of the exopolysaccharide cellulose via binding to the cyclic di-GMP receptors BcsE and a PilZ domain at the C-terminal end of the cellulose synthase BcsA.
A common ECM component of enterobacterial biofilms is the exopolysaccharide cellulose [59]. Thereby, the cellulose biosynthesis operon is present not only in environmental species, but also in human pathogens [60]. Cyclic di-GMP signalling (see below) was identified as a major post-translational regulatory mechanism leading to the activation of cellulose biosynthesis [60]. BcsZ, a cellulase of glycoside hydrolase (GH) family 8 is associated with cellulose biosynthesis operons, however, its function remains controversial. In the gastrointestinal pathogen Salmonella enterica serovar Typhimurium, in accordance with its enzymatic functionality, BcsZ negatively regulates cellulose biosynthesis [61]. Thereby, the periplasmic protein inversely regulates the major bacterial life styles biofilm formation and motility. Strikingly, although regulation of cellulose biosynthesis was not obvious under laboratory conditions at body temperature, bcsZ was required for efficient establishment of an infection in the mouse model of typhoid fever. Key virulence phenotypes of S. Typhimurium such as organ colonization and uptake and proliferation in macrophages were positively regulated by bcsZ. Most of the phenotypes mediated by bcsZ were relieved upon deletion of the cellulose synthase BcsA and/or the major biofilm activator CsgD, which indirectly regulates cellulose expression through activation of the diguanylate cyclase AdrA (Fig. 2b). Thus bcsZ effectively downregulates csgD mediated cellulose biosynthesis to enable Salmonella to efficiently cause acute infection [61]. Consequently, the BcsZ cellulase is the example of a periplasmic enzyme involved in the adjustment of biofilm formation versus motility and virulence downstream of major cytoplasmic biofilm hubs such as the cyclic di-GMP signaling system. Such a multilayer regulation might also occur in other bacteria, which produce cellulose as an extracellular matrix component of biofilms.
Elucidation of biofilm properties
Epithelial surfaces in the human body are very efficient in keeping microorganisms under surveillance. Artificial medical devices, however, which are obviously devoid of major innate immune defense mechanisms, become readily colonized by microbes. A prominent example is urinary tract catheters, which have a colonization probability of 5–10% per day. Experts working in networks such as “ipromedai”, “improved protection of medical devices against infection” (www.ipromedai.net), are developing novel antimicrobial and biocompatible surfaces. Biofilm formation on surfaces is multifactorial, influenced by the particular strain, the growth medium and, last but not least, surface characteristics. As such, the capability to form a biofilm, biofilm ECM components and the regulatory biofilm network can vary substantially between clinical isolates of the same species [62]. This biological variability has to be addressed in the development of anti-biofilm strategies. For example, the biofilm formed by Pseudomonas aeruginosa on a catheter surface in artificial urine is composed of a specific type of extracellular matrix partially dependent on eDNA [63]. This contrasts biofilm formation under alternative growth conditions where alginate, PEL and PSL exopolysaccharides are major extracellular matrix components. Remarkably, though, the second messenger cyclic di-GMP (see below) directs biofilm formation even under these substantially different environmental conditions [64] making this molecule a general anti-biofilm target. Surface characteristics that determine attachment and subsequent biofilm formation are hydrophobicity and surface charge; two parameters modulated, for example, by the application of polymer brushes. Thereby, a polycationic surface can dramatically enhance, while an anionic and zwitterionic surface can dramatically decrease biofilm formation, although leaving dense macrocolonies, roughly concomitant with the surface contact angle [65]. Furthermore, irregularities on a surface and surface patterning can affect biofilm formation [66]. Unconventional inclusion of anti-microbial substances in innovative surfaces also reduces bacterial adhesion [67]. Recently developed machine learning approaches might accelerate the design of effective antimicrobial surfaces [68].
It has to be pointed out, though, that a successful microbe-free implantation is not only determined by the implant surface characteristics, but also by the health status of the patient and disease parameters. For example, smoking, severe obesity and diabetes are well-known risk factors for post-operative infections, which automatically lead to an increase in the foreign body infection rate [69–71]. As such, the rate of infection can vary dramatically from 0.75% to 35% depending on implant characteristics and the patients’ health status.
Better understanding of biofilm properties requires advanced imaging technologies. The resolution of light microscopy is limited by the wavelength of light. The development of different live-cell nanoscopy techniques such as stimulated-emission depletion (STED) microscopy has overcome these physical constraints and thus has revolutionized the exploration of living cells at molecular resolution [72, 73]. In microbiology, nanoscopy has been applied, for example, to investigate the nucleoid structure and associated machineries [74]. Another technical development, atomic force microscopy (AFM), has facilitated new opportunities for imaging and manipulation of biological systems at the level of single cells and molecules [74, 75]. Originally developed for the scanning of technical surfaces, in force production between the tip and the surface enables AFM to unravel the surface architecture of single live cells at nanoscale in real time. The self-assembly of S-layer proteins and the effect of antimicrobial substances on cell walls has been observed with AFM [76]. AFM has also been used to investigate physical properties of biofilm communities such as an inverse correlation between the elasticity and cellulose production of an Aliivibrio fischeri biofilm [77]. Beyond imaging, AFM has been developed to probe forces in single-molecule spectroscopy to understand processes such as adhesion, unfolding and sugar recognition. Single-cell force spectroscopy is also used to assess mechanisms of cell adhesion. These analyses complement traditional methods to analyse the gene-function relationship of microbial cells at the nanoscale and structure-function relationships in the biological context [74].
Translational applications – from biofilms to treatment
Although first described as an allosteric regulator of the cellulose synthase, the bacterial cyclic dinucleotide second messenger cyclic di-GMP was early on suspected to be involved in interkingdom crosstalk [78, 79]. These first studies described effects of cyclic di-GMP on the retardation of growth of cancer cell and preliminary identified eukaryotic receptors. A number of follow-up studies have demonstrated that cyclic di-GMP and subsequently identified bacterial cyclic di-nucleotide second messengers indeed cause broad physiological responses, including the activation of a multifactorial innate and adaptive immune response [80, 81]. Furthermore, experimental data indicate that cyclic di-GMP and its nonhydrolizable analogues can efficiently induce shrinkage of tumours [82]. However, what are the molecular mechanisms by which cyclic di-GMP and other bacterial cyclic di-nucleotide second messengers cause such a broad response?
The most prominent cyclic di-GMP receptor is the immune adaptor STING that binds cyclic di-GMP with a dissociation constant of 5 lM and triggers a downstream signalling cascade with relocalization of STING to stimulate type 1 interferon production [83]. Indeed STING also binds the bacterial second messengers cyclic di-AMP and the cyclic 3′−3′ GAMP hybrid, but the physiological substrate is the intrinsic innate immune second messenger cyclic 2′−3′-GAMP [84]. Subsequent identification of additional human cyclic di-GMP receptors (Table 2) starts to explain the multi-faceted effects of cyclic di-nucleotide signalling that extend beyond the activation of the immune system and aid unravelling an intricate crosstalk between microbial and metazoan signalling systems.
Table 2.
Mammalian cyclic di-GMP receptors and their features
| Receptor | Specificity | Affinity | Function | C-dinucleotide effect | References |
|---|---|---|---|---|---|
| STimulator of INterferon Genes, STING | c-di-GMP, c-di-AMP, 3′3′-cGAMP, 2′3′-cGAMP | Kd = 5 μM (c-di-GMP) to C-terminal domain; 1:2 stoichiometry; Kd = 61 nM (2′3′-cGAMP) | Innate immune adaptor | type 1 IFN stimulation; some STING alleles have lost binding capacity | [102] |
| Helicase DDX41 | c-di-GMP; c-di-AMP | Kd = 5.7 μM (c-di-GMP) | Interacts with STING; STING activation | type 1 IFN stimulation | [103] |
| Hyperpolarization-activated cyclic nucleotide-gated channel 4 (HCN4) | c-di-GMP, c-di-AMP, 2′3′ and 3′3′-cGAMP | Ka = 1.8 μM (c-di-GMP) | Nonselective ligand-gated cation channel in heart and brain | Counteracts cAMP dependent HCN channel open probability, reduces heart rate | [86] |
| A2a adenosine receptor | c-di-AMP, 3′3′-cGAMP | IC50 = 2.5 μM (c-di-AMP) | Monocyte Gαs-coupled adenosine receptor | Apoptosis | [86] |
| Human siderocalin LCN2 | c-di-GMP | Kd = 1.63 μM | Siderophore receptor | Competition with ferric siderophores, antagonizes antibacterial activity | [104] |
| RECON | c-di-AMP, 3′3′-cGAMP | Kd = 87 nM (c-di-AMP) | Oxidoreductase; aldo-keto reductase family 1; member C13 (AKR1C13) | Inhibition of enzymatic activity; NF-κB activation; reduced bacterial survival | [105] |
| ERAdP | c-di-AMP | Kd = 76 nM | Endoplasmatic reticulum membrane adaptor | Promotes pro-inflammatory cytokine production through NF- κB | [106] |
In an unexpected twist of events, the hyperpolarization-activated cyclic nucleotide gated (HCN) ion channel 4 was identified as a cyclic di-GMP responsive protein. HCN4 ion channels are the molecular determinants of the cardiac pacemaker current responsible for the automaticity of the heart [85]. In the physiological setting, the autonomic regulation of the heart rate is affected by direct binding of cAMP to HCN channels, which increases channel current. Binding of cAMP to the cyclic nucleotide-binding domain (CNBD) of HCN channels subsequently promotes a conformational change that is transmitted via the C-linker to the pore and enhances the probability of channel opening. Unexpectedly, the C-linker contains a binding pocket for the bacterial and mammalian cyclic dinucleotides [86] that prevent cAMP modulation of the current in HCN4 and leads to heart rate reduction in mouse myocytes by about 30%. These and other findings indicate that cyclic di-GMP and other microbial derived cyclic (di) nucleotides can have a much wider impact on human physiology than what has been previously appreciated. It is therefore possible that these kind of second messenger molecules can have a therapeutic potential comparable to small chain fatty acids produced abundantly by the gut microbiome [87].
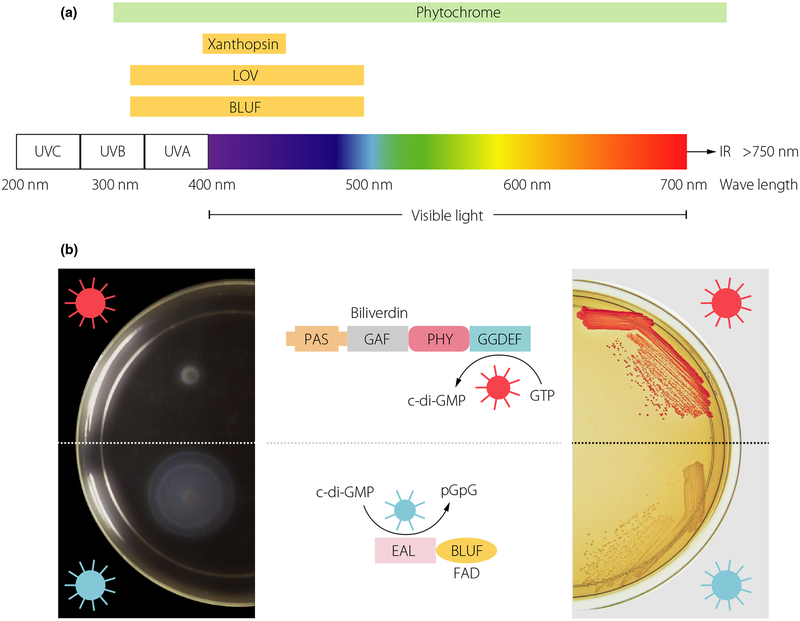
In its genuine context, the bacterial second messenger cyclic di-GMP plays a determinative role in motile-to-sessile and acute-to-chronic virulence lifestyle transitions in many, if not most bacteria. As the exposure to a regular light-dark cycle is a major determinant of life, it is not surprising that these life style transitions can be controlled by photoreceptor signalling domains covering the visible spectrum of light (Fig. 3; [88]). Thereby, photoreceptors that belong to several different classes such as the BLUF, the LOV or bacteriophytochrome domain are coupled to diguanylate cyclases and cyclic di-GMP phosphodiesterases in different bacterial phyla. Not surprisingly, many of the light-activated enzymes are found in phototrophic bacteria, including cyanobacteria [89]. Superior temporal and spatial restriction compared to chemical inducers suggests light as an attractive stimulus to manipulate bacterial and eukaryotic physiology by cyclic (di)nucleotide second messenger signalling through tailor-suited engineering of signal transduction pathways [90]. Proof-of-principle, first exemplified by an algal photoactivated adenylate cyclase [91], stimulated the subsequent development of synthetic light-regulated modules to control cyclic di-GMP levels in bacterial as well as animal cells. In bacteria, the combination of synthetic modules can be used to differentially manipulate intrinsic bacterial behaviour such as motility, biofilm formation and virulence with light of different wavelengths in in vitro and in vivo studies (Fig. 3b; [92]). In bacterial and animal cells that lack cyclic di-GMP signalling pathways, light-activated cyclic di-nucleotide modules can be linked to downstream effector modules for orthogonal regulation of biological processes through manipulation of e.g. gene expression, protein activities or protein–protein interactions [93]. Although the broad spectrum of light can be used for various therapeutic purposes, nondestructive low-energy near-infrared light penetrates most deeply into mammalian tissue. Photocontrol of cyclic (di)nucleotide-based modules can thus be used in the temporal and spatial precision manipulation in host-bacterial interactions in mammalian models with the aim to develop innovative therapeutics. These second messenger based modules can also be adapted for remote control of stem, immune or other mammalian cells with an intrinsic and/or engineered response, which eventually will lead to the development of novel treatment options [94].
Fig. 3.
Light directed control of bacterial behavior. (a) Absorption range of characterized photoreceptor domains that are coupled to downstream bacterial cyclic dinucleotide signalling domains [88]. Chromophores such as biliverdine/bilin derivatives, flavin derivatives such as FAD or cumarin that sense light of different wave length are covalently or noncovalently coupled to the protein scaffold of photoreceptors in phytochromes (including bacteriophytochromes and cyanobacteriochromes), LOV/BLUF and xanthopsin proteins, respectively [96–98]. Phytochrome/phytochrome-like proteins contain PAS/GAF/PHY domains in various combinations [99]. (b) Engineering of an optogenetic system to regulate cyclic di-GMP levels bidirectionally. A red light-activated diguanylate cyclase (based on a bacteriophytochrome photoreceptor with a biliverdine chromophore) and a blue light-activated phosphodiesterase (based on a BLUF domain photoreceptor with an FAD chromophore) to regulate motility (left) and Congo red stained biofilm formation (right) [92].
Conclusions
Biofilm-associated infections still remain a diagnostic and treatment challenge. However, besides the initiation of pipelines to discover novel antibiofilm compounds, even traditional antimicrobial compounds can provide a significant anti-biofilm effect and are (re)discovered for extended clinical use also in combination with physical and chemical treatment strategies. In-depth analyses of the ECM components of biofilms will provide the molecular basis to develop efficient cocktails of ECM degrading enzymes as powerful biofilm dispersal agents tailor-suited for a respective clinical biofilm. It is the combination of novel anti-biofilm strategies with traditional antibiotics that promises to master the challenge of eradication of biofilm infections. The regulatory molecules known to play key roles in biofilm regulation such as the second messenger cyclic di-GMP and related cyclic (di)nucleotide second messengers are targets for anti-biofilm strategies. Extending their regulatory context, light sensing domains coupled to cyclic (di)nucleotide turnover domains can be developed as signal-input amplifying modules to construct remote-controlled interkingdom cross-talk modules to be applied for innovative treatment options.
Acknowledgements
This review is based on contributions from the Biofilm conference held at the Birke Aula, Karolinska Institutet, Campus Flemingsberg in May 2015. The contribution of all speakers and participants to the success of the conference is highly appreciated.
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- 1.Römling U, Kjelleberg S, Normark S, Nyman L, Uhlin BE, Åkerlund B. Microbial biofilm formation: a need to act. J Intern Med 2014; 276: 98–110. [DOI] [PubMed] [Google Scholar]
- 2.Hoiby N A personal history of research on microbial biofilms and biofilm infections. Pathog Dis 2014; 70: 205–11. [DOI] [PubMed] [Google Scholar]
- 3.Elek SD. Experimental staphylococcal infections in the skin of man. Ann N Y Acad Sci 1956; 65: 85–90. [DOI] [PubMed] [Google Scholar]
- 4.Siemens N, Chakrakodi B, Shambat SM, et al. Biofilm in group A streptococcal necrotizing soft tissue infections. JCI Insight 2016; 1: e87882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu H, Johani K, Almatroudi A, et al. Bacterial biofilm infection detected in breast implant-associated anaplastic large-cell lymphoma. Plast Reconstr Surg 2016; 137: 1659–69. [DOI] [PubMed] [Google Scholar]
- 6.Kinnari TJ. The role of biofilm in chronic laryngitis and in head and neck cancer. Curr Opin Otolaryngol Head Neck Surg 2015; 23: 448–53. [DOI] [PubMed] [Google Scholar]
- 7.Dejea CM, Fathi P, Craig JM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018; 359: 592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoiby N, Bjarnsholt T, Moser C, et al. ESCMID guideline for the diagnosis and treatment of biofilm infections. Clin Microbiol Infect 2015; 21(Suppl 1): S1–25. [DOI] [PubMed] [Google Scholar]
- 9.Bjarnsholt T, Jensen PO, Fiandaca MJ, et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol 2009; 44: 547–58. [DOI] [PubMed] [Google Scholar]
- 10.Brooks BD, Brooks AE. Therapeutic strategies to combat antibiotic resistance. Adv Drug Deliv Rev 2014; 78: 14–27. [DOI] [PubMed] [Google Scholar]
- 11.Kolpen M, Appeldorff CF, Brandt S, et al. Increased bactericidal activity of colistin on Pseudomonas aeruginosa biofilms in anaerobic conditions. Pathog Dis 2016; 74: ftv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haagensen JA, Klausen M, Ernst RK et al. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J Bacteriol 2007; 189: 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 2008; 68: 223–40. [DOI] [PubMed] [Google Scholar]
- 14.Chua SL, Yam JK, Hao P, et al. Selective labelling and eradication of antibiotic-tolerant bacterial populations in Pseudomonas aeruginosa biofilms. Nat Commun 2016; 7: 10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banat IM, De Rienzo MA, Quinn GA. Microbial biofilms: biosurfactants as antibiofilm agents. Appl Microbiol Biotechnol 2014; 98: 9915–29. [DOI] [PubMed] [Google Scholar]
- 16.Barraud N, Kelso MJ, Rice SA, Kjelleberg S. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr Pharm Des 2015; 21: 31–42. [DOI] [PubMed] [Google Scholar]
- 17.Sakarya S, Gunay N, Karakulak M, Ozturk B, Ertugrul B. Hypochlorous acid: an ideal wound care agent with powerful microbicidal, antibiofilm, and wound healing potency. Wounds 2014; 26: 342–50. [PubMed] [Google Scholar]
- 18.Bjarnsholt T, Alhede M, Jensen PO, et al. Antibiofilm properties of acetic acid. Adv Wound Care (New Rochelle) 2015; 4: 363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pletzer D, Coleman SR, Hancock RE. Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr Opin Microbiol 2016; 33: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleming D, Rumbaugh KP. Approaches to dispersing medical biofilms. Microorganisms 2017; 5: pii: E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snarr BD, Baker P, Bamford NC, et al. Microbial glycoside hydrolases as antibiofilm agents with cross-kingdom activity. Proc Natl Acad Sci U S A 2017; 114: 7124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 2001; 39: 1452–63. [DOI] [PubMed] [Google Scholar]
- 23.Römling U Genetic and phenotypic analysis of multicellular behavior in Salmonella typhimurium. Methods Enzymol 2001; 336: 48–59. [DOI] [PubMed] [Google Scholar]
- 24.Chapman MR, Robinson LS, Pinkner JS, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002; 295: 851–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol 1989; 42: 872–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choong FX, Back M, Fahlen S et al. Real-time optotracing of curli and cellulose in live Salmonella biofilms using luminescent oligothiophenes. NPJ Biofilms and Microbiomes 2016; 2: 16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Wu X, Chen J, et al. Antimicrobial blue light inactivation of gram-negative pathogens in biofilms: in vitro and in vivo studies. J Infect Dis 2016; 213: 1380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourke P, Ziuzina D, Han L, Cullen PJ, Gilmore BF. Microbiological interactions with cold plasma. J Appl Microbiol 2017; 123: 308–24. [DOI] [PubMed] [Google Scholar]
- 29.Granick MS, Paribathan C, Shanmugam M, Ramasubbu N. Direct-contact low-frequency ultrasound clearance of biofilm from metallic implant materials. Eplasty 2017; 17: e13. [PMC free article] [PubMed] [Google Scholar]
- 30.Wollina U, Heinig B, Naumann G, Scheibe A, Schmidt WD, Neugebauer R. Effects of low-frequency ultrasound on microcirculation in venous leg ulcers. Indian J Dermatol 2011; 56: 174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai Y, Wang J, Liu X, Wang R, Xia L. A review of the combination therapy of low frequency ultrasound with antibiotics. Biomed Res Int 2017; 2017: 2317846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schatz V, Neubert P, Schröder A, et al. Elementary immunology: Na(+) as a regulator of immunity. Pediatr Nephrol 2017; 32: 201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahim K, Saleha S, Zhu X, Huo L, Basit A, Franco OL. Bacterial contribution in chronicity of wounds. Microb Ecol 2017; 73: 710–21. [DOI] [PubMed] [Google Scholar]
- 34.Hu FB. Globalization of diabetes: the role of diet, lifestyle, and genes. Diabetes Care 2011; 34: 1249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boulton AJ, Vileikyte L, Ragnarson-Tennvall G, Apelqvist J. The global burden of diabetic foot disease. Lancet 2005; 366: 1719–24. [DOI] [PubMed] [Google Scholar]
- 36.Smith AC, Rice A, Sutton B et al. Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect Immun 2017; 85: pii: e00116–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun 2014; 82: 4718–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watters C, Everett JA, Haley C, Clinton A, Rumbaugh KP. Insulin treatment modulates the host immune system to enhance Pseudomonas aeruginosa wound biofilms. Infect Immun 2014; 82: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet 2014; 10: e1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol 1998; 25: 134–44. [DOI] [PubMed] [Google Scholar]
- 41.Larsen T, Fiehn NE. Dental biofilm infections – an update. APMIS 2017; 125: 376–84. [DOI] [PubMed] [Google Scholar]
- 42.Charalampakis G, Belibasakis GN. Microbiome of periimplant infections: lessons from conventional, molecular and metagenomic analyses. Virulence 2015; 6: 183–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jakubovics NS, Kolenbrander PE. The road to ruin: the formation of disease-associated oral biofilms. Oral Dis 2010; 16: 729–39. [DOI] [PubMed] [Google Scholar]
- 44.Kallio KA, Buhlin K, Jauhiainen M, et al. Lipopolysaccharide associates with pro-atherogenic lipoproteins in periodontitis patients. Innate Immun 2008; 14: 247–53. [DOI] [PubMed] [Google Scholar]
- 45.Sahingur SE, Yeudall WA. Chemokine function in periodontal disease and oral cavity cancer. Front Immunol 2015; 6: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akar H, Akar GC, Carrero JJ, Stenvinkel P, Lindholm B. Systemic consequences of poor oral health in chronic kidney disease patients. Clin J Am Soc Nephrol 2011; 6: 218–26. [DOI] [PubMed] [Google Scholar]
- 47.Buhlin K, Holmer J, Gustafsson A, et al. Association of periodontitis with persistent, pro-atherogenic antibody responses. J Clin Periodontol 2015; 42: 1006–14. [DOI] [PubMed] [Google Scholar]
- 48.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol 2010; 8: 623–33. [DOI] [PubMed] [Google Scholar]
- 49.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 2013; 77: 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jennings LK, Storek KM, Ledvina HE, et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A 2015; 112: 11353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Schaik EJ, Giltner CL, Audette GF, et al. DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J Bacteriol 2005; 187: 1455–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeFrancesco AS, Masloboeva N, Syed AK, et al. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc Natl Acad Sci U S A 2017; 114: E5969–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science 2002; 295: 1487. [DOI] [PubMed] [Google Scholar]
- 54.Dengler V, Foulston L, DeFrancesco AS, Losick R. An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J Bacteriol 2015; 197: 3779–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Domenech M, Ruiz S, Moscoso M, Garcia E. In vitro biofilm development of Streptococcus pneumoniae and formation of choline-binding protein-DNA complexes. Environ Microbiol Rep 2015; 7: 715–27. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez CJ, Shivshankar P, Stol K, et al. The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog 2010; 6: e1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shivshankar P, Sanchez C, Rose LF, Orihuela CJ. The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol Microbiol 2009; 73: 663–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schulte T, Löfling J, Mikaelsson C, et al. The basic keratin 10-binding domain of the virulence-associated pneumococcal serine-rich protein PsrP adopts a novel MSCRAMM fold. Open Biol 2014; 4: 130090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zogaj X, Bokranz W, Nimtz M, Römling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun 2003; 71: 4151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Römling U, Galperin MY. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol 2015; 23: 545–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahmad I, Rouf SF, Sun L, et al. BcsZ inhibits biofilm phenotypes and promotes virulence by blocking cellulose production in Salmonella enterica serovar Typhimurium. Microb Cell Fact 2016; 15: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pannanusorn S, Ramirez-Zavala B, Lünsdorf H, Agerberth B, Morschhäuser J, Römling U. Characterization of biofilm formation and the role of BCR1 in clinical isolates of Candida parapsilosis. Eukaryot Cell 2014; 13: 438–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cole SJ, Records AR, Orr MW, Linden SB, Lee VT. Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect Immun 2014; 82: 2048–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cole SJ, Lee VT. Cyclic-di-GMP signaling contributes to Pseudomonas aeruginosa mediated catheter-associated urinary tract infection. J Bacteriol 2015; 198: 91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rzhepishevska O, Hakobyan S, Ruhal R, Gautrot J, Barbero D, Ramstedt M. The surface charge of anti-bacterial coatings alters motility and biofilm architecture. Biomater Sci 2013; 1: 589–602. [DOI] [PubMed] [Google Scholar]
- 66.Ruhal R, Antti H, Rzhepishevska O et al. A multivariate approach to correlate surface properties and biofilm formation by lipopolysaccharide mutants of P. aeruginosa. Colloids Surf, B 2015; 127: 182–91. [DOI] [PubMed] [Google Scholar]
- 67.Gomez-Carretero S, Nybom R, Richter-Dahlfors A. Electroenhanced antimicrobial coating based on conjugated polymers with covalently coupled silver nanoparticles prevents Staphylococcus aureus biofilm formation. Adv Healthc Mater 2017; 6: 1700435. [DOI] [PubMed] [Google Scholar]
- 68.Mikulskis P, Hook AL, Dundas A, et al. Prediction of broad-spectrum pathogen attachment to coating materials for biomedical devices. ACS Appl Mater Interfaces 2017; 10: 139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kapadia BH, Berg RA, Daley JA, Fritz J, Bhave A, Mont MA. Periprosthetic joint infection. Lancet 2016; 387: 386–94. [DOI] [PubMed] [Google Scholar]
- 70.Marmor S, Kerroumi Y. Patient-specific risk factors for infection in arthroplasty procedure. Orthop Traumatol Surg Res 2016; 102: S113–9. [DOI] [PubMed] [Google Scholar]
- 71.Pittet B, Montandon D, Pittet D. Infection in breast implants. Lancet Infect Dis 2005; 5: 94–106. [DOI] [PubMed] [Google Scholar]
- 72.Stelzer EH. Better imaging through chemistry. Cell 2014; 159: 1243–6. [DOI] [PubMed] [Google Scholar]
- 73.Hell SW. Far-field optical nanoscopy. Science 2007; 316: 1153–8. [DOI] [PubMed] [Google Scholar]
- 74.Xiao J, Dufrene YF. Optical and force nanoscopy in microbiology. Nat Microbiol 2016; 1: 16186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geoghegan JA, Foster TJ, Speziale P, Dufrene YF. Live-cell nanoscopy in antiadhesion therapy. Trends Microbiol 2017; 25: 512–4. [DOI] [PubMed] [Google Scholar]
- 76.Ozkan AD, Topal AE, Dana A, Guler MO, Tekinay AB. Atomic force microscopy for the investigation of molecular and cellular behavior. Micron 2016; 89: 60–76. [DOI] [PubMed] [Google Scholar]
- 77.Ziemba C, Shabtai Y, Piatkovsky M, Herzberg M. Cellulose effects on morphology and elasticity of Vibrio fischeri biofilms. NPJ Biofilms Microbiomes 2016; 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Steinberger O, Lapidot Z, Ben-Ishai Z, Amikam D. Elevated expression of the CD4 receptor and cell cycle arrest are induced in Jurkat cells by treatment with the novel cyclic dinucleotide 3′,5′-cyclic diguanylic acid. FEBS Lett 1999; 444: 125–9. [DOI] [PubMed] [Google Scholar]
- 79.Amikam D, Steinberger O, Shkolnik T, Ben-Ishai Z. The novel cyclic dinucleotide 3′−5′ cyclic diguanylic acid binds to p21ras and enhances DNA synthesis but not cell replication in the Molt 4 cell line. Biochem J 1995; 311: 921–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karaolis DK, Means TK, Yang D, et al. Bacterial c-di-GMP is an immunostimulatory molecule. J Immunol 2007; 178: 2171–81. [DOI] [PubMed] [Google Scholar]
- 81.Kobayashi H, Kobayashi CI, Nakamura-Ishizu A, et al. Bacterial c-di-GMP affects hematopoietic stem/progenitors and their niches through STING. Cell Rep 2015; 11: 71–84. [DOI] [PubMed] [Google Scholar]
- 82.Corrales L, Glickman LH, McWhirter SM, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep 2015; 11: 1018–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marinho FV, Benmerzoug S, Oliveira SC, Ryffel B, Quesniaux VFJ. The emerging roles of STING in bacterial infections. Trends Microbiol 2017; 25: 906–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ablasser A, Goldeck M, Cavlar T, et al. cGAS produces a 2′−5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013; 498: 380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DiFrancesco D Pacemaker mechanisms in cardiac tissue. Annu Rev Physiol 1993; 55: 455–72. [DOI] [PubMed] [Google Scholar]
- 86.Lolicato M, Bucchi A, Arrigoni C, et al. Cyclic dinucleotides bind the C-linker of HCN4 to control channel cAMP responsiveness. Nat Chem Biol 2014; 10: 457–62. [DOI] [PubMed] [Google Scholar]
- 87.Rios-Covian D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilan CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 2016; 7: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gomelsky M, Hoff WD. Light helps bacteria make important lifestyle decisions. Trends Microbiol 2011; 19: 441–8. [DOI] [PubMed] [Google Scholar]
- 89.Agostoni M, Waters CM, Montgomery BL. Regulation of biofilm formation and cellular buoyancy through modulating intracellular cyclic di-GMP levels in engineered cyanobacteria. Biotechnol Bioeng 2016; 113: 311–9. [DOI] [PubMed] [Google Scholar]
- 90.Gomelsky M Special issue on synthetic photobiology. ACS Synth Biol 2014; 3: 780–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schroder-Lang S, Schwarzel M, Seifert R, et al. Fast manipulation of cellular cAMP level by light in vivo. Nat Methods 2007; 4: 39–42. [DOI] [PubMed] [Google Scholar]
- 92.Ryu MH, Fomicheva A, Moskvin OV, Gomelsky M. Optogenetic module for dichromatic control of c-di-GMP signaling. J Bacteriol 2017; 199: pii: e00014–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Folcher M, Oesterle S, Zwicky K, et al. Mind-controlled transgene expression by a wireless-powered optogenetic designer cell implant. Nat Commun 2014; 5: 5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ryu MH, Gomelsky M. Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synth Biol 2014; 3: 802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sriramulu DD, Lünsdorf H, Lam JS, Römling U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol 2005; 54: 667–76. [DOI] [PubMed] [Google Scholar]
- 96.van der Horst MA, Hellingwerf KJ. Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res 2004; 37: 13–20. [DOI] [PubMed] [Google Scholar]
- 97.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu Rev Biochem 2015; 84: 519–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ikeuchi M, Ishizuka T. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci 2008; 7: 1159–67. [DOI] [PubMed] [Google Scholar]
- 99.Karniol B, Wagner JR, Walker JM, Vierstra RD. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem J 2005; 392: 103–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arora DP, Hossain S, Xu Y, Boon EM. Nitric oxide regulation of bacterial biofilms. Biochemistry 2015; 54: 3717–28. [DOI] [PubMed] [Google Scholar]
- 101.Hughson AG, Race B, Kraus A, et al. Inactivation of prions and amyloid seeds with hypochlorous acid. PLoS Pathog 2016; 12: e1005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burdette DL, Monroe KM, Sotelo-Troha K, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011; 478: 515–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Parvatiyar K, Zhang Z, Teles RM, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol 2012; 13: 1155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tosolini M, Pont F, Verhoeyen E, Fournie JJ. Cyclic dinucleotides modulate human T-cell response through monocyte cell death. Eur J Immunol 2015; 45: 3313–23. [DOI] [PubMed] [Google Scholar]
- 105.Li W, Cui T, Hu L, Wang Z, Li Z, He ZG. Cyclic diguanylate monophosphate directly binds to human siderocalin and inhibits its antibacterial activity. Nat Commun 2015; 6: 8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xia P, Wang S, Xiong Z, et al. The ER membrane adaptor ERAdP senses the bacterial second messenger c-di-AMP and initiates anti-bacterial immunity. Nat Immunol 2018; 19: 141–50. [DOI] [PubMed] [Google Scholar]