Abstract
Oral microbial communities are extraordinarily complex in taxonomic composition and comprise interdependent biological systems. The bacteria, archaea, fungi, and viruses that thrive within these communities engage in extensive cell-cell interactions, which are both beneficial and antagonistic. Direct physical interactions among individual cells mediate large-scale architectural biofilm arrangements and provide spatial proximity for chemical communication and metabolic cooperation. In this review, we summarize recent work in identifying specific molecular components that mediate cell-cell interactions and describe metabolic interactions, such as cross-feeding and exchange of electron acceptors and small molecules, that modify the growth and virulence of individual species. We argue, however, that although pairwise interaction models have provided useful information, complex community-like systems are needed to study the properties of oral communities. The networks of multiple synergistic and antagonistic interactions within oral biofilms give rise to the emergent properties of persistence, stability, and long-range spatial structure, with these properties mediating the dysbiotic transitions from health to oral diseases. A better understanding of the fundamental properties of interspecies networks will lead to the development of effective strategies to manipulate oral communities.
Keywords: biofilms, plaque/plaque biofilms, imaging, microbial ecology, microscopy, candidiasis
Introduction
Modern genomic analyses have identified approximately 700 species or phylotypes of bacteria as well as archaea, fungi, and viruses that comprise the human oral microbiome (Dewhirst et al. 2010; Abeles et al. 2014; Diaz et al. 2017). Sequencing studies of the different oral sites available for colonization (i.e., the teeth, tongue, buccal mucosa, soft and hard palate, and gingiva) have demonstrated that the different habitats support distinct microbial communities with taxon selection mediated by the characteristics of the surfaces available for attachment, oxygen availability, and exposure to host nutritional products delivered by saliva and gingival crevicular fluid (Human Microbiome Project Consortium 2012).
Oral microbial communities are biological systems, comprising individual cells whose coordinate interactions give rise to emergent properties (Marsh and Zaura 2017). The microbial communities that live on the nonshedding tooth surfaces, the supra- and subgingival plaque, assemble into biofilms. The cells that comprise these biofilms attach either to the tooth surface through the salivary pellicle or molecules derived from gingival crevicular fluid or to each other, and they are embedded in an extracellular matrix (Kolenbrander et al. 2010). In this review, we summarize recent work to elucidate the physical and chemical interactions among oral microbes, focusing on bacteria and fungi, the most studied components of the oral microbiome. We next consider the need to understand dental plaque emergent properties, which are characteristics that emerge only from the interactions of its components in a wider whole.
Physical Interactions among Bacteria
The binding of genetically distinct oral bacterial cells to each other was first reported by Gibbons and Nygaard in 1970 and was termed bacterial agglutination (Gibbons and Nygaard 1970). Coaggregation, defined as the specific molecularly mediated binding of genetically distinct organisms in solution, has been systematically tested for thousands of pairs of cultivable isolates (Kolenbrander et al. 1990). In this review, we consider coaggregation together with coadhesion, defined as the binding of a planktonic cell to another cell attached to a surface. Early work to characterize the molecular components that comprise the interbacterial adhesion apparati involved the use of carbohydrate inhibitors including lactate, protein-cleaving enzymes such as trypsin, and inhibitory concentrations of ions, which together shed light on the general nature of the surface molecules involved and revealed the ubiquity of lectin-like interactions (McIntire et al. 1978).
Cells of the genus Streptococcus as well as Actinomyces comprise the subset of oral microbes that act as the earliest colonizers, able to directly bind to components of the salivary pellicle that coats the teeth, including proline-rich proteins, albumin, sialic acid, alpha amylase, glycoproteins, and other proteins (Abranches et al. 2018). Streptococci comprise up to 80% of initial supragingival plaque bacteria (Nyvad and Kilian 1987; Diaz et al. 2006). Streptococcal adhesion is mediated by cell wall proteins as well as surface structures, including pili or fimbriae, which mediate binding to salivary gp340 as well as host extracellular matrix proteins, epithelial cells, and endothelial cells. CshA is a fibrillar cell surface protein in Mitis group streptococci that mediates binding to other microbes, including Actinomyces oris, as well as host fibronectin (Nobbs et al. 2015).
The serine-rich repeat glycoproteins (SRRPs), Hsa and GspB, on the surfaces of Streptococcus gordonii DL1 and S. gordonii M99 respectively bind α2,3-linked sialic acid of mucin and thereby mediate S. gordonii attachment to the salivary pellicle (Takahashi et al. 2002; Takamatsu et al. 2006). In an in vitro binding assay, the affinity of Hsa for mucin glycoproteins was shown to increase with increasing shear force (Ding et al. 2010). In general, low-affinity salivary glycan binding by bacterial adhesin proteins is synergized by the presence of multiple adhesins on the cell surface and the clustering of glycans in salivary mucins (Cross and Ruhl 2018). Recently, Hsa was demonstrated to mediate S. gordonii coaggregation with Veillonella spp., secondary early colonizers, through a mechanism that did not involve sialic acid recognition (Zhou et al. 2015). SRRPs have been identified in a number of oral streptococci, including Fap1 in Streptococcus parasanguinis, SrpA in Streptococcus sanguinis and Streptococcus cristatus, and Srp A, B, and C in Streptococcus salivarius (Nobbs et al. 2015).
The antigen I/II proteins of streptococci were among the first streptococcal surface antigens identified as adhesins. Various isoforms have been identified in most oral streptococci and include SpaP, P1, PAc, SsoA, and SspB (Abranches et al. 2018). AgI/II proteins are cell-wall anchored proteins that mediate binding to other bacteria and epithelial cells as well as the salivary glycoprotein gp340, and the AgI/II isoforms have recently been reported to function together as a supramolecular complex in Streptococcus mutans (Heim et al. 2015).
In coaggregation assays, Fusobacterium nucleatum demonstrates the unique ability to coaggregate with numerous species of oral microbes, including both early and late colonizers (Kolenbrander et al. 1989). Recently, several Fusobacterium genes have been identified that function as cell surface coaggregation mediators. The 350-kDa outer membrane protein, RadD, was initially identified in F. nucleatum subsp. polymorphum strain ATCC 23726 as an arginine-inhibitable, promiscuous adhesin that mediated coaggregation with Gram-positive organisms, including S. gordonii and Actinomyces naeslundii (Kaplan et al. 2009) (Fig. 1). RadD was recently shown to bind the S. mutans adhesin SpaP, which was previously implicated in S. mutans salivary glycoprotein binding (Guo et al. 2017). Aid1 (Adherence Inducing Determinant 1) was subsequently identified in a mutagenesis screen as a putative accessory protein that contributes to RadD-mediated coaggregation with streptococci (Kaplan et al. 2014). In another mutagenesis experiment, the F. nucleatum outer membrane autotransporter protein, Fap2, was identified as a mediator of coaggregation with Porphyromonas gingivalis as well as F. nucleatum binding to mammalian cells (Coppenhagen-Glazer et al. 2015). Mice infected with the fap2 knockout strain of F. nucleatum had reduced placental F. nucleatum colonization compared to controls, and strains of F. nucleatum lacking Fap2 had reduced binding to Gal-Gal-NAc-expressing colorectal adenocarcinoma tumors in mice, implicating this adhesin in the tumor-related virulence of F. nucleatum (Coppenhagen-Glazer et al. 2015; Abed et al. 2016). The F. nucleatum porin protein, FomA, has demonstrated binding to the salivary protein statherin as well as coaggregation with P. gingivalis (Kinder and Holt 1993). Recently, a second F. nucleatum ATCC 23726 outer membrane protein, CmpA (coaggregation mediating protein A), was identified as another mediator of F. nucleatum coaggregation with S. gordonii (Lima et al. 2017).
Figure 1.
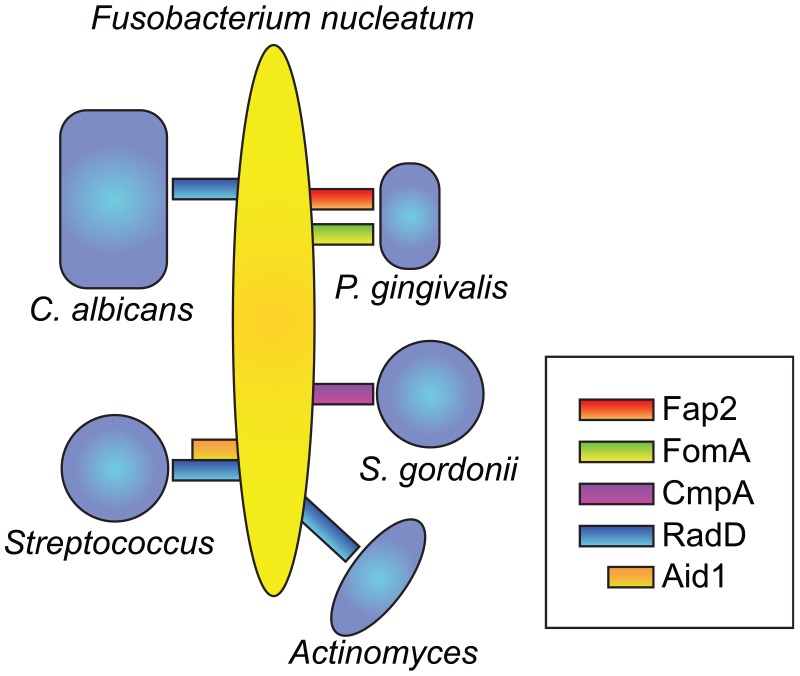
Fusobacterium nucleatum physical interactions and surface adhesion molecules. The surface adhesin RadD mediates F. nucleatum binding to Streptococcus spp., Actinomyces, and Candida albicans. Aid1 is an accessory adhesin that mediates F. nucleatum coaggregation with Streptococcus spp. The outer membrane protein Fap2 mediates coaggregation with Porphyromonas gingivalis as does the porin protein FomA, and the outer membrane protein CmpA mediates coaggregation with Streptococcus gordonii.
Interestingly, like F. nucleatum, P. gingivalis has also been observed to physically interact with a number of different oral community members, including both early and late colonizers. P. gingivalis coaggregation with S. gordonii has been demonstrated to be mediated by the fimbriae protein FimA specifically binding to S. gordonii surface-localized glyceraldehyde phosphate dehydrogenase (GAPDH) (Maeda et al. 2004; Ng et al. 2016), as well as the short fimbriae protein Mfa, which binds SspB proteins on S. gordonii. The P. gingivalis gingipains RgpA, RgpB, and Kgp were shown to play an essential role in synergistic biofilm formation with Treponema denticola in vitro (Zhu et al. 2013), and the T. denticola outer sheath dentilisin protein has been implicated in coaggregation with P. gingivalis and other bacteria, including Treponema forsythia (Sano et al. 2014).
As well as providing scaffolds for structuring the biofilm, direct physical interactions between oral microbes have been demonstrated to mediate changes in gene transcription. Differential regulation of 16 genes in F. nucleatum and 119 genes in S. gordonii were observed when these 2 organisms coaggregated in vitro as compared to mono-culture conditions (Mutha et al. 2018). More recently, transcriptional profiling revealed differential regulation of 69 genes in S. gordonii and 272 genes in Veillonella parvula when these organisms coaggregated in vitro (Mutha et al. 2019). Importantly, proximity in coculture was not sufficient to induce differential gene expression (i.e., coaggregation was required). Genes regulated included those related to oxidative stress and carbohydrate metabolism (Mutha et al. 2019).
Chemical Interactions
Synergistic Metabolic Interactions
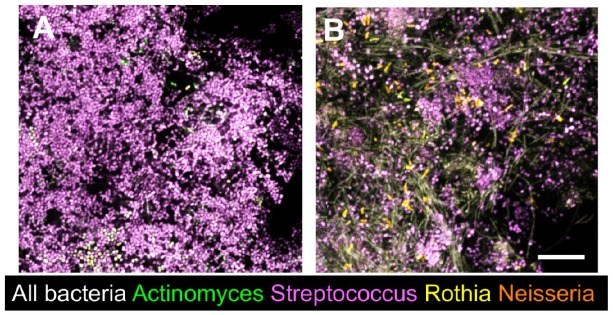
One of the strongest pieces of evidence for metabolic cooperation among oral bacteria comes from the observation of enhanced growth and biofilm formation when oral isolates are cocultured in saliva as the sole nutritional source (Periasamy and Kolenbrander 2009). When dental plaque from a healthy donor was used to seed a microcosm culture and grown in a rich medium, the community was dominated by streptococci, but when a dental plaque inoculum from the same donor was used to seed a microcosm community in dilute saliva, an extraordinarily diverse biofilm community resulted, as evidenced by microscopic observation after fluorescence in situ hybridization (Fig. 2) (previously unpublished).
Figure 2.
In vitro microcosm biofilms, seeded with dental plaque and labeled with fluorescence in situ hybridization probes. (A) When in vitro cultures are grown in a rich medium (in this case, fastidious anaerobe broth) for 7 d, the biofilm is dominated by streptococci. (B) A plaque inoculum from the same donor was used to seed an in vitro biofilm grown under the same conditions as in A, except 25% human saliva was used as the medium. A highly diverse community is present with multiple cells that are labeled with the bacterial domain probe but not any of the genus-level probes used. Scale bar = 10 µm. (Previously unpublished data from Valm lab.)
While the metabolic connectivity of oral microbial communities is undoubtedly complex and multifactorial, a number of specific interactions have been identified in simple in vitro coculture systems. S. gordonii ferments carbohydrate to form lactic acid, which is the preferred carbon substrate for Veillonella atypica. In an in vitro coculture system, V. atypica induces expression of the S. gordonii α-amylase gene amyB to induce hydrolysis of intracellular glycogen and secretion of lactate (Egland et al. 2004; Johnson et al. 2009).
Metabolic synergism and crosstalk have been observed between P. gingivalis and S. gordonii, mediated by streptococcal 4-aminobenzoate/para-amino benzoic acid (pABA), involved in folate biosynthesis (Kuboniwa et al. 2017). Exogenous pABA was shown to increase the colonization and fitness of P. gingivalis while reducing virulence in a mouse model (Kuboniwa et al. 2017). F. nucleatum has been shown to support the growth of P. gingivalis in oxygenated and CO2-depleted environments (Diaz et al. 2002), and recently, metabolic crosstalk in the form of increased electron acceptor bioavailability (O2) has been observed between S. gordonii and Aggregatibacter actinomycetemcomitans in a coinfection model (Stacy et al. 2016).
Antagonistic Interactions
Many streptococci produce bacteriocins (e.g., lantibiotics, among many others), which are small peptides with antimicrobial activity (Mathur et al. 2018). For example, S. mutans produces several mutacins with antimicrobial activity against many other streptococci (Merritt and Qi 2012). S. gordonii and S. sanguinis of the Mitis group streptococci produce hydrogen peroxide as a by-product of carbohydrate metabolism (Redanz et al. 2018; Chen et al. 2019). While some oral microbes possess catalase genes and therefore resistance to oxidative stress, streptococci do not, and strains of S. mutans display a range of susceptibility to H2O2 (Redanz et al. 2018). This antagonistic relationship may be clinically relevant in the context of dental caries (Giacaman et al. 2015). Moreover, some commensal oral microbes possess an arginine deaminase system (ADS) and produce ammonia through arginine metabolism, which raises the pH of the biofilm environment and inhibits growth of aciduric organisms, including S. mutans (Huang et al. 2018). H2O2 production by oral streptococci has also been shown to inhibit the growth of periodontopathogens (Hillman et al. 1985). Interestingly, A. actinomycetemcomitans was shown to reduce pyruvate oxidase (PoxL)–mediated H2O2 production by S. parasanguinis in coculture, which resulted in increased biofilm formation, a finding with implications for localized aggressive periodontitis (Duan et al. 2016). Recently, environmental arginine was demonstrated to have a direct effect on S. mutans growth and stress tolerance (Chakraborty and Burne 2017). Coaggregation of S. gordonii with A. oris resulted in downregulation of arginine biosynthesis genes and upregulation of biofilm genes, and recently, the ArcR arginine-dependent regulator of transcription in S. gordonii was shown to link arginine sensing with biofilm formation, further implicating arginine metabolism in community structure (Robinson et al. 2018).
An extreme form of potentially antagonistic metabolic cooperativity in the form of parasitism has been described among the oral microbiota. TM7x of the candidate phylum Saccharibacteria (formerly TM7) requires its bacterial host, Actinomyces odondolyticus, for survival because its greatly reduced genome is insufficient for independent growth (He et al. 2015). Although infection of naive hosts results in cell death, a subset of Actinomyces cells within infected populations survives with reduced growth rates, giving rise to long-term population stability and metabolic cooperativity (Bor et al. 2018). Taken together, antagonistic interactions in the oral community reflect the dynamic interconnectedness of oral biofilm microbes and represent potential avenues for therapeutic intervention.
Fungal Interactions with Other Oral Microbiome Members
Dozens of fungal taxa inhabit the oral cavity of humans. Molecular- and cultivation-based studies show yeasts such as Candida and Malassezia are common oral residents (Dupuy et al. 2014; Diaz et al. 2017; Abusleme et al. 2018). Other low-abundance fungi, such as Alternaria, Aspergillus, Fusarium, Rhodotorula, and Cryptococcus, have also been recovered from the oral cavity (Monteiro-da-Silva et al. 2014; Diaz et al. 2017). The function fungi play in oral microbiome communities, however, remains incompletely understood.
An overgrowth of Candida, especially Candida albicans, is associated with the development of oropharyngeal candidiasis (Abusleme et al. 2018; Diaz et al. 2019). Recent studies also suggest a role of Candida in other disruptions of oral homeostasis such as early childhood caries (Koo et al. 2018). Thus, due to its clinical relevance, Candida is the only fungal genus that has been evaluated in terms of its interactions with other microbiome members. In both oral candidiasis and caries, the interactions of C. albicans with other Candida species or with bacteria are important determinants of Candida virulence and appear to contribute to dysbiosis.
The interactions of Candida and Streptococcus are one of the best understood fungal-bacterial relationships. Candida and Mitis group streptococci have been shown to coaggregate and form synergistic biofilms (Bamford et al. 2009; Diaz et al. 2012). The specific binding between Candida and Mitis group streptococci is mediated by protein-protein interactions involving the streptococcal cell surface adhesins CshA, SspA, and SspB (Holmes et al. 1996; Silverman et al. 2010; Xu, Jenkinson, et al. 2014). The adhesins on C. albicans surface that recognize streptococci include members of the agglutinin-like sequence (ALS) and hyphal wall protein 1 (HWP1) families. Specifically, ALS3, ALS1, ALS5, and also HWP1 and EAP1 have been shown to mediate recognition of S. gordonii cells by C. albicans (Klotz et al. 2007; Nobbs et al. 2010; Silverman et al. 2010). Extracellular polysaccharide (EPS) produced by streptococcal glucosyl-transferases (Gtfs) also plays a role promoting adhesive interactions between S. gordonii and C. albicans (Ricker et al. 2014). Interactions between C. albicans and Mitis group streptococci have been shown to be important in oropharyngeal candidiasis. Mitis group streptococci appear enriched in mucosal samples of patients with chronic oral candidiasis (Abusleme et al. 2018). The relevance of this interaction has been demonstrated in animal and in vitro models, in which Streptococcus oralis enhances the virulence of C. albicans with coinoculation resulting in greater tissue invasion and damage (Diaz 2012; Xu, Sobue, et al. 2014).
C. albicans has also been shown to synergistically interact with S. mutans. Gtfs, which are produced by S. mutans under carbohydrate-rich environments, mediate its attachment to C. albicans and enhance the biofilm accretion of both microorganisms (Falsetta et al. 2014). On the Candida side, N- or O-linked mannans on the fungal cell wall appear to mediate binding to S. mutans GtfB (Hwang et al. 2017). The partnership of C. albicans and S. mutans has been shown to promote rampant carious lesions in a rodent model under high sucrose (Falsetta et al. 2014). The 2 microorganisms have been coisolated in children with caries (Xiao et al. 2018).
C. albicans has also been shown to coaggregate with F. nucleatum (Wu et al. 2015) and with the periodontitis-associated species P. gingivalis (Sztukowska et al. 2018). Candida spp. have been detected in periodontal pockets, and it is therefore possible that such interkingdom interactions are relevant in vivo (Dabdoub et al. 2016). C. albicans and P. gingivalis have been shown to physically interact through the InlJ internalin-family protein on the surface of P. gingivalis and ALS3 on the surface of Candida (Sztukowska et al. 2018). In a biofilm model, it has been shown that in the presence of oxygen, C. albicans creates a protective environment for P. gingivalis (Bartnicka et al. 2019).
Although less studied than fungal-bacterial interactions, fungal-fungal partnerships also influence oral homeostasis. For instance, Candida glabrata is often coisolated with C. albicans during oropharyngeal candidiasis (Redding et al. 1999). C. glabrata adheres to C. albicans hyphae, an interaction that seems important in an in vivo coinfection model (Tati et al. 2016).
In summary, fungal-bacterial and fungal-fungal interactions play a role in oral microbiome communities, mediating dysbiotic events associated with oral candidiasis, and are also possibly involved in caries. The function of other mycobiome members (e.g., Malassezia) that are dominant in certain individuals warrants further investigation.
Emergent Properties
The multitude of interactions that take place between individual cells gives rise to the emergent properties that are characteristic of oral biofilms, including persistence, stability, and resistance. By definition, the emergent properties of a system cannot be understood simply by analyzing the individual components in isolation. DNA sequencing has revealed the complex changes in community composition that take place during the multifactorial ecosystem transitions from health to dental caries and periodontal disease (Griffen et al. 2012; Abusleme et al. 2013; Koo et al. 2018; Tanner et al. 2018). Recent metatranscriptomic analyses have identified genes that are upregulated in subgingival communities in states of periodontal disease to include genes involved in iron uptake, among others, while longitudinal metatanscriptomic studies offer the possibility of identifying genes within communities that drive the transition from health to disease (Duran-Pinedo et al. 2014; Yost et al. 2015). However, these studies do not shed light into the assembly rules that operate in oral microbial communities and the events that mediate dysbiotic shifts.
The interspecies interactions described in previous sections have been observed for the most part in simple 2-species coculture models. Recent work on ecological theory of microbial communities shows that interactions that emerge only when 3 or more species are present determine stability, persistence, and maintenance of biodiversity in complex microbial systems (Levine et al. 2017; Goldford et al. 2018). These interactions include interaction chains, in which 1 species provides indirect benefits to another one, and higher-order interactions, in which the presence of 1 species modifies the way other species interact (Levine et al. 2017). Studying the properties of these networks could lead to the development of approaches to manipulate whole communities. For instance, there is certain predictability in the way nutrients determine community composition (Goldford et al. 2018). Also, network structure has been shown to influence the community resistance to perturbations (Levine et al. 2017).
Although it may not be possible to directly observe all of the individual interactions taking place in communities, the development of in vitro biofilm models with high species diversity offers promise for controlled laboratory testing of systems-level hypotheses (Thurnheer et al. 2016). Extraordinarily diverse human oral microbial communities have been grown in vitro by seeding cultures in minimal media with dental plaque inocula extracted from human volunteers (Fernandez et al. 2017). Metagenomic analysis of a previously well-characterized microcosm model system revealed the dynamics of community assembly during biofilm maturation to be sporadic while concomitant metatranscriptomic analysis revealed synergistic changes in gene expression that correlated more strongly with environmental changes in the community (i.e., pH) than with changes in community taxonomic structure (Edlund et al. 2018).
Through a process of ecological succession, mediated by dynamic intercelluar interactions and environmental inputs, tooth-associated oral microbes assemble into structures that may be orders of magnitude larger than the individual cells themselves. Multiplex imaging of subgingival plaque revealed the incorporation of C. albicans hyphae within biofilms, upon which streptococci assembled and could serve as scaffolds for long-range biofilm structure (Zijnge et al. 2010).
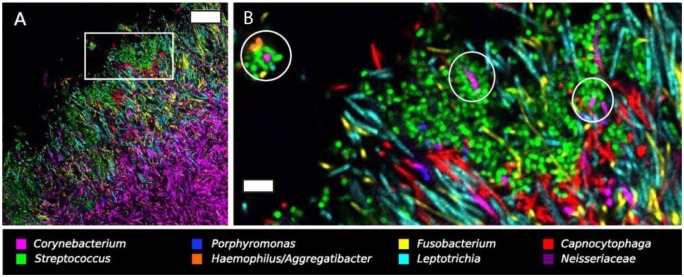
Systems-level imaging of plaque biofilms offers another omics-type approach for studying the emergent properties of oral communities in situ. Combinatorial labeling and spectral imaging fluorescence in situ hybridization (CLASI-FISH) was first used to quantify all of the physical interactions among 15 genera of microbes in extracted, semidispersed dental plaque (Valm et al. 2011). Analysis of these associations informed a network description of dental plaque communities and revealed the surprising finding that Fusobacterium spp. did not make physical associations with many other genera in situ (Valm et al. 2011). Subsequently, spectral imaging of semi-intact supragingival plaque biofilms carefully extracted from healthy donors revealed the presence of large multitaxon consortia of organisms in hedgehog structures (Mark Welch et al. 2016). In these structures, filamentous Corynebacterium spp. appear to form a bush-like scaffold, anchored to the presumed tooth surface through associations with Streptococcus and Actinomyces. The distal tips of the Corynebacterium filaments were often decorated with cocci, consisting of various combinations of Streptococcus, Haemophilus or Aggregatibacter, and Porphyromonas in “corncob” arrangements, which comprised an apical surface on the biofilm structure that functions to sequester oxygen and thereby create an anaerobic niche (Fig. 3).
Figure 3.
Systems imaging reveals long-range structure and corncob arrangements in supragingival plaque. (A) Hedgehog structure in supragingival plaque extracted from a healthy volunteer, labeled with fluorescent in situ hybridization probes for 8 genera. Scale bar = 20 µm. (B) Higher magnification view of the region of interest highlighted in A. Circles indicate variously comprised corncob arrangements in hedgehog structures, with coccoid cells (Streptococcus, Haemophilus/Aggregatibacter, and Porphyromonas) arranged around a central filamentous Corynebacterium cell. Scale bar = 5 µm. Reprinted with permission from Mark Welch et al. (2016).
The abundance and prevalence of Corynebacterium in supragingival plaque from multiple donors and the conspicuous arrangement of Corynebacterium filaments within hedgehog structures further led to the hypothesis that this organism plays a central role in structuring the biofilm community; however, the precise role that Corynebacterium plays in relation to that of Fusobacterium remains to be determined. Fusobacterium is highly abundant in supragingival plaque and has demonstrated an ability to coaggregate with numerous other species in vitro, and Fusobacterium cells are filamentous and centrally located within hedgehog structures alongside Corynebacterium (Mark Welch et al. 2016). F. nucleatum is aerotolerant, and its presence in oxygenated in vitro biofilms facilitates the growth of strict anaerobes within these communities (Bradshaw et al. 1998), suggesting that Fusobacterium and Corynebacterium may play different roles in structuring the biofilm at different times during community development. Because multiple biological explanations are consistent with many of the observations to date, the precise roles that Fusobacterium and Corynebacterium play in organizing supragingival plaque biofilms is therefore yet to be determined.
Conclusions and Future Directions
The human mouth is home to extraordinarily diverse microbial communities, which, especially on the hard, nonshedding surfaces of teeth, assemble into polymicrobial biofilms with defined spatial structure and community functions. Like many biological systems, dental plaque biofilms are composed of multiple compartments (microbial cells), each of which carries out repertoires of biochemical processes that sustain the individual cells. At the same time, the individual cells must interact with their environment and with each other. Collectively, the intercellular interactions among oral microbes, both physical and chemical, give rise to the emergent properties of plaque biofilms and the holistic functions of these microbial communities.
Under homeostatic conditions, plaque biofilms function to promote oral health; however, in states of dysbiosis, dental plaque biofilms mediate localized oral diseases, including dental caries and periodontal disease, and are further implicated in systemic diseases such as cardiovascular disease and others (Kholy et al. 2015; Lamont et al. 2018). The individuality of microbial communities as well as their persistence and regenerative capacities are emergent properties of the plaque biofilm. A greater understanding of the intercellular interactions that underlie community functions may provide an avenue to develop improved strategies to engineer plaque biofilms to promote health.
Although the study of pairwise interactions among oral microbes has unequivocally shown that species growth and virulence are modified by their community context, the field should also embrace complex community-like systems to study species networks in a more realistic setting. Defined-inoculum complex community models offer the possibility of studying the role of individual species or functional groups on the growth and stability of the whole community. Similarly, the use of model communities created with natural inocula could be useful to understand community assembly rules and response to perturbations in a more holistic context. The ultimate goal of studying oral microbial communities is to develop tools to engineer their composition to one compatible with oral health.
Author Contributions
P.I. Diaz, contributed to conception, design, and data acquisition, drafted and critically revised the manuscript; A.M. Valm, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. Both authors gave final approval and agree to be accountable for all aspects of the work.
Footnotes
The authors are supported by National Institutes of Health grant DE023967 (P.I.D.) and DE028042 (A.M.V.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: A.M. Valm  https://orcid.org/0000-0002-8286-080X
https://orcid.org/0000-0002-8286-080X
References
- Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, et al. 2016. Fap2 Mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 20(2):215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, Pride DT. 2014. Human oral viruses are personal, persistent and gender-consistent. ISME J. 8(9):1753–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J, Zeng L, Kajfasz JK, Palmer SR, Chakraborty B, Wen ZT, Richards VP, Brady LJ, Lemos JA. 2018. Biology of oral streptococci. Microbiol Spectr. 6(5). doi: 10.1128/microbiolspec.GPP3-0042-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusleme L, Diaz PI, Freeman AF, Greenwell-Wild T, Brenchley L, Desai JV, Ng W-I, Holland SM, Lionakis MS, Segre JA, et al. 2018. Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis. JCI Insight. 3(17): pii: 122061. doi: 10.1172/jci.insight.122061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7(5):1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford CV, d’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 77(9):3696–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnicka D, Karkowska-Kuleta J, Zawrotniak M, Satała D, Michalik K, Zielinska G, Bochenska O, Kozik A, Ciaston I, Koziel J, et al. 2019. Adhesive protein-mediated cross-talk between Candida albicans and Porphyromonas gingivalis in dual species biofilm protects the anaerobic bacterium in unfavorable oxic environment. Sci Rep. 9(1):4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor B, McLean JS, Foster KR, Cen L, To TT, Serrato-Guillen A, Dewhirst FE, Shi W, He X. 2018. Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc Natl Acad Sci USA. 115(48):12277–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw DJ, Marsh PD, Watson GK, Allison C. 1998. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. 66(10):4729–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty B, Burne RA. 2017. Effects of arginine on Streptococcus mutans growth, virulence gene expression, and stress tolerance. Appl Environ Microbiol. 83(15). pii: e00496-17. doi: 10.1128/AEM.00496-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chakraborty B, Zou J, Burne RA, Zeng L. 2019. Amino sugars modify antagonistic interactions between commensal oral streptococci and Streptococcus mutans. Appl Environ Microbiol. 85(10). pii: e00370-19. doi: 10.1128/AEM.00370-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, Bachrach G. 2015. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun. 83(3):1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BW, Ruhl S. 2018. Glycan recognition at the saliva–oral microbiome interface. Cell Immunol. 333:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub SM, Ganesan SM, Kumar PS. 2016. Comparative metagenomics reveals taxonomically idiosyncratic yet functionally congruent communities in periodontitis. Sci Rep. 6:38993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol. 192(19):5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI. 2012. Microbial diversity and interactions in subgingival biofilm communities. Front Oral Biol. 15:17–40. [DOI] [PubMed] [Google Scholar]
- Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ, Kolenbrander PE. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 72(4):2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Hong B-Y, Dupuy AK, Choquette L, Thompson A, Salner AL, Schauer PK, Hegde U, Burleson JA, Strausbaugh LD, et al. 2019. Integrated analysis of clinical and microbiome risk factors associated with the development of oral candidiasis during cancer chemotherapy. J Fungi (Basel). 5(2). pii: E49. doi: 10.3390/jof5020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Hong B-Y, Dupuy AK, Strausbaugh LD. 2017. Mining the oral mycobiome: methods, components, and meaning. Virulence. 8(3):313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, Ikonomou L, Dongari-Bagtzoglou A. 2012. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 80(2):620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Zilm PS, Rogers AH. 2002. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 148(Pt 2):467–472. [DOI] [PubMed] [Google Scholar]
- Ding AM, Palmer RJ, Cisar JO, Kolenbrander PE. 2010. Shear-enhanced oral microbial adhesion. Appl Environ Microbiol. 76(4):1294–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D, Scoffield JA, Zhou X, Wu H. 2016. Fine-tuned production of hydrogen peroxide promotes biofilm formation of Streptococcus parasanguinis by a pathogenic cohabitant Aggregatibacter actinomycetemcomitans. Environ Microbiol. 18(11):4023–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD. 2014. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 9(3):e90899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, Krishnan K, Frias-Lopez J. 2014. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 8(8):1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund A, Yang Y, Yooseph S, He X, Shi W, McLean JS. 2018. Uncovering complex microbiome activities via metatranscriptomics during 24 hours of oral biofilm assembly and maturation. Microbiome. 6(1):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egland PG, Palmer RJ, Kolenbrander PE. 2004. Interspecies communication in Streptococcus gordonii–Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci USA. 101(48):16917–16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai C-H, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, et al. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 82(5):1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Y, Mostajo M, Exterkate RAM, Buijs MJ, Beertsen W, van der Weijden GA, Zaura E, Crielaard W. 2017. A reproducible microcosm biofilm model of subgingival microbial communities. J Periodont Res. 43(Suppl. 11):1021–1031. [DOI] [PubMed] [Google Scholar]
- Giacaman RA, Torres S, Gómez Y, Muñoz-Sandoval C, Kreth J. 2015. Correlation of Streptococcus mutans and Streptococcus sanguinis colonization and ex vivo hydrogen peroxide production in carious lesion-free and high caries adults. Arch Oral Biol. 60(1):154–159. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Nygaard M. 1970. Interbacterial aggregation of plaque bacteria. Arch Oral Biol. 15(12):1397–1400. [DOI] [PubMed] [Google Scholar]
- Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, Segrè D, Mehta P, Sanchez A. 2018. Emergent simplicity in microbial community assembly. Science. 361(6401):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6(6):1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Shokeen B, He X, Shi W, Lux R. 2017. Streptococcus mutans SpaP binds to RadD of Fusobacterium nucleatum ssp. polymorphum. Mol Oral Microbiol. 32(5):355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu S-Y, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, et al. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA. 112(1):244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim KP, Sullan RM, Crowley PJ, El-Kirat-Chatel S, Beaussart A, Tang W, Besingi R, Dufrene YF, Brady LJ. 2015. Identification of a supramolecular functional architecture of Streptococcus mutans adhesin P1 on the bacterial cell surface. J Biol Chem. 290(14):9002–9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman JD, Socransky SS, Shivers M. 1985. The relationships between streptococcal species and periodontopathic bacteria in human dental plaque. Arch Oral Biol. 30(11–12):791–795. [DOI] [PubMed] [Google Scholar]
- Holmes AR, McNab R, Jenkinson HF. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect Immun. 64(11):4680–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Browngardt CM, Jiang M, Ahn S-J, Burne RA, Nascimento MM. 2018. Diversity in antagonistic interactions between commensal oral streptococci and Streptococcus mutans. Caries Res. 52(1–2):88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature. 486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G, Liu Y, Kim D, Li Y, Krysan DJ, Koo H. 2017. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 13(6):e1006407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BP, Jensen BJ, Ransom EM, Heinemann KA, Vannatta KM, Egland KA, Egland PG. 2009. Interspecies signaling between Veillonella atypica and Streptococcus gordonii requires the transcription factor CcpA. J Bacteriol. 191(17):5563–5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A, Kaplan CW, He X, McHardy I, Shi W, Lux R. 2014. Characterization of aid1, a novel gene involved in Fusobacterium nucleatum interspecies interactions. Microb Ecol. 68(2):379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CW, Lux R, Haake SK, Shi W. 2009. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol. 71(1):35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholy KE, Genco RJ, Van Dyke TE. 2015. Oral infections and cardiovascular disease. Trends Endocrinol Metab. 26(6):315–321. [DOI] [PubMed] [Google Scholar]
- Kinder SA, Holt SC. 1993. Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J Bacteriol. 175(3):840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz SA, Gaur NK, De Armond R, Sheppard D, Khardori N, Edwards JE, Lipke PN, El-Azizi M. 2007. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med Mycol. 45(4):363–370. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Moore LV. 1989. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect Immun. 57(10):3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Moore LV. 1990. Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl Environ Microbiol. 56(12):3890–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. 2010. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 8(7):471–480. [DOI] [PubMed] [Google Scholar]
- Koo H, Andes DR, Krysan DJ. 2018. Candida-streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog. 14(12):e1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, Houser JR, Hendrickson EL, Wang Q, Alghamdi SA, Sakanaka A, Miller DP, Hutcherson JA, Wang T, Beck DAC, et al. 2017. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol. 2(11):1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Koo H, Hajishengallis G. 2018. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 16(12):745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JM, Bascompte J, Adler PB, Allesina S. 2017. Beyond pairwise mechanisms of species coexistence in complex communities. Nature. 546(7656):56–64. [DOI] [PubMed] [Google Scholar]
- Lima BP, Shi W, Lux R. 2017. Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii. Microbiologyopen. 6(3):e00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Nagata H, Yamamoto Y, Tanaka M, Tanaka J, Minamino N, Shizukuishi S. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect Immun. 72(3):1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. 2016. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 113(6):E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD, Zaura E. 2017. Dental biofilm: ecological interactions in health and disease. J Clin Periodontol. 44(Suppl 18):S12–S22. [DOI] [PubMed] [Google Scholar]
- Mathur H, Field D, Rea MC, Cotter PD, Hill C, Ross RP. 2018. Fighting biofilms with lantibiotics and other groups of bacteriocins. NPJ Biofilms Microbiomes. 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire FC, Vatter AE, Baros J, Arnold J. 1978. Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34. Infect Immun. 21(3):978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Qi F. 2012. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol. 27(2):57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro-da-Silva F, Araujo R, Sampaio-Maia B. 2014. Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med Mycol. 52(5):498–505. [DOI] [PubMed] [Google Scholar]
- Mutha NVR, Mohammed WK, Krasnogor N, Tan GYA, Choo SW, Jakubovics NS. 2018. Transcriptional responses of Streptococcus gordonii and Fusobacterium nucleatum to coaggregation. Mol Oral Microbiol. 33(6):450–464. [DOI] [PubMed] [Google Scholar]
- Mutha NVR, Mohammed WK, Krasnogor N, Tan GYA, Wee WY, Li Y, Choo SW, Jakubovics NS. 2019. Transcriptional profiling of coaggregation interactions between Streptococcus gordonii and Veillonella parvula by dual RNA-Seq. Sci Rep. 9(1):7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HM, Kin LX, Dashper SG, Slakeski N, Butler CA, Reynolds EC. 2016. Bacterial interactions in pathogenic subgingival plaque. Microb Pathog. 94:60–69. [DOI] [PubMed] [Google Scholar]
- Nobbs AH, Jenkinson HF, Everett DB. 2015. Generic determinants of Streptococcus colonization and infection. Infect Genet Evol. 33:361–370. [DOI] [PubMed] [Google Scholar]
- Nobbs AH, Vickerman MM, Jenkinson HF. 2010. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell. 9(10):1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvad B, Kilian M. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 95(5):369–380. [DOI] [PubMed] [Google Scholar]
- Periasamy S, Kolenbrander PE. 2009. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infect Immun. 77(9):3542–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redanz S, Cheng X, Giacaman RA, Pfeifer CS, Merritt J, Kreth J. 2018. Live and let die: hydrogen peroxide production by the commensal flora and its role in maintaining a symbiotic microbiome. Mol Oral Microbiol. 33(5):337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding SW, Zellars RC, Kirkpatrick WR, McAtee RK, Caceres MA, Fothergill AW, Lopez-Ribot JL, Bailey CW, Rinaldi MG, Patterson TF. 1999. Epidemiology of oropharyngeal Candida colonization and infection in patients receiving radiation for head and neck cancer. J Clin Microbiol. 37(12):3896–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker A, Vickerman M, Dongari-Bagtzoglou A. 2014. Streptococcus gordonii glucosyltransferase promotes biofilm interactions with Candida albicans. J Oral Microbiol. 6. doi: 10.3402/jom.v6.23419. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JC, Rostami N, Casement J, Vollmer W, Rickard AH, Jakubovics NS. 2018. ArcR modulates biofilm formation in the dental plaque colonizer Streptococcus gordonii. Mol Oral Microbiol. 33(2):143–154. [DOI] [PubMed] [Google Scholar]
- Sano Y, Okamoto-Shibayama K, Tanaka K, Ito R, Shintani S, Yakushiji M, Ishihara K. 2014. Dentilisin involvement in coaggregation between Treponema denticola and Tannerella forsythia. Anaerobe. 30:45–50. [DOI] [PubMed] [Google Scholar]
- Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. 2010. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 78(11):4644–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy A, Fleming D, Lamont RJ, Rumbaugh KP, Whiteley M. 2016. A commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. MBio. 7(3): pii: e00782-16. doi: 10.1128/mBio.00782-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztukowska MN, Dutton LC, Delaney C, Ramsdale M, Ramage G, Jenkinson HF, Nobbs AH, Lamont RJ. 2018. Community development between Porphyromonas gingivalis and Candida albicans mediated by InlJ and Als3. MBio. 9(2). pii: e00202-18. doi: 10.1128/mBio.00202-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Konishi K, Cisar JO, Yoshikawa M. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect Immun. 70(3):1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Prakobphol A, Fisher SJ, Sullam PM. 2006. Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect Immun. 74(3):1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner ACR, Kressirer CA, Rothmiller S, Johansson I, Chalmers NI. 2018. The caries microbiome: implications for reversing dysbiosis. Adv Dent Res. 29(1):78–85. [DOI] [PubMed] [Google Scholar]
- Tati S, Davidow P, McCall A, Hwang-Wong E, Rojas IG, Cormack B, Edgerton M. 2016. Candida glabrata binding to Candida albicans hyphae enables its development in oropharyngeal candidiasis. PLoS Pathog. 12(3):e1005522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurnheer T, Bostanci N, Belibasakis GN. 2016. Microbial dynamics during conversion from supragingival to subgingival biofilms in an in vitro model. Mol Oral Microbiol. 31(2):125–135. [DOI] [PubMed] [Google Scholar]
- Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. 2011. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA. 108(10):4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Cen L, Kaplan C, Zhou X, Lux R, Shi W, He X. 2015. Cellular components mediating coadherence of Candida albicans and Fusobacterium nucleatum. J Dent Res. 94(10):1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Huang X, Alkhers N, Alzamil H, Alzoubi S, Wu TT, Castillo DA, Campbell F, Davis J, Herzog K, et al. 2018. Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res. 52(1–2):102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jenkinson HF, Dongari-Bagtzoglou A. 2014. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 29(3):99–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sobue T, Thompson A, Xie Z, Poon K, Ricker A, Cervantes J, Diaz PI, Dongari-Bagtzoglou A. 2014. Streptococcal co-infection augments candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 16(2):214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. 2015. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 7(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Liu J, Li X, Takahashi Y, Qi F. 2015. The sialic acid binding protein, Hsa, in Streptococcus gordonii DL1 also mediates intergeneric coaggregation with Veillonella species. PLoS One. 10(11):e0143898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Dashper SG, Chen Y-Y, Crawford S, Slakeski N, Reynolds EC. 2013. Porphyromonas gingivalis and Treponema denticola synergistic polymicrobial biofilm development. PLoS One. 8(8):e71727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, Gmür R, Harmsen HJM. 2010. Oral biofilm architecture on natural teeth. PLoS One. 5(2):e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]