Abstract
Regulation of iron (Fe) acquisition and homeostasis is critical for plant survival. In Arabidopsis, Fe deficiency‐induced bHLH039 forms a complex with the master regulator FIT and activates it to upregulate Fe acquisition genes. FIT is partitioned between cytoplasm and nucleus, whereby active FIT accumulates more in the nucleus than inactive FIT. At the same time, there is so far no information on the subcellular localization of bHLH039 protein and how it is controlled. We report here that the bHLH039 localization pattern changes depending on the presence of FIT in the cell. When expressed in cells lacking FIT, bHLH039 localizes predominantly in the cytoplasm, including cytoplasmic foci in close proximity to the plasma membrane. The presence of FIT enhances the mobility of bHLH039 and redirects the protein toward primarily nuclear localization, abolishing its accumulation in cytoplasmic foci. This FIT‐dependent change in localization of bHLH039 found in transient fluorescent protein expression experiments was confirmed in both leaves and roots of Arabidopsis transgenic plants, stably expressing hemagglutinin‐tagged bHLH039 in wild‐type or fit mutant background. This posttranslational mechanism for intracellular partitioning of Fe‐responsive transcription factors suggests a signaling cascade that translates Fe sensing at the plasma membrane to nuclear accumulation of the transcriptional regulators.
Keywords: bHLH039, Fer‐like iron deficiency‐induced transcription factor, iron deficiency, nucleocytoplasmic partitioning, transcription factor regulation
1. INTRODUCTION
Iron (Fe) is an essential element in plants as it participates in key redox reactions. Changes in metabolism due to execution of developmental programs or in response to environmental cues result in major rebalancing of the cellular and plant Fe homeostasis. Fe acquisition in Arabidopsis (Arabidopsis thaliana) and other Strategy I plants requires active proton extrusion to solubilize Fe in the soil, the reduction of Fe3+ to Fe2+ by the ferric reductase‐oxidase 2 (FRO2) and the subsequent uptake of Fe2+ in the root epidermis cells by the metal transporter iron‐regulated transporter 1 (IRT1) (Brumbarova, Bauer, & Ivanov, 2015; Jeong, Merkovich, Clyne, & Connolly, 2017). The execution of this strategy under Fe deficiency requires the essential basic helix–loop–helix (bHLH) transcription factor Fer‐like iron deficiency‐induced transcription factor (FIT) (Colangelo & Guerinot, 2004; Jakoby, Wang, Reidt, Weisshaar, & Bauer, 2004). Members of the bHLH subgroup Ib, BHLH038, BHLH039, BHLH100, and BHLH101 (Heim et al., 2003) are induced by Fe deficiency in roots and leaves (Vorwieger et al., 2007; Wang et al., 2007), controlled by a network of transcription factors (Gao, Robe, Gaymard, Izquierdo, & Dubos, 2019). BHLH039 induction serves as one of the robust Fe deficiency markers (Gratz, Manishankar, et al., 2019; Ivanov, Brumbarova, & Bauer, 2012; Khan et al., 2019). Each of the four members can interact with FIT, resulting in an active protein complex for upregulation of Fe uptake genes, with bHLH039 playing the most prominent role among them (Wang et al., 2013; Yuan et al., 2008).
The nucleocytoplasmic partitioning of proteins is an important regulatory aspect affecting their function, and, therefore, the signaling cascades in which they are involved (Meier & Somers, 2011). During Fe deficiency, the function of the plasma membrane‐localized FRO2 and IRT1 has to be synchronized with the transcriptional regulation of the Fe deficiency response in the nucleus, controlled by FIT and its activator interaction partner bHLH039. FIT undergoes strict posttranslational control and exists in two forms, active and inactive (Meiser, Lingam, & Bauer, 2011; Sivitz, Grinvalds, Barberon, Curie, & Vert, 2011), distinguishable based on the phosphorylation status (Gratz, Manishankar, et al., 2019). FIT is localized in the cytoplasm and nucleus, whereby active FIT shows greater accumulation in the nucleus versus the cytoplasm than inactive FIT (Gratz, Manishankar, et al., 2019). The FIT‐bHLH039 interaction is enhanced when FIT is activated by phosphorylation at Ser272 (Gratz, Manishankar, et al., 2019).
So far, studies on bHLH039, as representative of a subgroup Ib bHLH transcription factor, have remained focused on its transcriptional regulation and protein interaction with FIT. Therefore, this study was motivated by two significant gaps in understanding Fe acquisition regulation. First, the lack of information on the subcellular localization of bHLH039 and second, the lack of understanding whether post‐transcriptional events play a role in bHLH039 regulation. We report here a surprising pattern of bHLH039 localization that is not predominantly nuclear, as for the majority of studied transcription factors, and changes depending on the presence of another transcription factor in the cell, namely FIT. We analyze the localization of bHLH039 in two different biological systems, including Arabidopsis. Through a combination of standard and advanced imaging approaches together with biochemical analysis, we quantitatively assign the localization and subcellular dynamics of bHLH039 to the presence of FIT.
2. MATERIALS AND METHODS
2.1. Plant material and growth conditions
Tobacco (Nicotiana benthamiana) plants were grown on soil for 3–4 weeks in a greenhouse facility under long day conditions (16‐hr light/8‐hr dark). The Arabidopsis (Arabidopsis thaliana) ecotype Col‐0 was used as wild type (WT). FIT loss‐of‐function mutant plants, fit‐3 (GABI_108C10), were described previously (Jakoby et al., 2004). HA3‐bHLH039 transgenic plants overexpressing BHLH039, N‐terminally fused with a triple hemagglutinin tag, in WT (39/WT) or fit‐3 mutant background (39/fit) were described in Naranjo‐Arcos et al. (2017). For protoplast isolation, WT and fit‐3 plants were grown for three weeks upright on half‐strength Hoagland agar medium with sufficient (50 mM FeNaEDTA, +Fe) Fe supply before harvesting their shoots. For fractionation experiments, WT, 39/WT and 39/fit plants were grown for two weeks upright on half‐strength Hoagland agar medium with sufficient (50 mM FeNaEDTA, + Fe) Fe supply and then transferred to new plates with either sufficient or deficient (0 mM FeNaEDTA, −Fe) Fe supply for 3 days before harvesting, as described previously (2‐week system; Gratz, Manishankar, et al., 2019).
2.2. Generation of fluorescent protein fusions
The pABind vector system with XVE‐driven ß‐estradiol inducible promoter (Bleckmann, Weidtkamp‐Peters, Seidel, & Simon, 2010) was used for expression of fluorescently tagged FIT and bHLH039 proteins in tobacco leaf epidermis cells. The generation of pABind‐GFP:FIT, pABind‐mCherry:FIT, and pABind‐GFP:BHLH039 was previously described (Gratz, Manishankar, et al., 2019). Similarly, pDONR207:BHLH039 (Gratz, Manishankar, et al., 2019) was used to generate pABind‐mCherry:BHLH039. Free GFP was expressed using pMDC7:GFP (Khan et al., 2019), also under the control of the XVE‐driven ß‐estradiol inducible promoter. For transient expression in Arabidopsis protoplasts, FIT‐GFP was expressed using pMDC83:FIT (Gratz, Manishankar, et al., 2019), under the control of the 35S promoter. For bHLH039‐mCherry, the BHLH039 coding sequence was introduced in pDONR207 using primers 39 B1 (GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTGTGCATTAGTACCTCC) and 39ns B2 (GGGGACCACTTTGTACAAGAAAGCTGGGTTTATATATGAGTTTCCACATTC). Subsequently, the BHLH039‐containing cassette was recombined into the Gateway‐compatible vector pJNC1 under the control of the 35S promoter (Ivanov et al., 2014) to create pJNC1:bHLH039.
2.3. Transient transformation of tobacco leaf epidermis cells
The Agrobacterium (Rhizobium radiobacter) strain C58 (GV3101) carrying pABind constructs with GFP‐ or mCherry‐tagged FIT or BHLH039, or pMDC7:GFP were used for transient transformation of tobacco leaves. Agrobacterium cultures were grown to an OD600 = 0.4, centrifuged and resuspended in a solution containing 250 µM acetosyringone, 0.1% (w/v) glucose, 0.01% (v/v) Silwet, and 5% (w/v) sucrose. After one hour incubation on ice, the suspension was infiltrated into the abaxial side of tobacco leaves. p19 vector was co‐infiltrated to enhance gene expression. Expression was induced by application of a β‐estradiol solution (20 µM β‐estradiol, 0.1% Tween 20) 16 hr before imaging.
2.4. Confocal microscopy and cytoplasm‐to‐nucleus ratio determination
For cytoplasm‐to‐nucleus ratio determination, FIT‐GFP, FIT‐mCherry, bHLH039‐GFP, bHLH039‐mCherry, or free GFP proteins were transiently expressed in tobacco cells as described above. Full Z‐stacks of epidermal cells were recorded on LSM780 confocal microscope (Zeiss). GFP was imaged with an excitation wavelength of 488 nm and emission detection at 500–530 nm. mCherry and FM4‐64 membrane dye were imaged with an excitation of 561 nm with emission detection at 570–615 nm. C‐Apochromat 40×/1.20 W Korr M27 water immersion objective was used. Pinhole was set to 1 Airy unit, equivalent to optical slices of 0.8 µm, with a frame size of 1,024 × 1,024 pixels and a pixel dwell time of 0.79 µs. Full projections of Z‐stack images were generated in ZEN 2012 Blue Edition software (Zeiss), and densitometry on the resulting uncompressed images was performed in the Fiji distribution of ImageJ (fiji.sc) to calculate the cytoplasm‐to‐nucleus ratio of protein localization (Gratz, Manishankar, et al., 2019). Total intensities of the nucleus and the cytoplasm fluorescence were measured separately. The ratio was calculated for each individual cell. Fifteen to 20 cells were processed per fluorescent reporter under each condition.
2.5. Arabidopsis protoplast isolation, transformation, and imaging
Plant material was prepared as described in Dovzhenko, Bosco, Meurer, and Koop (2003). In short, shoots from 160 to 200 three‐week‐old Arabidopsis plants were chopped to fine pieces with a sterile scalpel in 5 ml of MMC medium (10 mM MES, 40 mM CaCl2, mannitol to 550 mOsm, pH 5.8), followed by an overnight enzymatic digestion in MMC medium supplemented with macerozyme R10 and cellulase Onozuka R10 (0.5% each, Duchefa Biochemie). The homogenized leaf material was filtered through a 70 µm mesh and centrifuged at 100 g for 20 min. The obtained pellet was resuspended in 10 ml MSC medium (10 mM MES, 20 mM MgCl2, mannitol to 550 mOsm, sucrose to 550 mOsm, pH 5.8) and overlaid with 3 ml MMM solution (5 mM MES, 15 mM MgCl2, mannitol to 600 mOsm, pH 5.8). After flotation for 10 min at 80 g, protoplasts were collected from the interphase and transferred to tubes containing W5 solution (2 mM MES, 154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, pH 5.8), adjusting the total volume to 10 ml. Protoplasts were counted, pelleted, and resuspended in MMM solution to a concentration of 500,000 protoplasts/ml. PEG‐mediated protoplast transformation was performed as follows: 15 µg of plasmid DNA in 20 µl of MMM solution were mixed with 100 µl protoplast suspension in 6‐well plates (Greiner Bio‐One, Germany). After 5 min, 120 μl of PEG4000 solution (62% w/v PEG4000, 0.3 M mannitol, 0.15 M CaCl2, prewarmed at 37°C) was carefully added to the transformation mixture and incubated for 8 min. Afterwards, 120 µl MMM solution and PCA regeneration medium (0.3% w/v B5 salts, 0.05% w/v MgSO4, 0.05% w/v CaCl2, 0.1% w/v MES, 2% w/v sucrose, 8% w/v glucose, 0.0002% w/v Ca‐panthotenate, 0.1% v/v Gamborg B5 Vitamin Mix, 0.02 ng/ml biotin, pH 5.8) were added to a final volume of 1.6 ml; the plate was sealed and incubated overnight at 21°C. The protoplasts were imaged using an AxioImager.M2 fluorescent microscope (Zeiss). Images were taken with a Plan‐Apochromat 40×/1.4 oil objective and recorded by an Axiocam 503 monochromatic camera. For GFP imaging, Filter set 38 HE eGFP shift free (E) (EX BP 470/40, BS FT 495, EM BP 525/50) was used. For mRFP/mCherry imaging, Filter set 43 HE Cy 3 shift free (E) (EX BP 550/25, BS FT 570, EM BP 605/70) was used.
2.6. Preparation of nuclear and cytoplasmic protein fractions
Cellular fractionation was performed as described in Li et al. (2019). Leaves and roots were collected separately from Arabidopsis plants grown in the 2‐weeks system described above. The plant material was ground in the presence of liquid nitrogen and resuspended in three volumes of buffer containing 20 mM Tris‐HCl (pH 7.4), 25% glycerol, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 250 mM sucrose, and 1x protease inhibitor cocktail (Roche). The samples were then sequentially filtered through 70 μm and 22–25 μm nylon mesh before being centrifuged at 1,500 x g at 4°C for 20 min to pellet the nuclei. The pellet was resuspended in a buffer containing 20 mM Tris‐HCl (pH 7.4), 25% glycerol, 2.5 mM MgCl2, 0.2% NP40, and 0.1% Triton X‐100, and the cycle was repeated twice, after which the last pellet was resuspended in 1x protein loading buffer (62 mM Tris‐HCl pH 6.8, 2.5% SDS, 2% 1,4‐dithiothreitol, 10% glycerol), producing the nuclear fraction. The first supernatant (after the first centrifugation) was centrifuged at 18,800 x g at 4°C for 20 min two times. This supernatant was mixed 1:1 (v/v) with 2× protein loading buffer, producing the cytosolic fraction.
2.7. SDS‐PAGE and protein detection by immunoblot
Protein samples were separated on a 4%–20% gradient polyacrylamide gel (Bio‐Rad) and transferred to a nitrocellulose membrane following the protocol described in Gratz, Manishankar, et al. (2019). The presence of target proteins on the membrane was verified using immunodetection and chemiluminescence. Images of signals on the membranes were digitally recorded in the dynamic range using FluorChem Q machine (Biozym). Band signal intensities were quantified in Fiji distribution of ImageJ (fiji.sc) and used to calculate cytoplasm‐to‐nucleus ratios.
The following antibodies were used for immunodetection: Rat monoclonal anti‐HA horseradish peroxidase conjugated (3F10; Roche) 1:5,000 was used to detect HA‐bHLH039; Rabbit anti‐UGPase (AS05 086; Agrisera) 1:1,000 together with goat anti‐rabbit IgG horseradish peroxidase (AS09 602; Agrisera) 1:5,000 were used to detect the cytoplasmic fraction marker protein UDP‐glucose pyrophosphorylase (UGPase); Rabbit anti‐histone H3 horseradish peroxidase (AS10710‐HRP; Agrisera) was used to detect the nuclear fraction marker protein Histone H3.
2.8. Fluorescence recovery after photobleaching (FRAP) analysis
Transient expression of pABind:bHLH039‐GFP, pABind:FIT‐GFP, pABind:FIT‐mCherry, and pMDC7:GFP was performed as described above. FRAP measurements in nucleus and at the cell periphery were taken with a frame size of 256 × 256 and pixel dwell time of 1.0 µs. A series of five images was taken before and 40 images after 50 iterations of photobleaching with full laser power of the argon laser at 488 nm. A non‐bleached region of interest with the same size as the bleached region was used to correct for acquisition bleaching. The fluorescence intensity values before the photobleach (pre‐bleach, F pre) were averaged, and together with the first value after the bleach (post‐bleach, F post) and the intensity after recovery (recovery, F end) were used to calculate the mobile fraction (M f) according to the following equation: M f = [(F end−F post)/(F pre−F post)] * 100 (Bancaud, Huet, Rabut, & Ellenberg, 2010). For calculating Fend, the average of the five last scans was used and was adjusted for the intensity loss due to the acquisition based on the intensity measured in the non‐bleached region. Ten to 15 cells were analyzed for each construct or combination.
2.9. Statistical analysis
Experimental data was processed using analysis of variance (ANOVA) and Fisher's least significant difference post hoc test. For signal colocalization, Pearson's correlation coefficient (PCC) was calculated using the JACoP plugin of ImageJ (Bolte & Cordelieres, 2006).
2.10. Accession numbers
Sequence data from this article can be found in the TAIR and GenBank data libraries under accession numbers: BHLH039 (TAIR: AT3G56980), FIT (TAIR: AT2G28160).
3. RESULTS
3.1. FIT is required for the nuclear localization of bHLH039
Partitioning of regulatory proteins between intracellular compartments represents a mean of controlling their activity. Since no information on the subcellular localization of bHLH039 was available, we determined the localization of this transcription factor, to assess whether partitioning between cytoplasm and nucleus is a potential target for its regulation, as is the case for FIT. Expressing FIT‐GFP in tobacco leaf epidermis cells showed the reported (Gratz, Manishankar, et al., 2019) dual localization in the cytoplasm and the nucleus (Figure 1a). bHLH039‐mCherry fusion, expressed in the same system, surprisingly showed strong signals at the cell periphery and only a rather weak presence in the nucleus (Figure 1b and Figure S1). However, coexpression of FIT and bHLH039 in the same cells led to a clear dual localization in nucleus and cytoplasm of bHLH039‐mCherry and a strong colocalization with FIT‐GFP in the nucleus (Figure 1c). When coexpressed with free GFP, bHLH039‐mCherry had strong cell periphery localization (Figure 1d), similar to bHLH039‐mCherry alone. We quantified the cytoplasm‐to‐nucleus ratio of the tested fluorescent proteins and found that indeed, only in the presence of FIT‐GFP, bHLH039‐mCherry showed a significant shift toward nuclear localization (Figure 1e), suggesting that the presence of FIT is important for the nuclear localization of bHLH039. At the same time, the localization ratio of FIT‐GFP remained unchanged by the presence of bHLH039, suggesting that bHLH039 does not influence the localization of FIT. As a control, we performed the reverse experiment, where the fluorescent tags of bHLH039 and FIT were swapped (Figure S2). The experiment yielded the same result, showing that the observed effect was independent of the fluorescent tag.
Figure 1.
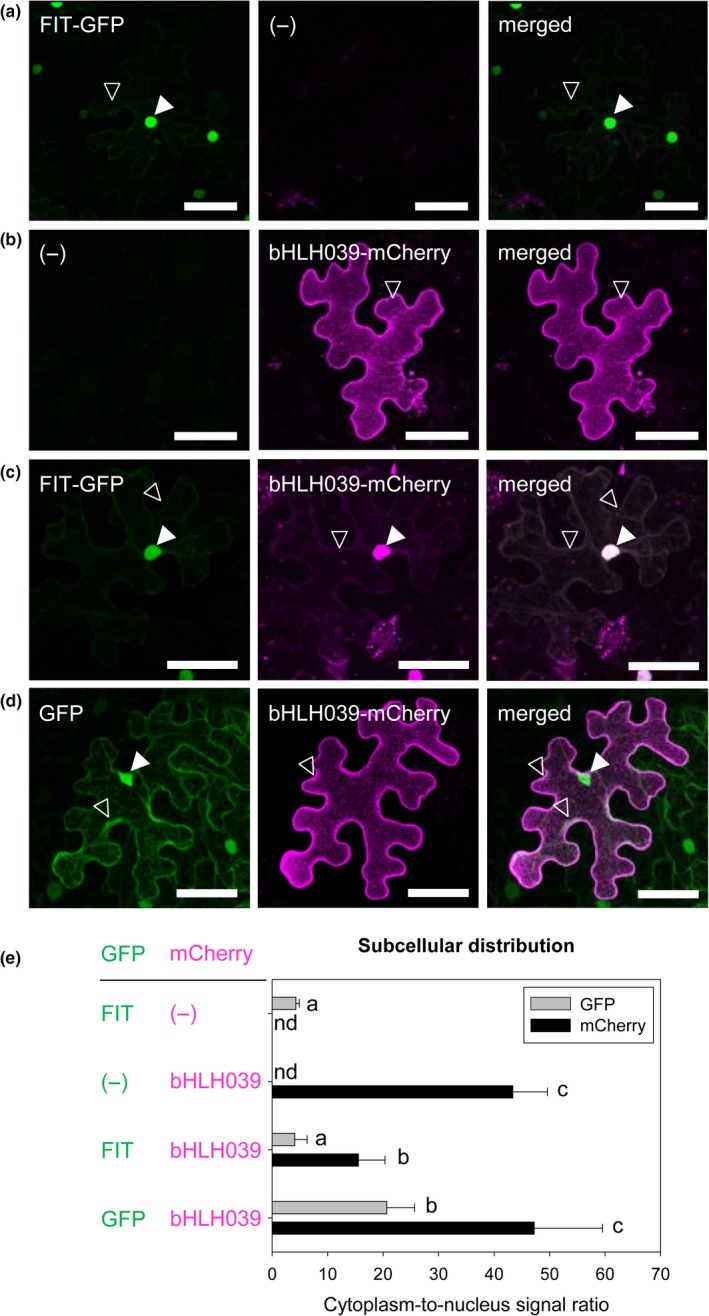
Subcellular localization of bHLH039 depends on FIT presence in tobacco leaf epidermis cells. (a‐d) Localization of FIT‐GFP, bHLH039‐mCherry, and free GFP, alone or in combinations. Representative full projections of laser‐scanning confocal images show GFP and mCherry fluorescence of FIT‐GFP alone (a), bHLH039‐mCherry alone (b), FIT‐GFP and bHLH039‐mCherry (c), and free GFP and bHLH039‐mCherry (d), following ß‐estradiol induction of transiently transformed tobacco leaf epidermis cells. Nuclear and cytoplasmic fluorescence signals are indicated by arrowheads (filled and empty, respectively). Bars: 50 µm. (e) Quantification of subcellular distribution of the fluorescently tagged protein combinations shown in (a‐d). Cytoplasm‐to‐nucleus signal ratio was calculated for the GFP (gray bars), and mCherry (black bars) signals obtained for each combination (n = 15–20). Data are represented as mean ± SD. Different letters indicate statistically significant differences (p < .05)
FIT is weakly expressed in leaves (Bauer et al., 2004; Colangelo & Guerinot, 2004; Jakoby et al., 2004), which offers the possibility to test the effect of low FIT levels on bHLH039 subcellular localization. Indeed, bHLH039 showed a dual, nuclear and cytoplasmic, localization in a wild‐type Arabidopsis leaf protoplast expression system (Figure 2a), similar to the subcellular localization of bHLH039‐mCherry in WT protoplasts cotransformed with FIT‐GFP (Figure 2b). In support of our previous observation in tobacco leaf cells, bHLH039 expressed alone in fit‐3 mutant protoplasts was mainly detectable at the cell periphery (Figure 2c), whereas cotransformation with FIT‐GFP restored bHLH039 nuclear accumulation (Figure 2d). Thus, colocalization experiments in both tobacco and Arabidopsis protoplast cells demonstrate the importance of FIT for full‐scale nuclear bHLH039 accumulation.
Figure 2.
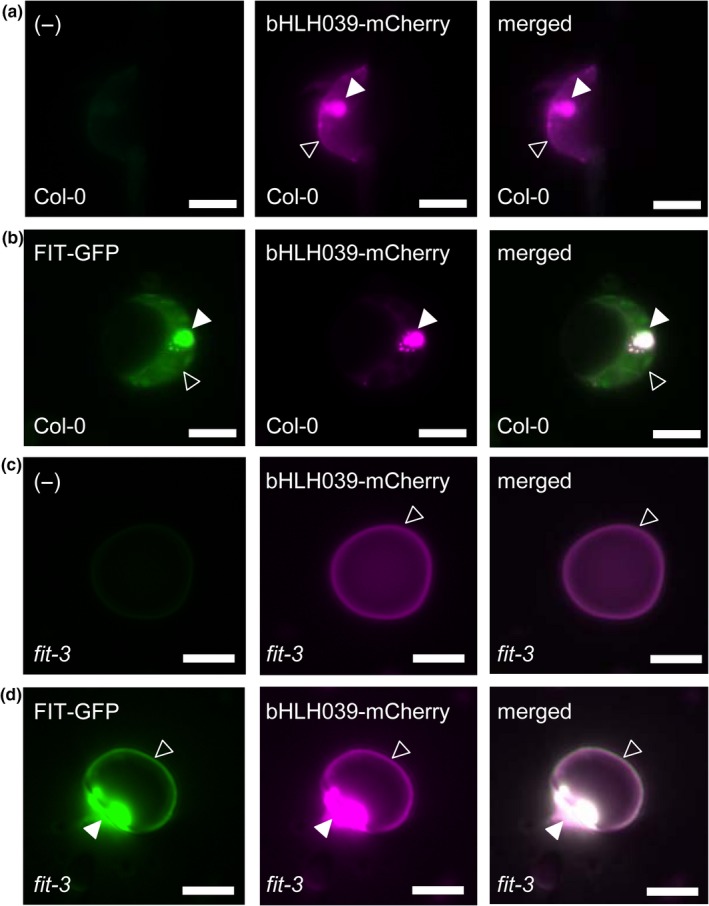
FIT‐dependent subcellular localization of bHLH039 in Arabidopsis protoplasts. Arabidopsis protoplasts from wild‐type Col‐0 (a,b) and fit‐3 mutant (c,d) plants were transformed with bHLH039‐mCherry alone (a,c) or FIT‐GFP together with bHLH039‐mCherry (b,d). Left panels, GFP channel; middle panels, mCherry channel; right panels, merged images. Nuclear and cytoplasmic fluorescence signals are indicated by arrowheads (filled and empty, respectively). Bars: 5 µm
3.2. bHLH039 presence in the nucleus is FIT‐dependent in Arabidopsis
Since FIT presence aids bHLH039 nuclear accumulation in our test systems, we asked whether this is also the case in leaves and roots of Arabidopsis plants grown under different Fe supply conditions. Under Fe deficiency, FIT has a strong expression in the root and much weaker in the leaves (Bauer et al., 2004; Colangelo & Guerinot, 2004; Jakoby et al., 2004), raising the question whether the subcellular distribution of bHLH039 would be differentially regulated between the two organs. To address this, we quantified bHLH039 protein amounts in cytoplasmic and nuclear fractions from leaves (Figure 3a‐c and Figure S3a‐c) and roots (Figure 3d‐f and Figure S3d‐f) of transgenic Arabidopsis plants constitutively expressing triple hemagglutinin‐tagged (HA3)‐bHLH039 in either wild‐type (39/WT) or fit‐3 loss‐of‐function (39/fit) background, grown under sufficient or deficient Fe supply. bHLH039 protein is active in enhancing Fe uptake in roots and leaves of 39/WT but not in 39/fit (Naranjo‐Arcos et al., 2017). HA3‐bHLH039 was found in both leaf and root fractions of 39/WT and 39/fit (Figure 3a,b,d,e and Figure S3a,b,d,e). Remarkably, in the absence of FIT, a clear increase of 1.7 to 3‐fold in cytoplasmic HA3‐bHLH039 (higher cytoplasm‐to‐nucleus signal ratio) is seen in both Fe supply conditions and both organs in 39/fit versus 39/WT (Figure 3c,f and Figure S3c,f). This shows that although a certain portion of bHLH039 can still accumulate in the nucleus in a FIT‐independent manner, the presence of FIT strongly promotes bHLH039 nuclear localization in Arabidopsis. Comparing leaves and roots, we observed a marked difference, depending on Fe supply. In leaves, the nuclear localization of HA3‐bHLH039 increased under Fe deficiency compared with sufficient Fe supply in both plant lines (lower cytoplasm‐to‐nucleus signal ratio at −Fe vs. +Fe, Figure 3c and Figure S3c), whereas in roots the cytoplasm‐to‐nucleus signal ratio was higher under Fe deficiency compared with Fe sufficiency, indicative of decreased nuclear localization of bHLH039 under Fe deficiency, despite the presence of active FIT (Figure 3f and Figure S3f). Taken together, bHLH039 nuclear accumulation is organ‐specific and Fe‐dependent, and, to a large extent, dependent on FIT.
Figure 3.
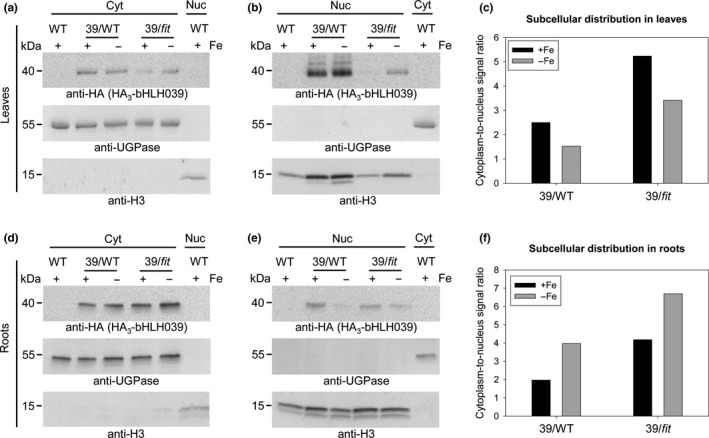
Nuclear accumulation of bHLH039 depends on FIT presence and Fe supply in Arabidopsis leaves and roots. Immunoblot analysis of HA3‐bHLH039 protein distribution in cytosolic (Cyt) (a,d) and nuclear (Nuc) (b,e) fractions of leaves (a,b) and roots (d,e) from plants overexpressing HA3‐bHLH039 in wild‐type (39/WT) and fit‐3 mutant background (39/fit). WT plants were used as a control. Fractionation was performed on 2‐week‐old plants transferred for 3 days on sufficient (+Fe) or deficient (−Fe) Fe supply. HA3‐bHLH039 was detected with anti‐HA‐HRP antibody. Anti‐UGPase and anti‐H3 antibodies were used as markers for the cytosolic and nuclear fractions, respectively. Arrows indicate the detection of the respective full‐length protein. Protein molecular weight (in kDa) is indicated. The intensity of each HA3‐bHLH039 protein band was normalized to the corresponding marker protein for the respective fraction. The obtained normalized values were used to calculate a cytoplasm‐to‐nucleus signal ratio representing the subcellular distribution of HA3‐bHLH039 in leaves (c) and in roots (f) of plants grown under sufficient (+Fe, black bars) and deficient (−Fe, gray bars) Fe supply. The assay was performed twice yielding comparable results (see also Figure S3)
3.3. In the presence of FIT, bHLH039 protein has increased mobility in the cytoplasm
Aiming to better understand the mechanism of bHLH039 localization and its FIT dependence, we measured the relative mobility of FIT and bHLH039 fluorescent protein fusions in the nucleus and the periphery of transformed tobacco cells. We applied fluorescence recovery after photobleaching (FRAP) to bHLH039‐GFP, FIT‐GFP, and free GFP as a control (Figure 4). In the absence of FIT, the bleached bHLH039‐GFP area in the nucleus showed fluorescence recovery, suggesting that in this region, apart from being very weakly present (Figure 4a and Figure S4), the bHLH039‐GFP protein is relatively mobile (Figure 4i). Outside the nucleus, however, the bleached area at the cell periphery remained almost completely devoid of signal, showing that bHLH039‐GFP was kept in an immobile state (Figure 4b,i). A closer inspection of these images revealed that the signal at the cell periphery was not uniformly distributed but rather concentrated in immobile punctate structures (foci) (Figure 4b). In the presence of FIT‐mCherry, bHLH039‐GFP remained mobile in the nucleus (Figure 4c,i). The cytoplasmic bHLH039‐GFP foci, however, were almost completely absent and were replaced by diffuse cytoplasmic signal that recovered quickly after photobleaching (Figure 4d,i). In comparison, FIT‐GFP (Figure 4e,f) and free GFP (Figure 4g,h) signals showed high mobility both in the nucleus and the cytoplasm (Figure 4i). Thus, in the presence of FIT, bHLH039 is kept in a mobile form, while without FIT, it is sequestered in immobile foci at the cell periphery, potentially preventing its entry in the nucleus.
Figure 4.
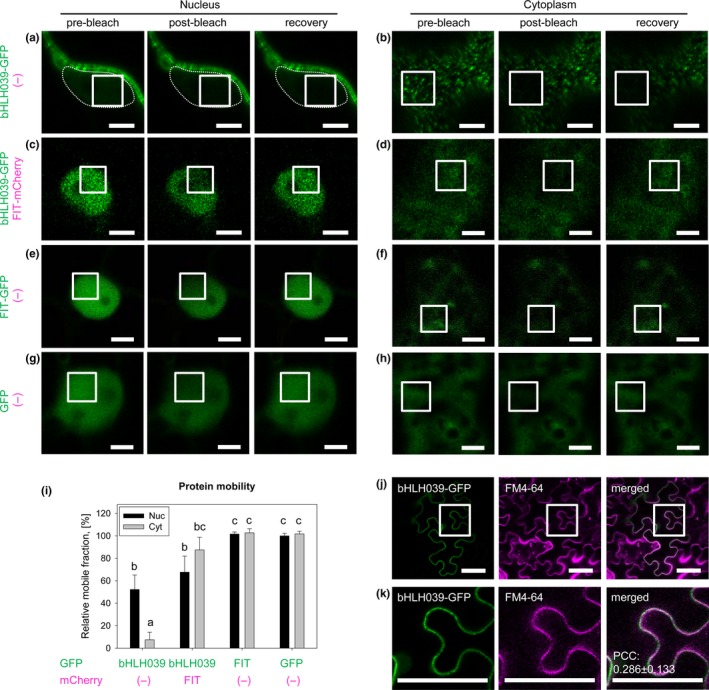
The intracellular mobility of bHLH039 depends on FIT. (a‐i) Fluorescence recovery after photobleaching (FRAP) analysis of the protein mobility of GFP‐tagged FIT and bHLH039. White rectangles in the laser‐scanning confocal images of transiently transformed tobacco leaf epidermis cells indicate the bleached areas in the nucleus (a,c,e,g; optical cross‐sections through the nucleus) and at the cell periphery (b,d,f,h; frontal images of the periphery of the cells). The protein mobility was assayed for bHLH039‐GFP alone (a,b), bHLH039‐GFP coexpressed with FIT‐mCherry (c,d), FIT‐GFP alone (e,f), and free GFP alone as a control (g,h). For each sample, representative pre‐bleach, post‐bleach and recovery images are shown. Due to the low intensity of the nuclear bHLH039‐GFP signal in (a), the nucleus contour is traced with a punctate line. For an enhanced intensity image of nuclear bHLH039‐GFP, see Figure S4. Bars: 5 µm. (i) Protein mobility quantification for the samples shown in (a‐h). The percentage of the mobile protein fraction for each protein is presented relative to the mobile fraction of nuclear free GFP alone (n = 10–15). Data are represented as mean ± SD. Different letters indicate statistically significant differences (p < .05). (j‐k) Colocalization analysis of GFP‐tagged bHLH039 and the plasma membrane labeled with FM4‐64. (k) Enlarged images of the areas in (j) indicated with white rectangles. bHLH039‐GFP signals are detectable in close proximity to the plasma membrane; however, statistical analysis indicates lack of significant colocalization. Pearson's correlation coefficient (PCC) = .286 ± .133 (n = 7). Bars: 50 µm
The intriguing localization of bHLH039‐GFP in static foci at the cell periphery in the absence of FIT raises the question on the nature of these structures and their relation to the plasma membrane. Using the lipophilic fluorescent dye FM4‐64 to label the plasma membrane, we investigated the degree of colocalization between bHLH039‐GFP and the marker staining. The bHLH039‐GFP and FM4‐64 signals were both located at the cell periphery, however, despite their proximity they were distinct from each other. In addition, FM4‐64 fluorescence was uniform across the membrane and not in foci (Figure 4j,k). Consistently, quantitative analysis showed relatively poor colocalization between the two signals, with an average Pearson's correlation coefficient (PCC) of .286 (while well‐colocalizing signals have a PCC of above .5) (Figure 4k). Therefore, the bHLH039‐GFP signals are in proximity to but not at the plasma membrane.
4. DISCUSSION
4.1. Nucleocytoplasmic partitioning of bHLH039 is FIT‐dependent
We report here a pattern of bHLH039 localization that changes depending on the presence of FIT in the cell. It is known that the effects of bHLH039 on the Fe deficiency response in Arabidopsis are FIT‐dependent and due to the dimerization of the two proteins (Naranjo‐Arcos et al., 2017; Wang et al., 2013). Our data adds a new level of complexity to the FIT‐bHLH039 regulation. bHLH039 accumulates to a low level in the nucleus in the absence of FIT, suggesting that additional factor(s), potentially other related transcription factors (Gao et al., 2019), may also influence this process. Still, FIT is essential for full‐scale bHLH039 nuclear accumulation. In the absence of FIT, bHLH039 is retained in immobile foci in close proximity to the plasma membrane. In the presence of FIT, bHLH039 is mobile and readily accumulates in the nucleus (Figure 5).
Figure 5.
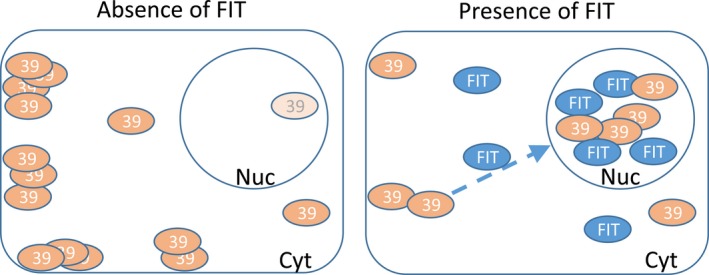
Different FIT‐dependent subcellular localization patterns of bHLH039. Model summarizing the need of FIT presence for full‐scale bHLH039 nuclear accumulation. Left, in the absence of FIT, bHLH039 is predominantly localized outside of the nucleus, concentrated in stable cytoplasmic foci in close proximity to the plasma membrane. Only a small portion of bHLH039 localizes to the nucleus. Right, in Fe‐deficient wild‐type root cells and/or in the presence of FIT, FIT aids the nuclear accumulation of bHLH039 preventing its sequestration in cytoplasmic foci. In the nucleus, the FIT‐bHLH039 protein complex triggers the expression of Fe response genes
4.2. Potential mechanisms of bHLH039 nuclear accumulation
The accumulation of bHLH039 in the nucleus in the presence of FIT may be achieved by different mechanisms. One possibility is that FIT helps maintain bHLH039 in a mobile form and/or shuttle together with it into the nucleus. Alternatively, FIT may prevent the nuclear export of bHLH039 and its sequestration in cytoplasmic foci. The physical exclusion of transcriptional regulators from the nucleus by sequestration in the cytoplasm is one strategy for regulating their activity. In animals, Bach1, STRA8, and the STAT family proteins are prominent examples of transcription factors translocating from the cytoplasm to the nucleus in response to different stimuli (Meyer & Vinkemeier, 2004; Tedesco, Sala, Barbagallo, Felici, & Farini, 2009; Yamasaki, Tashiro, Nishito, Sueda, & Igarashi, 2005). There are also several examples in plants, such as the transcription factors BES1 and BZR1 in response to brassinosteroid signaling (Yin et al., 2002), the PHR1 transcription factor in a phosphate status‐dependent manner (Osorio et al., 2019), and the chromatin remodeling factor ET2 during cell differentiation (Ivanov et al., 2008). Interestingly, when retained in the cytoplasm, ET2 also accumulates in punctate structures (Ivanov et al., 2008), which may thus represent a common mechanism for the sequestration of proteins not immediately required by the cell. The BHLH039 gene is strongly upregulated under Fe deficiency, suggesting that only minor levels of the protein are required at sufficient Fe supply, requiring a mechanism that keeps excess protein excluded from action. This notion is supported by the fact that overexpression of bHLH039 at +Fe probably overloads the Fe sequestration system and has drastic effects on Fe accumulation, since enough FIT is present to keep bHLH039 mobile and/or shuttle it to the nucleus (Naranjo‐Arcos et al., 2017). Further research will be needed to uncover the exact nature of the cytoplasmic foci, where bHLH039 accumulates in the absence of FIT, and the participating proteins that potentially anchor bHLH039.
Another aspect to consider is whether active FIT is needed for bHLH039 nuclear accumulation. We could recently show that FIT phosphorylation by CIPK11 at Ser272 is a major FIT‐activating process, whereby both Ser272‐phosphorylated and non‐phosphorylated FIT forms can interact with bHLH039, with the non‐phosphorylated S272AA FIT form showing a weaker interaction with bHLH039 (Gratz, Manishankar, et al., 2019). Additionally, phosphorylation events at Ser221, Tyr238, and Tyr278 also affect FIT activity (Gratz, Brumbarova, et al., 2019). Therefore, it is possible that differential FIT phosphorylation may modulate FIT activity levels and, in turn, the effect of FIT on bHLH039 nuclear accumulation. In the future, it will be crucial to understand the mechanism by which FIT influences bHLH039 cellular mobility, its nuclear accumulation, and the role of their direct interaction in this process.
4.3. Biological significance of FIT‐dependent bHLH039 nuclear accumulation
Our finding has interesting implications regarding the Fe deficiency‐induced signaling cascade. It is tempting to speculate that the cytoplasmic retention of bHLH039 could be linked to direct crosstalk at protein level with other Fe deficiency‐related proteins that are localized at the plasma membrane, such as the effectors FRO2 and IRT1, and/or the Fe response modulators CBL1/9 (Gratz, Manishankar, et al., 2019) and EHB1 (Khan et al., 2019). Posttranslational control plays a prominent role in the functioning of bHLH039 and potentially the other three group Ib bHLH proteins involved in Fe acquisition and homeostasis. It is possible that fit mutant plants are functional knock‐out mutants of the four Ib bHLH proteins due to their reduced nuclear accumulation in the absence of FIT, which may explain the very strong Fe deficiency phenotype of the fit mutant.
The differences in subcellular partitioning of bHLH039 between leaves and roots at different Fe supply may reflect different organ‐specific functions of this transcription factor (Andriankaja et al., 2014). Proteasomal degradation, similar to the stability control shown for FIT (Lingam et al., 2011; Meiser et al., 2011; Sivitz et al., 2011), and/or other factor(s) that negatively regulate(s) bHLH039 nuclear accumulation, may account for the observed differential subcellular partitioning.
An important aspect of the phenomenon that we describe here is its potential conservation across the plant kingdom. A recent study in rice identified OsbHLH156 as a rice ortholog of FIT, albeit with low sequence similarity and controlling Strategy II, instead of Strategy I, Fe uptake. Interestingly, this transcription factor was found necessary for the nuclear localization of OsIRO2, the rice ortholog of bHLH039 (Wang et al., 2019). This underlines the importance of the here described phenomenon in Arabidopsis as an integral part of the plant Fe deficiency response.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
K.T., R.I., P.B., and T.B. designed the research. K.T., R.I., and T.B. designed experiments. K.T., R.I., M.E., B.A., I.M., and T.B. performed research. K.T., R.I., and T.B. analyzed data. R.I. and T.B. wrote the manuscript. K.T., R.I., I.M., P.B., and T.B. revised the manuscript. P.B. acquired funding. All authors participated in discussion of the manuscript.
Supporting information
ACKNOWLEDGMENTS
R.I., I.M., and P.B. are members of CRC 1208; I.M. is member of the MB Train graduate school, integrated in CRC 1208. This study was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 267205415—SFB 1208, project B05 to P.B. This work received funding from Germany´s Excellence Strategy, EXC 2048/1, Project ID: 390686111.
Trofimov K, Ivanov R, Eutebach M, et al. Mobility and localization of the iron deficiency‐induced transcription factor bHLH039 change in the presence of FIT. Plant Direct. 2019;3:1–11. 10.1002/pld3.190
Trofimov and Ivanov contributed equally to this article.
Contributor Information
Petra Bauer, Email: Petra.Bauer@uni-duesseldorf.de.
Tzvetina Brumbarova, Email: Tzvetina.Brumbarova@uni-duesseldorf.de.
REFERENCES
- Andriankaja, M. E. , Danisman, S. , Mignolet‐Spruyt, L. F. , Claeys, H. , Kochanke, I. , Vermeersch, M. , … Inzé, D. (2014). Transcriptional coordination between leaf cell differentiation and chloroplast development established by TCP20 and the subgroup Ib bHLH transcription factors. Plant Molecular Biology, 85, 233–245. [DOI] [PubMed] [Google Scholar]
- Bancaud, A. , Huet, S. , Rabut, G. , & Ellenberg, J. (2010). Fluorescence perturbation techniques to study mobility and molecular dynamics of proteins in live cells: FRAP, photoactivation, photoconversion, and FLIP. Cold Spring Harbor Protocols, 2010(12), pdb top90. [DOI] [PubMed] [Google Scholar]
- Bauer, P. , Thiel, T. , Klatte, M. , Bereczky, Z. , Brumbarova, T. , Hell, R. , & Grosse, I. (2004). Analysis of sequence, map position, and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiology, 136, 4169–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann, A. , Weidtkamp‐Peters, S. , Seidel, C. A. , & Simon, R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiology, 152, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte, S. , & Cordelieres, F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. Journal of Microscopy, 224, 213–232. [DOI] [PubMed] [Google Scholar]
- Brumbarova, T. , Bauer, P. , & Ivanov, R. (2015). Molecular mechanisms governing Arabidopsis iron uptake. Trends in Plant Science, 20, 124–133. [DOI] [PubMed] [Google Scholar]
- Colangelo, E. P. , & Guerinot, M. L. (2004). The essential basic helix‐loop‐helix protein FIT1 is required for the iron deficiency response. The Plant Cell, 16, 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovzhenko, A. , Dal Bosco, C. , Meurer, J. , & Koop, H. U. (2003). Efficient regeneration from cotyledon protoplasts in Arabidopsis thaliana. Protoplasma, 222, 107–111. [DOI] [PubMed] [Google Scholar]
- Gao, F. , Robe, K. , Gaymard, F. , Izquierdo, E. , & Dubos, C. (2019). The transcriptional control of iron homeostasis in plants: A tale of bHLH transcription factors? Frontiers in Plant Science, 10, 6 10.3389/fpls.2019.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz, R. , Brumbarova, T. , Ivanov, R. , Trofimov, K. , Tunnermann, L. , Ochoa‐Fernandez, R. , … Bauer, P. (2019). Phospho‐mutant activity assays provide evidence for alternative phospho‐regulation pathways of the transcription factor FER‐LIKE IRON DEFICIENCY‐INDUCED TRANSCRIPTION FACTOR. New Phytologist. 10.1111/nph.16168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz, R. , Manishankar, P. , Ivanov, R. , Köster, P. , Mohr, I. , Trofimov, K. , … Brumbarova, T. (2019). CIPK11‐dependent phosphorylation modulates FIT activity to promote arabidopsis iron acquisition in response to calcium signaling. Developmental Cell, 48(5), 726–740. 10.1016/j.devcel.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Heim, M. A. , Jakoby, M. , Werber, M. , Martin, C. , Weisshaar, B. , & Bailey, P. C. (2003). The basic helix‐loop‐helix transcription factor family in plants: A genome‐wide study of protein structure and functional diversity. Molecular Biology and Evolution, 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Ivanov, R. , Brumbarova, T. , & Bauer, P. (2012). Fitting into the harsh reality: Regulation of iron deficiency responses in dicotyledonous plants. Molecular Plant, 5, 27–42. 10.1093/mp/ssr065 [DOI] [PubMed] [Google Scholar]
- Ivanov, R. , Brumbarova, T. , Blum, A. , Jantke, A. M. , Fink‐Straube, C. , & Bauer, P. (2014). SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON‐REGULATED TRANSPORTER1. The Plant Cell, 26, 1294–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, R. , Tiedemann, J. , Czihal, A. , Schallau, A. , le Diep, H. , Mock, H. P. , … Baumlein, H. (2008). EFFECTOR OF TRANSCRIPTION2 is involved in xylem differentiation and includes a functional DNA single strand cutting domain. Developmental Biology, 313, 93–106. [DOI] [PubMed] [Google Scholar]
- Jakoby, M. , Wang, H. Y. , Reidt, W. , Weisshaar, B. , & Bauer, P. (2004). FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana . FEBS Letters, 577, 528–534. [DOI] [PubMed] [Google Scholar]
- Jeong, J. , Merkovich, A. , Clyne, M. , & Connolly, E. L. (2017). Directing iron transport in dicots: Regulation of iron acquisition and translocation. Current Opinion in Plant Biology, 39, 106–113. [DOI] [PubMed] [Google Scholar]
- Khan, I. , Gratz, R. , Denezhkin, P. , Schott‐Verdugo, S. N. , Angrand, K. , Genders, L. , … Ivanov, R. (2019). Calcium‐promoted interaction between the C2‐domain protein EHB1 and metal transporter IRT1 inhibits Arabidopsis iron acquisition. Plant Physiology, 180, 1564–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Li, Y. , Zhao, Q. , Li, T. , Wei, J. , Li, B. , … Gao, C. (2019). The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nature Plants, 5, 512–524. [DOI] [PubMed] [Google Scholar]
- Lingam, S. , Mohrbacher, J. , Brumbarova, T. , Potuschak, T. , Fink-Straube, C. , Blondet, E. , … Bauer, P. (2011). Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. The Plant Cell, 23, 1815–1829. 10.1105/tpc.111.084715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, I. , & Somers, D. E. (2011). Regulation of nucleocytoplasmic trafficking in plants. Current Opinion in Plant Biology, 14, 538–546. [DOI] [PubMed] [Google Scholar]
- Meiser, J. , Lingam, S. , & Bauer, P. (2011). Posttranslational regulation of the iron deficiency basic helix‐loop‐helix transcription factor FIT is affected by iron and nitric oxide. Plant Physiology, 157, 2154–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, T. , & Vinkemeier, U. (2004). Nucleocytoplasmic shuttling of STAT transcription factors. European Journal of Biochemistry, 271, 4606–4612. [DOI] [PubMed] [Google Scholar]
- Naranjo‐Arcos, M. A. , Maurer, F. , Meiser, J. , Pateyron, S. , Fink‐Straube, C. , & Bauer, P. (2017). Dissection of iron signaling and iron accumulation by overexpression of subgroup Ib bHLH039 protein. Scientific Reports, 7(1), 10911 10.1038/s41598-017-11171-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio, M. B. , Ng, S. , Berkowitz, O. , De Clercq, I. , Mao, C. , Shou, H. , … Jost, R. (2019). SPX4 acts on PHR1‐dependent and ‐independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiology, 181(1), 332–352. 10.1104/pp.18.00594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz, A. , Grinvalds, C. , Barberon, M. , Curie, C. , & Vert, G. (2011). Proteasome‐mediated turnover of the transcriptional activator FIT is required for plant iron‐deficiency responses. The Plant Journal, 66(6), 1044–1052. 10.1111/j.1365-313X.2011.04565.x [DOI] [PubMed] [Google Scholar]
- Tedesco, M. , La Sala, G. , Barbagallo, F. , De Felici, M. , & Farini, D. (2009). STRA8 shuttles between nucleus and cytoplasm and displays transcriptional activity. Journal of Biological Chemistry, 284, 35781–35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwieger, A. , Gryczka, C. , Czihal, A. , Douchkov, D. , Tiedemann, J. , Mock, H. P. , … Baumlein, H. (2007). Iron assimilation and transcription factor controlled synthesis of riboflavin in plants. Planta, 226, 147–158. [DOI] [PubMed] [Google Scholar]
- Wang, H. Y. , Klatte, M. , Jakoby, M. , Baumlein, H. , Weisshaar, B. , & Bauer, P. (2007). Iron deficiency‐mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana . Planta, 226, 897–908. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Cui, Y. , Liu, Y. , Fan, H. , Du, J. , Huang, Z. , … Ling, H. Q. (2013). Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana . Molecular Plant, 6, 503–513. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Li, L. , Ying, Y. , Wang, J. , Shao, J. F. , Yamaji, N. , … Shou, H. (2019). A transcription factor OsbHLH156 regulates Strategy II iron acquisition through localizing IRO2 to the nucleus in rice. New Phytologist. 10.1111/nph.16232 [DOI] [PubMed] [Google Scholar]
- Yamasaki, C. , Tashiro, S. , Nishito, Y. , Sueda, T. , & Igarashi, K. (2005). Dynamic cytoplasmic anchoring of the transcription factor Bach1 by intracellular hyaluronic acid binding protein IHABP. Journal of Biochemistry, 137, 287–296. [DOI] [PubMed] [Google Scholar]
- Yin, Y. , Wang, Z. Y. , Mora‐Garcia, S. , Li, J. , Yoshida, S. , Asami, T. , & Chory, J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell, 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Yuan, Y. , Wu, H. , Wang, N. , Li, J. , Zhao, W. , Du, J. , … Ling, H. Q. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Research, 18, 385–397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


