Abstract
Spatial regulation of microtubule catastrophe is important for controlling microtubule length and consequently contributes to the proper establishment of cell polarity and cell growth. The +TIP proteins including Tip1/CLIP-170, Klp5/Kinesin-8, and Alp14/XMAP215 reside at microtubule plus ends to regulate microtubule dynamics. In the fission yeast Schizosaccharomyces pombe, Tip1 and Alp14 serve as microtubule-stabilizing factors, while Klp5 functions oppositely as a catastrophe-promoting factor. Despite that Tip1 has been shown to play a key role in restricting microtubule catastrophe to the cell end, how Tip1 fulfills the role remains to be determined. Employing live-cell microscopy, we showed that the absence of Tip1 impairs the localization of both Klp5 and Alp14 at microtubule plus ends, but the absence of Klp5 prolongs the residence time of Tip1 at microtubule plus ends. We further revealed that Klp5 accumulates behind Tip1 at microtubule plus ends in a Tip1-dependent manner. In addition, artificially tethering Klp5 to microtubule plus ends promotes premature microtubule catastrophe, while tethering Alp14 to microtubule plus ends in the cells lacking Tip1 rescues the phenotype of short microtubules. These findings establish that Tip1 restricts microtubule catastrophe to the cell end likely by spatially restricting the microtubule catastrophe activity of Klp5 and stabilizing Alp14 at microtubule plus ends. Thus, the work demonstrates the orchestration of Tip1, Alp14, and Klp5 in ensuring microtubule catastrophe at the cell end.
Keywords: microtubule, microtubule catastrophe, kinesin-8, Tip1, Alp14
Introduction
The microtubule cytoskeleton is involved in a wide range of cellular activities and displays dynamic instability (Brouhard and Rice, 2018). Within the cell, microtubule minus ends are generally capped by the gamma-tubulin ring complex and thus exhibit less dynamic than microtubule plus ends (Petry and Vale, 2015). At microtubule plus ends reside a large number of proteins named +TIPs, which possess various abilities in modulating microtubule dynamics to meet the demands of cells in response to the internal and external environmental changes (Kumar and Wittmann, 2012). In the fission yeast Schizosaccharomyces pombe, proper extension of microtubule plus ends to the cell ends is important for delivering polarity factors to direct cell growth (Chang and Martin, 2009), and similar roles of microtubule plus ends in establishing cell polarity and directing cell migration are found in mammalian cells (Jiang et al., 2009; Adams et al., 2016).
Among +TIPs, the evolutionarily conserved protein EB1 serves as a hub for recruiting many other +TIPs to microtubule plus ends (Kumar and Wittmann, 2012; Xia et al., 2014). In the fission yeast S. pombe, the EB1 counterpart Mal3 forms a complex with Tip1 (CLIP-170 in human) and Tea2 (Kinesin) at the plus ends of growing microtubules (Browning et al., 2003; Busch and Brunner, 2004; Busch et al., 2004). The Mal3/Tip1/Tea2 complex plays important roles in inhibiting microtubule catastrophe and in regulating cell polarity (Browning et al., 2003; Busch and Brunner, 2004). In cells lacking any component of the Mal3/Tip1/Tea2 complex, microtubules are short and undergo premature catastrophe before reaching the cell end (Brunner and Nurse, 2000; Busch and Brunner, 2004; Busch et al., 2004). In contrast, microtubules in wild-type (WT) cells generally undergo catastrophe only after reaching the cell end. Despite the fact that Tip1 has been shown to play a critical role in restricting microtubule catastrophe to the cell end (Busch and Brunner, 2004), how Tip1 fulfills the role remains elusive.
Multiple reports have demonstrated the important role of kinesin-8 family proteins in promoting microtubule catastrophe (Gupta et al., 2006; Varga et al., 2006; Unsworth et al., 2008; Tischer et al., 2009; Varga et al., 2009; Gardner et al., 2011). The kinesin-8 family proteins are highly processive and are able to accumulate at microtubule plus ends (Leduc et al., 2012; Zheng et al., 2014; Arellano-Santoyo et al., 2017). Similarly, in fission yeast, two kinesin-8 proteins, Klp5 and Klp6, work as a heterodimeric kinesin at microtubule plus ends to promote microtubule catastrophe (Garcia et al., 2002). In the absence of either Klp5 or Klp6, microtubules are unusually long and curve around the cell end (West et al., 2001; Garcia et al., 2002; Unsworth et al., 2008). The interplay between Mal3/Tip1/Tea2 and Klp5/Klp6 in regulating microtubule dynamics is unclear. Similar to Mal3/Tip1/Tea2 and Klp5/Klp6, the evolutionarily conserved tumor overexpressed gene (TOG) domain containing proteins XMAP215/Stu2/Alp14 also localizes to microtubule plus ends, presumably at the extreme end, where it functions as a polymerase to promote microtubule growth (Slep and Vale, 2007; Widlund et al., 2011; Al-Bassam et al., 2012; Ayaz et al., 2012). Although both Alp14 and Tip1 functions similarly to prevent microtubule catastrophe, the relationship between the two proteins is poorly understood.
In this work, we delineated the interplay between Tip1, Klp5, and Alp14 in regulating microtubule catastrophe and found that Tip1 plays critical roles in restricting the microtubule catastrophe activity of Klp5 to the cell end and in stabilizing Alp14 at microtubule plus ends. Thus, the work demonstrates the concerted actions of the microtubule stabilizing and destabilizing factors in the spatial regulation of microtubule catastrophe.
Results
Deletion of klp5 in cells lacking tip1 partially restores WT microtubule properties
In the fission yeast S. pombe, microtubules generally depolymerize after reaching the cell end, and it has been shown that Tip1 functions as a catastrophe-inhibiting factor to restrict microtubule catastrophe to the cell end (Busch and Brunner, 2004). In contrast, the heterodimeric kinesin Klp5/6 (Klp5 and Klp6) promotes microtubule catastrophe (Unsworth et al., 2008; Tischer et al., 2009). Since the absence of either Klp5 or Klp6 affects microtubule dynamics similarly, we focused on Klp5 in the present work. Microtubules are unusually long and often curve around the cell end in klp5Δ cells, whereas microtubules in tip1Δ cells are very short (Brunner and Nurse, 2000; West et al., 2001; Garcia et al., 2002). These opposite effects of klp5 and tip1 deletion on microtubules prompted us to ask whether deletion of both tip1 and klp5 could restore WT microtubule properties.
To address this question, we first imaged WT, tip1Δ, klp5Δ, and tip1Δklp5Δ cells expressing mCherry-Atb2 (α-tubulin). Consistent with previous works (Brunner and Nurse, 2000; West et al., 2001; Garcia et al., 2002), our imaging data showed that short and long buckled microtubules were present in tip1Δ and klp5Δ cells, respectively (Figure 1A). Interestingly, the microtubule length of tip1Δklp5Δ cells was indeed longer than the microtubule length of tip1Δ cells but shorter than the microtubule length of klp5Δ cells (Figure 1A). Quantitative measurements of microtubule length also confirmed the findings (Figure 1B). Such restoration effect on the microtubule length by the double deletion was partial since the microtubule length of tip1Δklp5Δ cells was still shorter than the microtubule length of WT cells (Figure 1B). We noticed that cell length appeared to be altered in the mutant cells. To exclude the effect of the altered cell length on the quantification, we performed Pearson correlation analysis of microtubule length and cell length. As shown in Supplementary Figure S1B, the correlation coefficients (R) are less than 0.35, suggesting almost no linear relationship between the tested variables. Therefore, it is unlikely that the altered cell length affects the quantification of microtubule length.
Figure 1.
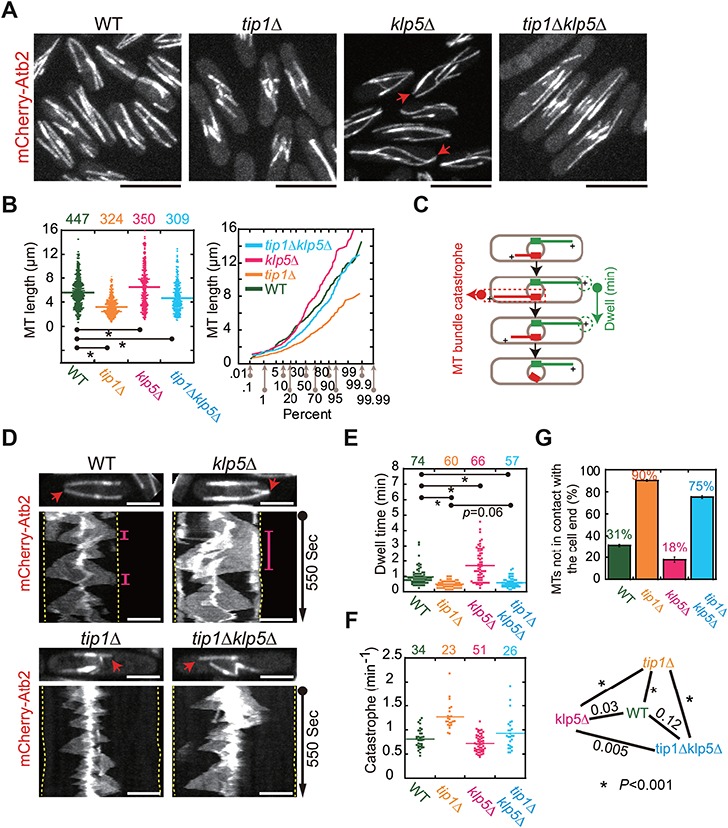
Microtubule length and dynamics in WT, tip1Δ, klp5Δ, and tip1Δklp5Δ cells. (A) Maximum projection images of WT, tip1-deletion (tip1Δ), klp5-deletion (klp5Δ), and double deletion (tip1Δklp5Δ) cells expressing mCherry-Atb2 (α-tubulin). Red arrows mark the microtubules that curve around the cell end. Scale bar, 10 μm. (B) Dot and probability plots of microtubule length measured for the four types of cells in A. The measured cell number is shown on the top of the graph. P-values were calculated by student’s t-test, and the stars indicate P < 0.001. The X-axis of the probability plot shows the percentage of the microtubule length that is less than the data point. (C) Diagram illustrating microtubule dynamics within a cell. The dwell time of a microtubule at the cell end is the time during which the microtubule plus end is in contact with the cell end (green microtubule). Due to the resolution limit (~300 nm) of conventional confocal microscopy, a single microtubule is unresolvable. The catastrophe frequency is the frequency of microtubule bundles as indicated (in red). (D) Kymograph analysis of WT, tip1Δ, klp5Δ, and tip1Δklp5Δ cells expressing mCherry-Atb2. Red arrows mark the microtubules used to create the kymograph graphs. The pink lines indicate the period of time when the microtubules were in contact with the cell ends (marked by dashed lines). Scale bar, 5 μm. (E) Dot plots of the dwell time of the microtubules in contact with the cell end. Statistical analysis was performed by student’s t-test (stars, P < 0.001), and the measured cell number is shown on the top of the graph. (F) Dot plots of the catastrophe frequency of microtubule bundles. Statistical analysis was performed by student’s t-test, and the stars on the right indicate P < 0.001. The measured cell number is shown on the top of the graph. (G) The percentage of microtubules not able to contact the cell end. A triplet of live-cell imaging experiments were performed for each type of the indicated cells in parallel. The top of the column is the mean of the triplet quantifications (see Supplementary Figure S1A for each individual quantification), and error bars represent standard deviation (SD).
Given the important roles of Tip1 and Klp5 in regulating microtubule catastrophe, we then analyzed the catastrophe frequency of microtubules and the dwelling time of microtubule plus ends at the cell end (Figure 1C). The dwelling time was measured for only the microtubules that reached the cell end, while the catastrophe frequency was determined for all microtubules. Due to the resolution limit of conventional confocal microscopy (~300 nm), a single microtubule is unresolvable. Therefore, the catastrophe frequency in this present work was determined with microtubule bundles, and thus the catastrophe frequency reflected the dynamics of microtubule bundles. To visualize microtubule dynamics, we employed live-cell spinning disk microscopy and took stack images containing three planes with a 0.5-μm step size every 5 sec for WT, tip1Δ, klp5Δ, and tip1Δklp5Δ cells. Consistent with the role of Klp5 in promoting microtubule catastrophe, microtubules in klp5Δ cells were more stable at the cell end than the microtubules in other cells (Figure 1D). This was further supported by the quantitative measurements; the dwelling time of microtubule plus ends at the cell end increased significantly in klp5Δ cells, while the catastrophe frequency of microtubules decreased significantly in klp5Δ cells (Figure 1E and F). By contrast, microtubules in tip1Δ cells were more dynamic since the catastrophe frequency increased significantly and the dwelling time decreased significantly (Figure 1E and F). It is unlikely that the altered cell length of mutant cells affected the quantification because Pearson correlation analysis showed almost no linear relationship between dwell time, as well as catastrophe frequency, and cell length (Supplementary Figure S1C and D). It was apparent that the majority of the microtubules in tip1Δ cells underwent premature catastrophe before touching the cell end (Figure 1D and G). Similar to the microtubules in tip1Δ cells, the majority of the microtubules in tip1Δklp5Δ cells underwent premature catastrophe (Figure 1D and G). Although the microtubules in tip1Δklp5Δ cells were able to grow longer than the microtubules in tip1Δ cells (Figure 1A and B), the catastrophe frequency of the microtubules in tip1Δklp5Δ and WT cells was comparable (Figure 1F). Taken together, the findings suggest that Tip1 and Klp5 regulate microtubule catastrophe in an antagonistic manner.
Tip1 is required for the proper accumulation of Klp5 at microtubule plus ends
To understand the interplay between Tip1 and Klp5 in regulating microtubule catastrophe, we imaged WT cells expressing Tip1-tdTomato and Klp5-2mNeonGreen at their own loci by high-temporal live-cell microscopy (single plane, 1-sec intervals). As shown in Figure 2A, three characteristic phases of Klp5 dynamics were identified. Initially, multiple Klp5-2mNeonGreen foci emerged on the growing microtubule and moved towards the microtubule plus end decorated by Tip1-tdTomato; the Klp5 traffic appeared to halt upon encountering Tip1 and did not pass Tip1 (a phase of following Tip1). During the second phase, the microtubule was in contact with the cell end, and the Klp5 intensity at the microtubule plus end continuously rose (a phase of Klp5 accumulation). During the third phase, Tip1 colocalized with Klp5 at the microtubule plus end, and upon Tip1 dissociation, the microtubule began to depolymerize (a phase of Tip1 and Klp5 colocalization). The adjacent nature of Tip1 and Klp5 localization was also evident by kymograph analysis in which the trajectories of Tip1 and Klp5 adjoined each other (Figure 2B). It is therefore conceivable that Tip1 may serve as a tip blocker to halt Klp5 traffic at microtubule plus ends. It is also possible that Tip1 simply restricts Klp5 from microtubule plus ends.
Figure 2.

The absence of Tip1 affects the microtubule localization of Klp5. (A) Maximum time-lapse images of a WT cell expressing Tip1-tdTomato and Klp5-2mNeonGreen. Line-scan intensity measurements were performed along the highlighted microtubule (dashed rectangle). The intensity plot for each frame is shown below, and the corresponding frame time is indicated. The three phases of Klp5 dynamic behaviors are indicated. Scale bar, 5 μm. (B) Kymograph analysis of Tip1-tdTomato and Klp5-2mNeonGreen for the microtubule marked by the red arrow. Dashed lines indicate cell ends, and the magnified images indicated by the dashed rectangle are shown on the right. Scale bar, 5 μm. (C) Maximum projection images of WT and tip1Δ cells expressing Klp5-2mNeonGreen and mCherry-Atb2. Note that the majority of Klp5-2mNeonGreen was present in the nucleus, and apparent Klp5-2mNeonGreen signals (highlighted by red arrows) were also seen at microtubule plus ends in WT cells but not in tip1Δ cells. Scale bar, 10 μm. (D) Kymograph analysis of Klp5-2mNeonGreen and mCherry-Atb2 in WT and tip1Δ cells. Dashed lines mark the cell ends, and red arrows indicate the analyzed microtubules. Note that microtubules underwent catastrophe before reaching the cell ends in tip1Δ cells in which Klp5-2mNeonGreen was still present at the microtubule plus ends. Scale bar, 5 μm. (E) Intensity measurements of the microtubules indicated by the dashed arrows. Note that the frame of images taken from time-lapse movies for intensity measurement was selected based on the criterion that the measured microtubule was about to depolymerize. The intensity plots are shown under each cell. Red arrows indicate the peak Klp5 signals at the tips of the microtubules. The corresponding quantification data are included in Supplementary Figure S2. Scale bar, 10 μm. (F) Maximum projection images of WT and klp5Δ cells expressing Tip1-GFP. Scale bar, 10 μm. (G) Kymograph analysis of the marked microtubules (red arrows) in WT and klp5Δ cells expressing mCherry-Atb2 and Tip1-GFP. The dashed lines mark the cell ends. Scale bar, 5 μm.
To test the hypothesis, we went on to assess the effect of losing Tip1 on Klp5 localization. Maximum projection imaging (11 optical planes with 0.5 μm spacing) showed that the accumulation of Klp5 to microtubule plus ends was impaired in the absence of Tip1 (Figure 2C). These data demonstrate the important role of Tip1 in maintaining Klp5 at microtubule plus ends. We further performed high-temporal live-cell imaging to monitor Klp5 dynamics in WT and tip1Δ cells. As shown in the kymographs (Figure 2D), the absence of Tip1 did not abolish the accumulation of Klp5 at microtubule plus ends but appeared to compromise the accumulation of Klp5 there. Evidently, intensity measurements of the microtubules in the selected image frames, in which the microtubules were about to depolymerize, showed that Klp5 intensity at the microtubule plus ends in tip1Δ cells was attenuated (Figure 2E and Supplementary Figure S2). The residual Klp5 at microtubule plus ends in tip1Δ cells is still capable of promoting microtubule catastrophe because the catastrophe frequency of microtubules in tip1Δ cells is significantly higher than the one in tip1Δklp5Δ cells (Figure 1F).
Additionally, we assessed the effect of losing Klp5 on Tip1 localization. We first examined WT and klp5Δ cells expressing only Tip1-GFP. As shown in Figure 2F, the localization patterns of Tip1-GFP in both cell types were indistinguishable except that slightly stronger Tip1 staining at the cell ends in klp5Δ cells was detected. We then performed live-cell microscopy to monitor Tip-GFP dynamics in WT and klp5Δ cells expressing mCherry-Atb2. As shown in Figure 2G, Tip1-GFP was present at microtubule plus ends in the two types of cells but stayed longer at the microtubule plus end that was contacting the cell tip in klp5Δ cells. Therefore, Klp5 accumulation may contribute to the dissociation of Tip1 from microtubule plus ends. Together, the results suggest that Tip1 is required for the proper accumulation of Klp5 at microtubule plus ends, but the localization of Tip1 at microtubule plus ends is independent of Klp5.
Tethering Klp5 to microtubule plus ends results in premature microtubule catastrophe
It has been reported that the absence of Klp5 results in long and buckled microtubules (West et al., 2001; Garcia et al., 2002). This leads to the notion that Klp5 is a catastrophe factor in promoting microtubule depolymerization (Unsworth et al., 2008; Tischer et al., 2009). Our present work further demonstrated that Klp5 and Tip1 work in an antagonistic manner to control microtubule catastrophe (Figure 1F and G) and that Tip1 likely serves as a tip blocker to accumulate Klp5 at microtubule plus ends (Figure 2). It is then important to determine whether Klp5 functions directly or indirectly to promote microtubule catastrophe.
If Klp5 is a direct catastrophe factor, we expect that tethering Klp5 to microtubule plus ends could overcome the inhibitory effect of Tip1 on microtubule depolymerization and promotes microtubule catastrophe. To test the hypothesis, we employed the green fluorescent protein (GFP)-binding protein (GBP)-targeting approach to tether Klp5 to microtubule plus ends (Rothbauer et al., 2006, 2008; Chen et al., 2017). Green fluorescent protein (GFP)-binding protein (GBP) is a GFP-binding nanobody, developed for biochemically purifying GFP-fusion proteins and targeting GFP-fusion proteins to a subcellular location of interest (Rothbauer et al., 2008; Chen et al., 2017). Specifically, we expressed Tip1-GBP at the leu1 locus from the ase1 promoter in WT cells expressing mCherry-Atb2 and Klp5-3GFP. In a control strain, Tip1-VC (the C-terminal half of the Venus fluorescent protein), but not Tip1-GBP, was expressed. In addition, we created a strain expressing Tip1-GBP, mCherry-Atb2, and Rga7-3GFP (Rga7 is a GAP protein localizing to the cell cortex), in which Rga7-3GFP was tethered to microtubule plus ends to assess the possible effect of the tethering on Tip1 function. As shown in Figure 3A, Tip1-GBP, instead of Tip1-VC, successfully recruited Klp5-3GFP and Rga7-3GFP to microtubule plus ends, and GFP signals were also present at the cell ends given the presence of Tip1 there. Microtubule length measurements showed that tethering Klp5-3GFP, but not Rga7-3GFP, to microtubule plus ends shortened microtubules (Figure 3B), indicative of the direct role of Klp5 in promoting microtubule catastrophe. To show the catastrophe effect clearly, we performed live-cell imaging for the three types of cells. As shown in Figure 3C, frequent premature microtubule catastrophe was apparent in the Tip-GBP Klp5-3GFP cells. Evidently, quantification showed that ~64% of the microtubules in the Tip1-GBP Klp5-3GFP cells were not able to reach the cell end, whereas only ~33% and ~39% of microtubules in Tip1-VC Klp5-3GFP and Tip1-GBP Rga7-3GFP cells, respectively, were not able to reach the cell end (Figure 3D and Supplementary Figure S3). Thus, these data support that Klp5 plays a direct role in promoting microtubule catastrophe.
Figure 3.

Forced localization of Klp5 to microtubule plus ends promotes microtubule catastrophe. (A) Maximum projection images of Tip1-VC and Tip1-GBP cells expressing mCherry-Atb2 and Klp5-3GFP. Tip1-VC and Tip1-GBP under the control of the ase1 promoter were integrated at the leu1 locus. Tip1-GBP cells expressing mCherry-Atb2 and Rga7-3GFP (a GTPase-activating protein localizing to the cell cortex) was used as a control. Note that both Klp5-3GFP and Rga7-3GFP in Tip1-GBP cells, but not in Tip1-VC cells, were successfully forced to localize to the cell ends (red arrows) and to the microtubule plus ends. Scale bar, 5 μm. (B) Measurements of the microtubule length in the three indicated cells. Statistical analysis was performed by student’s t-test (stars, P < 0.001), and the measured cell number is shown on the top of the graph. (C) Kymograph graphs of microtubule and the indicated GFP-fusion proteins. Red arrows indicate the analyzed microtubules. Note that tethering Klp5-3GFP to microtubule plus ends in the Tip1-GBP cells resulted in frequent microtubule catastrophes during the observation period. Scale bar, 10 μm. (D) Quantification of the microtubules not able to reach the cell end for the three types of cells indicated in A. A triplet of live-cell imaging experiments were performed for each type of cells in parallel. The top of the column is the mean of the triplet quantifications (see Supplementary Figure S3 for each individual quantification), and error bars represent SD. Note that the percentage (~64%) of the microtubules that underwent premature catastrophe increased significantly in Tip1-GBP cells expressing Klp5-3GFP.
Tip1 is required for the proper localization of Alp14 to microtubule plus ends
The microtubule length of tip1Δklp5Δ cells is not completely restored (Figure 1B). Moreover, similar to the microtubules of tip1Δ cells, the majority of the microtubules in tip1Δklp5Δ cells underwent catastrophe prematurely (Figure 1G). These findings implied that other factor(s) might be involved in regulating microtubule length in a Tip1-dependent manner. A possible candidate is the microtubule polymerase Alp14, which localizes to the extreme microtubule plus end to promote microtubule growth. Therefore, we carefully examined the localization of Alp14 in WT, tip1Δ, klp5Δ, and tip1Δklp5Δ cells expressing mCherry-Atb2 and Alp14-GFP. As shown in Figure 4A, Alp14 clearly decorated microtubule plus ends and displayed as multiple foci along microtubules both in WT and in klp5Δ cells. In contrast, the Alp14 staining at microtubule plus ends became weaker in tip1Δ and tip1Δklp5Δ cells. Approximate 50% and 36% of tip1Δ and tip1Δklp5Δ cells, respectively, did not display Alp14 staining at microtubule plus ends in the still images, compared to ~11% and ~7% of WT and klp5Δ cells (Figure 4B and Supplementary Figure S4). Intensity measurements confirmed that Alp14 signals at microtubule plus ends were weaker in tip1Δ and tip1Δklp5Δ cells than in WT and klp5Δ cells (Figure 4C). Kymograph analysis of Alp14 dynamics further revealed that the absence of Tip1 caused Alp14 to dissociate prematurely before the growing microtubules initiated catastrophe (Figure 4D). Next, we determined the residence time of Alp14 at microtubule plus ends. As shown in Figure 4E, klp5Δ cells displayed the longest residence time of Alp14 at microtubule plus ends, then WT, tip1Δklp5Δ, and tip1Δ cells. These results suggest an important role of Tip1 in stabilizing the localization of Alp14 at microtubule plus ends. It is likely that Tip1 functions by spatially separating Alp14 from the microtubule depolymerization promoting factor Klp5 at microtubule plus ends.
Figure 4.
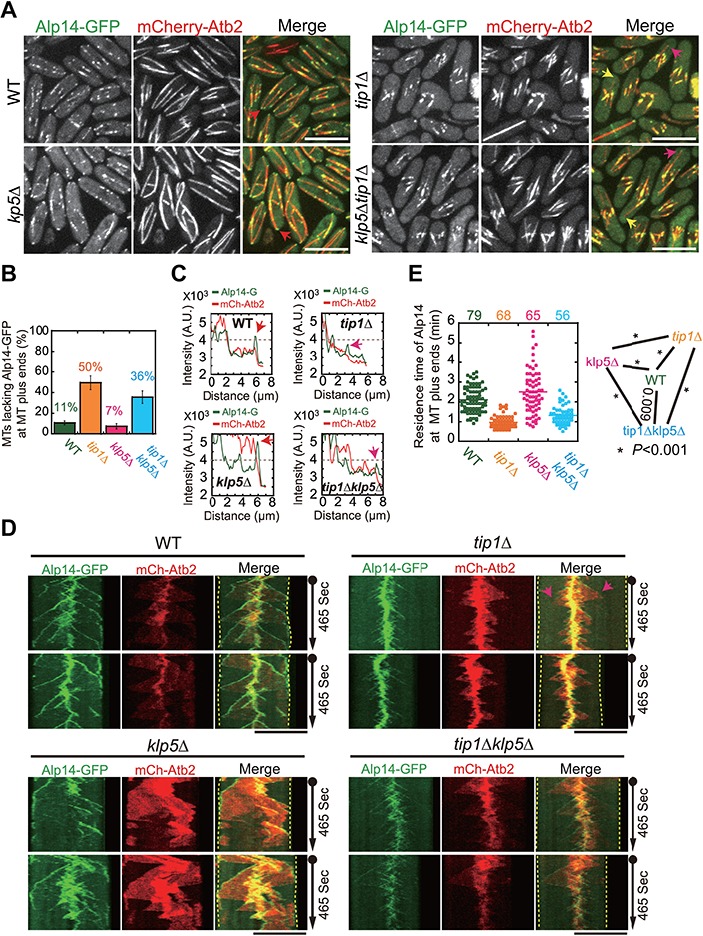
Alp14-GFP localization in WT, tip1Δ, klp5Δ, and tip1Δklp5Δ cells. (A) Maximum projection images of WT, tip1Δ, klp5Δ, and tip1Δklp5Δ cells expressing Alp14-GFP and mCherry-Atb2. Red, pink, and yellow arrows mark Alp14 staining at microtubule plus ends (red, pink, and yellow indicate strong, weak, and none Alp14 signals, respectively). Scale bar, 10 μm. (B) Quantification of the microtubules lacking Alp14-GFP at their plus ends. A triplet of live-cell imaging experiments were performed for each type of cells in parallel. The top of the column is the mean of the triplet quantifications (see Supplementary Figure S4 for each individual quantification), and error bars represent SD. Note that the absence of Tip1 (tip1Δ and tip1Δklp5Δ cells) increased the number of the microtubules that lack Alp14-GFP at the microtubule plus end. (C) Intensity measurement of the indicated microtubule and Alp14 in A. Arrows mark the peak Alp14-GFP signals at the microtubule plus tips. (D) Kymograph analysis of microtubule and Alp14-GFP for the four types of cells indicated in A. Note that the arrows highlight premature dissociation of Alp14-GFP from the growing microtubule plus end. Dashed lines mark the cell ends. Scale bar, 10 μm. (E) Quantification of the residence time of Alp14-GFP at microtubule plus ends in the four types of cells. Statistical analysis was performed by student’s t-test (stars, P < 0.001). Note that the absence of Tip1 decreased the Alp14 residence time, whereas the absence of Klp5 increased the Alp14 residence time.
Tethering Alp14 to microtubule plus ends in tip1Δ cells lengthens microtubules
Next, we tested if stabilizing Alp14 at microtubule plus ends could rescue the short microtubule phenotype in tip1Δ cells. To stabilize the localization of Alp14 at microtubule plus ends in tip1Δ cells, we similarly employed the GBP-targeting approach, by which Alp14-GFP was tethered to microtubule plus ends by Klp2-GBP (a kinesin localizing at microtubule plus ends) expressed at the leu1 locus from the ase1 promoter in tip1Δ cells. In a control tip1Δ strain, Klp2 (without tags), instead of Klp2-GBP, was expressed. Imaging data showed that Alp14 signals at the microtubule plus ends were much more apparent in the Klp2-GBP-expressing cells than in the Klp2-expressing cells, indicative of successful recruitment of Alp14 to microtubule plus ends by Klp2-GBP (Figure 5A). Indeed, Alp14 in the Klp2-GBP-expressing cells resided significantly longer at microtubule plus ends than in the Klp2-expressing cells (Figure 5B), and the microtubules in the Klp2-GBP-expressing cells tended to be longer (Figure 5C). Importantly, ~40% of the microtubules in the Klp2-GBP-expressing cells were now able to reach the cell end, compared to ~28% of the microtubules in the Klp2-expressing cells (Figure 5D and Supplementary Figure S5). Thus, these results support our idea that tethering Alp14 to microtubule plus ends sustains microtubule growth.
Figure 5.

Increasing the residence time of Alp14-GFP at microtubule plus ends in tip1Δ cells lengthens microtubules. (A) Maximum projection images of tip1Δ Klp2 (without a tag) and tip1Δ Klp2-GBP cells expressing mCherry-Atb2 and Alp14-GFP. Klp2 and Klp2-GBP under the control of the ase1 promoter were integrated at the leu1 locus. Note that Alp14-GFP in Klp2-GBP cells, but not in Klp2 cells, was successfully forced to localize to the microtubule plus ends. Scale bar, 10 μm. (B) Quantification of the residence time of Alp14-GFP at microtubule plus ends in the indicated cells in A. Statistical analysis was performed by student’s t-test (star, P < 0.001). (C) Quantification of the microtubule length in the indicated cells in A. Statistical analysis was performed by student’s t-test (star, P < 0.001). (D) Quantification of cell end-contacted microtubules for the two types of cells indicated in A. A triplet of live-cell imaging experiments were performed for each type of cells in parallel. The top of the column is the mean of the triplet quantifications (see Supplementary Figure S5 for each individual quantification), and error bars represent SD. Note that ~40% of the microtubules were able to reach the cell end in Klp2-GBP cells, compared to ~28% of the Klp2 cells. (E) Working model illustrating the concerted actions of Tip1, Alp14, and Klp5 in regulating microtubule catastrophe. Tip1 restricts the access of Klp5 to microtubule plus ends until Tip1 dissociation at the cell end and also stabilizes Alp14 at microtubule plus ends to promote microtubule growth.
Discussion
Tip1 and Klp5 have been shown to be critical factors in inhibiting and promoting microtubule catastrophe, respectively, and Alp14 functions as a microtubule polymerase at microtubule plus ends to promote microtubule growth (Busch and Brunner, 2004; Unsworth et al., 2008; Tischer et al., 2009; Al-Bassam et al., 2012). Here we delineated the interplay of Tip1, Klp5, and Alp14 in regulating microtubule dynamics. We demonstrated that Klp5 functions directly to promote microtubule catastrophe, but the catastrophe activity of Klp5 is restricted by Tip1 to the cell end through accumulating Klp5 behind Tip1 at microtubule plus ends (Figures 2 and 3). We further demonstrated that Tip1 and Klp5 function antagonistically to regulate the localization of Alp14 to microtubule plus ends (Figure 4). These data suggest a possible model explaining the concerted actions of Tip1, Klp5, and Alp14 in spatially controlling microtubule catastrophe. At the microtubule plus end of a growing microtubule, Tip1 stabilizes the localization of Alp14, while it blocks and accumulates Klp5, creating a microenvironment favoring microtubule growth; once the microtubule reaches the cell end, Tip1 and Alp14 dissociate from the microtubule plus end, enabling Klp5 to promote microtubule depolymerization (Figure 5E).
Tip1 plays a critical role in regulating microtubule catastrophe because most of the microtubules in tip1Δ and tip1Δklp5Δ cells depolymerize before reaching the cell end (i.e. premature microtubule catastrophe) (Figure 1D and G), and the absence of Tip1 significantly increases the microtubule catastrophe frequency (Figure 1F). As a result, frequent premature microtubule catastrophe could then lead to short microtubules (Figure 1A, B, and D). It has been established that Tip1 functions to restrict microtubule catastrophe to the cell end and that Klp5 is a critical factor in promoting microtubule catastrophe (Busch and Brunner, 2004; Unsworth et al., 2008; Tischer et al., 2009). Our GBP-targeting experiments further demonstrated that Klp5 plays a direct role in promoting microtubule catastrophe since artificially tethering Klp5 to microtubule plus ends causes premature microtubule catastrophe (Figure 3). Moreover, our high-temporal live-cell imaging data clearly showed that Klp5 moves towards and accumulates at microtubule plus ends where Tip1 resides (Figure 2A and B). Klp5 accumulation at microtubule plus ends is most pronounced once microtubules reach the cell end (Figure 2C and D). It is therefore conceivable that Tip1 may function as a microtubule tip blocker to halt and accumulate Klp5 at microtubule plus ends. Alternatively, the accumulation of Klp5 at microtubule plus ends could be in a Tip1-independent manner given the fact that the Klp5 counterpart Kip3 itself in budding yeast has a high affinity to microtubule plus ends (Leduc et al., 2012; Arellano-Santoyo et al., 2017). If the latter was true, we would expect the same degree of Klp5 accumulation at microtubule plus ends in WT and tip1Δ cells. However, Klp5 accumulation at microtubule plus ends is attenuated by the absence of Tip1 (Figure 2C–E). In addition, trajectory analysis of Tip1 and Klp5 clearly showed that Klp5 localizes behind Tip1 at microtubule plus ends (Figure 2A and B). Hence, the tip1 blocker model is favorable.
The attenuated accumulation of Klp5 to microtubule plus ends in tip1Δ cells could also be interpreted as an indirect effect of the shortened microtubules (Figure 2C–E), which bind less Klp5 than long microtubules. Despite the attenuated accumulation, Klp5 is able to promote microtubule catastrophe in tip1Δ cells because the microtubule catastrophe frequency of tip1Δklp5Δ cells is significantly lower than the microtubule catastrophe frequency of tip1Δ cells (Figure 1F). It is likely that the absence of Tip1 enables Klp5 to function freely to promote microtubule catastrophe and/or that the absence of Tip1 significantly decreases the threshold of Klp5 molecules required for initiating microtubule depolymerization. Whichever interpretation prevails, Tip1 plays a critical role in preventing the access of Klp5 to the extreme microtubule plus ends until Tip1 dissociation at the cell end. It has been demonstrated, by a cryo-electron microscopy work (Maurer et al., 2012), that EB1/Mal3 localizes at the corners of four tubulin subunits to bridge protofilaments at the GTP cap regions. Such localization arrangement may enable the Mal3/Tip1/Tea2 complex to set up a physical barrier to restrict Klp5 from microtubule plus tips.
Although Tip1 has a great effect on Klp5 accumulation at microtubule plus ends, we are not clear whether Klp5 accumulation, on the other hand, contributes to Tip1 dissociation from microtubules at the cell end. Interestingly, the myosin V Myo52 has been shown to enhance the dissociation of Tip1 from microtubule plus ends (Martin-Garcia and Mulvihill, 2009). Nevertheless, we observed a prolonged residence time of Tip1 at microtubule plus ends in klp5Δ cells (Figure 2G). This could be due to the absence of Klp5 that positively contributes to Tip1 dissociation. Alternatively, it could be indirectly due to the over-stabilization of microtubules caused by the absence of Klp5. To clarify these possibilities, it is worthwhile to tackle the interplay between Klp5 and Tip1 with the in vitro microtubule reconstitution system as previously reported (Bieling et al., 2007). In our live-cell imaging experiments, we observed that ~30% of the microtubules in even WT cells undergo premature catastrophe before reaching the cell end (Figure 1G). Preliminary analysis showed that such microtubules display a similar percentage of Tip1 dissociation as the microtubules that are able to reach the cell end and that Klp5 intensity at the plus tips of the microtubules are not able to reach the cell end appears to decrease (data not shown). This further highlights the complex interplay between Klp5 and Tip1 in regulating microtubule dynamics, awaiting further investigation.
In addition to demonstrating the role of Tip1 in regulating Klp5, this present work establishes that Tip1 is required for stabilizing the localization of Alp14 to microtubule plus ends (Figure 4). Alp14 is the conserved XMAP215/Stu2 family protein and has been shown to be a microtubule polymerase (Al-Bassam et al., 2012). The absence of Tip1 does not abolish, but attenuates, the localization of Alp14 at microtubule plus ends (Figures 4A–C). This is consistent with the previous claim that Alp14 does not depend on Mal3/Tip1 for localizing to microtubule plus ends but underscores the fact that Tip1 is a critical factor in stabilizing the localization of Alp14 to microtubule plus ends (Al-Bassam et al., 2012). Not only is the signal intensity of Alp14 at microtubule plus ends largely reduced in tip1Δ cells but also the residence time (Figure 4C and E). Moreover, Alp14 occasionally dissociates from growing microtubules before reaching the cell end in tip1Δ cells (Figure 4D). Artificially stabilizing Alp14 at the microtubule plus ends of tip1Δ cells lengthens microtubule length (Figure 5A–C). These results support our idea that Tip1 is required for stabilizing Alp14 at microtubule plus ends and that the stabilization of Alp14 is important for sustaining microtubule growth. Interestingly, Klp5 destabilizes Alp14 at microtubule plus ends in the absence of Tip1 since klp5 removal in tip1Δ cells increases the residence time of Alp14 at microtubule plus ends (Figure 4E). This finding suggests an antagonistic action of Klp5 and Tip1 in regulating Alp14 stability.
In conclusion, this work demonstrates the new roles of Tip1 in restricting the microtubule catastrophe activity of Klp5 to the cell end and in stabilizing the localization of Alp14 at microtubule plus ends. The concerted actions of the microtubule-stabilizing factors Tip1 and Alp14 and the microtubule catastrophe factor Klp5 ensure proper spatial regulation of microtubule catastrophe. Given the conserved nature of the proteins, the findings may also be conserved in other organisms.
Materials and methods
Yeast genetics
The strains and plasmids used in this study are listed in Supplementary Tables S1 and S2, respectively. Strains were created by random spore analysis or tetra-dissection analysis. Plasmids were constructed either by the one-step cloning technique based on homologous recombination (Vazyme Biotech) or by the traditional cloning method (NEB). Yeast and Escherichia coli culture media were purchased from ForMedium. For live-cell imaging, strains were inoculated into Edinburgh minimal medium with the five supplements: adenine, leucine, uracil, histidine, and lysine (EMM5S).
Microscopy
All imaging experiments were carried out with a PerkinElmer Ultraview Spinning Disk confocal microscope equipped with a Nikon Apochromat TIRF 100X 1.49NA objective and a Hamamatasu C9100-23B EMCCD camera (PerkinElmer). Images were acquired at room temperature with log-phase cells sandwiched between an EMM5S agarose pad and a coverslip. For high-temporal resolution imaging, stack images containing three planes with 0.5 μm spacing were taken every 5 sec by Volocity (PerkinElmer) unless otherwise specified; for single time-point imaging, stack images containing 11 planes with 0.5 μm spacing were acquired.
Data analysis
Imaging data was analyzed with MetaMorph 7.7 (Molecular Devices) and ImageJ. Microtubule length was measured with maximum projection images generated from raw stack images and was presented as dot and probability plots using KaleidaGraph (Synergy Software). Quantification of the microtubules that contacted the cell end was carried out with live-cell microscopy movies; the same set of imaging were performed three times, and the quantification results were presented by KaleidaGraph as summary column graphs, in which the top of the column is the mean of the quantifications and the error bar represents standard deviation. The residence time of microtubule plus ends at the cell end was referred to as dwell time and was determined with live-cell microscopy movies. The catastrophe frequency of microtubule bundles was calculated by dividing the number of catastrophe events by the total time of the microtubule bundle in growth as previously described (Walker et al., 1988). Intensity measurements were carried out with the line-scan function in MetaMorph along the selected targets in maximum projection images. The residence time of Alp14 at microtubule plus ends was determined by calculating the duration between the appearance and disappearance time of Alp14 at microtubule plus ends. Statistical analysis was performed with Microsoft Excel, and the illustration diagram was generated with Adobe Illustrator.
Supplementary Material
Acknowledgements
We thank Drs Phong Tran (University of Pennsylvania) and Quan-wen Jin (Xiamen University) for providing yeast strains. We also thank members of the Fu Laboratory for helpful discussions.
Conflict of interest
none declared.
Funding
This work is supported by grants from the National Key Research and Development Program of China (2018YFC1004700), the National Natural Science Foundation of China (91754106, 31671406, 31871350, and 31601095), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19040101), the Major/Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology (2017FXCX008), and China’s 1000 Young Talents Recruitment Program.
References
- Adams G. Jr, Zhou J., Wang W., et al. (2016). The microtubule plus end tracking protein TIP150 interacts with cortactin to steer directional cell migration. J. Biol. Chem. 291, 20692–20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., Kim H., Flor-Parra I., et al. (2012). Fission yeast Alp14 is a dose-dependent plus end-tracking microtubule polymerase. Mol. Biol. Cell 23, 2878–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano-Santoyo H., Geyer E.A., Stokasimov E., et al. (2017). A tubulin binding switch underlies Kip3/Kinesin-8 depolymerase activity. Dev. Cell 42, 37–51.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P., Ye X., Huddleston P., et al. (2012). A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science 337, 857–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., et al. (2007). Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450, 1100–1105. [DOI] [PubMed] [Google Scholar]
- Brouhard G.J., and Rice L.M. (2018). Microtubule dynamics: an interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol. 19, 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H., Hackney D.D., and Nurse P. (2003). Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 5, 812–818. [DOI] [PubMed] [Google Scholar]
- Brunner D., and Nurse P. (2000). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695–704. [DOI] [PubMed] [Google Scholar]
- Busch K.E., and Brunner D. (2004). The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 14, 548–559. [DOI] [PubMed] [Google Scholar]
- Busch K.E., Hayles J., Nurse P., et al. (2004). Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev. Cell 6, 831–843. [DOI] [PubMed] [Google Scholar]
- Chang F., and Martin S.G. (2009). Shaping fission yeast with microtubules. Cold Spring Harb. Perspect. Biol. 1, a001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., Wang G.Y., Hao H.C., et al. (2017). Facile manipulation of protein localization in fission yeast through binding of GFP-binding protein to GFP. J. Cell Sci. 130, 1003–1015. [DOI] [PubMed] [Google Scholar]
- Garcia M.A., Koonrugsa N., and Toda T. (2002). Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 12, 610–621. [DOI] [PubMed] [Google Scholar]
- Gardner M.K., Zanic M., Gell C., et al. (2011). Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell 147, 1092–1103. [DOI] [PubMed] [Google Scholar]
- Gupta M.L. Jr., Carvalho P., Roof D.M., et al. (2006). Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8, 913–923. [DOI] [PubMed] [Google Scholar]
- Jiang K., Wang J., Liu J., et al. (2009). TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 10, 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., and Wittmann T. (2012). +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 22, 418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc C., Padberg-Gehle K., Varga V., et al. (2012). Molecular crowding creates traffic jams of kinesin motors on microtubules. Proc. Natl Acad. Sci. USA 109, 6100–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Garcia R., and Mulvihill D.P. (2009). Myosin V spatially regulates microtubule dynamics and promotes the ubiquitin-dependent degradation of the fission yeast CLIP-170 homologue, Tip1. J. Cell Sci. 122, 3862–3872. [DOI] [PubMed] [Google Scholar]
- Maurer S.P., Fourniol F.J., Bohner G., et al. (2012). EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S., and Vale R.D. (2015). Microtubule nucleation at the centrosome and beyond. Nat. Cell Biol. 17, 1089–1093. [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., et al. (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusionproteins. Mol. Cell Proteomics 7, 282–289. [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Tillib S., et al. (2006). Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3,887–889. [DOI] [PubMed] [Google Scholar]
- Slep K.C., and Vale R.D. (2007). Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell 27, 976–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer C., Brunner D., and Dogterom M. (2009). Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Mol. Syst. Biol. 5, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth A., Masuda H., Dhut S., et al. (2008). Fission yeast kinesin-8 Klp5 and Klp6 are interdependent for mitotic nuclear retention and required for proper microtubule dynamics. Mol. Biol. Cell 19, 5104–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., et al. (2006). Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8,957–962. [DOI] [PubMed] [Google Scholar]
- Varga V., Leduc C., Bormuth V., et al. (2009). Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138, 1174–1183. [DOI] [PubMed] [Google Scholar]
- Walker R.A., O'Brien E.T., Pryer N.K., et al. (1988). Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 107, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Malmstrom T., Troxell C.L., et al. (2001). Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell 12, 3919–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund P.O., Stear J.H., Pozniakovsky A., et al. (2011). XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc. Natl Acad. Sci. USA 108, 2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Liu X., Wu B., et al. (2014). Superresolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol. Biol. Cell 25, 4166–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F., Li T., Cheung M., et al. (2014). Mcp1p tracks microtubule plus ends to destabilize microtubules at cell tips. FEBS Lett. 588,859–865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


