Abstract
The inhibitory effect of large tumor suppressor kinase (LATS1/2) on the activity of the oncoprotein yes-associated protein (YAP) is crucial to maintain tissue homeostasis. Proteomic studies have identified several new regulators of this process. Recently, citron kinase (CIT) was listed as a potential binding candidate of Hippo-related components, suggesting a new connection between CIT and the Hippo pathway. Aside from CIT’s role in cytokinesis, the molecular crosstalk between CIT and the Hippo pathway is largely unknown. Here, we demonstrate a role for CIT as a scaffold protein linking LATS2 and YAP. More importantly, CIT interacts with LATS2 to directly suppress LATS2 phosphorylation at the hydrophobic motif—targeted by MST1, leading to LATS2 inactivation and YAP activation. By studying their genetic interactions, we found that Sticky, the CIT homolog in Drosophila melanogaster, functions with Warts to control Drosophila eye development. Together, our study confirms citron kinase as a novel regulator of the Hippo pathway.
Keywords: citron kinase, LATS2–YAP interaction, LATS2 inhibition, sticky–warts, Hippo pathway
Introduction
The Hippo pathway was initially discovered in Drosophila melanogaster more than a decade ago. Since its discovery, the conservation of the Hippo pathway’s function in mammals has motivated intense study of its components (Huang et al., 2005; Yu and Guan, 2013; Meng et al., 2016). Canonical Hippo signaling begins with the activation of large tumor suppressor kinase LATS1/2 (homologs of Drosophila Warts). This occurs via phosphorylation of hydrophobic motifs in LATS1/2 (threonine 1079 in LATS1, threonine 1049 in LATS2) by the upstream mammalian Ste20-like kinase 1/2 (MST1/2, homolog of Drosophila Hpo), and in its activation loop (serine 909 in LATS1, serine 872 in LATS2) by subsequent auto-phosphorylation. Active LATS1/2 phosphorylates the two paralogous transcriptional co-activators Yes-associated protein (YAP) and WW domain-containing transcription regulator 1 (WWTR1/TAZ) (homologs of Drosophila Yorkie) at their HxRxxS/T consensus motifs (Hao et al., 2008; Ni et al., 2015; Hoa et al., 2016). In particular, phosphorylation of YAP at serine 127 by LATS1/2 is an indicator of inactive YAP. Phosphorylated YAP is shuttled from the nucleus to the cytoplasm, where it is eventually degraded by the proteasome (Dong et al., 2007). This core Hippo pathway cascade is essential in translating a wide range of external cues (e.g. cell-cell contact, mechanical stress, extracellular matrix stiffness, and nutrient availability) into appropriate cellular responses (e.g. proliferation, differentiation, and cell fate determination) (Hong et al., 2005; Dupont et al., 2011; Zhao et al., 2011, 2012; Yu et al., 2012, 2013; Aragona et al., 2013; Azzolin et al., 2014; Park et al., 2015). Malfunctions of the Hippo pathway in vivo alter tissue development, affect tissue regeneration, and accelerate cancer progression in organs like the liver, pancreas, stomach, and intestine (Harvey et al., 2013; Moroishi et al., 2015; Panciera et al., 2017). It is, therefore, important to better understand the intrinsic cellular regulation of the Hippo pathway, especially the activity of LATS1/2 and YAP/TAZ.
Citron kinase (CIT), a serine/threonine kinase, was first identified through a yeast two-hybrid experiment using GTP-bound Rho and Rac (Madaule et al., 1995). CIT protein contains multi-functional domains including a kinase domain, a coiled-coil domain, a Rho-binding domain, a zinc finger, a pleckstrin homology domain, and a citron homology domain (CNH) (Madaule et al., 1998; D’Avino, 2017). The most well-studied function of CIT is its role as a scaffold protein for the recruitment of the core cytokinetic machinery (Madaule et al., 1998; Gruneberg et al., 2006; Gai et al., 2011; Bassi et al., 2013). Depletion of CIT in mitotic cells prevents the formation of the mid-body and leads to the formation of multinucleated cells (D’Avino, 2017). Supporting an essential role for CIT in vivo, the loss of Sticky, the Drosophila CIT homolog, significantly reduces the size of the imaginal discs and induces hyperploidy in the larval brain (Shandala et al., 2004). Mice lacking CIT have shorter lifespans and suffer from severe brain defects due mainly to pronounced cell death triggered by abnormal cytokinesis (Di Cunto et al., 2000). More importantly, two groups have independently uncovered several pathogenic variants in the gene encoding CIT from microcephaly patients (Harding et al., 2016; Li et al., 2016). This suggests that the function of CIT is conserved in Drosophila and humans.
Despite these published in vivo data that reveal the function of CIT, the details of its molecular mechanisms in cellular phenomena other than cytokinesis remain poorly understood. While searching for novel regulators of the canonical Hippo pathway, we noticed a potential connection to CIT (Moya and Halder, 2014). Here, we demonstrate that CIT is, indeed, a novel component of the Hippo signaling network. CIT serves as a scaffold protein that facilitates the interaction of LATS2 and YAP. CIT interacts with LATS2 to directly inhibit its kinase activity by hindering MST1’s phosphorylation of the LATS2 hydrophobic motif. This inactivates LATS2 and activates YAP. We confirmed with genetic interaction assays in Drosophila melanogaster that Sticky (the Drosophila CIT homolog) and Warts (the Drosophila LATS1/2 homolog) function in a common pathway to control eye development.
Results
CIT physically interacts with YAP and LATS2
In a protein–protein interaction analysis, we identified CIT as a candidate binding partner for purified YAP tagged with streptavidin-binding peptide (SBP) (Supplementary Figure S1A and Table S1) (Kim et al., 2016). Since CIT appeared in the lists of overlapping ‘prey’ proteins baited by other Hippo components in other Hippo proteomic studies (Kwon et al., 2013; Wang et al., 2014; Toloczko et al., 2017), we wanted to confirm any interactions between CIT and the Hippo pathway protein network. First, we checked CIT and YAP, finding that HA-tagged YAP is co-immunoprecipitated with Myc-tagged CIT (Figure 1A). We next attempted to identify the binding domain for each respective protein. After obtaining all available CIT and YAP truncation mutants, we used them to repeat the co-immunoprecipitation experiments (Supplementary Figure S1B and C). We found that loss of a conserved PPxY motif within the CIT C-terminus is sufficient to abolish the binding of CIT and YAP (Figure 1B and C). We also found that YAP lacking its WW domain can no longer pull-down CIT (Figure 1D). Next, we tested the interaction between CIT and LATS1/2, upstream regulators of YAP. Interestingly, we found that CIT binds strongly to LATS2 and weakly to LATS1 (Figure 1E) (Furth and Aylon, 2017). Equally important, we confirmed the endogenous interaction between CIT, LATS2 under attachment (AT) and detachment (DE) conditions (Supplementary Figure S1D). Also, we used immunoprecipitation with a set of serially truncated CIT mutants to map the domains required for CIT binding to LATS2. As shown in Figure 1F, we narrowed this region down to the area containing the Rho binding domain (RBD) and the zinc finger pleckstrin homology domain (ZF-PH). Since the deletion of each of these domains alone is insufficient to alter the interaction between CIT and LATS2 (Supplementary Figure S1E), we conclude that the CIT RBD and ZF-PH domains act redundantly in the binding of LATS2. Collectively, CIT interacts strongly with YAP and LATS2, weakly with LATS1, at two different sites: the PPxY motif for YAP and the RBD and ZF-PH domains for LATS2.
Figure 1.
CIT physically interacts with YAP and LATS2. (A) CIT was co-immunoprecipitated with YAP. Immuno-blotting for the HA tag showed the presence of HA-YAP along with Myc-CIT. (B) Schematic describing the structure of full-length Myc-CIT with the PPxY motif, Myc-CIT C-terminus (Myc-CIT Ct) from amino acid 955 to the last amino acid, and the Myc-CIT C-terminus with a deletion of its PPxY motif (Myc-CIT CtΔPY). There is a conserved PPxY motif at the C-terminus of CIT in zebrafish (D. rerio), mouse (Mus. musculus), and human (H. sapiens). (C) YAP interacted with CIT through the PPxY motif. The DNA constructs in B were used in a co-immunoprecipitation assay with Flag-YAP. Western blot showed that Myc-CIT CtΔPY failed to interact with YAP. (D) CIT interacted with YAP through its WW domain. Full-length YAP (Flag-YAP FL) and YAP with a deleted WW domain (Flag-YAPΔWW) were subjected to co-immunoprecipitation. Only Flag-YAP FL was pulled down by Myc-CIT. (E) CIT interacted strongly with LATS2 and weakly with LATS1. Myc-CIT was co-transfected together with HA-LATS1 or HA-LATS2 into HEK293T cells. Immunoprecipitation with Myc tag pulled down both HA-LATS1 and HA-LATS2. s.e., short exposure; l.e., long exposure. (F) LATS2 bound to CIT through its RBD and ZF-PH. Serial truncated mutants of Myc-CIT including Ct, Nt, and ΔC were subjected to co-immunoprecipitation with HA-LATS2. CIT fragments Ct, Nt, and ΔC were generated by deleting the sequence encoding the following amino acids (0–955), (955–2055), and (1746–2055), respectively.
Full-length CIT functions as a scaffold that promotes the YAP–LATS2 interaction
Given that all three proteins—YAP, LATS2, and CIT—are interconnected, we asked how the interaction of two of the three proteins is affected when the third component is added. We observed that adding Flag-YAP enhances the association between Myc-CIT and HA-LATS2 (Figure 2A Lanes 1 and 4, and 2B-i). Similarly, additional expression of HA-LATS2 increases the level of Flag-YAP immunoprecipitated by Myc-CIT and vice versa (Figure 2A Lanes 2 and 4, and 2B-ii). Also, Flag-YAP pulls down HA-LATS2 more efficiently in the presence of Myc-CIT (Figure 2A Lanes 3 and 4, and 2B-iii). This result suggests adding the missing third component to a complex of two of the three proteins strengthens the association of all three. Alternatively, we examined the endogenous interaction of LATS2 and YAP in the absence of CIT. We employed the CRISPR/Cas9 system to stably knock out full-length CIT in retinal pigment epithelial cells (RPE). Hereafter, we will refer to the resulting RPE CIT-null cells as sgCIT and to the RPE control cells as sgCTR. We performed experiments under two conditions—attached (AT) and detached (DE). Consistent with the effect of CIT overexpression, we found that knockout of CIT reduces the binding affinity of YAP to LATS2 under both attached and detached conditions (Figure 2C).
Figure 2.
Full-length CIT functions as a scaffold for the YAP–LATS2 interaction. (A) Comparison of the binding affinity between bipartite and tripartite complexes of Flag-YAP, HA-LATS2, and Myc-CIT. The indicated combinations were expressed in HEK293T cells. Antibodies against the Myc tag and Flag tag were used for analysis. (B) Quantification of the immunoprecipitation experiment in A. (i) Protein level of LATS2 pulled down by CIT in the presence or absence of YAP. (ii) Protein level of YAP precipitated by CIT in the presence or absence of LATS2. (iii) Protein level of LATS2 bound to YAP in the presence or absence of CIT. n = 3, unpaired student’s t-test, *P < 0.05, **P < 0.01, error bars indicate the SEM. (C) Endogenous immunoprecipitation of YAP and LATS2 in RPE cells under the attached condition (AT, cells normally attached to culture dishes) and the detached condition (DE, cells trypsinized and suspended for 1 h before the experiment). Endogenous immunoprecipitation were performed in RPE control cells (sgCTR) and RPE CIT-null cells (sgCIT). (D) Sequential immunoprecipitation scheme (sequential immunoprecipitation) of CIT–LATS2–YAP. (E) The western blot showing the result from D.
Next, we hypothesized that CIT, YAP, and LATS2 form a stable tripartite complex. To prove this, we designed a sequential immunoprecipitation experiment with exogenous expression of Flag-YAP, Myc-CIT, and HA-LATS2 in HEK293T cells (Figure 2D). We reasoned that upon initial immunoprecipitation with Flag-YAP, followed by elution with Flag peptide, three potential complexes may exist in the eluate: (i) a bipartite complex of Myc-CIT and Flag-YAP, (ii) a bipartite complex of HA-LATS2 and Flag-YAP, and (iii) a tripartite complex of Myc-CIT, Flag-YAP, and HA-LATS2. After the second immunoprecipitation with Myc-CIT, if HA-LATS2 forms a tripartite complex with Flag-YAP and Myc-CIT, we should detect HA-LATS2 in the final eluate (Figure 2D). Indeed, we detected HA-LATS2 together with Myc-CIT and Flag-YAP among the products of the second immunoprecipitation (Figure 2E). Furthermore, we checked the subcellular localization of Flag-tagged YAP and HA-tagged LATS2 while co-expressing them with either GFP-tagged CIT or a GFP empty vector control (Supplementary Figure S2). As previously reported, CIT shows a discrete punctuate appearance (Supplementary Figure S2i and ii) (Di Cunto et al., 1998). Interestingly, when YAP-LATS2 is co-expressed with GFP-CIT, the resulting staining pattern consists of a few dotted structures overlapping with CIT-positive punctate. We did not observe this pattern in the case of co-expression with a GFP empty vector control (Supplementary Figure S2iii–viii). These data further confirmed the existence of the CIT–LATS2–YAP complex. Together, our data support a model in which full-length CIT functions as a scaffold protein for the formation of a stable CIT–LATS2–YAP complex.
CIT promotes YAP activity by inhibiting LATS2 kinase
So far, our data show CIT enhances the association between YAP and LATS2. While LATS2 is known to phosphorylate and inhibit YAP activity, we next asked how CIT affects YAP activity. Before analyzing YAP activity, it is important to note that the morphologies of RPE CIT-null cells and RPE control cells are different. RPE CIT-null cells show significantly more multinucleation than RPE control cells (Supplementary Figure S3A and B). RPE CIT-null cells also grow more slowly than control cells (Supplementary Figure S3C) and incorporate less BrdU (Supplementary Figure S3D). These characteristics may be due to CIT-depletion-induced cytokinesis failure (D’Avino, 2017).
Next, we measured YAP activity in RPE CIT-null cells using three common assays: measurement of YAP phosphorylation at serine 127, observation of YAP localization, and measurement of the expression of YAP target genes. First, we found RPE CIT-null cells (sgCIT) show higher levels of YAP phosphorylation at serine 127 than RPE control cells (sgCTR) (Figure 3A and B). Given that depletion of CIT reduces YAP–LATS2 interaction under attachment and detachment conditions, we measured the levels of phosphorylated YAP under the same conditions. We saw a consistent increase in the level of YAP phosphorylation in CIT-null attached cells; but when YAP was fully phosphorylated under cell-detached conditions, CIT knockout did not further elevate YAP phosphorylation (Figure 3C). These data suggest that in the presence of LATS activating signals, the reduced LATS–YAP binding caused by CIT knockout may not affect LATS-enzymatic catalysis of YAP. This result raises the additional question of when CIT knockout starts to affect YAP phosphorylation. Thus, we re-plated suspended RPE cells and measured phosphorylated YAP at different time points. Interestingly, we observed an increase in YAP phosphorylation induced by CIT knockout at the middle stage of cell re-attachment (i.e. 4–8 h after plating) (Figure 3D). We saw a similar pattern in the experiment using Latrunculins B (LatB) to activate LATS kinase (Supplementary Figure S4A). Based on these data, we reasoned that CIT promotes YAP de-phosphorylation in the presence of YAP-activating signals (Supplementary Figure S4B). Also, the scaffolding activity of Citron for YAP and LATS2 is of unknown functional significance, and does not seem connected to YAP phosphorylation by LATS. Second, given that highly phosphorylated YAP is excluded from the cytoplasm, we analyzed YAP subcellular localization through immuno-florescent staining. We found that rather than completely excluding YAP from the cytoplasm, CIT depletion equalizes the amount of YAP in the nuclear and cytoplasmic compartments, especially in multinucleated cells (Figure 3E). We, therefore, compared the number of cells with predominantly nuclear YAP (Nu) to the number of cells with equal YAP distribution in the nucleus and cytoplasm (Nu = Cyto). Among the RPE CIT-null cells, we found fewer Nu cells than Nu = Cyto cells (Figure 3F). Third, we measured the mRNA level of YAP-regulated genes (e.g. CTGF, CYR61, ANKRD1, AMOTL2) in the absence of CIT and found significant reduction in their expression (Figure 3G). Together, these data suggest CIT promotes YAP activity.
Figure 3.
CIT promotes YAP activity by inhibiting LATS2 kinase. (A) Western blot comparing the level of phosphorylated YAP at serine 127 between RPE control cells (sgCTR) and RPE CIT-null cells (sgCIT). (B) The intensities of the YAP and phosphorylated YAP bands were quantified with the ImageJ software, and the ratio between them was calculated. n = 3, error bars indicate SEM, **P < 0.01, unpaired student’s t-test. (C) Western blot showing the level of YAP phosphorylated at serine 127 in RPE attached cells compared to RPE detached cells. (D) The level of YAP phosphorylated at serine 127 when RPE detached cells were re-plated and harvested at the indicated time points. (E) Representative image displaying YAP localization in RPE control cells (sgCTR) and RPE CIT-null cells (sgCIT). YAP, red. DAPI, blue. Scale bar, 50 μm. (F) Percentage of RPE cells showing nuclear YAP (Nu) or YAP equally distributed between the nucleus and cytoplasm (Nu = Cyto) quantified by the ImageJ software. At least 100 cells in total were taken from 5 to 6 random fields. n = 3, error bars indicate SEM, ***P < 0.001, unpaired student’s t-test. (G) mRNA level of YAP target genes: CTGF and CYR61 (n = 3), AMOTL2, and ANKRD1 (n = 4). Error bars indicate SEM, ***P < 0.001, unpaired student’s t-test. (H) Western blot showed that siRNA-mediated knockdown of LATS1/2 rescued the effect of CIT knockout on YAP phosphorylation. (I and J) Kinase assay measuring LATS2 and YAP as substrates in the absence of endogenous CIT (I) or upon overexpression of full-length CIT (J). LATS2 was immunoprecipitated with an HA tag antibody and subjected to a kinase assay with His-YAP as a substrate and cold ATP.
The phosphorylation of YAP at serine 127 is mediated by LATS1/2, as knockdown of LATS1/2 rescues the effect of CIT on YAP phosphorylation (Figure 3H). A study from Ganem et al. (2014) indicated that tetraploid cells generated by dihydrocytochalasin B treatment or actin polymerization also show enhanced LATS kinase activity and reduced YAP activity. Thus, the decrease in YAP activity we observed upon CIT depletion may result from a mitotic catastrophe. To rule out this possiblity, we peformed a kinase assay to directly examine LATS2’s ability to phosphorylate YAP. When we immunoprecipitated exogenous HA-LATS2 from HEK293T cells with transient depletion of endogenous CIT, we found increased phosphorylation of LATS2 at its hydrophobic motif. After adding YAP as substrate to the LATS2 kinase reaction in vitro, we observed an increase in YAP phosphorylated at serine 127 (Figure 3I). In contrast, HA-LATS2 precipitated from HEK293T cells overexpressing full-length CIT leads to less LATS2 phosphorylation at the hydrophobic motif and less phosphorylation of YAP at serine 127 (Figure 3J). These results support our hypothesis that CIT directly interferes with the ability of LATS2 kinase to phosphorylate YAP.
The CIT N-terminus interferes with MST1 phosphorylation of the LATS2 hydrophobic motif
The above data allowed us to devise a model in which CIT interacts with LATS2 to weaken its ability to phosphorylate YAP. Next, we asked how CIT regulates the activity of LATS2. First, we reasoned that the CIT interacting domain may be important for CIT’s inhibition of LATS2 kinase. Therefore, we performed a LATS2 kinase assay with a CIT N-terminus mutant lacking both the YAP and LATS2 binding domains. Unexpectedly, we found that this N-terminus mutant reduces the phosphorylation of LATS2 at its hydrophobic motif and YAP at serine 127 to an extent similar to full-length CIT (Figure 4A). Next, we purified the CIT N-terminus and LATS2 separately and then combined them in different ratios before re-evaluating LATS2 kinase activity in vitro. Since the CIT N-terminus itself contains a kinase domain, we confirmed that the CIT kinase domain does not phosphorylate YAP at serine 127 by itself (Figure 4B). Also, we found that a CIT N-terminus with an Alanine in place of lysine 126 (CIT Nt K/A mut), a mutation known to produce a CIT kinase-dead protein, failed to reduce the phosphorylation of LATS2 at its hydrophobic motif (Supplementary Figure S5A) (Di Cunto et al., 1998). We then observed that the wild-type CIT N-terminus suppresses the phosphorylation of the LATS2 hydrophobic motif and the phosphorylation of YAP at serine 127 (Figure 4B). These data strongly support the conclusion that the CIT N-terminus, which contains a kinase domain and a coiled-coil domain, is the main factor that hinders LATS2 kinase activity, and this inhibition may require CIT kinase activity.
Figure 4.
The CIT N-terminus interferes with MST1’s phosphorylation of the LATS2 hydrophobic motif. (A) Kinase assay using LATS2 and YAP as substrates when the CIT N-terminus (Myc-CIT Nt) and HA-LATS2 were co-expressed. HA-LATS2 was immunoprecipitated via its HA tag, and then subjected to a kinase assay with His-YAP and cold ATP. (B) Kinase assay using LATS2 and YAP as substrates when LATS2 was combined with the CIT N-terminus in vitro. SBP-LATS2 was pulled down using streptavidin beads, while Flag-CIT Nt was precipitated via its Flag tag. The two proteins were combined and subjected to a kinase assay with His-YAP and cold ATP. (C) MST1-mediated phosphorylation of the LATS2 hydrophobic motif in the presence of MST1 alone or MST1 with the CIT N-terminus (Myc-CIT Nt). HA-LATS2 was immunoprecipitated via its HA tag, and LATS2 hydrophobic motif phosphorylation was assessed using a specific antibody. (D) The interaction between MST1 and LATS2 when MST1 was expressed alone or in combination with the CIT N-terminus (Myc-CIT Nt). Flag-LATS2 was immunoprecipitated via its Flag tag, and the level of bound MST1 was checked using a specific antibody. (E) Schematic describing the proposed mechanism by which CIT inhibits LATS2 kinase and promotes YAP activity.
The phosphorylation of the LATS1/2 hydrophobic motif by MST1/2 is a prerequisite for full activation of LATS1/2 kinase (Ni et al., 2015). Thus, we next asked whether the CIT N-terminus prevents the phosphorylation of LATS2 by MST1/2. To answer this question, we co-expressed the CIT N-terminus with MST1 and immunoprecipitated LATS2 to assess its phosphorylation status. Before this, we confirmed that the expression of the CIT N-terminus alone reduces phosphorylation at the LATS2 hydrophobic motif and that the expression of MST alone increases LATS2 phosphorylation at this site in a dose-dependent manner (Figure 4C Lanes 1–5). We found that the additional expression of the CIT N-terminus interferes with MST1-mediated phosphorylation of the LATS2 hydrophobic motif (Figure 4C Lanes 6 and 7). Of note, this inhibition does not disrupt the interaction between LATS2 and MST1 (Figure 4D). Since LATS1 is MST1’s substrate, we asked whether the CIT N-terminus has a similar effect on LATS1. Indeed, we found the CIT N-terminus strongly inhibited the MST1-mediated phosphorylation of LATS1 at its hydrophobic motif (Supplementary Figure S5B and C). From these results, we reasoned that the CIT N-terminus is rendering the hydrophobic motif of LATS2 inaccessible to MST1. Based on our molecular data, we propose the following model for CIT’s regulation of LATS2 and YAP (Figure 4E). CIT has two independent functions: full-length CIT mediates the interaction between LATS2 and YAP, while the CIT N-terminus inhibits the MST1-mediated phosphorylation of the LATS2 hydrophobic motif, inactivating LATS2 and eventually blocking YAP suppression. In summary, we conclude that CIT promotes YAP activity by inhibiting the MST1-induced activation of LATS2 kinase.
CIT/sticky (sti) shows genetic interaction with warts (wts) in Drosophila
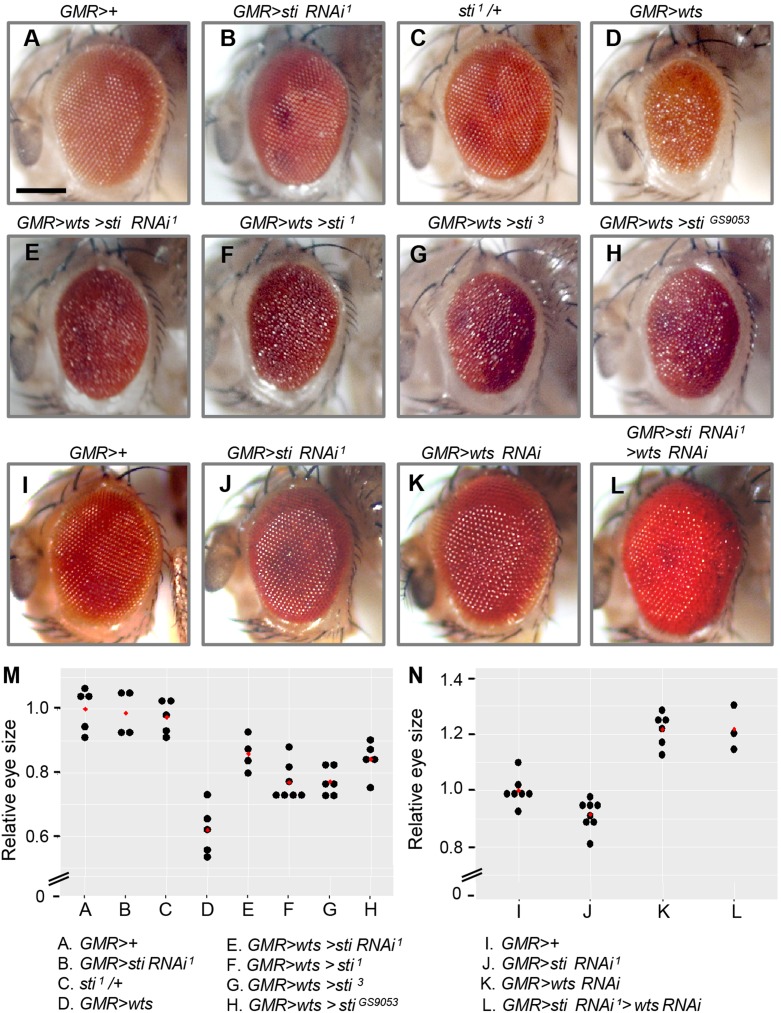
The Drosophila homologs of LATS1/2 and CIT are encoded by warts (wts) and sticky (sti), respectively. To determine whether wts and sti are functionally related in vivo, we analyzed the genetic interaction between them. We used the GAL4-UAS system to examine the effects of sti RNAi in targeted tissues. Knockdown of sti by UAS-sti RNAi1 and GMR-GAL4 in the developing eye induces a slight reduction in eye size (Figure 5A and B). GMR-GAL4-mediated overexpression of wts (GMR>wts) induces a strong reduction in eye size and roughening of the eye surface (Figure 5D). Under this condition, sti RNAi1 considerably suppresses the wts overexpression phenotype (Figure 5E and M). Two other UAS-sti RNAi lines (sti RNAi2 and sti RNAi3) also produce similar suppression of the wts overexpression phenotype (Supplementary Figure S7B, C, and G). This suppression by sti RNAi is not due to a titration of the GAL4 by an additional copy of UAS, because UAS-GFP does not affect the wts overexpression phenotype (Supplementary Figure S6C and D). The rough eye phenotype caused by wts overexpression is also suppressed by sti1/+ heterozygous mutation (Figure 5F and M). We obtained similar results with two other heterozygous sti mutants (sti3, stiGS9053) (Figure 5G, H, and M). Such genetic interactions suggest Wts function may, at least in part, depend on Sti. We then examined the epistatic relationship between the sti and wts RNAi conditions. wts RNAi knockdown using GMR-GAL4 increases eye size (Figure 5K), whereas sti RNAi1 slightly decreases it (Figure 5J). Interestingly, double knockdown of sti and wts (GMR>sti RNAi1, wts RNAi) results in similar phenotype as wts RNAi alone (Figure 5L and N). This implies wts RNAi is epistatic to the sti RNAi phenotype. Together, our results show that sti genetically interacts with both gain and loss of wts function.
Figure 5.
Genetic interaction between sti and wts in Drosophila eye. (A–D) Genotypes used for the genetic interaction between sti and wts. (A) GMR>+ control. (B) Sti knockdown by sti RNAi1 causes a slight reduction in eye size. (C) sti1/+ heterozygotes show normal eyes. (D) Wts overexpression reduces eye size and causes severe roughness. (E) sti RNAi1 partially suppresses the Wts overexpression phenotypes. (F–H) The Wts overexpression phenotypes are partially suppressed by sti1/+ (F), sti3/+ (G), or stiGS9053/+ (H). (I–L) Genetic interaction between sti RNAi1 and wts RNAi. (I) GMR>+ control. (J) sti RNAi1 weakly reduces eye size. (K) wts RNAi increases eye size. (L) sti RNAi does not suppress the wts RNAi phenotype. Scale bar, 100 μm. (M) Dot plot quantification of relative eye size in A–H. Red diamond spot indicates the average size of eye. (N) Dot plot quantification of relative eye size in I–L. Red diamond spot indicates the average size of eye.
Discussion
In this study, we reveal a new function for citron kinase in the regulation of the Hippo pathway. We first demonstrate CIT physically interacts with the core Hippo components YAP and LATS2. CIT binds to YAP through its PPxY motif and to LATS2 through its RBD and ZF-PH. CIT forms a complex with YAP and LATS2, and the co-expression of YAP–LATS2 together with CIT enhances the interaction between YAP and LATS2. Furthermore, CIT interacts with LATS2 to restrain its kinase activity toward YAP. This occurs as the CIT N-terminus acts to inhibit the MST1-mediated phosphorylation of LATS2 at its hydrophobic motif without perturbing the physical interaction between MST1 and LATS2. We propose the model (Figure 4E) in which CIT’s inhibition of LATS2 kinase activity relies mainly on the CIT N-terminus because overexpression of the CIT N-terminus alone is sufficient to reduce LATS2 kinase activity (Figure 4A). Although, the scaffolding function of CIT seems to be dispensable for YAP phosphorylation by LATS2, our results do not exclude a role for the CIT C-terminus as an important binding domain. We reasoned that in one hand, enzyme-catalyzed reaction do not require strong and stable interaction between enzyme and substrate; on the other hand, CIT’s dual role as a scaffold protein and a LATS2 inhibitor may mean it acts differently over time. In conditions that favor LATS2 kinase activity (e.g. cell detachment or serum starvation), CIT may function solely as a scaffold protein for YAP and LATS2 interaction. In conditions that disfavor LATS2 kinase activity (e.g. cell re-attachment or serum stimulation), CIT inhibits LATS2 to promote YAP activity.
Drosophila Sti and mammalian CIT proteins consist of conserved N-terminal kinase domain, coiled-coiled domain, and C-terminal region (D’Avino et al., 2004). Hence we also examined the relationship between CIT and the Hippo pathway in vivo using the Drosophila eye model. We found that sticky (sti) and warts (wts), the Drosophila homologs of CIT and LATS2, respectively, function in a common pathway to control Drosophila eye size. Our genetic data seem to be consistent with the proposed dual functions of CIT, that is, direct inhibition of LATS2 phosphorylation by CIT N-terminus and stabilization of LATS2–YAP interaction by CIT C-terminus (Figure 4E). According to this model, Drosophila Sti may antagonize Wts to promote eye growth when there is a normal level of Wts. In the case of Wts overexpression, a high level of ectopic Wts may override the inhibitory effects of Sti/CIT N-terminus, hence dominantly inhibiting Yki activity. However, when Sti is depleted, the Wts–Yki interaction may be impaired due to loss of Sti/CIT C-terminal function, leading to the suppression of wts overexpression effects. Conversely, when wts is knocked down, Yki activity will be increased independent of the level of Sti, consistent with the observation that wts is epistatic to sti. These data suggest that functional relationships between Sti and Wts are influenced by the level and activity of Wts. Several external cellular stimuli are known to affect the kinase activity of LATS2/Wts. Thus, depending on the cellular context, different protein-protein networks might control LATS2/Wts kinase activity to maintain cellular homeostasis (Meng et al., 2016). We will further examine the effects of CIT/Sti on LATS2/Wts in different cellular contexts in the future.
Materials and methods
Cell culture
Retinal pigment epithelial cell (RPE) and Human embryonic kidney 293T cell (HEK293T) were cultured in DMEM/F12 and DMEM, respectively, supplemented with 10% FBS and 1% penicillin/streptomycin.
Plasmid constructs
The following CIT plasmids Myc-CIT, Myc.mcherry.CIT FL, Myc.mcherry.CITΔRBD, Myc-CIT-ΔN, Myc-CIT Ct, Myc-CIT Nt, Myc-CITΔZF-PH, Myc-CITΔC, and Myc.mcherry.CIT KD were kindly provided by Prof. Ferdinado Di Cunto (Gai et al., 2011). Based on these constructs, we additionally generated Flag-CIT Nt, Myc-CIT CtΔPY, and Flag-CIT Nt K/A mut. The GFP-CIT plasmid was kindly provided by Prof. Shuh Narumiya (Madaule et al., 1998).
CRISPR/Cas9 knockout of CIT
A single guide RNA (sgRNA) was designed according to protocol (Hsu et al., 2013) and cloned into the lentiCRIPRv2 vector (Sanjana et al., 2014). sgRNA plasmids were transfected into HEK293T cells together with the packaging vectors pMD2.G and psPAX2. Lentiviral supernatant was collected after two days and used to infect RPE cells for 24 h. Infected RPE cells were selected using 3 μg/ml puromycin. To obtain RPE CIT-null cells, a single clone was isolated and tested for CIT depletion by western blot. The single guide sequence used for CIT is 5′-GTCAGAAGTCTTGTAGGTTG-3′ (human).
siRNA transfection
siRNA oligos were annealed and diluted to a final concentration of 20 μM. Cells were prepared at sparse conditions (100000/60-cm dish) and transfected with either control siRNA or siRNA against a target gene of interest using Lipofectamine RNAiMax (Invitrogen) with a ratio of 6 μl siRNA:6 μl reagent in 400 μl Opti-MEM medium and incubated at room temperature for 10 min. Transfected cells were analyzed after two days. The siRNA oligo sequences used for control, CIT, LATS1, and LATS2 were: CIT, 5′-ACGATGAGCTGCTAGAAAA-3′ (human); LATS2, 5′-AAAGGCGTATGGCGAGTAG-3′ (human); universal control, 5′-CGTACGCGGAATACTTCGA-3′; LATS1, 5′-GTCTGCTTCATACATTCCTAA-3′.
RNA isolation and analysis
Cells were lysed using Easy-Blue (Intron Biotechnology) and their RNA was purified according to the manufacturer’s instructions. Briefly, 2 μg RNA was heated to 70°C for 5 min and then subjected to reverse transcription for 2 h using M-MLV reverse transcriptase, oligo-dT, and random hexamer primers. Then, 1 μl of cDNA was used as a template for quantitative PCR. Target mRNA levels were normalized with the mRNA level of GAPDH. The qPCR primers used for the target genes were: (human) hGAPDH qF, 5′-CTTCGCTCTCTGCTCCTCCT-3′; hGAPDH qR, 5′-GTTAAAAGCAGCCCTGGTGA-3′; (human) hCTGF qF, 5′-AAAAGTGCATCCGTACTCCCA-3′; hCTGF qR, 5′-CCGTCGGTACATACTCCACAG-3′; (human) hCYR61 qF, 5′-CCAATGACAACGCCTCCTG-3′; hCYR61 qR, 5′-TGGTGCAGCCAGAAAGCTC-3′; (human) hAMOTL2 qF, 5′-GCATTGAGAAGCTGGAAAGC-3′; hAMOTL2 qR, 5′-CTTGTTCCGCATGGTCTTCT-3′; (human) hANKRD1 qF, 5′-AGTAGAGGAACTGGTCACTGG-3′; hANKRD1 qR, 5′-TGGGCTAGAAGTGTCTTCAGAT-3′.
Co-immunoprecipitation
The indicated DNA combinations were transiently transfected into HEK293T cells with polyethylenimine (PEI) at a ratio of 1 μg DNA:3 μl PEI. After 2 days, the cells were harvested and lysed with lysis buffer (0.5% Triton X-100, 50 mM Tris-Cl, pH 8.0, 150 mM NaCl, and 1 mM EDTA). The extracted proteins were diluted to 1 mg in 1 ml lysis buffer and incubated with 1–2 μg antibodies overnight. Then, 20 μl protein A/G agarose beads were added the next day, and the resulting mixture was rotated for 1–2 h at 4°C, followed by three washes in lysis buffer. Finally, the samples were boiled with Laemmli buffer and subjected to western blotting.
Endogenous immunoprecipitation
RPE cells were trypsinized and suspended one hour prior to immunoprecipitation using 1 mg of extracted protein in 1 ml lysis buffer (0.5% Triton X-100, 50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA). In the case of endogenous immunoprecipitation, the lysate was pre-cleared with 10 μl protein A/G agarose beads for 1 h before incubating with the antibody. After this, the samples were centrifuged at 12000 rpm for 3 min to pellet all the beads. Then, the supernatant was collected and incubated with 2 μg antibody overnight. The next day, 20 μl protein A/G agarose beads were added, rotated for 1 h, followed by three washes with lysis buffer. Finally, the samples were boiled with Laemmli buffer and subjected to western blotting.
Sequential immunoprecipitation
The indicated combinations were expressed in HEK293T cells. The first immunoprecipitation step was performed according to the co-immunoprecipitation protocol above. The immunoprecipitation samples were eluted using elution buffer containing 20 μl 3× Flag peptide from a stock concentration of 5 mg/ml (Sigma) in 2 ml washing buffer (0.1% Triton X-100, 50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1× protease inhibitors, and a phosphatase inhibitor). Then 100 μl of elution buffer was added to the samples, incubated for 30 min on ice, and gently mixed every 10 min (or rotating at mode 15 speed 18 rpm). Then, the samples were centrifuged at maximum speed for 1 min, the supernatants were collected, and this was repeated several times until all supernatant was collected for each immunoprecipitation sample. The second round of immunoprecipitation was carried out using 1 μg antibody in 0.1% Triton X-100 buffer overnight, followed by 1 h of 20 μl protein A/G bead incubation, and three washes with 0.1% Triton X-100 buffer. Finally, the samples were boiled with Laemmli buffer.
BrdU staining and counting
Cells were split one day prior to collection at 10000–15000 cells/35 cm dish. The day after, 5 μg/ml BrdU (stock 10 mg/ml, 2000×) was added, incubated for 2 h, then fixed with 4% PFA. The nucleus was permeabilized with 2 N HCl. Cells were stained with an antibody against BrdU, and their nuclei were stained with DAPI. Seven different areas were photographed per coverslip, for which the number of cells positive for DAPI and BrdU was counted. The percentage of cells positive for BrdU (BrdU+) was calculated by dividing the number of BrdU-positive cells by the number of DAPI-positive cells and multiplying by 100.
Kinase assay
HA-LATS2 and the indicated plasmid combinations were transfected into HEK293T cells. Cells were lysed with 1× cell lysis buffer (0.5% Triton X-100, 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA). Then, lysate was subjected to immunoprecipitation with HA antibody overnight, followed by incubation with 20 μl protein A/G agarose beads for 1 h the next day. The immunoprecipitation samples were washed twice with 1× washing buffer (0.5% Triton X-100, 50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA), and twice with 1× kinase buffer (25 mM Tris, pH 7.5, 2 mM DTT, 10 mM MgCl2, 1× PAFS). For the kinase reaction, 500 μM cold ATP, 10 μl purified his-YAP, and a volume of 1× kinase buffer was added to the 20 μl protein A/G agarose beads to bring the final volume to 40 μl. Then, the samples were incubated at 30°C for 30 min. Finally, the kinase reaction was terminated by boiling with Laemmli buffer.
Antibodies
The following antibodies were used in our study: HA (Covance, MMS-101P), Flag (Sigma, F1804), Myc (Santa Cruz, sc-40), Myc (ascite), IgG Mouse (Santa Cruz, sc-2025), CIT (Santa Cruz, sc-390437), and YAP (Novus H00010431-M01) for immunostaining and immunoprecipitation; Yap (Cell Signaling 4912 s) for western blotting; pYAP S127 (Cell Signaling, 4911 s), LATS1 (Benthyl, A300-477A), LATS2 (Cell Signaling, 5888 s), β-actin (Sigma, A5361), pLATS 1079 (Cell Signaling, 8654), pLATS 909 (Cell Signaling, 9157 s), MST1 (Cell Signaling, 3682), MST2 (Cell Signaling, 3952 s), and BrdU (BD555627).
Mass spectrometry for identification of YAP binding proteins
RPE cells stably expressing YAP-tagged streptavidin binding protein (SBP) or the SBP tag alone (control) were trypsinzed and suspended for 1 h before being harvested and lysed with 0.5% Triton X-100. The lysates were pulled down and resolved by SDS-PAGE before being subjected to silver staining.
Drosophila genetics
Drosophila stocks were maintained at 25°C. The GAL4-UAS system (Brand and Perrimon, 1993) was used for gene overexpression or knockdown. Genetic crosses were made at 25°C unless stated otherwise. w1118 was used as wild-type. GAL4 and UAS lines were: GMR-GAL4 (Bloomington Drosophila Stock Center), UAS-sti RNAiHMJ03025 (Bloomington Stock Center, BL64012, sti RNAi1 in short), UAS-sti RNAiNIG10522R-1 (National Institute of Genetics, NIG 10522R-1, sti RNAi2), UAS-sti RNAiNIG10522R-2 (NIG 10522R-2, sti RNAi3), UAS-wts RNAi (Vienna Drosophila Research Center, v9928), and UAS-wts (a gift from G. Halder). sti1, sti3, and stiGS9053 mutants were obtained from the Bloomington stock center (BL4366, BL4871) and the Kyoto stock Center (DGRC201488), respectively. Eye sizes were measured using the Image J software. Dot plot graphs were generated using the R language in the RStudio IDE with the ggplot2 package.
Statistical analysis
Graphs were created using GraphPad Prism 6. Statistical analysis was performed using two-tailed student’s t-tests with a confidence level of 95%.
Supplementary Material
Acknowledgements
We thank Prof. Shuh Narumiya for providing the GFP-CIT plasmid. We thank all our lab members for critical comments on the manuscript, especially DaeHee Hwang, Wonyul Jang, David Eisenbarth, Sun-hye Jeong, and Quoc Duy Tran.
Funding
This work was supported by grants from National Creative Research Initiative Program (2010–0018277 to D.-S.L.), Individual Basic Science & Engineering Research Program (NRF-2016R1D1A1B03935764 to D.-H.L), Global Research Laboratory Program (NRF-014K1A1A2042982 to K.-W.C.), Mid-Career Research Program (NRF-2017R1A2B3007516 to K.-W.C.), Ministry of Education of the Republic of Korea and the National Research Foundation of Korea (NRF-2018R1A5A1024261) Korean Advanced Institute of Science and Technology (N11180249).
Conflict of interest
none declared.
Author contributions
M.K. performed the mass spectrometry experiment and the analysis. F.D.C. and M.G. provided CIT-related DNA constructs. T.H.Y.T. and D.-W.Y. conceived the project, performed the experiments, and analyzed the data. K.-W.C. supervised the fly work. D.-S.L. supervised the project and wrote the manuscript with K.-W.C. and D.-H.L.
References
- Aragona M., Panciera T., Manfrin A., et al. (2013). A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059. [DOI] [PubMed] [Google Scholar]
- Azzolin L., Panciera T., Soligo S., et al. (2014). YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 158, 157–170. [DOI] [PubMed] [Google Scholar]
- Bassi Z.I., Audusseau M., Riparbelli M.G., et al. (2013). Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc. Natl Acad. Sci. USA 110, 9782–9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cunto F., Calautti E., Hsiao J., et al. (1998). Citron Rho-interacting kinase, a novel tissue-specific Ser/Thr kinase encompassing the Rho–Rac-binding protein citron. J. Biol. Chem. 273, 29706–29711. [DOI] [PubMed] [Google Scholar]
- Di Cunto F., Imarisio S., Hirsch E., et al. (2000). Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron 28, 115–127. [DOI] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., et al. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183. [DOI] [PubMed] [Google Scholar]
- D’Avino P.P. (2017). Citron kinase—renaissance of a neglected mitotic kinase. J. Cell Sci. 130, 1701–1708. [DOI] [PubMed] [Google Scholar]
- D’Avino P.P., Savoian M.S., and Glover D.M. (2004). Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J. Cell Biol. 166, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N., and Aylon Y. (2017). The LATS1 and LATS2 tumor suppressors: beyond the Hippo pathway. Cell Death Differ. 24, 1488–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai M., Camera P., Dema A., et al. (2011). Citron kinase controls abscission through RhoA and anillin. Mol. Biol. Cell 22, 3768–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Cornils H., Chiu S.-Y., et al. (2014). Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell 158, 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Li X., et al. (2006). KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 172, 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Chun A., Cheung K., et al. (2008). Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509. [DOI] [PubMed] [Google Scholar]
- Harding B.N., Moccia A., Drunat S., et al. (2016). Mutations in citron kinase cause recessive microlissencephaly with multinucleated neurons. Am. J. Hum. Genet. 99, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K.F., Zhang X., and Thomas D.M. (2013). The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246. [DOI] [PubMed] [Google Scholar]
- Hoa L., Kulaberoglu Y., Gundogdu R., et al. (2016). The characterisation of LATS2 kinase regulation in Hippo-YAP signalling. Cell. Signal. 28, 488–497. [DOI] [PubMed] [Google Scholar]
- Hong J.-H., Hwang E.S., McManus M.T., et al. (2005). TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309, 1074–1078. [DOI] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., et al. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434. [DOI] [PubMed] [Google Scholar]
- Kim M., Kim M., Park S.J., et al. (2016). Role of Angiomotin-like 2 mono-ubiquitination on YAP inhibition. EMBO Rep. 17, 64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y., Vinayagam A., Sun X., et al. (2013). The hippo signaling pathway interactome. Science 342, 737–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Bielas S.L., Zaki M.S., et al. (2016). Biallelic mutations in citron kinase link mitotic cytokinesis to human primary microcephaly. Am. J. Hum. Genet. 99, 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P., Eda M., Watanabe N., et al. (1998). Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature 394, 491–494. [DOI] [PubMed] [Google Scholar]
- Madaule P., Furuyashiki T., Reid T., et al. (1995). A novel partner for the GTP-bound forms of rho and rac. FEBS Lett. 377, 243–248. [DOI] [PubMed] [Google Scholar]
- Meng Z., Moroishi T., and Guan K.-L. (2016). Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T., Hansen C.G., and Guan K.-L. (2015). The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya I.M., and Halder G. (2014). Discovering the Hippo pathway protein-protein interactome. Cell Res. 24, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Zheng Y., Hara M., et al. (2015). Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 29, 1416–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panciera T., Azzolin L., Cordenonsi M., et al. (2017). Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 18, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.W., Kim Y.C., Yu B., et al. (2015). Alternative Wnt signaling activates YAP/TAZ. Cell 162, 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandala T., Gregory S.L., Dalton H.E., et al. (2004). Citron kinase is an essential effector of the Pbl-activated Rho signalling pathway in Drosophila melanogaster. Development 131, 5053–5063. [DOI] [PubMed] [Google Scholar]
- Toloczko A., Guo F., Yuen H.-F., et al. (2017). Deubiquitinating enzyme USP9X suppresses tumor growth via LATS kinase and core components of the Hippo pathway. Cancer Res. 77, 4921–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Li X., Huang J., et al. (2014). Defining the protein-protein interaction network of the human hippo pathway. Mol. Cell. Proteomics 13, 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., and Guan K.L. (2013). The Hippo pathway: regulators and regulations. Genes Dev. 27, 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.-X., Zhang Y., Park H.W., et al. (2013). Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 27, 1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Zhao B., Panupinthu N., et al. (2012). Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Lu Q., et al. (2011). Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Wang L., et al. (2012). Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 26, 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.