Abstract
CASP (Critical Assessment of Structure Prediction) assesses the state of the art in modeling protein structure from amino acid sequence. The most recent experiment (CASP13 held in 2018) saw dramatic progress in structure modeling without use of structural templates (historically ‘ab initio’ modeling). Progress was driven by the successful application of deep learning techniques to predict inter-residue distances. In turn, these results drove dramatic improvements in three-dimensional structure accuracy: With the proviso that there are an adequate number of sequences known for the protein family, the new methods essentially solve the long-standing problem of predicting the fold topology of monomeric proteins. Further, the number of sequences required in the alignment has fallen substantially. There is also substantial improvement in the accuracy of template-based models. Other areas - model refinement, accuracy estimation, and the structure of protein assemblies - have again yielded interesting results. CASP13 placed increased emphasis on the use of sparse data together with modeling and chemical crosslinking, SAXS, and NMR all yielded more mature results. This paper summarizes the key outcomes of CASP13. The special issue of PROTEINS contains papers describing the CASP13 assessments in each modeling category and contributions from the participants.
Keywords: Protein Structure Prediction, Community Wide Experiment, CASP
INTRODUCTION
CASP (Critical Assessment of Structure Prediction) is a biennial community experiment to determine the state of the art in modeling protein structure. Participants are provided with amino acid sequences of target proteins, and build models of the corresponding three-dimensional structures. Submissions are compared with experiment by independent assessors. The experiment is double blinded - participants have no access to the experimental structures and assessors do not know the identity of those making the submissions. In addition to structure models, a number of other aspects of protein modeling are assessed as well: refinement of an approximate structure closer to the experimental one, estimates of the accuracy of an overall structure model and of each residue, modeling the structure of protein oligomers, the ability to improve models using a variety of sparse data types, and the accuracy of protein structure features related to deducing aspects of function. Here we summarize the current state of the art in each of these areas as determined in the CASP13 experiment (2018). Papers in this special issue of PROTEINS provide detailed analysis by the independent assessors in each modeling area and contributions from some of the more successful participants.
In CASP13, a total of 98 research groups from 21 countries tested 185 modeling methods and submitted over 57,000 predictions in six prediction categories, maintaining the previous high level of participation. There were 90 modeling targets for tertiary structure prediction (80 assessed), and 45 for quaternary structure prediction (42 assessed), including 13 hetero-complexes (12 assessed). The 80 tertiary structure modeling targets were parsed into 111 evaluation units, which were assessed as separate targets1. Models are solicited in two initial stages. First on a short (72 hour) time scale, intended for automated model building servers, then on a three-week time scale, allowing time for more complex procedures and human input (though the latter now appears to be rare). Relatively easy targets are only released for the server phase. For evaluation, targets are divided into two main categories: template based (TBM), for those where one or more structural templates can be identified by sequence search, and template free (FM), for targets with no sequence detectable template. Some targets fall into a grey area between these categories, and are labeled TBM/FM. One significant change in target composition in CASP13 was from the ongoing revolution in high resolution cryo electron microscopy (EM)2. There are EM targets for a total of six complexes (four heteromeric) and one protein monomer. These targets tend to be considerably larger than typical in CASP, but once parsed into evaluation domains are less unusual3. A fuller account of the procedures used in CASP is available in4
PROGRESS IN CASP13
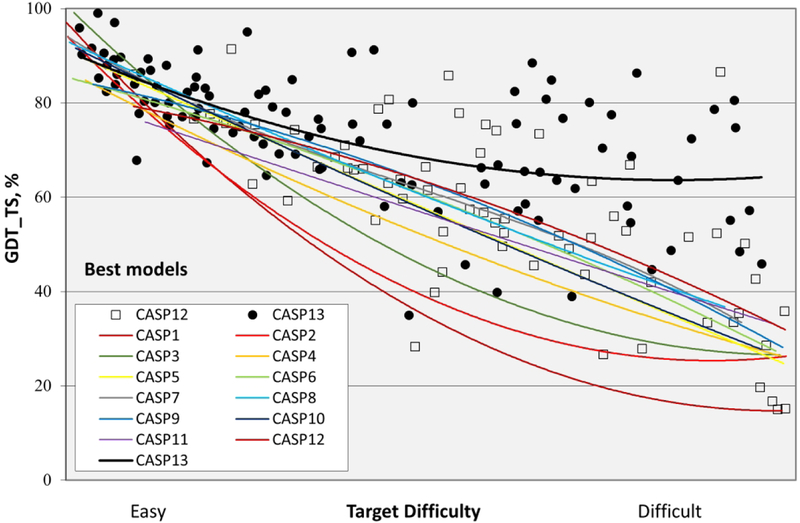
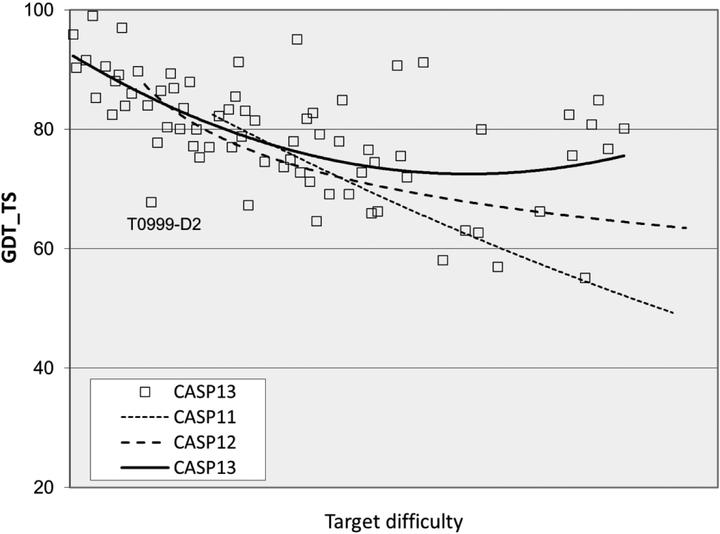
The overall accuracy of models improved dramatically in CASP13, especially for the more difficult targets where comparative modeling cannot be used. Figure 1 shows the trends in backbone accuracy for the best models received in each CASP, as a function of target difficulty (the extent to which a target or target domain is related to the sequence and structure of other proteins with already known structures5 - supplementary figure S1 gives more data on target difficulty). The vertical axis shows backbone accuracy in terms of GDT_TS6,7). With this measure, 100% is exact agreement of the Cα co-ordinates of a model with those of the experimental structure, and a random model typically has a GDT_TS of between 20 and 30%. As a rule of thumb, models with values greater than about 50% have correct overall topology, and models with values greater than ~75% have many correct atomic level details. As the trend lines show, early CASPs saw rapid improvement, but started from very low accuracy. Until CASP13, most recent CASPs have shown very limited overall improvement by this measure (though more fine-grained analysis shows improvement in specific areas4). Dramatically, the CASP13 trend line, instead of plunging downwards, continues horizontally to the most difficult targets, with a sustained GDT_TS greater than 60. Supplementary figure S2 shows a similar (though not quite so pronounced) CASP13 trend for average GDT_TS over the six best performing groups on each target, indicating that multiple groups have improved substantially. Below, we discuss the methodological advances that drove this progress.
Figure 1:
Trend lines of backbone accuracy for the best models in each of the 13 CASP experiments. Individual target points are shown for the two most recent experiments. The accuracy metric, GDT_TS, is a multiscale indicator of the closeness of the Cα atoms in a model to those in the corresponding experimental structure. Target difficulty is based on sequence and structure similarity to other proteins with known experimental structures (see5 for details). There is a striking improvement in model accuracy in CASP13 (top black line), particularly for the more difficult targets.
PREDICTING CONTACTS IN PROTEIN STRUCTURES
For a quarter of a century8, attempts have been made to predict three-dimensional contacts between residues in proteins, based on correlations in amino-acid substitutions found in protein family protein sequence alignments9.
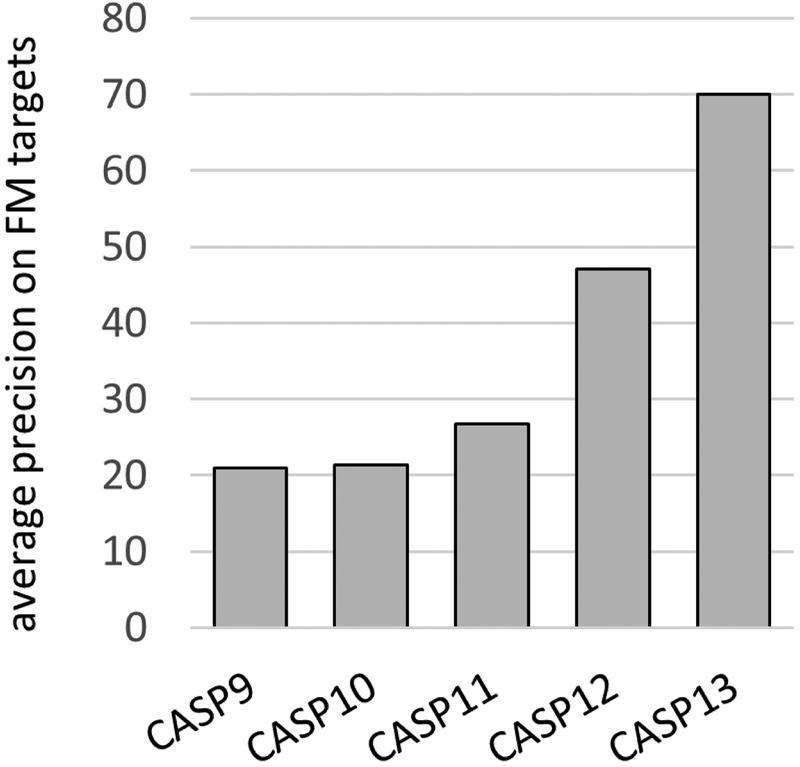
For many years, the precision of these methods as measured in CASP was stalled at 20% or a little higher. Figure 2 summarizes progress in recent CASPs. Starting in CASP11 (2014), and much more successfully in CASP12, statistical methods that consider all pairs of residues simultaneously to address transitivity effects9 began to improve accuracy, resulting a best overall precision of 47% in CASP12 - almost doubling in one CASP round - one of the biggest single improvements in any metric seen in any CASP. Some predictors combined the statistical models with machine learning, for instance10,11. But the key algorithmic advance appears to be the proper treatment of transitivity. A limitation of those methods is that at least several hundred appropriate sequences are needed to produce accurate predictions.
Figure 2:
Best contact prediction precision in recent CASPs. CASPs 9 and 10 continued a long trend of low precision. CASP11 shows a small advance, while the two most recent, CASP12 and 13, show dramatic improvements. In CASPs 11 and 12 progress is the result of more sophisticated statistical models, together with largely conventional machine learning. The further jump in CASP13 is the result of the effective deployment of deep learning methods. (Average fraction of correctly predicted contacts for the most confidently predicted L/5 contacts 24 or more residues apart in the sequence, where L is target length. Free modeling targets, average for the best performing group in each CASP. Contacting residue pairs defined as those with less than 8 Angstroms between Cβ atoms).
In CASP13, there is another large advance in precision, to 70%, again with several groups delivering similar performance. This time it is clear the improvement came from the use of deep neural network methods (discussed further in a CASP13 special issue paper12). These techniques have of course been very effective in other areas, particularly image analysis13,14 and speech recognition15. Contact prediction uses a similar methodology, treating the contact matrix (an L by L matrix for a sequence length L, with 1 for elements representing contacting residues pairs and 0 for non-contacting ones) as an image. The network is trained on a large set of known structures, typically with multiple sequence alignment information, secondary structure prediction, co-evolution analysis, and related features as input and the contact matrices as output. Input of information for a new protein then generates an approximate contact map. These methods were already being tested in CASP12 and promising benchmarking has since been published16. But as is often the case, they took some time to mature to the point where improvements in performance are clearly measurable (very clearly in this instance!).
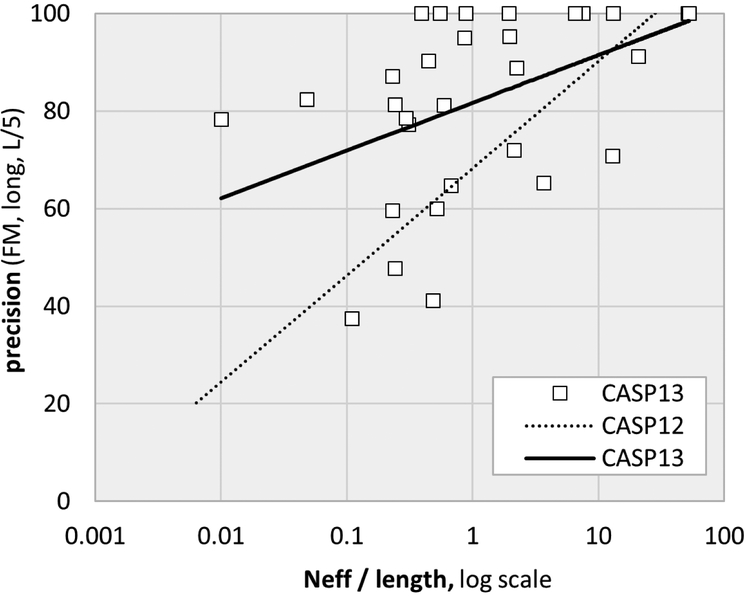
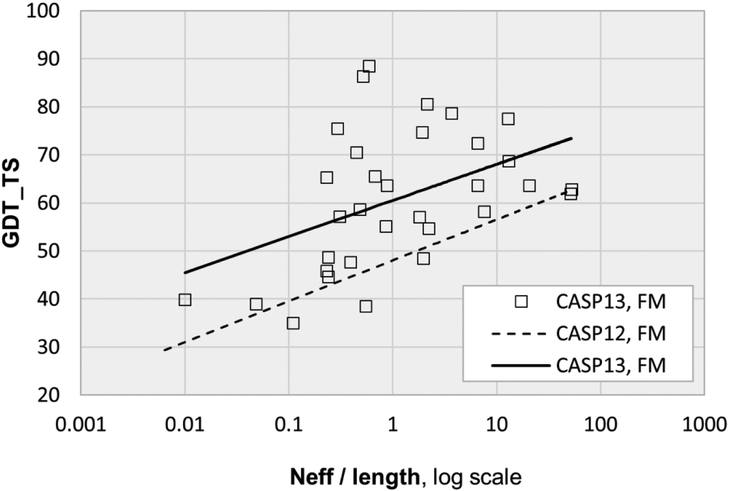
Although the data representation in the advanced statistical methods and deep learning approaches are very different, both rely on correlations in amino acid substitutions for contacting residue pairs. As a result, a limitation of both is the need for a substantial depth of sequence alignment. The effect of this can be seen in figure 3, where trend lines for contact precision slope upwards as a function of normalized alignment depth. But this dependency is greatly reduced with the CASP13 deep learning methods, resulting in higher accuracy over a wide range of alignment depths. In CASP13, inclusion of metagenomics sequence data increased alignment depth for some targets. For example, metagenomics data as described in17,18 increases alignment depth for two free modeling targets from marginally adequate (less than 1L) to greater than 2L. But generally, addition of these data has had only a modest impact so far.
Figure 3:
Contact prediction precision trend lines as a function of sequence alignment depth and target length. In CASP13 there is a reduced dependency on alignment depth, resulting in more accurate results for shallow alignments as well as higher precision overall. Strikingly, for ten out of the 31 free modeling targets, the best predictions achieved 100% precision for this subset of contacts (see figure 2 for definitions). The effective alignment depth, Neff, includes metagenomic sequences compiled as described in17,18 Neff was calculated using a 90% sequence identity cutoff and a minimum of 60% sequence coverage (details in44).
TEMPLATE FREE MODELING
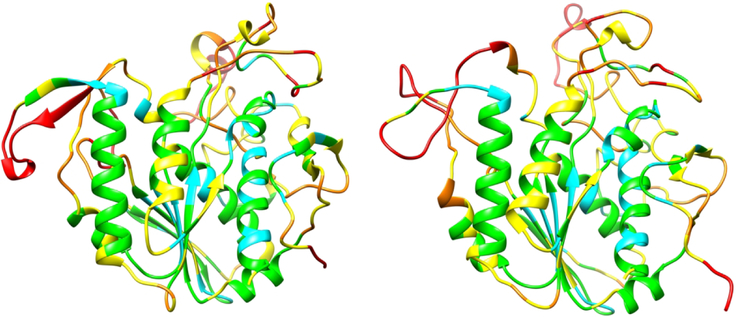
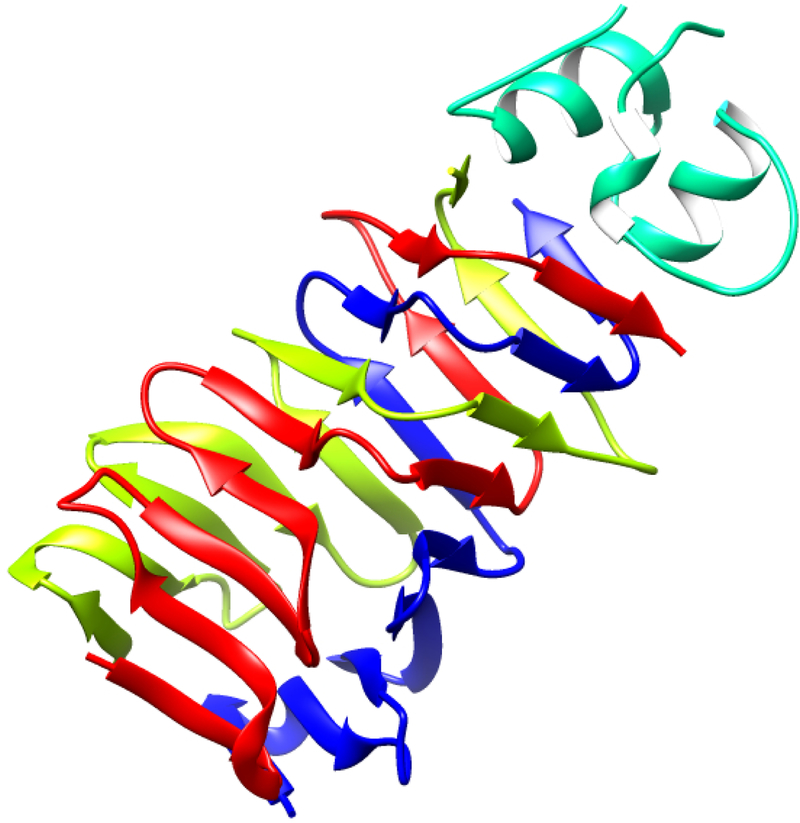
In CASP13, the largest improvement in model accuracy is for the most difficult, free modeling, targets (Figure 1, right hand side) where no structural template could be detected using sequence. Figure 4 shows an example for a free modeling target where a number of groups produced good models.
Figure 4:
Crystal structure of a 354 residue domain of a free modeling target (T0969-D1), ESKIMO 1, a probable xylan acetyltransferase, PDB 6CCI (left panel) and the most accurate CASP model (right panel). Most of the structure core is modeled to a Cα accuracy of better than 1 (cyan) or 2 Angstroms (green). Irregular loop regions are less accurate (yellow, better than 4 Angstroms or orange, up to 8 Angstroms error. Some residues (red) in external loops have larger errors.
If a sufficiently reliable set of contacts are predicted, these can be used as restraints to obtain more accurate three-dimensional models. Figure 5 shows the relationship between main chain accuracy and normalized alignment depth for template free modeling targets in the most recent CASPs. There is a strong dependency of accuracy on alignment depth, consistent with the major jumps in contact performance driving the 3D improvement for free modeling targets. The trend line for CASP13 is well above that for CASP12, consistent with the more accurate contact predictions from deep learning. In CASP13, all FM targets with Neff/L greater than 1 (effective sequence alignment depth equal or greater than the length of the target) have a GDT_TS greater than 50, indicating a correct topology. The majority of targets with Neff/L > 0.1 also have GDT_TS >50. As discussed later, a number of the less accurate models are affected by inter-molecular protein interactions, something current methods are not able to handle. (Earlier CASPs already showed a link between contacts and 3D structure accuracy4, but not nearly to this extent).
Figure 5:
Best model main chain accuracy (GDT_TS) as a function of sequence alignment depth and target length for CASPs 12 and 13. Accuracy depends on alignment depth, as is expected if the result is dominated by contact prediction accuracy and related advances. Across all alignment depths, CASP13 models are on average more accurate than those in CASP12.
Part of the three-dimensional accuracy improvement in CASP13 comes from not only more accurate prediction of contacts but also prediction of inter-residue distances at a range of thresholds, something deep neural networks are capable of and the statistical methods are not. Approaches vary19,20, but in essence, ‘contact’ maps are predicted for each of a set of inter-residue distances - say atoms within 6, 8, 10…20… Angstroms. Properly normalized, these predictions allow an effective potential of mean force to be derived between every pair of residues in a structure (that is, up to L*L/2 - L potentials for an L residue long sequence). These potentials can then be used to drive a structure folding procedure. One group, A7D from DeepMind20, appear to have very successfully deployed this technique, and had the most accurate results overall. It is not fully clear what current deep learning procedures are ‘learning’ about protein architecture. The ability to predict inter-residue distance probabilities as well as contacts suggests that the topology of helices and beta-sheets and inter-secondary structure packing are captured in some form. But so far there is no published analysis and indeed such an analysis may not be meaningfully possible. There are many potential variations on the type of residual networks currently being deployed, as well as other variables that have yet to be evaluated, such as the best use of dilation and dropout21. This and other aspects of the methods will likely be further developed and refined by the next CASP and it will very interesting to see how much further improvement can be made.
By definition, all free modeling targets are cases where no template structure can be easily detected from sequence. But there may nevertheless be similar folds already known. An alternative approach to using predicted contacts as restraints is to survey a library of known structures, assessing which, if any, are most compatible with the contact set. Supplementary Figure S3 shows the dependency of backbone accuracy on the nearness of structural templates. Both CASP 12 and 13 show clear dependency, but it is substantially reduced in CASP13, suggesting that template searches were less competitive with folding algorithms, probably because greater contact accuracy and the use of more general inter-residue distance prediction made the latter approach more effective.
As always in CASP, care is needed to make sure that apparent progress is not an artifact of different target difficulty in successive rounds. The insert in supplementary figure S1 shows only very small differences in average free modeling (FM) target difficulty in the most recent three CASPs. Additionally, supplementary figure S3 shows that the average similarities of CASP 12 and 13 FM targets to structural templates are also nearly identical. The figure also shows the underlying CASP12 and 13 distributions of target/template similarity values are close, further supporting similar target difficulty.
TEMPLATE BASED MODELING
As the number of experimentally determined structures grows, so does the number of sequences for which it is possible to directly use structural templates to build a model, using comparative modeling techniques. Figure 6 shows the relationship between backbone accuracy of best models received for the template-based modeling category in the three most recent CASPs. For the easiest targets (left hand side) with a high level of sequence identity to a known structure there is no apparent improvement by this measure. For harder targets, CASP12 is improved over CASP11, and CASP13 is substantially further improved. Given the major accuracy advance in template-free modeling from improved inter-residue distance predictions, an obvious question is whether those methods are contributing here too. Supplementary Figure S4 shows the relationship between backbone accuracy and alignment depth for the template-based targets. CASP13 shows a mild dependence of accuracy on alignment depth, suggesting that contacts are also playing some role in this regime. However, as expected for these targets, almost all the alignments are deep enough for good contact prediction, which may obscure a larger signal. Conversely, there may be a tendency for targets with deeper alignment to have more useful templates, which would also tend to contribute to the signal seen in the figure. Further support for contact prediction contributing to the TBM improvement comes from a post-CASP analysis comparing the performance of one method with and without contact prediction included - contact information leads to higher accuracy for a number of targets (Yang Zhang, personal communication).
Figure 6:
Best model backbone accuracy (GDT_TS) as a function of target difficult for template-based models in recent CASPs. CASP13 shows a marked improvement in accuracy compared to previous CASP. Targets are those where there is clear sequence relationship to a known structure (termed TBM) and those with a marginal relationship (TBM/FM).
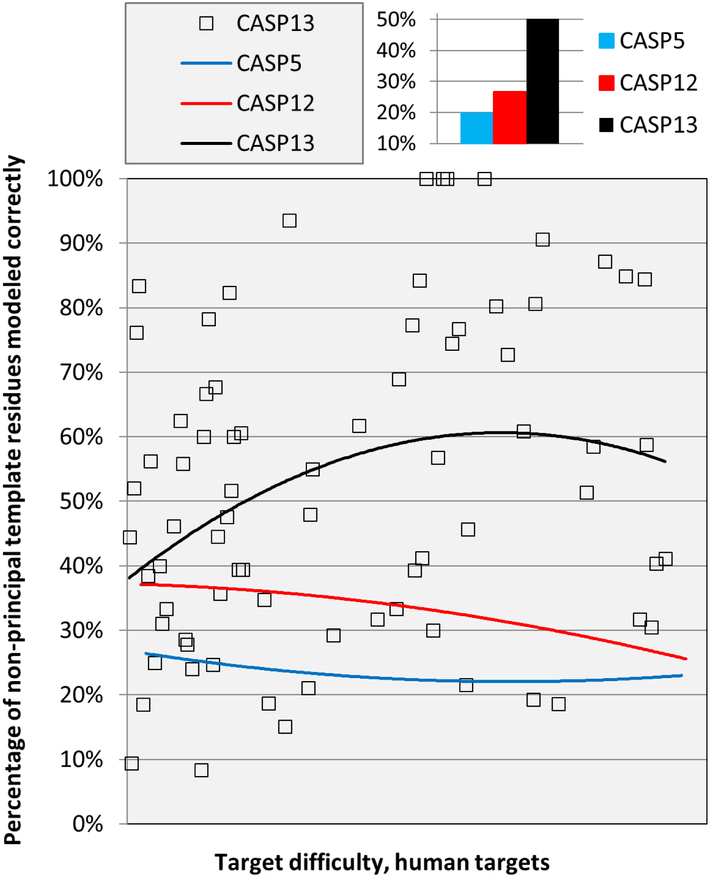
Typically, structure templates do not provide complete coverage of a target structure, and overall accuracy depends not only on the appropriateness of the templates but also on modeling of regions with no template. As Figure 7 shows, by this measure, there was modest improvement between CASP5 in 2002 and CASP12, but a much larger improvement between CASP12 and CASP13. As we have discussed before4, earlier improvements resulted from two principal modeling strategies: identification of other templates with the correct structure in these regions or in some sense building these substructures from scratch. Note that one would not expect the improvement to come from better contact prediction: by definition these are regions that are not structurally conserved within the protein family, and contact prediction generally relies on such conservation. Though it is possible that more accurate modeling of the structurally conserved regions creates a more accurate context for modeling non-template regions.
Figure 7:
Trend lines for the fraction of non-principal template (‘loop’) residues correctly modeled. There is a substantial improvement in CASP13. (Best models received for each target, 3.8 Angstrom Cα atom agreement or better considered correct, TBM and TBM/FM targets).
REFINEMENT
Models generated in both the template-free and template-based modeling sections of CASP are approximate, and there is an end-game problem of further improving agreement with experiment. To address this challenge, CASP includes a section on refinement, where participants are provided with an initial model and asked to submit a more accurate version. Performance has improved enormously over the succession of experiments, from initial attempts that marginally improved some of the targets22 to impressive examples of structure correction in recent rounds23,24,25. But it is still the case that no single method improves every target. In the three most recent CASPs23,24,25, the best groups have returned improvements for 60 to 70% of the targets. One probable reason for limited performance is that the area suffers from a serious Red Queen problem. Refinement methods that have been shown to be useful are increasingly incorporated in initial modeling pipelines so that the starting point structures supplied are already partly refined. Thus, methods must improve every round just to appear as effective as previously. This particularly affects those who participate in both initial structure modeling and refinement, as their models may be selected as starting structures for refinement. As a consequence, metrics of improved structure accuracy may not be very useful for measuring refinement performance. Nevertheless, the three groups who have been consistently successful in recent CASPs do show modest improvement in performance over successive rounds25. A more qualitative judgement of progress is to analyze the type of structural features that are corrected. A few CASPs ago, success with minor repositioning of secondary structure elements became common, for example26. More recently, and especially in this CASP25, larger range corrections (for example a 7.5 Angstrom helix shift in target R0981-D4) and significant repacking (for example in R0974s1) have been achieved.
A persistent feature of refinement performance is that some targets are more refine-able than others, and there are always some for which no group achieves a significant improvement (10 to 15% of targets in recent CASPs). This has been a puzzle. The most recent assessment provides partial insight into that phenomenon, with clear examples where the non-inclusion of interactions with other protein domains or binding partners limits accuracy25. As will be apparent again later, the modeling field has now advanced to the point where there is a critical need for methods that effectively include the full molecular environment.
As in previous CASPs, there are differences among the most successful refinement methods. These range from a major focus on molecular dynamics27 to hybrid Monte Carlo/sampling method28, to methods dominated by sampling29. But overall, there is an increasing emphasis on the importance of conformational sampling.
ACCURACY ESTIMATION
Although modeling methods have improved enormously, models still greatly vary in accuracy, both globally and in different parts of a structure. For any application it is critical to know the accuracy of a model, and so CASP includes a section on estimating model accuracy. As detailed in30, these predictions are very useful, and have been for some rounds of CASP. The methods roughly fall into two categories - consensus methods that rely on the degree to which a model is similar both overall and in detail to others, and so-called ‘single model’ methods that use some form of structure-based scoring function, often together with machine learning. Both approaches have performed well and comparably in recent CASPs. In CASP13, the assessor observed a relationship between the reliability of single model accuracy estimates and the methods used to generate a model, particularly for models created with high reliance on contact prediction related methods31, apparently because of method-specific characteristics of the models. To address this, some groups are now developing method-specific accuracy estimation approaches. CASP already requires that models are accompanied by detailed accuracy estimates, and in future, more emphasis may be placed on these.
PROTEIN ASSEMBLIES
As noted earlier, the enormous progress in domain and monomeric protein structure modeling has highlighted the next bottleneck - limitations on initial model accuracy and on refinement imposed by no or inadequate inclusion of the larger molecular environment. More generally, most proteins exist as part of complexes, and function is often dependent on the assembly. One aspect of the problem, the ability to dock subunits of proteins to each other, has been evaluated by CASP’s sister organization, CAPRI (http://www.capri-docking.org/) since 2001. In the three most recent CASPs, CAPRI and CASP have worked together to engage both communities in the broader problem of protein assembly, including the use of protein models. Assessment papers from both organizations are included32,33 in the CASP13 special issue. Participation from both communities increased over CASP12, showing growing interest in this important problem. The CASP13 assessor found evidence of some improvement compared to CASP1233.
Assembly is most successful when there is a template for the complex, presenting an assembly comparative modeling problem34, and that was again demonstrated in this CASP, where the CASP13 assessor concluded that availability of good assembly templates usually results in good models. Next most successful is assembly of complexes where the experimental structure of all the components is known, and there is little or no conformational change associated with assembly. ‘Free’ docking methods can be effective for these35, but these targets do not generally occur in CASP. Relatively few complexes do not involve significant conformational changes of at least side chains and local regions of structure, and without an assembly template, current methods are unable to cope with these situations. Add to this the complications of working with approximate models for assembly components, and the problem is daunting. As the CASP13 assembly assessor points out33, because of the importance of conformational changes on assembly formation, the current standard procedure of first building monomer structures in isolation and then attempting to dock them is flawed. The assessor found seven of the heteromeric CASP13 assembly targets have substantial interdependences between monomers, in a variety of ways. Figure 8 shows an example for target H0953, an A3B1 multimer, where the trimer assembly generates strong subunit interdependencies. In other targets (T0973, 991, 998), a helix is swapped between subunits.
Figure 8:
Part of the experimental structure of target H0953 (PDB 6F45), the adhesin tip complex of a bacteriophage tail fiber, illustrating subunit structure interdependence. One of the two protein chains contributing to this assembly forms a trimer (colored red, green and blue), with the N terminal five strand beta sheets of the three monomers packing against each other. The C terminal three beta strands of each monomer inter-digitate with each other. The C terminal stands also form an interface with the helical end of another subunit (green). Impressively, in spite of the apparent interdependency of the five-strand beta-sheets, accurate models were returned for that part of the structure. But failure to consider the even more intimate subunit interactions of the three N terminal strands resulted in incorrect models for that subdomain.
The obvious message is that successful assembly methods will have to take subunit interdependences into account and not rely exclusively on modeling isolated subunits. Following the emergence of powerful deep learning methods for monomers in CASP13, there is intense interest in whether these approaches may be adaptable to the problem. Of note in this regard is that the assembly assessor found prediction of inter-subunit contacts in homo-assemblies was ‘surprisingly successful’33. It’s not clear how these predictions were made, but they may be useful in identifying first, which regions are in contact even in the absence of a good three-dimensional model of the monomers, and second, possibly where there are conformational interdependencies. Deep learning methods for image analysis have been shown to be very robust to noise12, for example see https://clarifai.com/demo. It will be very interesting to see in the next CASP whether any examples of contact driven non-trivial assembly can be achieved, particularly with the use of deep learning.
DATA ASSISTED MODELING
Even high-resolution experimental structures incorporate aspects of modeling, making use of bond length and bond angle restrictions, avoidance of steric clashes, and sometimes imposing reasonable electrostatic interactions. Lower resolution methods - SAXS, chemical cross-linking coupled with mass spectrometry, and sparse NMR - depend critically on modeling to make maximum use of the experimental data. Since CASP11, CASP has experimented with providing these types of data to participants after data-free models have been obtained, and assessing whether the sparse data can be effective in increasing model accuracy. This area has great promise, but is proving challenging to successfully implement. First, additional data must be generated by the experimental community. A number of groups have been very co-operative and supportive, but still, protein samples are only available for a few targets, and those targets may not be ideal. Second, it requires specialized expertise to make optimum use of these types of data. In spite of vigorous efforts to provide webinars and other material in CASP13, rather few predictors have so far moved into this area. Third, because of low participation, the newer contact prediction and deep learning methods were not used together with the sparse data. As a result, more accurate models were obtained without use of the experimental information.
NMR:
The Gaetano Montelione and Antonio Rosato groups produced simulated sparse NMR data for 12 proteins or protein domains, in the form of ambiguous interatomic contact lists, chemical shifts, and RDCs. They also provided real sparse NMR data for one CASP target, N1008. The data provided are intended be similar to that available for large structures, and are insufficient for structure solution by standard NMR techniques36. Nine groups took part in NMR-assisted modeling, three of whom were controls from the Montelione lab. Generally, the models submitted are of similar accuracy to the best unassisted models received, but for one target, N0981-D2, a model built using the simulated data is over 30 GDT_TS units better than the best unassisted, a notable success. The target with experimental NMR data, N1008, is a designed protein37, and even though there are no sequence homologs, was very accurately modeled by a number of groups, without the use of the data. As a result, the NMR assisted models were not as accurate. That outcome illustrates how tricky it is to choose targets in which to invest experimental effort. Of the nine groups submitting NMR assisted structures, two (Laufer and Meiler) had markedly better results than the controls. Laufer used molecular dynamics with a filtering technique to remove non-consistent restraints38. We hope the encouraging results will lead to larger scale participation in this category in CASP14.
SAXS:
Data were generated for 11 targets in all, including 7 complexes. This was a very impressive contribution from the experimental group (Susan Tsutakawa, Greg Hura and John Tainer). 13 groups submitted models using these data. A number of teething troubles that plagued the first SAXS experiment in CASP1239 were avoided or greatly reduced, so that a more meaningful assessment of the contribution from the SAXS data could be made.
For no target was the best data assisted structure as accurate as the best unassisted, although there are some examples of improved inter-subunit relationships. Again, the issue here may be the relatively low participation, so that the results are not necessarily representative of the newest unassisted methods. Several groups did develop interesting pipelines incorporating SAXS data, and as is often the case in CASP, it may take further iterations before the power of these can be properly assessed. Methods typically take the full set of server models available for a target and evaluate the fit of these the SAXS data, often also using additional accuracy estimations. One group also investigated the use of normal mode driven structure changes.
Comparison of the SAXS envelopes with the X-ray structures suggests that for up to half the targets there could be differences between the solution and crystal structures. Such differences may in principle limit the accuracy that modeling can achieve using the crystal structure as the gold standard. But there could be a number of explanations for the discrepancies, including sequence differences between crystal and SAXS samples, more disorder in solution, and the inherent difficulties of interpreting SAXS data.
Chemical crosslinking and mass spectrometry:
Experimental data derived from two different cross-linking chemistries were provided. Alexander Leitner and Ruedi Abersold (ETH, Zurich) used a predominantly Lys primary amine-oriented (BS3) chemistry and Adam Belsom and Juri Rappsilber (University of Edinburgh and Berlin Technical University) employed the heterobifunctional, photoactivable cross-linking chemistry. One data set was also provided by Marcus Hartmann, using disuccinimidyl suberate (DSS) chemistry. Altogether data were collected for eight different protein samples, including three hetero-multimers, two homo-multimers, and three single chain proteins. Based on these data, five heteromeric targets and 17 single-sequence targets (monomers or subunits of multimers) were released for prediction. An analysis by the assessor40 shows that a surprisingly high fraction of cross-links appear not to be compatible with the targets’ X-ray structures (27–47%). In total, 14 prediction groups participated. For the monomeric protein domains and subunits, there are many instances where the data-assisted models are more accurate than the corresponding un-assisted models from the same participants. But this comparison likely provides too optimistic a view, since the groups with the greatest improvement started from scratch in utilizing the cross-link data, ignoring their initial submissions, and instead making use of the full set of server models available for each target. A more stringent criterion, comparing the data-assisted models to the best received for each target from any group generally shows all the cross-link assisted models are less accurate. For the complete complexes, there is one instance of a significant improvement, for X0957, a bacterial toxin/immunity protein complex, where several inter-subunit crosslinks helped select a more appropriate overall assembly33, an encouraging result. The results illustrate both the promise and the challenges of using cross-link information to improve models. Many cross-links are misleading in that they conflict with the corresponding X-ray structure and some can be false positives. Further, the large variation in distance between crosslinked residues40 makes the technique inherently low-resolution, and so likely best suited to complexes, as the result for X0957 illustrates.
Pilot experiments were also conducted with FRET data on one target (generously provided by Claus Seidel and Mykola Dimura) and SANS on another (provided by Anne Martel). We expect to include more data of these types in CASP14.
DISCUSSION
Successful use of relatively standard deep learning techniques for predicting not only three-dimensional contacts but more general inter-residue distance distributions was the outstanding development of CASP13 and caused much excitement and creative thinking at the CASP meeting. There is an expectation that similar approaches can be applied to other areas of structure modeling, particularly improved estimates of both global and local model accuracy, improved model refinement by allowing focus on regions of maximum error, and recognition and prediction of protein-protein interfaces. We will have to wait until CASP14 in 2020 to see which of these bears fruit. CASP14 is also likely to see further progress in 3D structure modeling based on deep learning approaches. Several modeling groups are developing servers that will make the new methods available to broader community. It’s likely that the impact on the usefulness of modeling will be large.
The major progress in modeling domains and monomeric proteins without direct use of a structural template is a very significant break-through: for these proteins, the long-standing problem of ‘protein folding’ (generating a model with the correct topology) is essentially solved, albeit it in way that early work in the field never imagined. An alignment with of at least a few dozen sequences is usually needed for the methods to work, but most protein families are now that large. Success with topology prediction has increased focus on the remaining problems - we are still a long way from the accuracy of X-ray structures or from enabling structure-based drug design, and more complex structures are the norm in biology. CASP already has well-established categories in the relevant areas, particularly refinement and protein assemblies, and as already noted it will be exciting to see what impact deep learning and related approaches have on those. Other areas, such as conformational change in response to ligand binding and environmental conditions, remain future challenges.
CASP continues to experiment with other aspects of modeling. Of note this round was the expanded number of targets for which sparse experimental data were available. Although the results in terms of more accurate models are not impressive, it is clear that much more development is possible, and we have already seen several groups introduce methods specifically tailored to particular data types. CASP continues to investigate the best ways of assessing how effectively functional information can be derived from models41,42, and in this round, solicited assessment comments from those who provided the prediction targets43. An interesting development during CASP13 was the introduction of a CASP commons experiment (http://predictioncenter.org/caspcommons/). Biologists were canvased to identify a total of 35 small proteins for which structure would be particularly useful for their research. The Montelione group cloned and expressed these, with the goal of determining which are suitable for NMR structure determination, and in parallel the CASP community was invited to submit models. So far one experimental structure has been obtained37. A new round of modeling is now beginning, using the new free modeling methods from CASP13.
We plan to hold CASP14 in 2020, with a similar timetable to previous rounds. The prediction season will be spring and summer, and the conference will be at the end of the year. Details will be posted on the Prediction Center web site as they become available.
Supplementary Material
ACKNOWLEDGMENTS
CASP is only possible through the generosity and support of three groups of people: the data providers, the assessors, and the participants. A total of 36 experimental groups from 14 countries provided targets for CASP13. Nine of these groups also provided material for the collection of SAXS, SANS, or cross-linking experimental data: Owen Davies (Newcastle University); Petr Leiman (UTMB, Galveston, TX) and Matthew Dunne (ETH Zurich); Andrew Lovering (University of Birmingham); Karolina Michalska and Andrzej Joachimiak (Argonne National Lab, Chicago, IL); Anne Martel (Institute Laue-Langevin, Grenoble); Jose Henrique Pereira (LBNL, Berkeley, CA); Mark van Raaij (CNB-CSIS, Madrid); Lindsey Spiegelman (University of California, San Diego, CA); Chin-Lin Tsai (MD Anderson Cancer Center, University of Texas), making that aspect of the experiment possible. SAXS data were acquired for ten protein samples by Susan Tsutakawa, Greg Hura and John Tainer (LBNL, Berkeley). Cross-link data were acquired for seven samples by Esben Trabjerg and Alexander Leitner (Institute of Molecular Systems Biology, ETH, Zurich), and for four samples by Adam Belsom and Juri Rappsilber (Technische Universitat, Berlin; University of Edinburgh). Marcus Hartmann (Max Planck Institute for Developmental Biology, Tuebingen) provided SAXS and cross-linking data for a target they had submitted. The data for the NMR study were generated by Antonio Rosatto (University of Florence), Janet Huang and Gaetano Montelione (Rutgers). The material for the FRET study was produced by Claus Seidel and Mykola Dimura (University of Dusseldorf), who also performed the FRET analysis. We are grateful to Emily Tai (NCI/NIH) for her major contributions to the CASP commons experiment.
We once again thank the assessment teams for their thorough and insightful analyses: Randy Read and Tristan Croll (Cambridge University) for template-based modeling and refinement assessment; Luciano Abriata and Matteo Dal Peraro (EPFL, Lausanne) for free modeling assessment; Jose Duarte (RCSB, San Diego, CA) for assembly assessment; Andras Fiser (Albert Einstein College of Medicine, New York, NY) for contact and cross-link assisted assessment; Greg Hura and Susan Tsutakawa (LBNL, Berkeley, CA) for SAXS-assisted assessment; Gaetano Montelione (Rutgers, NJ) for NMR-assisted assessment; Chaok Seok (Seoul National University, South Korea) for model accuracy assessment; and Lisa Kinch for her work on target structure analysis. As always, for participants, it takes courage to expose their methods to such intense and public scrutiny. We greatly appreciate the 98 research groups who submitted their work to this CASP. We again thank PROTEINS for providing a mechanism for peer reviewed publication of the outcome of the experiment.
The CASP Prediction Center at UC Davis is supported by a grant from the US National Institute of General Medical Sciences (NIGMS/NIH), R01GM100482 to KF.
REFERENCES
- 1.Kinch LN, Kryshtafovych A, Monastyrskyy B, Grishin NV. CASP13 target classification into tertiary structure prediction categories. Proteins 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egelman EH. The Current Revolution in Cryo-EM. Biophys J 2016;110(5):1008–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kryshtafovych A, et al. Cryo-EM targets in CASP13: overview and evaluation of results. [CASP13 special issue] 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)-Round XII. Proteins 2018;86 Suppl 1:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kryshtafovych A, Fidelis K, Moult J. CASP10 results compared to those of previous CASP experiments. Proteins 2014;82 Suppl 2:164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemla A, Venclovas, Moult J, Fidelis K. Processing and evaluation of predictions in CASP4. Proteins 2001;Suppl 5:13–21. [DOI] [PubMed] [Google Scholar]
- 7.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res 2003;31(13):3370–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gobel U, Sander C, Schneider R, Valencia A. Correlated mutations and residue contacts in proteins. Proteins 1994;18(4):309–317. [DOI] [PubMed] [Google Scholar]
- 9.de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet 2013;14(4):249–261. [DOI] [PubMed] [Google Scholar]
- 10.Kosciolek T, Jones DT. Accurate contact predictions using covariation techniques and machine learning. Proteins 2016;84 Suppl 1:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Sun S, Xu J. Analysis of deep learning methods for blind protein contact prediction in CASP12. Proteins 2018;86 Suppl 1:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadathil S, Greener J, Jones D. Recent developments in deep learning applied to protein structure prediction. [CASP13 special issue] 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, van Ginneken B, Sanchez CI. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 14.Biswas M, Kuppili V, Saba L, Edla DR, Suri HS, Cuadrado-Godia E, Laird JR, Marinhoe RT, Sanches JM, Nicolaides A, Suri JS. State-of-theart review on deep learning in medical imaging. Front Biosci (Landmark Ed) 2019;24:392–426. [DOI] [PubMed] [Google Scholar]
- 15.Afouras T, Chung J, Senior A, Vinyals O, Zisserman A. Deep audio-visual speech recognition. IEEE transactions on pattern analysis and machine intelligence 2018. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Sun S, Li Z, Zhang R, Xu J. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLoS Comput Biol 2017;13(1):e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinegger M, Mirdita M, Soding J. Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nat Methods 2019;16(7):603–606. [DOI] [PubMed] [Google Scholar]
- 18.Kandathil SM, Greener JG, Jones DT. Prediction of interresidue contacts with DeepMetaPSICOV in CASP13. Proteins 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J. Distance-based protein folding powered by deep learning. Proc Natl Acad Sci U S A 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senior A, et al. Protein structure prediction using multiple deep neural networks in CASP13. PROTEINS 2019;[CASP13 special issue]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adhikari B. DEEPCON: Protein Contact Prediction using Dilated Convolutional Neural Networks with Dropout. bioRxiv 2019. [DOI] [PubMed] [Google Scholar]
- 22.MacCallum JL, Hua L, Schnieders MJ, Pande VS, Jacobson MP, Dill KA. Assessment of the protein-structure refinement category in CASP8. Proteins 2009;77 Suppl 9:66–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modi V, Dunbrack RL, Jr. Assessment of refinement of template-based models in CASP11. Proteins 2016;84 Suppl 1:260–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovan L, Oleinikovas V, Yalinca H, Kryshtafovych A, Saladino G, Gervasio FL. Assessment of the model refinement category in CASP12. Proteins 2018;86 Suppl 1:152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Read RJ, Sammito MD, Kryshtafovych A, Croll TI. Evaluation of model refinement in CASP13. Proteins 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nugent T, Cozzetto D, Jones DT. Evaluation of predictions in the CASP10 model refinement category. Proteins 2014;82 Suppl 2:98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heo L, Arbour CF, Feig M. Driven to near-experimental accuracy by refinement via molecular dynamics simulations. Proteins 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park H, Lee GR, Kim DE, Anishchenko I, Cong Q, Baker D. High-accuracy refinement using Rosetta in CASP13. Proteins 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee GR, Won J, Heo L, Seok C. GalaxyRefine2: simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res 2019;47(W1):W451–W455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kryshtafovych A, Monastyrskyy B, Fidelis K, Schwede T, Tramontano A. Assessment of model accuracy estimations in CASP12. Proteins 2018;86 Suppl 1:345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Won J, Baek M, Monastyrskyy B, Kryshtafovych A, Seok C. Assessment of Protein Model Structure Accuracy Estimation in CASP13: Challenges in the Era of Deep Learning. PROTEINS 2019;[CASP13 special issue]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L M., et al. Blind prediction of homo- and hetero-protein complexes: The CASP13-CAPRI experiment. PROTEINS 2019;[CASP13 special issue]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guzenko D, Lafita A, Monastyrskyy B, Kryshtafovych A, Duarte JM. Assessment of protein assembly prediction in CASP13. Proteins 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anishchenko I, Kundrotas PJ, Vakser IA. Modeling complexes of modeled proteins. Proteins 2017;85(3):470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vakser IA. Protein-protein docking: from interaction to interactome. Biophys J 2014;107(8):1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montelione G, Others. Assessment of Guided Protein Structure Prediction using Sparse NMR Data in CASP13. [CASP13 special issue] 2019. [Google Scholar]
- 37.Koepnick B, Flatten J, Husain T, Ford A, Silva DA, Bick MJ, Bauer A, Liu G, Ishida Y, Boykov A, Estep RD, Kleinfelter S, Norgard-Solano T, Wei L, Players F, Montelione GT, DiMaio F, Popovic Z, Khatib F, Cooper S, Baker D. De novo protein design by citizen scientists. Nature 2019;570(7761):390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson JC, Nassar R, Liu C, Brini E, Dill KA, Perez A. NMR-assisted protein structure prediction with MELDxMD. Proteins 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogorzalek TL, Hura GL, Belsom A, Burnett KH, Kryshtafovych A, Tainer JA, Rappsilber J, Tsutakawa SE, Fidelis K. Small angle X-ray scattering and cross-linking for data assisted protein structure prediction in CASP 12 with prospects for improved accuracy. Proteins 2018;86 Suppl 1:202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fajardo J, et al. Assessment of chemical-crosslink-assisted protein structure modeling in CASP13. [CASP13 special issue] 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu T, Ish-Shalom S, Torng W, Lafita A, Bock C, Mort M, Cooper DN, Bliven S, Capitani G, Mooney SD, Altman RB. Biological and functional relevance of CASP predictions. Proteins 2018;86 Suppl 1:374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huwe PJ, Xu Q, Shapovalov MV, Modi V, Andrake MD, Dunbrack RL, Jr. Biological function derived from predicted structures in CASP11. Proteins 2016;84 Suppl 1:370–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lepore A, others. Target highlights in CASP13: experimental target structures through the eyes of their authors. [CASP13 special issue] 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monastyrskyy B, D’Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. New encouraging developments in contact prediction: Assessment of the CASP11 results. Proteins 2016;84 Suppl 1:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.