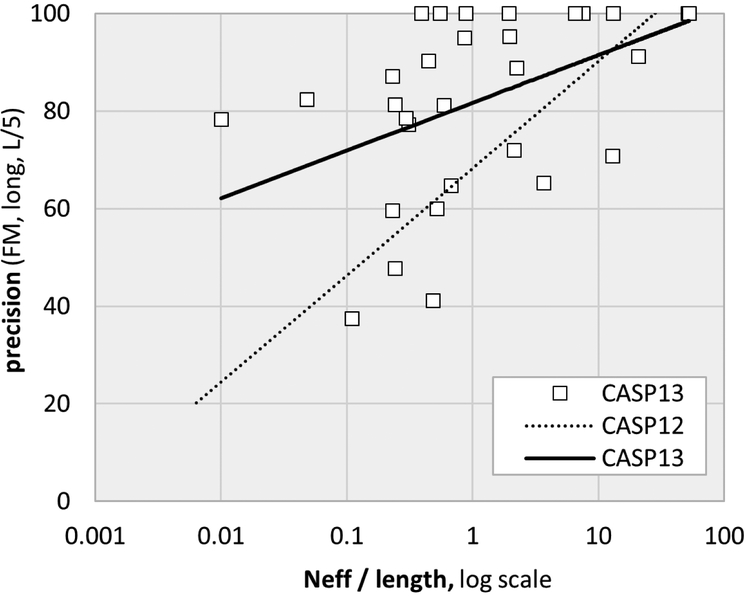
Figure 3:
Contact prediction precision trend lines as a function of sequence alignment depth and target length. In CASP13 there is a reduced dependency on alignment depth, resulting in more accurate results for shallow alignments as well as higher precision overall. Strikingly, for ten out of the 31 free modeling targets, the best predictions achieved 100% precision for this subset of contacts (see figure 2 for definitions). The effective alignment depth, Neff, includes metagenomic sequences compiled as described in17,18 Neff was calculated using a 90% sequence identity cutoff and a minimum of 60% sequence coverage (details in44).