Abstract
Background
To investigate the distribution of urine isolates and antibiotic resistance patterns in the predominant uropathogen Escherichia coli in migrant and non-migrant individuals.
Methods
We linked a cohort consisting of all migrants obtaining residence as refugees or family-reunited migrants in Denmark between January 1993 and December 2015 to hospital urine samples examined from January 2000 to December 2015 at the Department of Microbiology, University Hospital Hvidovre, Denmark. Samples from non-migrant individuals, Danish-born from Danish parents, were included as comparison. Analysis was carried out using multivariate logistic regression.
Results
There were 14 561 first-time urine samples included, with E. coli being the most prevalent bacterial pathogen. Of the identified isolates, 4686/11 737 were E. coli among non-migrants and 1032/2824 among migrants.
Sulfamethoxazol–Trimethoprim (SXT) resistance was found in 34.3% (350/1020) of E. coli isolates among migrants and 23.2% (1070/4619) among non-migrant patients [odds ratio (OR) 1.73, 95% confidence interval (CI): 1.47–2.03]. Ciprofloxacin resistance was found in 5.8% (36/618) of isolates among migrants and 2.2% (67/3092) among non-migrants (OR 2.20, 95% CI: 1.37–3.53). Gentamicin (GEN) resistance was seen in 10.8% (61/565) and 4.7% (110/2328) of isolates (OR 2.33, 95% CI:1.63–3.34), Cefuroxime resistance in 8.5% (87/1019) and 3.4% (158/4618) (OR 2.40, 95% CI:1.77–3.24), Ampicillin (AMP) resistance in 51.4% and 40.8% (OR 1.65, 95% CI: 1.42–1.92) and Piperacillin–Tazobactam resistance in 6.9% (30/432) and 4.2% (65/1532) for migrant and non-migrant patients, respectively. When stratifying according to migrant status, family-reunited had higher odds of resistance than refugees for SXT, GEN and AMP.
Conclusions
Prevalence of antibiotic resistance was significantly higher in E. coli isolates among migrants, both refugees and family-reunited, than non-migrant patients. Differences could not be explained by comorbidity or income. The results emphasize the importance of urine sample testing in both local-born and migrants before antibiotic start-up and point to the benefit of considering migration to secure individual treatment and equal health outcomes.
Keywords: Antimicrobial resistance, antibiotic susceptibility, ciprofloxacin, gentamycin, refugees, Enterobacteriaceae, Enterobacterales
Introduction
Antibiotic resistance represents one of the largest threats to human health and causes significant challenges in many aspects of our society including prevention and treatment of infection.1 Antibiotic resistance is the ability of microbes to withstand the effects of the antimicrobial therapies that were previously effective in treating infections caused by such microbes.2 It results in longer duration of illness, increased risk of mortality and increased costs.3 Several factors may contribute to antibiotic resistance, including mutations in bacteria with antibiotic use being the main driver.4,5 In addition, increased global migration and international travel may impact epidemiological patterns of antibiotic resistance worldwide.2,6–8 Prevalence of antibiotic resistance differs globally with relevative low prevalences in Scandinavian countries compared with other European countries.9 Antibiotic resistance in Escherichia coli isolates is found to be high in the Indian subcontinent, China and Southeast Asia, as well as Middle East.2,10 Migrants constitute an increasing part of the population in European countries.11 In April 2019, migrants comprised 11% of the Danish population, of whom 58% originated from non-Western countries.12 Migrants are a diverse group covering many types of migrant groups as migration overall means movement of people.13
Bacteriuria is common,14 with symptomatic urinary tract infection (UTI) being the most common health care-associated infection.15 The predominant bacterial pathogen found in urine is E. coli. However, very few studies have examined the resistance patterns of E. coli in urine among migrants. The few studies examining antibiotic resistance in E. coli isolates in both rectal swaps and urine samples among adult migrants have found a prevalence of 8.1–58.7%.8,16–22 The studies examine different migrant groups often with few included individuals and all without a comparison group. A previous study looking at bacteriuria with a comparison group and found that patients of South Asian ethnicity, with unknown migrant status, were more likely to have bacteriuria with extended spectrum beta-lactamase (ESBL)-producing Enterobacterales than patients of caucasian ethnic background.23 Besides this, no previous study has, to our knowledge, examined the prevalence of antibiotic resistance in bacteriuria in migrants compared with a non-migrant population. To shed light on this issue, we studied the differences in distribution of urine isolates and antibiotic resistance patterns in E. coli, in migrant and non-migrant individuals.
Methods
Study population
This study reports data from urine cultures taken from migrant and non-migrant patients from a part of the Capital Region of Denmark. Patient data were linked to a nationwide migrant cohort using the unique Patient Identification Number (PIN) provided to all Danish-born individuals at birth and assigned to all migrants upon obtaining residence status. This enabled us to look at urine samples from individuals with known migrant status and compare them with Danish-born individuals with Danish-born parents in a cross-sectional study design. For clarification, we use the term ‘non-migrants’ for Danish-born individuals with Danish-born parents.
For the migrant sample, the study utilizes register data from an existing migrant cohort. The migrant cohort includes all foreign-born individuals who obtained residence permission as refugees or through family reunification in Denmark between January 1993 and December 2015, and for whom a PIN was available. The migrant cohort was obtained through the Aliens Register at the Danish Immigration Service, which provided data on age, sex, ethnicity and reason for obtaining residence permission. We have described this cohort in detail previously.24 Refugees obtained residence permits as quota refugees or as spontaneous asylum seekers. Family-reunified migrants were reunified with refugees, Danish and Nordic individuals or other immigrants. Family reunification may be granted to spouses, children and other family members and consists of a more heterogenous subgroup of migrants, originating from high-, middle- and low-income countries. In this paper, the term ‘migrant’ is used as the combined term for refugees and family-reunified migrants.
We used residence permit as a proxy for migrant status as refugees or family-reunited migrants. Nationality upon arrival was recorded by the Danish Immigration Service. Nationality was grouped into seven regions according to the World Bank Country and Lending Groups: Central Asia and Eastern Europe, East Asia and Pacific, Middle East and North Africa, Sub-Saharan Africa, Latin America and Caribbean, Western (Central Europe and the Baltics, Canada, USA, Australia and New Zealand) and stateless.25
Patients for whom no PIN was available were excluded, as were statelessness migrants, migrants with a Western country of origin and patients under 18 years old.
Outcome
Retrospective clinical data from the analysis of urine samples taken from January 2000 to December 2015 were extracted through the laboratory information system (Department of Microbiology at Hvidovre Hospital), which covers patients across hospitals in the Capital Region of Denmark.
Significant bacteriuria was set at ≥103 CFU/ml for E. coli and ≥104 CFU/ml for other bacterial pathogens according to local guidelines. Among the samples we analysed, bacterial identification had been performed using standard laboratory methods [growth on chromogenic agar, p-Dimethylaminocinnamaldehyde-spot indole test for E. coli, Analytical Profile Index and VITEC systems (bioMerieux), MALDI-TOF MS (Bruker) for other bacterial pathogens]. Urine isolates were grouped into seven groups of pathogens: E. coli, Klebsiella spp., Enterobacterales other than E. coli and Klebsiella spp., Pseudomonas spp., other Gram-negative bacteria, Gram-positive bacteria and other.
Antibiotic susceptibility tests were performed by the laboratory using disc diffusion according to guidelines of the Swedish Reference Group for Antibiotics, now NordicAST26 and in the later years EUCAST.27 We analysed antibiotics that may be used for UTIs and urosepsis: Sulfamethoxazol–Trimethoprim (SXT), Ciprofloxacin (CIP), Gentamicin (GEN), Cefuroxim (CXM), Piperacillin–Tazobactam (TZP) and Ampicillin (AMP).
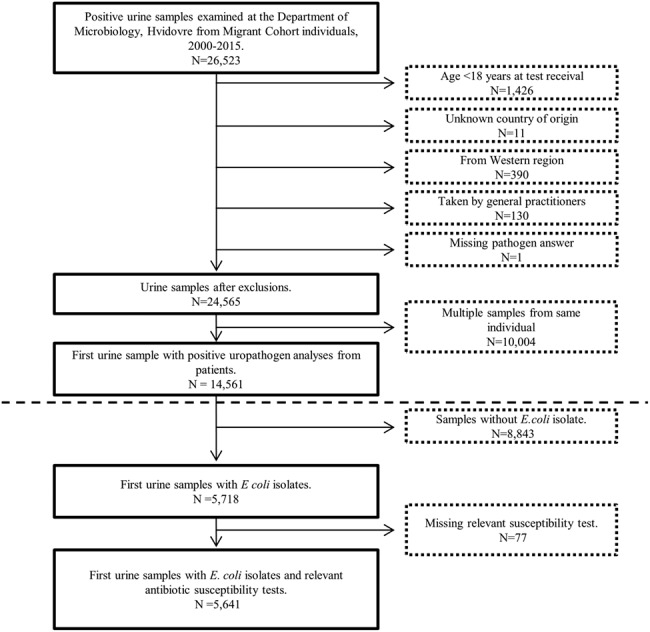
Date of test is when the Department of Microbiology received the urine sample. We excluded (i) urine samples with negative, no or missing isolate analysis; (ii) urine samples considered contaminated; and (iii) urine samples that were not first-time samples including only first-time urine samples and excluding any following urine samples from the same individual in the study period (Figure 1).
Figure 1. Flowchart showing the selection process.
Adjustment variables
Information on annual income was derived from socioeconomic registries at Statistics Denmark registered each year from 1998 to 2015 and grouped into four categories (in Euros): <15 000, 15 000–30 000, 30 000–40 000 and >40 000 based on an average from the past 2 years at urine sample year.
Through the Danish National Patient Registry, we derived information on acquired hospital diagnoses from 2000 to 2015 using WHO’s ICD-10. Applying the Canadian Charlson Comorbidity Index,28,29 we calculated a yearly Charlson Index Score (CIS) for each individual. Four comorbidity levels were defined: 0, 1–2, 3–4 and 5 or more based on the maximum CIS in the past 5 years prior to test year.
Test year was considered a potential confounder, as antibiotic resistance and laboratory methods change over time. To account for the small variations of sample distribution as well as the changes over time, we adjusted for test year in our fully adjusted models.
Interaction analyses showed no interactions for the potential confounders (data not shown).
Statistical analyses
Main analyses were stratified according to migrant status. Sub-analyses were done stratifying by region of origin, by sex and by time from residence permission.
We calculated odds ratios (ORs) for urine isolates and antibiotic resistance patterns of E. coli using multivariate logistic regression, with non-migrants as the reference group. Fully adjusted models included the following: test year, sex, annual income and CIS as categorical variables and age at urine sample test as a continuous variable. For the categorical variables income and CIS, individuals with missing information were included in analyses as a separate group. P values were calculated from two-tailed analyses, with P < 0.05 considered statistically significant. Differences between descriptive characteristics were analysed with the use of χ2 tests for categorical characteristics and one sample t-test for continuous variables.
Analyses were done using SAS 9.4 and forest plots using STATA 13.1.
Ethics
The study was approved by the Danish Data Protections Agency (approval number: 2016-41-4576) and was conducted according to the Declaration of Helsinki. Individual participant data are subject to protection from the national Danish Data Protection Agency and we are not allowed to share the data. L.B.S. and M.N. had full access to all data in the study and had final responsibility for the decision to submit for publication.
Results
Study population
In total, 14 561 first-time urine samples were included. Of the identified isolates, 4686 of 11 737 isolates were E. coli among non-migrants and 1032 of 2824 were E. coli isolates among migrants, with 1873 from family-reunited migrants and 951 from refugees. Baseline characteristics are illustrated in Table 1 and differed across migrants and non-migrant patients for income, CIS and sex (for further characteristics, see Table S1).
Table 1. Description of demographic and clinical characteristics for adult patients with positive uropathogen analyses.
| All migrants | Family-reunited migrants | Refugees | Non-migrants | P value a | |
|---|---|---|---|---|---|
| Number % (n) | 19.4 (2824) | 66.3 (1873) | 33.7 (951) | 80.6 (11 737) | |
| Age at urine sample test yrs, median (25–75 Pctl) | 35.7 (29.9–46.1) | 34.6 (29.6–41.2) | 40.9 (30.5–58.9) | 34.5 (29.2–47.6) | 0.459 |
| Sex | 0.023 | ||||
| Male % (n) | 11.1 (314) | 5.9 (110) | 21.5 (204) | 12.7 (1489) | |
| Female % (n) | 88.9 (2510) | 94.1 (1763) | 78.6 (747) | 87.3 (10 248) | |
| Mean length of stay at sample test yrs, median (25–75 Pctl) | 9.1 (4.5–14.4) | 7.2 (3.3–12.3) | 12.8 (7.7–16.7) | ||
| Region of originb % (n) | |||||
| Middle East and North Africa | 32.1 (906) | 28.9 (541) | 38.4 (365) | ||
| East Asia and Pacific | 23.7 (670) | 30.8 (577) | 9.8 (93) | ||
| Central Asia and Eastern Europe | 23.4 (662) | 20.3 (380) | 29.7 (282) | ||
| Sub-Sahara | 16.1 (454) | 13.2 (248) | 21.7 (206) | ||
| Latin America and Caribbean | 4.7 (132) | 6.8 (127) | 0.5 (5) | ||
| Income, Euro % (n) | <0.001 | ||||
| <15 000 Euro | 34.8 (982) | 33.5 (627) | 37.3 (355) | 20.9 (2456) | |
| 15 000–30 000 Euro | 41.0 (1158) | 37.3 (698) | 48.4 (460) | 45.0 (5277) | |
| 30 000–40 000 Euro | 5.9 (167) | 5.2 (97) | 7.4 (79) | 20.2 (2373) | |
| >40 000 Euro | 2.4 (67) | 2.1 (39) | 2.9 (28) | 13.1 (1536) | |
| No information on income | 15.9 (450) | 22.0 (412) | 4.0 (38) | 0.8 (96) | |
| Charlson score % (n) | <0.001 | ||||
| 0 | 67.6 (1908) | 69.5 (1302) | 63.7 (606) | 69.7 (8179) | |
| 1–2 | 13.5 (382) | 10.4 (194) | 19.8 (188) | 15.5 (1816) | |
| 3–4 | 1.9 (54) | 1.2 (22) | 3.4 (32) | 2.5 (287) | |
| ≥5 | 1.6 (45) | 1.2 (23) | 2.3 (22) | 1.3 (154) | |
| No information on diagnoses | 15.4 (435) | 17.7 (332) | 10.8 (103) | 11.1 (1301) |
aAll statistical tests were performed between all non-Western migrants and non-migrants.
bRegions according to the World Bank Grouping.
25–75 Pctl, 25th and 75th percentile; Yrs, years.
Pathogen distribution
Escherichia coli was the most prevalent bacterial pathogen found, accounting for 79.9% of Gram-negative bacteria.
There were 4686 E. coli isolates from non-migrant patients and 1032 from migrants (39.9% vs 36.5%, P < 0.001, Table 2). When stratifying according to sex, this difference was seen in female patients only (Table S2).
Table 2. Distribution of bacterial pathogen in first-time urine samples from migrants and non-migrants from 2000 to 2015 grouped according to migrant status.
| Non-migrants | All migrants | Refugees | Family-reunited migrants | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Urine isolate | No./total | % | OR (95% CI) | No./total | % | OR (95% CI) | P value | No./total | % | OR (95% CI) | P value | No./total | % | OR (95% CI) | P value |
| Gram-positive | 6715/11 737 | 57.2 | 1.00 (ref) | 1692/2824 | 59.9 | 1.25 (1.14–1.38) | <0.001 | 562/951 | 59.1 | 1.36 (1.18–1.56) | <0.001 | 1130/1873 | 60.3 | 1.20 (1.07–1.34) | 0.002 |
| Gram-negative | 5867/11 737 | 50.0 | 1.00 (ref) | 1334/2824 | 47.2 | 0.80 (0.73–0.88) | <0.001 | 482/951 | 50.7 | 0.81 (0.70–0.93) | 0.003 | 852/1873 | 45.5 | 0.80 (0.71–0.89) | <0.001 |
| E. coli | 4686/11 737 | 39.9 | 1.00 (ref) | 1032/2824 | 36.5 | 0.78 (0.71–0.86) | <0.001 | 354/951 | 37.2 | 0.77 (0.67–0.89) | <0.001 | 678/1873 | 36.2 | 0.78 (0.70–0.88) | <0.001 |
| Klebsiella spp. | 413/11 737 | 3.5 | 1.00 (ref) | 114/2824 | 4.0 | 1.21 (0.96–1.53) | 0.109 | 45/951 | 4.7 | 1.09 (0.78–1.51) | 0.629 | 69/1873 | 3.7 | 1.36 (1.01–1.82) | 0.041 |
| Enterobacterales other than E. coli and Klebsiella spp. | 420/11 737 | 3.6 | 1.00 (ref) | 93/2824 | 3.3 | 0.92 (0.71–1.18) | 0.490 | 48/951 | 5.0 | 1.08 (0.78–1.48) | 0.659 | 45/1873 | 2.4 | 0.79 (0.55–1.13) | 0.192 |
| Pseudomonas spp. | 143/11737 | 1.2 | 1.00 (ref) | 29/2824 | 1.0 | 1.01 (0.65–1.55) | 0.980 | 15/951 | 1.6 | 1.05 (0.60–1.84) | 0.864 | 14/1873 | 0.7 | 0.96 (0.52–1.76) | 0.886 |
| Other Gram-negative bacteria | 434/11737 | 3.7 | 1.00 (ref) | 119/2824 | 4.2 | 1.15 (0.92–1.44) | 0.230 | 51/951 | 5.4 | 1.35 (0.99–1.83) | 0.059 | 68/1873 | 3.6 | 1.00 (0.75–1.34) | 0.988 |
| Other | 461/11 737 | 3.9 | 1.00 (ref) | 129/2824 | 4.6 | 1.15 (0.92–1.43) | 0.214 | 37/951 | 3.9 | 1.01 (0.71–1.44) | 0.949 | 92/1873 | 4.9 | 1.24 (0.96–1.60) | 0.106 |
OR adjusted for test year, Charlson Index, income, sex and age at test.
Total; Total number of urine samples. Total number of isolates exceed total number of urine samples as multiple isolates can be identified in one sample.
As shown in Table 2, 57.5% of the positive urine samples had Gram-positive isolates, which were found to comprise a greater proportion of positive urine samples among migrants with 1692 isolates compared with non-migrant patients with 6715 isolates (59.9% vs 57.2%, P < 0.001).
Antibiotic resistance in E. coli isolates
A total of 34.3% (350/1020) of E. coli isolates with susceptibility test were resistant to SXT among migrants compared with 23.2% (1070/4619) of isolates among non-migrant patients [OR 1.73, 95% confidence interval (CI): 1.47–2.03]. 5.8% (36/618) of isolates from migrants were resistant to CIP compared with 2.2% (67/3092) for non-migrants (OR 2.20, 95% CI: 1.37–3.53). GEN resistance was found in 10.8% (61/565) of isolates from migrants and 4.7% (110/2328) of isolates from non-migrants (OR 2.33, 95% CI: 1.63–3.34). CXM resistance was found in 8.5% (87/1019) of isolates from migrants and 3.4% (158/4618) from non-migrants (OR 2.40, 95% CI: 1.77–3.24). AMP resistance was found in 51.4% (523/1018) of isolates from migrants and 40.8% (1882/4616) from non-migrants (OR 1.65, 95% CI: 1.42–1.92) (Figure 2).
Figure 2. Antibiotic susceptibility among E. coli isolates in first-time urine samples from migrants and non-migrants from 2000 to 2015. OR adjusted for test year, Charlson index, income, sex and age at test. R/No. total, resistant isolates/total number of isolates tested. *Significant difference between OR for family-reunited and refugees (SXT P < 0.001, GEN P = 0.004, AMP P = 0.002).
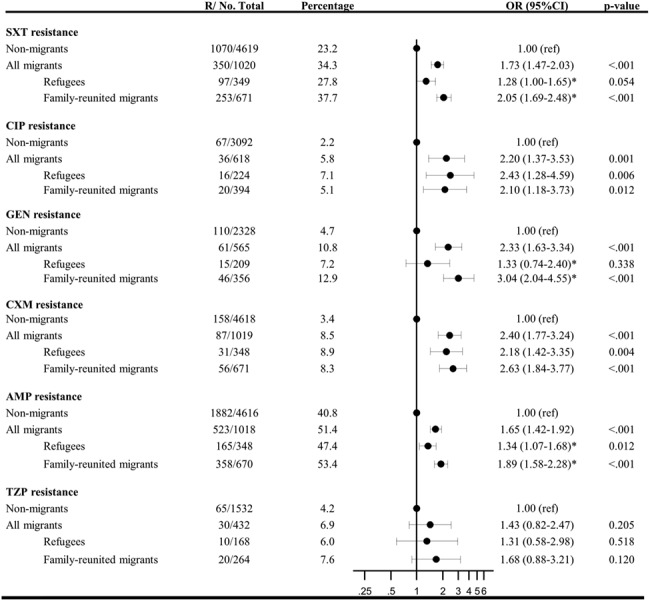
No significant difference between migrants and non-migrant patients was seen for TZP with 6.9% (30/432) and 4.2% (65/1532) among non-migrants (OR 1.43, 95% CI: 0.82–2.47). However, when stratifying according to sex, a higher prevalence of antibiotic resistance was seen for male migrants compared with male non-migrants with 19.4% vs 6.9% (P = 0.006, Figure S1).
In general, when stratifying by sex, a tendency of higher antibiotic resistance was found for male compared with female, subject to few observations (Figure S1).
Although multi-resistant E. coli isolates were rare among both migrants and non-migrants, there was a significant difference (4.1%. vs 1.4% migrants and non-migrants, respectively, P < 0.001. Table S3).
Migrant status
Figure 2 shows antibiotic resistance in E. coli isolates stratified according to migrant status. Among patients with E. coli isolates in their urine samples, we found higher odds of E. coli isolates resistant to SXT, CIP, GEN, CXM or AMP among family-reunited migrants compared with non-migrant patients and for CIP, CXM or AMP among refugees compared with non-migrant patients.
Further, SXT, GEN and AMP resistance was more prevalent in family-reunited migrants compared with refugees with 37.7% (253/671) vs 27.8% (97/349) (P < 0.001), 12.9% (46/356) vs 7.2% (15/209) (P = 0.004) and 53.4% (358/670) vs 47.4% (165/348) resistant isolates (P = 0.002), respectively. Sub-analyses were further carried out classifying the subgroup ‘family-reunited to refugees’ as ‘refugees’ instead of ‘family reunified’ showing a similar trend (Table S4).
Region of origin
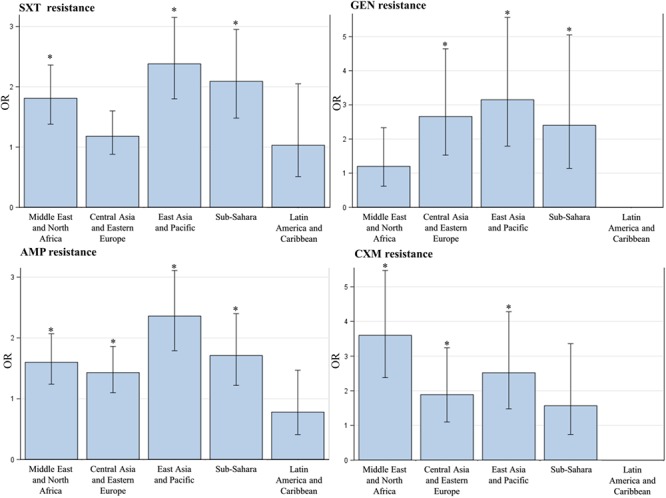
When stratifying according to region of origin and comparing to non-migrant individuals, higher odds of resistant E. coli isolates were seen among migrants from all five regions except Latin America and the Caribbean concerning different antibiotics, showing that resistance did not only occur in migrants from specific regions (Figure 3, Table S5).
Figure 3. Antibiotic susceptibility among E. coli isolates in first-time urine samples from migrants and non-migrants grouped by region of origin. Non-migrant individuals as the reference group with odds ratio 1. Odds ratio adjusted for test year, Charlson index, income, sex and age at test. *Significantly different from non-migrants.
Length of stay
Sub-analyses with stratification of migrants according to time in host country found the prevalence of overall antibiotic resistance among E. coli resistance to be lower in migrants who had been in the host country more than 10 years after gaining a residence permit compared with urine samples from migrants being in the host country under 5 years with an overall resistance of 57.6% compared with 62.6% (Table S6).
Discussion
Our study represents the first description of antibiotic resistance among E. coli isolates in urine samples in migrants compared with the non-migrant population and presents unique and substantial data on antibiotic resistance in migrant communities.
Our findings that a greater proportion of E. coli isolates were resistant to antibiotics in migrants compared with non-migrant patients support previous studies that have found a higher occurrence of ESBL-producing Enterobacterales in urine samples from individuals of South Asian ethnicity,23 as well as a higher prevalence of multi-drug resistant Gram-negative bacteria in rectal swabs from refugees and asylum seekers compared with the overall prevalence in the population.19,30 A recent systematic review and meta-analysis examining antibiotic resistance in migrants to Europe found an increased prevalence of antibiotic resistance carriage or infections with higher prevalence of resistant bacteria in refugees and asylum seekers compared with other migrant groups.8
In our study, we only looked at first-time urine samples to facilitate simple presentation of data and to avoid bias from individuals with repeated positive urine samples.
Our study is one of the most comprehensive datasets to date on antibiotic resistance in migrant populations, and due to the unique migrant cohort, we had accurate information on migrant status and country of origin. Furthermore, we examined susceptibility in specific antibiotics, for which previous data are limited.
Denmark is a country with relative low occurrence of antibiotic resistance and the differences between migrants and the Danish non-migrant population may be higher compared with differences in other countries with a high occurrence of antibiotic resistance. However, we believe our results to be valid for other high-income countries with a low occurrence of antibiotic resistance.
There are several limitations to the data that should be acknowledged. First, occurrence of antibiotic resistance is rising over time, and the types of antibiotic resistance routinely tested for are subject to constant change in the period from 2000 to 2015. Further, interpretation of the result of susceptibility testing may have changed over time (e.g. fluoroquinolone resistance was defined using either CIP or Nalidixin). However, the urine samples were all tested at the same Department of Microbiology, and changes in interpretation of antibiotic susceptibility therefore affected both migrants and non-migrants, meaning the data are consistent over time across the two groups. Further, we adjusted for test year. Second, even though antibiotic use facilitates antibiotic resistance and travel is found to amplify resistance,2 we do not have information about previous travel or antibiotic use in our study population and cannot account for recent antibiotic use or recent travel at urine sample test. Danish register data do not allow us to investigate international travel patterns of the population further, and further research such as health surveys are needed to strengthen data in this area.
Third, a major limitation is the lack of clinical data as we cannot distinguish asymptomatic bacteriuria from UTI.
Conclusion
We found a higher proportion of E. coli isolates among non-migrant individuals and higher rates of antibiotic resistance in E. coli isolates among migrants, compared with non-migrant individuals. Differences could not be explained by comorbidity or income.
Further, our findings point to a higher prevalence of antibiotic resistance among family-reunited migrants compared with refugees, which is novel, indicating that the migration process alone cannot explain the higher occurrence of antibiotic resistance.
The findings point to the value of urine sampling and drug susceptibility testing in both local-born and migrant patients before antibiotic start-up to reduce poor and costly health outcomes associated with drug resistant infections. Our study emphasizes the benefit of considering migration when treating patients in the interest of public health and helps identify groups who may be at increased risk of drug resistant infections to improve health outcomes.
Supplementary Material
Acknowledgements
We thank Thomas Benfield and Bjarne Ørskov at the Department of Infectious Diseases, University Hospital Hvidovre, Denmark, for funding this project as well as the employees at Section of Immigrant Medicine.
Funding
This work was supported by a research grant from the Department of Infectious Diseases, Hvidovre Hospital, Denmark, and funding from The Wellcome Trust (209993/Z/17/Z) and the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group Research Funding for the ESCMID Study Group for Infections in Travellers and Migrants to L.B.N., S.H. and J.S.F. The funders had no role in study design, data collection, data analysis, data interpretation or writing of the report.
Authors’ contributions
M.N., L.B.N. and J.S.F. conceived the idea. L.B.S., M.N., R.T.N., C.Ø., L.B.N. and J.S.F. contributed to the design of the study. C.Ø. and M.N. provided study material. L.B.S analysed data. L.B.S. wrote the first draft in collaboration with R.T.N. and M.N. All authors contributed to the interpretation of data and revision of the manuscript.
Conflict of interest:
None declared.
References
- 1. World Economic Forum Global Risks Report 2013 Eighth Edition. Accessed 22 May 2019 http://reports.weforum.org/global-risks-2013/risk-case-1/digital-wildfires-in-a-hyperconnected-world/ .
- 2. Dphil IF, Van Boeckel TP, Pires J et al. Global geographic trends in antimicrobial resistance: the role of international travel. J Travel Med 2019; 26:1–13. [DOI] [PubMed] [Google Scholar]
- 3. Laxminarayan R, Duse A, Wattal C et al. Antibiotic resistance—the need for global solutions. Lancet Infect Dis 2013; 13:1057–98. [DOI] [PubMed] [Google Scholar]
- 4. Søgaard M, Vandenbroucke JP, Schønheyder HC. Risk factors for extended-spectrum β-lactamase-producing Escherichia coli urinary tract infection in the community in Denmark: a case-control study. Clin Microbiol Infect 2017; 23:952–60. [DOI] [PubMed] [Google Scholar]
- 5. European Centre for Disease Prevention and Control (ECDC), European Food Safety Authority (EFSA), European Medicines Agency (EMA) . Joint Interagency Antimicrobial Consumption and Resistance Analysis (JIACRA) Report. ECDC, EFSA and EMA for the European Commission in EFSA Journal; 2017; 15(7):4872–135. https://efsa.onlinelibrary.wiley.com/doi/epdf/10.2903/j.efsa.2017.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holmes AH, Moore L, Sundsfjord A et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016; 387:176–87. [DOI] [PubMed] [Google Scholar]
- 7. Van der Bij AK, JDD P. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother 2019; 74:1469–72. [DOI] [PubMed] [Google Scholar]
- 8. Nellums LB, Thompson H, Holmes A et al. Antimicrobial resistance among migrants in Europe: a systematic review and meta-analysis. Lancet Infect Dis 2018; 3099:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. European Centre for Disease Prevention and Control Surveillance of antimicrobial resistance in Europe 2017. Accessed 9 October 2019 https://www.ecdc.europa.eu/sites/default/files/documents/EARS-Net-report-2017-update-jan-2019.pdf.
- 10. Center for Disease Dynamics, Economics & Policy (CDDEP) Resistance map. Accessed 9 October 2019 https://resistancemap.cddep.org/.
- 11. Rechel B, Mladovsky P, Ingleby D et al. Migration and health in an increasingly diverse Europe. Lancet 2013; 381:1235–45. [DOI] [PubMed] [Google Scholar]
- 12. Statistics Denmark Population at the first day of the quarter by ancestry and time. 2019Q1. Accessed 5 May 2019 https://www.statistikbanken.dk/statbank5a/selectvarval/saveselections.asp?MainTable=FOLK1E&PLanguage =1&TableStyle=&Buttons=&PXSId=2019111221524264216036FOLK1E&IQY=&TC=&ST=ST&rvar0=&rvar1=&rvar2=&rvar3=&rvar4=&rvar5=&rvar6=&rvar7=&rvar8=&rvar9=&rvar10=&rvar11=&rvar12=&rvar13=&rvar14=.
- 13. Douglas P, Cetron M, Spiegel P. Definitions matter: migrants, immigrants, asylum seekers and refugees. J Travel Med 2019; 26:1–3. [DOI] [PubMed] [Google Scholar]
- 14. Ipe DS, Sundac L, Benjamin WH et al. Asymptomatic bacteriuria: prevalence rates of causal microorganisms, etiology of infection in different patient populations, and recent advances in molecular detection. FEMS Microbiol Lett 2013; 346:1–10. [DOI] [PubMed] [Google Scholar]
- 15. Eriksen HM, Iversen BG, Aavitsland P. Prevalence of nosocomial infections in hospitals in Norway, 2002 and 2003. J Hosp Infect 2005; 60:40–5. [DOI] [PubMed] [Google Scholar]
- 16. Angeletti S, Ceccarelli G, Vita S et al. Unusual microorganisms and antimicrobial resistances in a group of Syrian migrants: sentinel surveillance data from an asylum seekers centre in Italy. Travel Med Infect Dis 2016; 14:115–22. [DOI] [PubMed] [Google Scholar]
- 17. Georgakopoulou T, Mandilara G, Mellou K et al. Resistant Shigella strains in refugees, August–October 2015, Greece. Epidemiol Infect 2016; 144:2415–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heudorf U, Albert-Braun S, Hunfeld KP et al. Multidrug-resistant organisms in refugees: prevalences and impact on infection control in hospitals. GMS Hyg Infect Control 2016; 11:Doc16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ravensbergen SJ, Berends M, Stienstra Y et al. High prevalence of MRSA and ESBL among asylum seekers in the Netherlands. PLoS One 2017; 12:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reinheimer C, Kempf VAJ, Göttig S et al. Multidrug-resistant organisms detected in refugee patients admitted to a university hospital, Germany June–December 2015. Euro Surveill 2016; 21:1–5. [DOI] [PubMed] [Google Scholar]
- 21. Pop R, Zillig D, Schibli U et al. A cross-sectional study of colonization rates with methicillin-resistant Staphylococcus aureus (MRSA) and extended-spectrum beta-lactamase (ESBL) and carbapenemase-producing Enterobacteriaceae in four Swiss refugee centres. PLoS 2017; 261:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Valverde A, Turrientes MC, Norman F et al. CTX-M-15-non-ST131 Escherichia coli isolates are mainly responsible of faecal carriage with ESBL-producing Enterobacteriaceae in travellers, immigrants and those visiting friends and relatives. Clin Microbiol Infect 2015; 21:252e1–4. [DOI] [PubMed] [Google Scholar]
- 23. Gopal Rao G, Batura D, Batura N et al. Key demographic characteristics of patients with bacteriuria due to extended spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in a multiethnic community, in North West London. Infect Dis (Lond) 2015; 47:719–24. [DOI] [PubMed] [Google Scholar]
- 24. Nørredam M. Migration and health: exploring the role of migrant status through register-based studies. Dan Med J 2015; 62:B5068–8. [PubMed] [Google Scholar]
- 25. The World Bank Country and Lending Groups Accessed 7 October 2019 Countries and Economies. https://data.worldbank.org/country.
- 26. The Nordic Committee on Antimicrobial Susceptibility Testing (NordicAST) Methods: Explanatory methods documents. Accessed 5 October 2019 http://www.nordicast.org/.
- 27. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint tables for interpretation of MICs and zone diameters. 2019. Accessed 5 October 2019 http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf.
- 28. Charlson ME, Pompei P, Ales KL et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373–83. [DOI] [PubMed] [Google Scholar]
- 29. Sundararajan V, Quan H, Halfan P et al. Cross-national comparative performance of three versions of the ICD-10 Charlson index. Med Care 2007; 45:1210–5. [DOI] [PubMed] [Google Scholar]
- 30. Reinheimer C, Kempf VAJ, Jozsa K et al. Prevalence of multidrug-resistant organisms in refugee patients, medical tourists and domestic patients admitted to a German university hospital. BMC Infect Dis 2017; 17:4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


