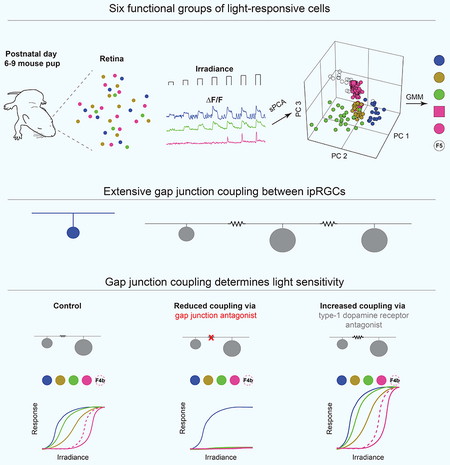
SUMMARY
Detection of ambient illumination in the developing retina prior to maturation of conventional photoreceptors is mediated by intrinsically photosensitive retinal ganglion cells (ipRGCs) and is critical for driving several physiological processes, including light-aversion, pupillary light reflexes, and photoentrainment of circadian rhythms. The strategies by which ipRGCs encode variations in ambient light intensity at these early ages are not known. Using unsupervised clustering of two-photon calcium responses followed by inspection of anatomical features, we found that the population activity of the neonatal retina could be modeled as six functional groups that were comprised of mixtures of ipRGC subtypes and non-ipRGC cell types. By combining imaging, whole-cell recording, pharmacology, and anatomical techniques, we found that functional mixing of cell types is mediated in part by gap junction coupling. Together, these data show that both cell-autonomous intrinsic light responses and gap junction coupling among ipRGCs contribute to the proper encoding of light intensity in the developing retina.
Graphical Abstract
Blurb
Prior to the maturation of rods and cones, mammals detect light with intrinsically photosensitive retinal ganglion cells (ipRGCs). At this age, ipRGCs are extensively gap junction coupled. Caval-Holme and Feller find that gap junctions contribute to ipRGC functional diversity and determine the light sensitivity of the developing retina.
INTRODUCTION
Prior to the maturation of retinal circuits that mediate image-forming vision, light can still trigger behaviors in neonatal rodents. Neonates have a pupillary light reflex [1], entrain their circadian rhythms to the solar day independent of cues from their parents [2], and exhibit aversion to bright light [3]. Neonatal light exposure also sets the intrinsic period length of the circadian clock [4]. These behaviors occur before synapse formation between photoreceptors and bipolar cells [5], implying that they are not mediated by conventional rod and cone inputs to retinal circuitry.
Light’s influence on neonatal behavior is instead attributed to a population of intrinsically photosensitive retinal ganglion cells (ipRGCs) that express the light sensitive protein melanopsin [6] and exhibit intrinsic light responses [7–9] at an embryonic age. Deletion of melanopsin prevents light aversion [3], while ablation of the ipRGCs themselves prevents the light influencing the circadian period length [4].
In the adult retina, ipRGCs come in multiple subtypes (M1-M6) distinguished by their morphology, dendritic stratification, expression of molecular markers, and projection targets in the brain (M1-M5 Schmidt et al., 2011; M6 Quattrochi et al., 2019). These ipRGC subtypes differ in their encoding of irradiance because of differences in their bipolar cell inputs, melanopsin expression levels, intrinsic membrane conductances, and the effector channels of the melanopsin phototransduction cascade [12–17].
Similarly, ipRGC subtypes described thus far in the neonatal retina exhibit distinctive cell-intrinsic properties [18]. In addition, developing ipRGCs are gap junction coupled both to other ipRGCs and to non-ipRGCs [19]. Pharmacological blockade of these networks reduced the overall number of light-responsive cells [19]. Conversely, blockade of dopamine receptor (D1R) signaling increased the number of light-responsive cells [19,20], raising the possibility that neuromodulation by dopamine could contribute to the functional properties of ipRGCs by changing the extent of their gap junction coupling, analogous to dopamine’s well-established modulatory role in gap junction networks in the adult retina [21–23]. The relative important of cell intrinsic properties and network connectivity in generating the neonatal light response is not known.
In the mouse, synapse formation between photoreceptors and bipolar cells occurs at postnatal day 11 (P11) [5], so recording before P11 offers the unique opportunity to understand how the neonatal retina uses ipRGCs to encode the ~8 log unit range of irradiance encountered over a typical day-night cycle [24] in the absence of input from rod and cone photoreceptors. Multielectrode array (MEA) recordings reveal three functionally distinct groups of light-responsive cells present at P8 [8], but it is unclear how these functional groups map to ipRGC subtypes.
The goals of this study were threefold: first, establish a complete description of ipRGC encoding of irradiance during development; secondly, determine the relative contributions of ipRGC subtype versus network connectivity via gap junctions to the functional diversity of light responses; and thirdly, determine if gap junction plasticity mediated by dopamine could regulate the light responses of ipRGCs.
RESULTS
Functional groups of light-responsive cells represent mixtures of ipRGC subtypes
We characterized light-evoked activity in the developing retina before synaptic inputs from conventional photoreceptors form at P10 by dissecting retinas from P6-P9 mice and loading the ganglion cell layer with the red-shifted calcium indicator Cal 590 [25](Figure 1A; see STAR Methods). Calibration experiments with Cal 590 indicated that fluorescence transient amplitudes scaled linearly with action potential number and did not saturate at the amplitudes we measured during population imaging experiments (Figure S1). During two photon imaging we presented a series of light stimuli approximating the range of effective irradiance for melanopsin from predawn to daylight [26]. Experiments were carried out in the presence of Di-hydro-β-erythroidine (DHβE, 8 μM, see STAR Methods) to prevent spontaneous cholinergic retinal waves that would otherwise interfere with measurements of light responses (Tu et al, 2005).
Figure 1. A survey of light-evoked activity in the neonatal retina.
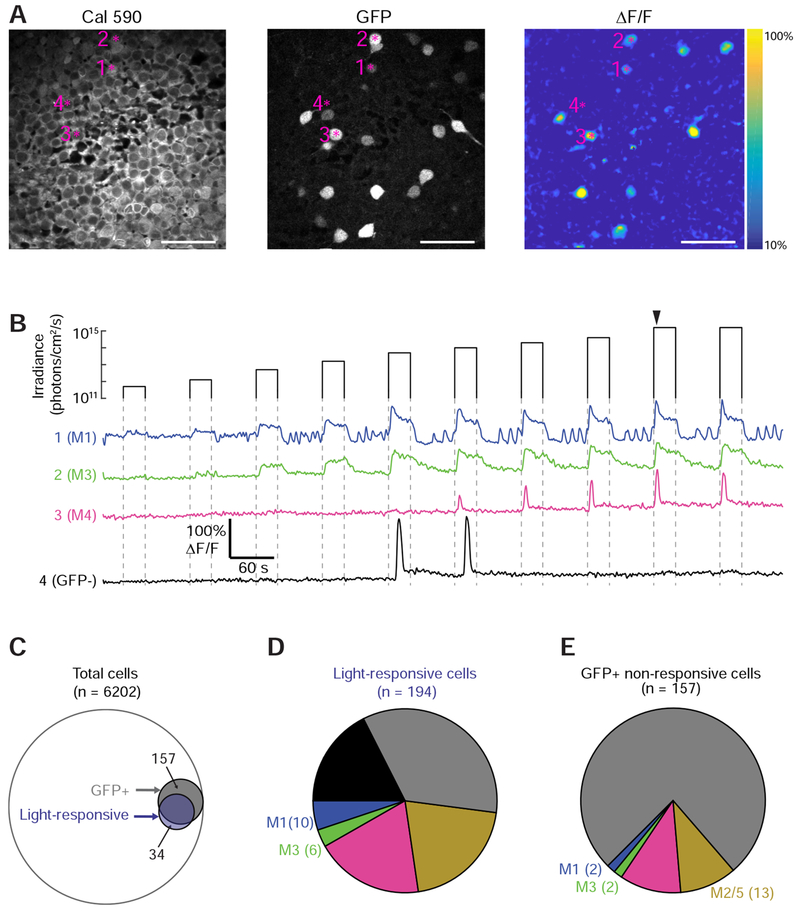
(A) Left: Ganglion cell layer of a retina dissected from a P6 Opn4::eGFP mouse after bolus loading with Cal 590 calcium indicator. Numbered asterisks correspond to cells with fluorescence transients in (B). Middle: GFP expression. Right: Fluorescence response two seconds after a light stimulus.
(B) Fluorescence traces from four cells in (A). Arrowhead: timing of the fluorescence response in (A). See Figure S1 for calibration of fluorescence transients with action potential firing.
(C) Intersections between sets of all cells, GFP+ cells, and light-responsive cells.
(D) Light-responsive cell types. “GFP+” cells were not identifiable as specific ipRGC subtypes. See Figure S2 details of ipRGC subtype identification.
(E) GFP+ cells without responses to light. See Figure S2B for details on these cells.
Putative ipRGCs were identified using the Opn4::eGFP transgenic mouse, which expresses enhanced GFP under the control of the melanopsin promoter [7]. A typical field of view contained a sparse set of light-responsive cells, with most individual cells responding to a sub-section of the irradiance range (Figure 1B). We classified light-responsive cells as GFP+ (putative ipRGCs) or GFP− based on the intensity of their green fluorescence relative to the background (Figure S2A; see STAR Methods). GFP+ cells comprised 82.5% (160/194) of light responsive cells, while GFP− cells accounted for 17.5% (34/194) of light responsive cells (Figure 1C). Surprisingly, only 50.5% (160/317) of GFP+ cells had detectable light responses. In a subset of recordings conducted in the absence of DHβE, we observed GFP+ cells that did not respond to light nevertheless had fluorescence transients during cholinergic retinal waves, indicating that they could produce detectable fluorescence transients during robust action potential firing (Figure S2C).
The ipRGC subtype of the light responsive cells was determined by fixing the retinas after imaging experiments and examining dendritic stratification and immunofluorescence in the field of view in which we had performed two-photon imaging (Figure S2D–G). Note that the dendritic trees of some GFP+ cells could not be classified, because they were physically overlapping with those of neighboring GFP+ cells or they had little fluorescence signal. Though we were only able to classify a subset of light-responsive cells, we could distinguish M1-M5 ipRGC subtypes (Figure 1D, E). We noted that there were few M3 ipRGCs, consistent with the observation of sparsely distributed M3s in the adult mouse retina [27]. We did not identify any GFP+ cells as M6 ipRGCs, either because they had not yet developed their adult morphology, or because of their low melanopsin expression levels and thin dendrites, as described in the adult retina [12]. It is important to note the GFP− cells do not necessarily lack melanopsin since, in the Opn4::eGFP mouse line, GFP expression is thought to be proportional to melanopsin expression [7], which, in the adult retina, is low in M5 and M6 ipRGCs [11,13].
We found that light-responsive cells differed considerably in their light sensitivity and the timing of their responses (Figure 1B). To characterize functional light responses independent of anatomical and molecular criteria, we used an unsupervised learning approach [28] to classify light responsive cells based solely on their fluorescence transients (Figure 2A–B; see also Methods and Figure S3). This approach yielded six functional groups of light-responsive cells (Figure 2C) that we subsequently refer to as F1, F2, F3, F4a, F4b, and F5.
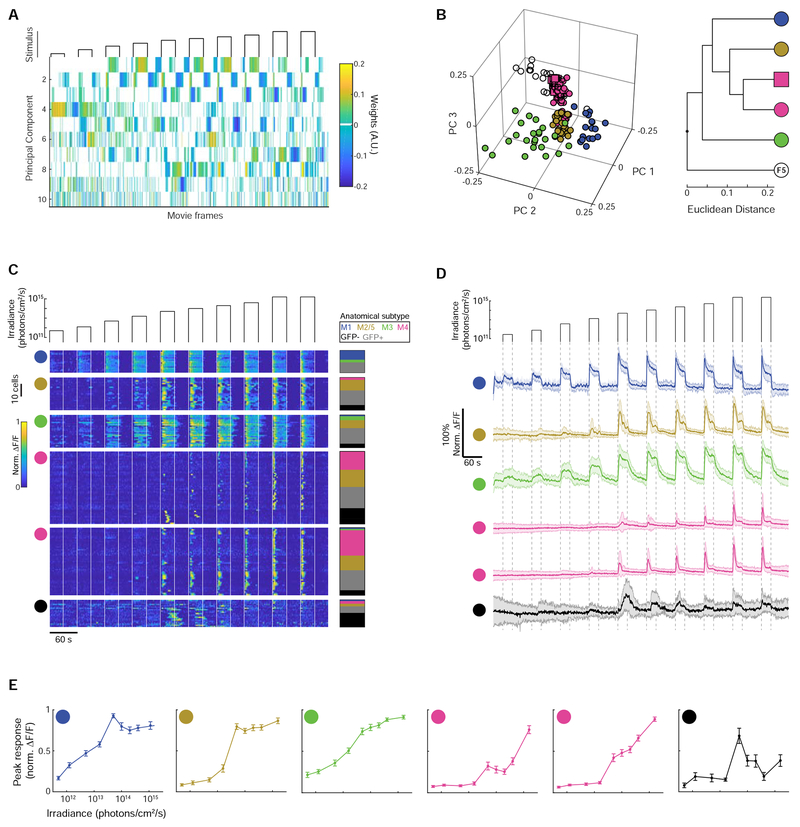
Figure 2. Unsupervised clustering divides light-responsive cells into six functional groups.
(A) Weights of the first ten principal components from sparse principal components analysis of fluorescence transients in (C). See Figure S3 for further algorithmic details.
(B) Left: Feature vectors for each cell in the space of the first three principal components. Colors correspond to functional cluster assignments. Right: hierarchical relationship between functional groups.
(C) Left: functional groups. Data were truncated so that only the first 30 s following stimulus offset were included (the latter 30 s of the interstimulus interval were excluded) and high-pass filtered to remove slow baseline drifts. Right: Color-coded cell type of each cell matched by row. See Figure S4 for details on the light responses of GFP− cells.
(D) Mean fluorescence transients for each functional group. Shading indicates standard deviation.
(E) Mean irradiance-response functions for each functional group. Error bars are standard error of the mean.
We characterized the light responses of cells within each functional group by examining their mean normalized light-evoked fluorescence transients (Figure 2D) and their irradiance-response functions (Figure 2E). Cells in the F1 functional group had sustained responses and the highest light sensitivity of all the groups. These cells often responded directly to the onset of imaging and exhibited periodic oscillations in fluorescence during the intervals between light stimuli (for example the F1-classified M1 cell in Figure 1B). F2 cells were less light sensitive than F1 cells and had a sustained, biphasic light response. F3 cells were intermediate in sensitivity between F1 and F2 and had especially sustained responses. Cells in the F4a and F4b groups had transient responses and low sensitivity, encoding irradiance over the range for which the responses of F1-F3 cells saturated. Finally, F5 cells, which were predominantly GFP− cells, had variable responses in which the majority (81%, 17/21) of the cells had maximum responses to an intermediate irradiance, after which they strongly adapted (Figure S4).
To determine whether the functional groups of light-responsive cells corresponded to anatomically defined ipRGC subtypes, we mapped the subset of cells for which we obtained an unambiguous cell type classification onto these functional groups (Figure 2C). Note, we did not anatomically distinguish M2 and M5 ipRGCs (which both have ON-stratifying dendrites) and M6 ipRGCs (which we did not detect). Some ipRGCs subtypes displayed clear functional similarities (Figure S3G–H). For example, M1 cells accounted for 78% (7/9) of the anatomically identified cells in functional group F1, and functional group F1 captured 70% (7/10) of the total anatomically identified M1 cells, indicating that the properties of this functional group were determined by the M1 ipRGC cell type.
In contrast, functional groups F2-F6 exhibited considerable mixing of ipRGC subtypes and GFP− cells. M2 ipRGCs were likely present in the F2 and F3 functional groups, given that their level of light sensitivity observed in the adult retina is intermediate between that of M1 ipRGCs and the much less sensitive M4 [17,29]. M5 and M6 ipRGCs may have been present in some of the functional groups with low light sensitivity, given their low light sensitivity observed in the adult retina [11,13]. However, the abundance of M3 and M4 cells in the functional groups occupied by these putative M2, M5, and M6 ipRGCs implies that functional groups, except for F1, are comprised of mixtures of cell types. Interestingly, GFP− cells, which may be GFP-negative RGCs or the wide field amacrine cells, that are gap junction coupled coupled to ipRGCs in the adult retina [30,31], comprised 52% (11/21) of functional group F5.
Taken together, these data indicate that the developing retina has functional groups of cells that encode distinct features of light stimuli, including absolute irradiance, the amplitude of rapid changes in irradiance, and recent light history. Interestingly, apart from the correspondence of M1 ipRGCs with functional group F1, there was considerable mixing of ipRGC subtypes and GFP− cells within functional groups. Below we explore the role of gap junction coupling in this heterogeneity.
Gap junctions coupling dictates light-evoked depolarization in a subset of ipRGCs
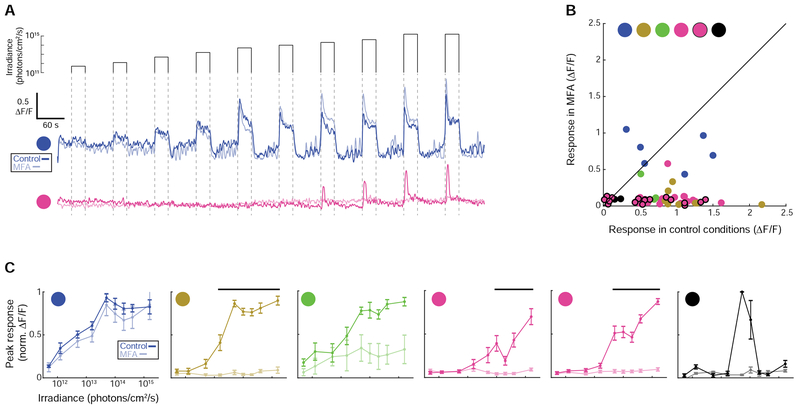
We bath applied the gap junction antagonist meclofenamic acid (MFA), which was previously shown to reversibly block junctional conductance [32] and dye coupling [33] between gap junction coupled neurons in the retina, during a subset of imaging trials to determine if gap junction circuits contribute to the light responses in putative ipRGCs or light-responsive GFP− cells (Figure 3A, Video S1). MFA caused a complete loss of light sensitivity in GFP− cells (Figure S7), consistent with the hypothesis that they inherit their light response from ipRGCs via gap junction coupling [19]. Surprisingly, MFA also caused a loss of light sensitivity in F2-F5 cells, while leaving F1 cells unaffected (Figure 3B, C, Figure S7). These data suggest the unexpected hypothesis that ipRGCs that are not in the F1 functional group require gap junction circuits to generate detectable light-evoked calcium transients.
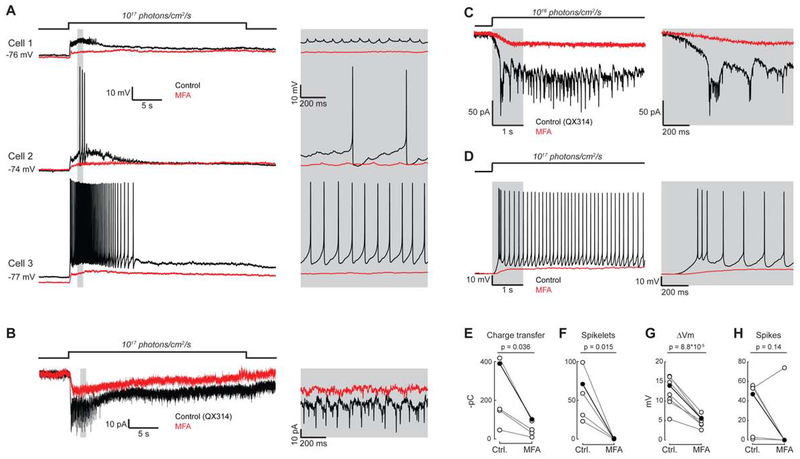
Figure 3. Cells in functional groups F2-F5 require gap junctions to generate a detectable light response.
(A) Fluorescence transients before (dark traces) and after (faded traces) 30-minute bath application of the gap junction antagonist meclofenamic acid (MFA, 50 μM). See Video S1 for an example experiment and Figure S7 for a full characterization of light responses. Figure S5 describes controls for off-target effects of MFA.
(B) Peak fluorescence response of each cell to the maximum irradiance light stimulus before and after bath application of MFA.
(C) Mean irradiance-response functions for each functional group before (dark lines) and after (faded lines) bath application of MFA. Responses are normalized to the maximum response in control conditions. Thick black lines indicate irradiances for which responses were significantly different in control and drug conditions (Wilcoxon Signed Rank Test, p < 0.05).
To understand the role of gap junction coupling in generating light-evoked depolarizations, we conducted whole cell patch-clamp recordings from M4 cells, which were easily targeted by their large, GFP+ soma, in the absence and presence of MFA (Figure 4). Recordings were conducted in the presence of DHβE to block cholinergic retinal waves. Some retinas from mice older than P7 also generated glutamatergic retinal waves, which we blocked by bath application of D-2-amino-5-phosphonovalerate (D-AP5, 50 μM). Light stimuli depolarized all M4 cells but the extent of these depolarizations varied, ranging from subthreshold depolarizations to a variety of spiking responses (Figure 4A). This variation in the spiking responses paralleled the variation in the calcium responses of M4s in functional group F4a and F4b (Figure 2C–E). Similarly, M4 cells that had little to no spiking in response to light (25%; 2/8 cells) offer a potential explanation for the lack of calcium responses in 27% (14/51) of the M4 cells recorded in calcium imaging experiments. Voltage clamp experiments revealed that the small, fast voltage fluctuations we observed during current clamp recordings were correlated with spikelets (Figure 4B), inward currents induced by the spiking activity of prejunctional light-responsive cells [19]. Bath application of MFA eliminated the spikelet currents in voltage clamp (Figure 4C, F), and the spikelet-generated depolarizations in current clamp (Figure 4D). MFA reduced the amplitude of the subthreshold depolarization in current clamp (Figure 4D, G) and the slow light-evoked current in voltage clamp (Figure 4C, E), indicating that the depolarization in M4 cells is due to cell-intrinsic photocurrent combined with junctional currents. Five of the six M4 cells that fired action potentials in control conditions failed to fire action potentials in MFA (Figure 4H), indicating that intrinsic photocurrents are insufficient for supra-threshold depolarizations in most M4s.
Figure 4. Gap junctions are required for supra-threshold light responses in M4 ipRGCs.
(A) Left: current clamp recordings before (black traces) and after (red traces) 20-minute bath application of the gap junction antagonist meclofenamic acid (MFA, 50 μM). Grey boxes in A-D correspond to expanded regions of traces on the right. Figure S5 describes controls for off-target effects of MFA.
(B) Left: Voltage clamp recording from an M4 cell before (black trace) and after (red trace) bath application of MFA. QX314 is a voltage-gated sodium channel antagonist.
(C) Left: Voltage clamp recording during the first five seconds of the light stimulus.
(D) Left: Current clamp recording during the first five seconds of the light stimulus.
(E) Charge transfer (in negative pico-Coulombs). Charge transfer was computed during the first five seconds of the light stimulus. P values in E-H are from paired t tests. Filled circles and dark lines in E-F correspond to example cells shown in C (E-F) and D (G-H)
(F) Number of spikelets (n = 5 cells from 5 mice). Spikelets were counted during the first five seconds of the light stimulus.
(G) Light-evoked depolarization three seconds after the light stimulus (n = 8 cells from 3 mice). Traces were low pass filtered before taking this measurement to remove action potential waveforms.
(H) Number of spikes during the light stimulus. (n = 6 cells from 3 mice; 2 cells that did not spike in control conditions were excluded).
We conducted several experiments to control for off-target effects of MFA on ipRGCs. First, M1 ipRGC calcium transients were unaffected by MFA, as were depolarization-induced calcium transients in M4 cells (Figure S5A–E), indicating that the loss of light responses was not due to a general degradation of cellular health [34], or an interaction with voltage-gated calcium channels, as described for the gap junction antagonist carbenoxolone [35]. MFA significantly increased the excitability of M4s (Figure S5F, I), indicating that gap junction coupling contributed to their resting conductance. Finally MFA did not reduce the sag depolarization or change the resting potential of M4s (Figure S5H–L), indicating that it did not affect the hyperpolarization-activated cyclic nucleotide-gated (HCN) or two-pore potassium conductances implicated in phototransduction in M4s [36,37]. The data are therefore most consistent with MFA acting on the light responses of ipRGCs through disruption of gap junction coupling.
Motivated by our finding that gap junctions contribute differentially to light responses in different ipRGC subtypes, we mapped gap junction circuits using an anatomical approach in which we filled GFP+ ipRGCs in the Opn4::eGFP mouse with Neurobiotin, a tracer molecule that diffuses through gap junctions to label somas of post-junctional cells. We used antibody staining against GFP and the neurofilament protein SMI32—which labels alpha RGCs, of which M4s are a subset—to characterize the somas of post-junctional cells as M4 ipRGCs, other ipRGC subtypes, or GFP− cells (Figure 5A). M2-M5 ipRGCs were tracer-coupled to many nearby cells, while M1 ipRGCs were coupled to very few or no nearby cells (Figure 5B–F). The tracer-coupled neighbors of M2-M5 cells typically included M4 ipRGCs, ipRGC subtypes that were not M4, and GFP− cells. ‘Heterotypic’ coupling between different ipRGC subtypes was therefore the rule rather than the exception. Together, these data are consistent with a circuit in which M2-M5 ipRGCs are extensively coupled and rely on coupling to generate a detectable light response, while M1 ipRGCs are isolated and generate their light response from intrinsic phototransduction.
Figure 5. Anatomical map of ipRGC gap junction circuitry.
(A) Tracer coupling of an M4 ipRGC filled with Neurobiotin (Nb).
(B) Tracer coupling patterns for M1-M5 ipRGCs. The identities of tracer coupled cells (color-coded arrowheads) were determined by antibody staining against GFP and SMI32, as in (A).
(C) Total number of tracer-coupled cells. Each point represents the number of cells found to be tracer coupled to Neurobiotin-injected ipRGC. N = 33 injected cells (5 M1s, 11 M2s, 1 M3, 10 M4s, 5 M5s) from 17 mice.
(D) Coupling to other ipRGCs (GFP+ & SMI32− or GFP+ and SMI32+).
(E) Coupling to GFP− cells that are either non-intrinsically photosensitive RGCs or amacrine cell interneurons.
(F) Coupling to M4 ipRGCs (GFP+ & SMI32+).
Increasing extent of gap junction circuits via blockade of type-1 dopamine receptors increases light sensitivity
Our studies so far suggested a model in which ipRGCs depolarize in response to light because of the summation of intrinsic melanopsin-mediated photocurrents with junctional currents arriving from other ipRGCs. In this model, ipRGCs also provide light-evoked excitatory currents to non-ipRGCs through gap junctions. We hypothesized that increasing gap junction coupling would (1) increase the light sensitivity of ipRGCs and (2) increase the extent of light-evoked activity in cells that are not ipRGCs or ipRGCs with very low melanopsin expression, e.g. GFP− cells in the Opn4::eGFP mouse.
To test these hypotheses, we took advantage of the fact that dopamine is a potent modulator of gap junction coupling in the retina [23,38]. Blockade of D1R signaling was previously shown to increase the number of light-responsive cells in the neonatal retina [39] and enhance the influence of light stimulation on the initiation rate of gap junction-mediated spontaneous activity observed in mice lacking cholinergic retinal waves [20].
To determine if D1R signaling regulates the extent of gap junction coupling in ipRGC networks, we performed Neurobiotin tracer coupling experiments in M4 ipRGCs. M4s filled with Neurobiotin in a bath solution containing SCH23390, a specific antagonist of D1Rs, were tracer coupled to more neurons than M4s filled under baseline conditions (Figure S6). These data suggest that SCH23390 relieves the suppression of gap junction coupling that occurs under baseline levels of dopamine signaling.
We then used two-photon calcium imaging to record light-evoked fluorescence before and after bath application of SCH23390 (Figure 6A–C, Video S2). Within each functional group, ipRGCs had significantly larger light responses in the presence of SCH23390 (Figure 6C), particularly to the highest irradiance stimuli. SCH23390 also increased both the charge transferred by the photocurrent and the number of spikelets (Figure 6D–F). Furthermore, many cells that did not have a detectable light response in control conditions gained one in SCH23390. Of the cells that gained light responses, 56% (31/55) were GFP− while the remaining were GFP+. These GFP+ cells may have been ipRGCs that had subthreshold depolarizations in control conditions but received enough current through gap junctions in the presence of SCH23390 to reach spike threshold (see Figure 4 and Discussion).
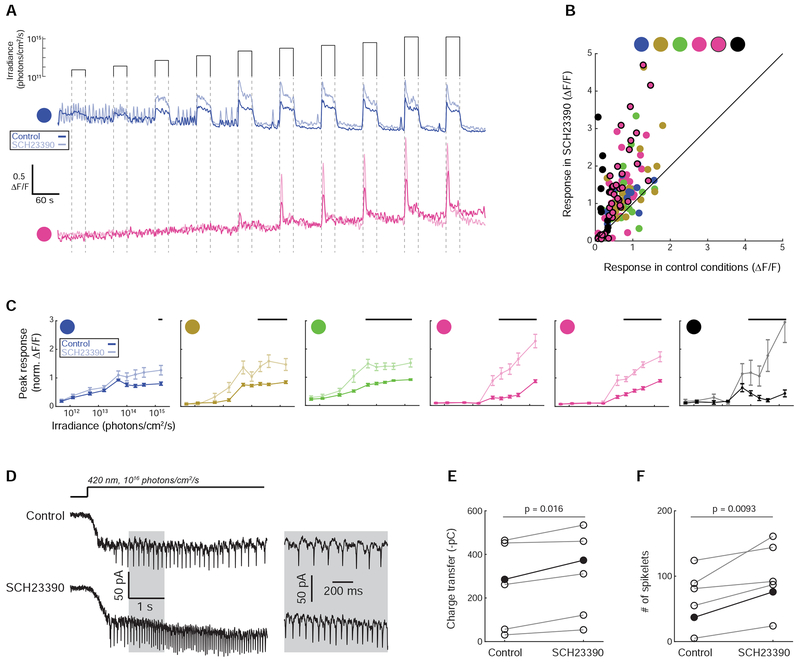
Figure 6. Blockade of type-1 dopamine receptors increases the extent of gap junction circuits and enhances light sensitivity.
(A) Fluorescence traces before (dark trace) and after (faded trace) 10-minute bath application of a type-1 dopamine receptor antagonist SCH23390 (8 μM) that increased the extent of ipRGC gap junction coupling (Figure S6). See Video S2 for an example experiment and Figure S7 for a full characterization of light responses.
(B) Peak fluorescence response of each cell to the maximum irradiance light stimulus before and after bath application of SCH23390.
(C) Mean irradiance-response functions for each functional group. Responses are normalized to the maximum response in control conditions. Thick black lines indicate irradiances for which responses were significantly different in control and drug conditions (Wilcoxon Signed Rank Test, p < 0.05).
(D) Left: Voltage clamp recording from an M4 cell before and after bath application of SCH23390. Right: a portion of the traces expanded in time to show details of spikelets.
(E) Charge transfer (in negative pico-Coulombs) during the light stimulus. Charge transfer was computed during the five second light stimulus, as shown in (D). N = 6 cells from 4 mice. Filled circles and dark line indicates data from the example cell shown in (D). The p value is from a paired t test.
(F) Number of spikelets. Spikelets were counted during the five second light stimulus, as shown in (D). Filled circles and dark line indicates data from the example cell shown in (D). The p value is from a paired t test.
We generated an ipRGC-specific D1R knockout mouse to determine if D1Rs on ipRGCs are necessary for modulation of light sensitivity by SCH23390 (Figure 7). SCH23390 caused a significantly smaller change in light-response amplitudes (Figure 7A–E) and failed to induce many cells to gain light responses in D1R KO retinas compared to retinas from heterozygous and wild type littermates (Figure 7F, G).
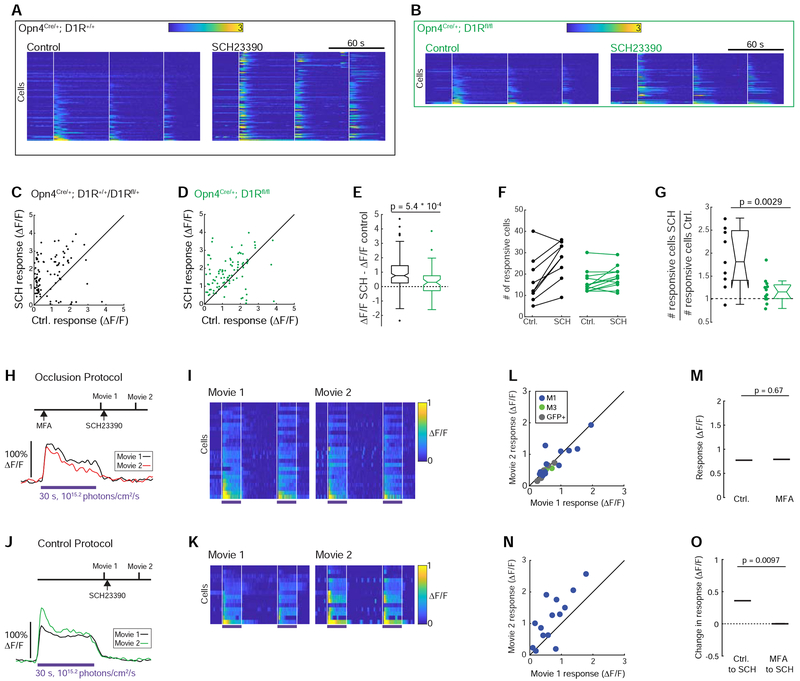
Figure 7. Knockout of type-1 dopamine receptors and blockade of gap junction circuitry occlude the effect of D1R blockade.
(A) Light-evoked fluorescence transients before (left heatmap) and after (right heatmap) 10-minute bath application of SCH23390 (8 μM). Retinas were from an Opn4Cre/+; D1R+/+ mouse. The light stimulus (420 nm, 1015.2 photons/cm2/s) consisted of three one-second pulses (vertical white lines).
(B) Same as (A), except that retinas were from an Opn4Cre/+; D1Rfl/fl mouse, in which the Drd1a gene was deleted specifically in ipRGCs.
(C) Peak fluorescence response of each cell before and after bath application of SCH23390. Retinas were from the Opn4Cre/+; D1R+/+ and Opn4Cre/+; D1Rfl/+ mice that had two or one functional copy of the Drd1a gene, respectively.
(D) Same as in (C), except that retinas were from an Opn4Cre/+; D1Rfl/fl mouse.
(E) Box and whisker plots of the change in peak fluorescence in each cell after bath application of SCH23390. The bottom and top edges of the box extend from the 25th to the 75th percentile. The whiskers extend to the most extreme data points not considered outliers, while outliers are individually plotted. The notches are 95% confidence intervals about the median value, indicated by a horizontal line. N = 175 cells from two Opn4Cre/+; D1R+/+ mice and 68 cells from one Opn4Cre/+; D1Rfl/fl mouse. The p value is from a two-sample t-test. Note that there was a small but significant increase in peak fluorescence for cells from the Opn4Cre/+; D1Rfl/fl mouse (p = 0.011, Student’s t-test), indicating either functionally relevant D1Rs in non-ipRGCs or that some copies of the Drd1a gene were not knocked out in some ipRGCs.
(F) Comparison of the number of light-responsive cells in fields of view before and after bath application of SCH23390.
(G) Comparison of the fold-change in the number of light-responsive cells following bath application of SCH23390. Each point represents one field of view from (F). N = 9 fields of view from three Opn4Cre/+; D1R+/+ mice and one Opn4Cre/+; D1Rfl/+ mouse and 12 fields of view from four Opn4Cre/+; D1Rfl/fl mice. Box and whisker plots were constructed as described in (E). The p value is from a two-sample t-test.
(H) Top: experimental design. Bottom: fluorescence response of an M1 ipRGC.
(I) Heatmaps of the fluorescence responses of all the M1 ipRGCs during the Occlusion Protocol shown in (H).
(J) Top: experimental design. Bottom: fluorescence response of an M1 ipRGC to a light.
(K) Heatmaps of fluorescence responses of M1 ipRGCs during the Control Protocol in (J). These are the same M1 cells that are shown in Figure 6. Mostly only M1 cells were light-responsive at any point during the Occlusion Protocol shown in (H), so M1 cells in the Control Protocol were the control group used in statistical comparisons in (N) and (O).
(L) Comparison of the peak fluorescence amplitudes in each cell during Movie 1 and Movie 2 of the Occlusion Protocol. Only cells that were light-responsive in at least one of the two movies were included.
(M) Comparison of peak fluorescence amplitudes in Movie 1 during the Control Protocol to those from Movie 1 during the Occlusion Protocol. Only M1 cells were included in this analysis. The p value is from an unpaired t test. For the Occlusion protocol, n = 13 M1 cells from 3 mice. For the Control Protocol, n = 14 M1 cells from 5 mice.
(N) Comparison of peak fluorescence amplitudes in each cell during Movie 1 and Movie 2 of the Occlusion Protocol.
(O) Comparison of changes in peak fluorescence amplitudes across Control and Occlusion protocols. Only M1 cells were included in this analysis. The p value is from an unpaired t test. For the Occlusion protocol, n = 13 M1 cells from 3 mice. For the Control Protocol, n = 14 M1 cells from 5 mice.
To distinguish between an effect of SCH23390 on light sensitivity via the gap junction network as opposed to a cell autonomous effect, such as an impact on the D1R interaction with the phototransduction cascade reported in rat ipRGCs [40], we performed a pharmacological occlusion experiment in which gap junctions were blocked with MFA prior to bath application of SCH23390 (Figure 7H, I). The effect of SCH23390 on light-response amplitudes was prevented by the presence of the MFA (Figure 7J–O). Note that bath application of MFA did not change the light response amplitude of M1 cells (Figure 7L, M), consistent with Figure 3. These data indicate that the increase in light sensitivity in the presence of SCH23990 is via modulation of the gap junction network.
DISCUSSION
Irradiance encoding is a fundamental property of the developing retina, even prior to the maturation of the circuits that mediate image-forming vision. We found that light-responsive cells in the developing retina could be functionally classified into six groups. Importantly, the mapping of these functional groups onto known anatomically-defined ipRGC subtypes was structured, in that certain subtypes mapped to certain functional groups (e.g. M1 cells to F1), but also mixed, in that multiple subtypes (e.g. M2-M5) coexisted in other functional groups. Several lines of evidence indicate that this mixing of subtypes into different functional groups was mediated by gap junctions. First, tracer coupling revealed that M2-M5 ipRGC subtypes were coupled to one another while M1s had little to no tracer coupling with other cells. Second, blockade of gap junctions not only eliminated light responses in GFP− cells but, surprisingly, also significantly reduced light responses in most non-M1 ipRGCs. Finally, blockade of type-1 dopamine receptor signaling led to an increase in the extent of gap junction coupling and the amplitude of light responses, indicating that plasticity in the gap junction circuits regulates light sensitivity. These results provide a new understanding of how the developing retina encodes light stimuli and a mechanistic basis for light-driven processes and behaviors during development. Moreover, they show that circuit connectivity, in addition to cell-type specific intrinsic properties, determines light sensitivity of the neonatal retina.
During development, ipRGC subtype does not completely specify function
There has been rapid progress in methods for classifying retinal cell types based on RNA-seq profiles [41]. However, it is critically important to have independent functional classification methods for comparison of cell types [42] and functional properties [28,43,44]. Here we functionally classify ipRGCs for the first time via an unsupervised learning approach. Interestingly, unlike a previous study in the adult retina [28], we find that these functional classes do not map onto molecularly identified cell types.
Our functional analysis of ipRGC subtypes yielded several key findings. First, we identified multiple subtypes of ipRGCs and characterized their functional properties, consistent with previous studies that identified M1s [4,45], M2s [18], M4s [46], and other ON-stratifying ipRGCs [47] in neonatal mice. In contrast to our finding of six functional groups, an MEA study identified 3 functional groups [8]. This is likely because of differences in recording methods— extracellularly recorded spikes vs. intracellular calcium transients—or because this previous classification relied on two measurements—latency and irradiance that evoked a half-maximal firing rate—while our unsupervised approach used additional structure in the fluorescence transients for classification. Finally, studies that used an MEA to sample multi-unit activity [8] and cell-attached electrophysiology to record spikes in M4 cells [46] in neonatal retina did not reveal cells with transient spiking responses to light stimuli, in contrast to our finding of transient responses in F4a and F4b cells, many of which were M4 cells. We confirmed that M4 cells had transient spiking responses with current clamp recordings. The origin of this discrepancy is unknown.
Second, we found that functional groups did not map directly to ipRGC subtypes. While we found that ipRGCs belonging to specific anatomically defined subtypes have clear functional similarities, an unsupervised clustering analysis revealed considerable mixing of ipRGC subtypes within functional groups. Thus, cell type only partially accounted for functional properties. Whether the population of ipRGCs in the adult retina comprises mixed functional groups, as we found here in the developing retina, is unknown. IpRGCs subtypes in the adult rat [48] and mouse [17] retina have distinct physiological properties, but is unclear if they are sufficient to functionally define these subtypes. A previous study on functional classification of RGC subtypes excluded most ipRGCs from analysis as the stimulus set was not suited for evoking responses from ipRGCs [28]. However, a functional classification of ipRGCs in the adult mouse retina based on MEA recordings found just two functional groups [8], implying that some of the six subtypes must have been either combined into or excluded from the functional groups.
Third, our findings suggest that the presence of multiple ipRGC subtypes within each functional group is partially mediated by gap junction coupling. Ml cells—the only subtype with exclusively OFF-stratifying dendrites and therefore a limited set of gap junction partners—were not tracer coupled to other ipRGCs and occupied primarily a single functional group (F1). Moreover, the light responses of M1 cells were not lost when gap junctions were blocked, indicating that their light response are primarily intrinsically generated. By contrast, M2-M5 cells—the subtypes with ON-stratifying dendrites and therefore many potential gap junction partners—were extensively coupled, including to other ipRGC subtypes, and their light responses were strongly modulated by gap junction coupling. This heterotypic coupling could produce functional groups that derive their light response properties both from intrinsic photocurrents and from prejunctional neighbors. For example, cells in F2 had biphasic light responses, potentially due to a sustained intrinsic response combined with an initial transient response inherited from prejunctional F4a/b cells. To completely understand how interactions via gap junction coupling contribute to light responses would likely require additional tools to specifically manipulate ipRGC subtypes.
Gap junctions increase the amplitude and extent of light responses in the developing retina
Gap junctions impact light responses in the developing retina in three ways. First, gap junction coupling enhances light sensitivity across the population of ipRGCs (Figure 3, 4). Second, gap junctions conduct light-evoked currents from ipRGCs to cells that lack intrinsic phototransduction (Figure 5, S7). Third, gap junction network exhibits plasticity that regulates the light sensitivity of ipRGCs (Figure 6).
How do gap junctions enhance light sensitivity in ipRGCs? One might expect a more coupled network to produce weaker light responses, due to the addition of non-photosensitive cells acting as current sinks. On the other hand, gap junctions could enhance light sensitivity by contributing depolarization from the prejunctional cells via spikelets to the postjunctional cell. There is the additional possibility that interactions between gap junction currents and nonlinear conductances enhance responses to light. There are two examples of such synergistic interactions in retinal circuits. First, gap junction coupling in the bipolar cell circuitry enhances responses to light stimuli in the receptive fields of RGCs [34]. Second, gap junctions between the dendrites of Hb9 RGCs conduct spikelets, which can evoke dendritic spikes and action potentials when they coincide with chemical synaptic input [49]. Both examples are based on synergistic interactions between chemical and electrical synapses, but chemical synapses do not contribute to the light response in the neonatal retina. Whether synergistic interactions, in this case between voltage-dependent conductances and gap junction conductances, enhance light responses in ipRGCs is unknown.
We found that non-ipRGCs exhibited strongly adapting light responses via gap junction coupling with ipRGCs. Consistent with this functional result, we found that ipRGCs were tracer coupled to non-ipRGCs (Figure 5, S6). This may reflect early establishment of a circuit also found in the adult retina, in which ipRGCs are tracer coupled [50] and electrically coupled to amacrine cells [31,51]. Interestingly, in macaque, there is some evidence that ipRGCs are coupled to other RGCs in addition to amacrine cells [52].
In conclusion, we have provided a complete characterization of encoding of ambient light in the neonatal retina and reveal for the first time that gap junction coupling significantly contributes to the heterogeneity of ipRGC light responses. The strength of gap junction coupling is modulated by dopamine, providing a powerful source of modulation of light responses prior to maturation of intraretinal circuits.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
This study did not generate new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marla Feller (mfeller@berkeley.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments were performed on mice aged postnatal day P6–P9 of either sex. Animals used in experiments had not previously been involved in other experiments or exposed to any drugs. Animal health was monitored daily and only healthy animals were used in experiments. WT mice were from the C57BL/6J strain. The Opn4::EGFP mouse strain is a BAC transgenic in which the promoter of Opn4 drives expression of enhanced GFP, as previously described [7]. Mice with the Drd1a gene deleted specifically in ipRGCs were generated by crossing mice from the Opn4Cre line, in which one of the copies of melanopsin is replaced by Cre [53], with mice from the Drd1tm2.1Stl strain, in which the Drd1a gene is flanked by loxP sites. Animal procedures were approved by the University of California, Berkeley Institutional Animal Care and Use Committees and conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals, the Public Health Service Policy, and the Society for Neuroscience Policy on the Use of Animals in Neuroscience Research.
METHOD DETAILS
Retina preparation
Mice were deeply anesthetized with isoflurane inhalation and euthanized by decapitation. Eyes were immediately enucleated and retinas were dissected in oxygenated (95% O2/5% CO2) ACSF (in mM, 119 NaCl, 2.5 KCl, 1.3 MgCl2, 1 K2HPO4, 26.2 NaHCO3, 11 D-glucose, and 2.5 CaCl2) at room-temperature under white light. Each isolated retina was cut into two pieces that were mounted over a 1–2 mm2 hole in nitrocellulose filter paper (Millipore) with the photoreceptor layer side down, dark-adapted for one hour, and transferred to the recording chamber of a two-photon microscope for imaging or electrophysiological recording. The whole-mount retinas were continuously perfused (3 ml/min) with oxygenated ACSF warmed to 32-34°C by a regulated inline heater (TC-344B, Warner Instruments) for the duration of the experiment. Additional retina pieces were kept in the dark at room temperature in ACSF bubbled with 95% O2, 5% CO2 until use (maximum 8 h).
Two-photon calcium imaging
Retinas were bulk loaded with the calcium indicator Cal 590 AM (AAT Bioquest) using the multicell bolus loading technique described previously [54,55]. Two-photon fluorescence measurements were obtained with a modified movable objective microscope (MOM, Sutter instruments) using an Olympus 60X, 1.00 NA, LUMPlanFLN objective (Olympus America) for single cell resolution imaging (field of view: 203 x 203 μm) or a Nikon 16X, 0.80 NA, N16XLWD-PF objective for large field of view (850 x 850 μm) imaging. Two-photon excitation was evoked with an ultrafast pulsed laser (Chameleon Ultra II; Coherent) tuned to 1040 nm to image Cal590 and tuned to 920 nm to image GFP. Di-hydro-β-erythroidine (DHβE, 8 μM, Tocris) was added to the perfusion system immediately before imaging to block spontaneous retinal waves that would otherwise interfere with measurements of light responses. Previously, we showed that extended blockade of waves with DHβE leads to an increase in the number of light sensitive cells [19]. We carried out most of our experiments within the first 20 minutes of wave blockade, during which time we did not see an increase in number of light sensitive cells (n = 5, data not shown). Following calcium imaging, a high resolution z-stack of GFP expression was collected to characterize dendritic structure. Laser power was set to 6.5 –W for imaging of Cal590 and single planes of GFP expression and set to 9-12 μW for z-stacks of GFP expression. The microscope system was controlled by ScanImage software [56]. Scan parameters were [pixels/line x lines/frame (frame rate in Hz)]: [256 x 256 (1.48 Hz)], at 2 ms/line for imaging of Cal590 and single planes of GFP and [1024x1024] at 8-16 ms/line for z stacks. This MOM was equipped with through-the-objective light stimulation and two detection channels for fluorescence imaging.
Visual stimulation
Visual stimuli were delivered by a violet LED (420 nm) coupled to a digital micromirror device (Digital Light Innovations Cel 5500). To decrease the background signal entering the photomultiplier tubes due to phosphorescence of the objective glass and violet stimulation of the Cal 590, the stimulus was delivered on the flyback of the fast-axis scanning mirror during a unidirectional scan to interleave the stimulus with the imaging. The intensity of the violet stimulus ranged from 1011.7 to 1015.2 photons/cm2/s, replicating the range of light intensities present at the retina from twilight to full sunlight [26]. Each light stimulus lasted 30 seconds and was separated from the previous light stimulus by 60 s. In all cases, the stimuli had a 100% positive contrast (bright on dark background). To minimize light adaptation, we waited at least ten minutes between presentations of a series of light stimuli.
Calibration of two-photon calcium imaging
To calibrate fluorescence measurements with action potential firing, we performed targeted current clamp recordings of several GFP+ cells, filled them with Cal590 at a concentration that produced approximately the same baseline fluorescence intensity as cells that had been bulk-loaded (37 μM in internal solution), and imaged fluorescence transients while evoking action potentials (Figure S1). Peak amplitudes of fluorescence transients correlated linearly with the number of action potentials, indicating that irradiance-response relations do not reflect nonlinearities in Cal590’s reporting of intracellular calcium concentrations.
Electrophysiology and Neurobiotin fills
Retinas were visualized through a window cut in the filter paper using a Köhler illumination system mounted below the objective of the microscope. ipRGCs were identified by their GFP signal in Opn4::EGFP mice during brief (1-5 s) of 920 nm two-photon excitation. A hole was pierced in the inner limiting membrane of the retina using a glass recording pipette to access the RGC layer. ipRGCs were targeted under control of a micromanipulator (MP-225, Sutter Instruments). Recording pipettes were fabricated using a vertical puller (Narishige PC-10) and had a tip resistance of 6–7 MΩ (for Neurobiotin fills) or 4–5 MΩ (for voltage clamp and current clamp). Internal solution for current clamp recordings was, in mM: 116 K+ D-Gluconate, 6 KCl, 2 NaCl, 20 HEPES, 0.5 EGTA, 4 ATP-Mg2+, 0.3 GTP-Na3, 10 phosphocreatine-Na2. Internal solution for voltage clamp recordings was identical to the internal solution used in current clamp recordings, except that it also contained 5 mM QX 314-Br. Internal solution for Neurobiotin fills was identical to the internal solution used in current clamp recordings, except that it also contained 7.5 mM Neurobiotin (Vector Laboratories SP-1120). Immediately before performing electrophysiological recordings, we bath applied Di-hydro-β-erythroidine (DHβE) to block cholinergic retinal waves and D-2-amino-5-phosphonovalerate (D-AP5) and 6,7-Dinitroquinoxaline-2,3-dione (DNQX) to block glutamatergic retinal waves. During voltage clamp experiments, M4 cells were held at −75 mV, near the mean resting potential of current-clamped M4 cells. All reported voltages are corrected for a liquid junction potential of +15 mV, calculated using the ‘Calculate Junction Potentials’ tool in Clampex 10.3 (Molecular Devices). Data were acquired using Clampex 10.3 recording software with a Multiclamp 700B amplifier (Molecular Devices) and a Digidata 1322A digitizer (Axon Instruments). Voltage clamp recordings were sampled at 10 kHz, while current clamp recordings were sampled at 33.3 kHz and low pass filtered at 10 kHz. Spikes and spikelets were identified using custom algorithms implemented in MATLAB. Spikes were defined as peaks in voltage with a prominence greater than 20 mV located at least 2 ms apart from another spike. Spikelets were defined as negative peaks of inward current with a prominence between −20 and −40 pA.
For tracer coupling experiments, cells were voltage-clamped and pipettes were removed after a 5-minute diffusion of Neurobiotin internal solution. Retinas were incubated for 25 minutes in the recording chamber after pipette removal. Cell morphology was assessed after pipette removal to confirm good cell health. Tissue was subsequently fixed, stained for Neurobiotin, immunolabeled for immunomarkers of interest (e.g. GFP and SMI32, Figure 5, S6), and imaged on a confocal scanning microscope (Zeiss LSM 880 NLO AxioExaminer, Molecular Imaging Center at UC Berkeley). Z stacks of optical slices (1 μm between slices) were acquired using a Zeiss 20x water-immersion objective. Two observers performed cell counts by hand on each optical slice. If the counts differed by more than five cells, the cells that only one observer had noticed were discarded. The observers performed their counts blind to the condition under which the cell was filled. Stacks were reconstructed offline using FIJI [57] maximum intensity projections for figure presentation.
Immunoassays
Whole-mount retinas were removed from the recording chamber and transferred to a 4% paraformaldehyde solution for 20 minutes at room temperature. Following fixation, retinas were washed in blocking buffer (1.5% BSA, 0.2% Na-Azide, 0.2% Triton X-100) (3 times, 10 minutes each time). Retinas were then incubated in a primary immunoreaction solution for 1-3 days at 4°C. Primary immunoreaction solution consisted of blocking buffer that contained 1:600 streptavidin-594 (Invitrogen S11227) to label Neurobiotin and one or more of the following primary antibodies: 1:1000 rabbit anti-GFP (Invitrogen A11122) and 1:250 mouse anti-SMI32 (Sigma Aldrich NE1023). After incubation in the primary immunoreaction solution, retinas were washed in PBS (3 times, 10 minutes each time), and then incubated for 2 hours at room temperature in secondary immunoreactive solution containing one or more of the following secondary antibodies: 1:1000 donkey anti-rabbit conjugated to Alexa 488 (Invitrogen A21206) and 1:200 goat anti-mouse conjugated to Alexa 647 (Invitrogen A21235). Retinas were washed again in PBS and then mounted on slides with an anti-fade agent (Vectashield H-1400, Vector Laboratories).
Pharmacology
Di-hydro-β-erythroidine (DHβE, 8 μM, Tocris), Meclofenamic acid (MFA, 50 μM, Sigma Aldrich), SCH23390 hydrochloride (8 μM, Tocris), D-2-amino-5-phosphonovalerate (D-AP5, 50 μM, Tocris), and 6,7-Dinitroquinoxaline-2,3-dione (DNQX, 20 μM, Tocris) were added to perfusion media as stock solutions prepared in distilled water. QX 314 Bromide (5 mM, Tocris) was added to the internal solution.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Tests
Details of statistical tests, number of replicates, and p values are indicated in the figures and figure captions. P values less than 0.05 were considered significant.
Image analysis of population calcium imaging movies
Movies were preprocessed using a custom FIJI macro written in ImageJ version 1.52g. Movies were spatially median-filtered to remove high-frequency outlier noise and then registered relative to a frame in the middle of the movie to remove mechanical drift. The baseline movie frame (F0) was computed by taking the temporal median projection of all the movie frames. Each movie frame (F) was normalized by dividing its difference from the baseline frame (F-F0) by the baseline frame ((F-F0)/F0) to produce a ΔF/F0 movie. Circular regions of interest (ROIs) were drawn on cells that were GFP+ (see next section) or displayed >20% increases in AF/F0 during at least one of the light stimuli. The ROIs and the ΔF/F0 movie was then imported into MATLAB for further analysis using custom algorithms, which are available on GitHub at https://github.com/FellerLabCodeShare/ipRGC-development-project. Traces for each ROI were computed as the mean value of the pixels enclosed by the ROI in each frame of the ΔF/F0 movie. For each ROI, the amplitude of the response to each light stimulus was computed as the difference between the peak ΔF/F0 value during the light stimulus and the ΔF/F0 value during the movie frame before the light stimulus. ROIs were defined as ‘light-responsive’ if 1) the maximum amplitude of their ΔF/F0 signal during any light stimulus exceeded the mean ΔF/F0 in the 30 second interval preceding the light stimulus by more than six standard deviations and 2) the signal-to-noise ratio between the ΔF/F0 signal during the light stimulus that evoked the maximum amplitude response and the ΔF/F0 signal during the 30 second interval preceding the light stimulus exceeded 5.0. Cells were defined as ‘non-responsive’ if the maximum amplitude of their ΔF/F0 signal during all light stimuli was within five standard deviations of the mean ΔF/F0 in the 30 second interval preceding each light stimulus. Cells that did not meet the set of criteria for being called ‘light-responsive’ or ‘non-responsive’ were deemed ‘ambiguous’. Only cells that were light-responsive were used for functional classification of cells and only cells that were light-responsive in at least one experimental condition were used for analysis of light response amplitudes.
Identification of GFP+ and GFP− cells
We compared the distribution of GFP fluorescence intensity in manually-selected GFP+ ROIs to the distribution of fluorescence intensity within ROIs automatically sampled from the background region of the image (Figure S2A). GFP+ cells were defined as cells whose fluorescence intensity matched or exceeded either the 99th percentile of background intensity or the minimum GFP+ ROI intensity, whichever was greater. The remaining cells were classified as GFP−. Note for Figure S4 that we used a more conservative criterion to rule out a contribution from very weak melanopsin expression: a subset of GFP− cells were analyzed whose fluorescence intensity was less than or equal to the 75th percentile of background intensity.
Identification of ipRGC subtypes
The procedure for classifying a GFP+ cell as a specific ipRGC subtype depended on classifying its dendritic stratification in the inner plexiform layer (IPL) as ON, OFF, or ON/OFF and determining if it was immunoreactive for SMI32 (Figure S2). Fields of view from calcium imaging experiments were registered with those in the fixed immunolabeled retinas by manually locating the field of view in the immunolabeled retina that contained GFP+ cells in the same spatial arrangement and with the same individual morphology. To classify dendritic stratification, we inspected z stacks of GFP expression taken in live and fixed immunolabeled tissue. A dendritic tree that could be manually traced back to the soma of a GFP+ cell was used to classify that cell. Dendritic stratification was assessed according to the depth of the dendrites below the axon bundles of RGCs that run just under the inner limiting membrane (ILM) of the retina. Dendritic trees were considered stratified if they had segments of >10 μm running parallel to the ILM (e.g. within the strata of the IPL). Dendritic trees were classified as ON if they stratified within 20 μm of the axon bundles, OFF if they stratified >25 μm below the axon bundles, and ON/OFF if they contained both ON-stratifying and OFF-stratifying dendrites. These threshold measurements were determined using neonatal mice from the mGluR2-GFP line [58], where a membrane-tethered human interleukin-2α/GFP fusion protein is expressed specifically in starburst amacrine cells in the retina, whose processes demarcate the boundaries between the ON and OFF layers of the IPL. Note that the dendritic trees of some GFP+ cells could not be classified, because they were physically overlapping with those of neighboring GFP+ cells or they had little to no fluorescent signal. To distinguish M2 ipRGCs from M5 ipRGCs in our tracer coupling experiments, we classified GFP+, SMI32− cells with ON-stratifying dendritic trees as M2s if their dendritic arbor diameters were > 200 μm (when viewed in the x-y plane parallel to the ILM) and as M5s if their dendritic arbor diameters were < 200 μm.
Unsupervised clustering of light-evoked fluorescence transients
Fluorescence traces for all light-responsive cells were combined into a single matrix (cells x movie frames). The traces were high pass filtered at 0.01 Hz to remove slow changes in fluorescence caused by mechanical drift in the z axis and the samples corresponding to the 30 seconds preceding each light stimulus were removed. Each trace was then normalized to its own maximum value. Note that subsequent steps were inspired by a previous study that performed unsupervised functional clustering of fluorescence transients from RGCs in the adult retina [28]. The columns of the matrix were normalized so that each column had a mean of zero and a Euclidean length of one. This normalization ensured that each timepoint (column) of the fluorescence traces in the population of cells contributed equally to the overall variance, which means, for example, that small fluorescence responses during the first few dim stimuli contributed as much variance as large fluorescence responses during the last few bright stimuli. We used a Sparse Principal Components (sPCA) algorithm implemented in the SpaSM Matlab toolbox [59] to find the Sparse Principal Components (sPCs) of the normalized data matrix. For subsequent steps in the analysis, we selected only the sPCs that individually explained >1% of the variance in the data matrix, which resulted in 10 sPCs. Matrix multiplication of the data matrix (cells x movie frames) with the sPC weights (movie frames x 10 sPCs) resulted in a feature matrix (cells x 10 sPCs). To cluster cells within this 10-dimensional space, we fit Gaussian Mixture Models (GMMs) to the feature matrix. For each candidate number of functional clusters (1-11), we fit 500 GMMs, which differed due to the dependence of a GMM’s final parameters on the randomization of its initial parameters at the start of model fitting. For each candidate number of functional clusters, we selected the GMM with the lowest Bayesian Information Criterion (BIC), a metric that rewards models for fitting data while penalizing them in proportion to their complexity. BIC declined rapidly from 1–6 clusters, plateaued, and then rose again (Figure S3B). To determine whether to split the dataset into more than six functional clusters, we computed the Bayes Factor, which is the ratio of the likelihood of a model with n+1 clusters to the likelihood of a model with n clusters, given the observed data. A heuristic interpretation of the value of the Bayes Factor indicated that there was not strong evidence for splitting the dataset into more than 6 clusters (Figure S3C). The code-base for this analysis is available on GitHub at https://github.com/FellerLabCodeShare/ipRGC-development-project.
Previous classification of RGC subtypes relied upon white noise stimulation [44] or stimuli with more spatial and temporal complexity [28]. Here, our functional classification is based on responses to increases in full-field irradiance. A more complex set of stimuli could potentially distinguish cells based upon their distinct biophysical properties [60]. We used a relatively simple stimulus in this study for several reasons. First, it is most closely associated with ipRGC function as irradiance detectors. Second, ipRGCs have slow temporal responses and receptive fields with little spatial structure since they are stimulated by melanopsin that is distributed throughout their dendrites. Hence, stimuli with more spatial and temporal complexity may not be effective at distinguishing subtypes. Third, this stimulus set based on impulse response was successful in distinguishing six different functional classes of light-responsive cells. More complex stimuli may therefore have broken these groups into more subgroups but would likely not be informative in the context of the physiological role of ipRGCs in encoding irradiance.
DATA AND SOFTWARE AVAILABILITY
The calcium imaging dataset is available on Mendeley Data at: https://data.mendeley.com/datasets/ddsj9pjhf6. The dataset is organized as a spreadsheet that contains a row for each cell with columns for fluorescence traces for each imaging trial, summary statistics computed from the fluorescence traces, functional cluster assignments, morphological criteria, and experimental metadata.
The code for extracting traces for calcium imaging analysis and unsupervised clustering is available on GitHub at https://github.com/FellerLabCodeShare/ipRGC-development-project. Other datasets generated during and/or analyzed during the current study and all custom scripts and functions generated or used during the current study are available from the Lead Contact (mfeller@berkeley.edu) on request.
Supplementary Material
Video S1: Blockade of gap junctions reduces light sensitivity in the neonatal retina. Related to Figure 3 and Figure S7.
Scalebar is 50 μm. Original video acquired at 1.5 Hz. Playback is at 15 frames per second. Red arrowheads indicate ipRGCs that retained their light response after bath application of 50 μm MFA.
Video S2: Blockade of type-1 dopamine receptors enhances light sensitivity in the neonatal retina. Related to Figure 6 and Figure S7.
Scalebar is 50 μm. Original video acquired at 1.5 Hz. Playback is at 15 frames per second. Red arrowheads indicate cells that gained light responses after bath application of 8 μm SCH23390.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti GFP | Thermo Fisher Scientific | Cat# A-11122; RRID: AB_221569 |
| Mouse monoclonal anti SMI32 | Millipore | Cat# NE1023-100UL; RRID: AB_10682557 |
| Donkey anti rabbit Alexa Fluor 488 | Molecular Probes | Cat# A-21206, RRID:AB_141708 |
| Goat anti mouse Alexa Fluor 647 | Molecular Probes | Cat# A-21235, RRID:AB_141693 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Vectashield | Vector Laboratories | Cat# H-1400, RRID:AB_2336787 |
| Cal-590 AM | AAT Bioquest | Cat# 20510 |
| Cal-590 potassium salt | AAT Bioquest | Cat# 20518 |
| Neurobiotin | Vector Laboratories | Cat# SP-1120, RRID:AB_2313575 |
| Streptavidin Alexa Fluor 594 conjugate | Thermo Fisher Scientific | Cat# S11227 |
| QX 314 bromide | Tocris | Cat# 1014 |
| Di-hydro-β-erythroidine (DHβE) | Tocris | Cat# 2349 |
| Meclofenamic acid (MFA) | Sigma Aldrich | Cat# M4531-1G |
| SCH23390 Hydrochloride (SCH23390) | Tocris | Cat# 0925 |
| D-2-amino-5-phosphonovalerate (D-AP5) | Tocris | Cat# 0106 |
| 6,7-Dinitroquinoxaline-2,3-dione (DNQX) | Tocris | Cat# 0189/10 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Calcium imaging of light responses in the neonatal retina | This paper | https://data.mendeley.com/datasets/ddsj9pjhf6 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Opn4 eGFP mice | [7] | Paulo Kofuji |
| Opn4 Cre mice | [53] | Tiffany Schmidt |
| Drd1tm2.1Stl mice | Jax | Cat# JAX:025700, RRID:IMSR_JAX:025700 |
| C57Bl/6J | Jax | Cat# JAX:000664, RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| ScanImage 4 | [56] |
http://scanimage.vidriotechnologies.com/display/SIH/ScanImage+Home RRID:SCR_014307 |
| Clampex 10.3 | Molecular Devices | RRID:SCR_011323 |
| MATLAB 2019a | MathWorks |
https://www.mathworks.com/products/matlab.html RRID:SCR_001622 |
| FIJI | [57] |
https://imagej.net/ RRID:SCR_003070 |
| Calcium imaging analysis and functional clustering code | This paper | https://github.com/FellerLabCodeShare/ipRGC-development-project |
| SpaSM | [59] | http://www2.imm.dtu.dk/projects/spasm/ |
| Other | ||
Highlights.
Six functional groups of light-responsive cells correspond to mixed ipRGC subtypes
Gap junctions transmit slow photocurrents and spikelets
Blocking gap junctions reduces light sensitivity in most functional groups
Increased gap junction coupling enhances light sensitivity of all functional groups
AKNOWLEDGEMENTS
We thank Yizhen Zhang, Christiane Voufo, and Amanda Gonzalez for assistance with gathering preliminary data and Yizhen Zhang for assistance with data analysis. Benjamin Smith, Mathew Summers, Alexandre Tiriac, and Ignas Cerniauskas commented on the manuscript. Tiffany Schmidt provided Opn4 Cre mice. Confocal images were obtained on a microscope at the Molecular Imaging Center at UC Berkeley. FSCH was supported by F31EY028022-03. MBF was supported by NIH RO1EY019498, NIH RO1EY013528, and NIH P30EY003176.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.McNeill DS, Sheely CJ, Ecker JL, Badea TC, Morhardt D, Guido W, Hattar S, Hatori M, Le H, Vollmers C, et al. (2011). Development of melanopsin-based irradiance detecting circuitry. Neural Dev. 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncan MJ, Banister MJ, and Reppert SM (1986). Developmental appearance of light-dark entrainment in the rat. Brain Res. 369, 326–330. [DOI] [PubMed] [Google Scholar]
- 3.Johnson J, Wu V, Donovan M, Majumdar S, Rentería RC, Porco T, Van Gelder RN, and Copenhagen DR (2010). Melanopsin-dependent light avoidance in neonatal mice. Proc. Natl. Acad. Sci. U. S. A 107, 17374–17378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi JS, Hughes H, Chew KS, Renna JM, Mcneill DS, Fernandez DC, Keenan WT, Thomsen MB, Ecker JL, Loevinsohn GS, et al. (2017). A subset of ipRGCs regulates both maturation of the circadian clock and segregation of retinogeniculate projections in mice. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoon M, Okawa H, Della Santina L, and Wong ROL (2014). Functional architecture of the retina: Development and disease. Prog. Retin. Eye Res 42, 44–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarttelin EE, Bellingham J, Bibb LC, Foster RG, Hankins MW, Gregory-Evans K, Gregory-Evans CY, Wells DJ, and Lucas RJ (2003). Expression of opsin genes early in ocular development of humans and mice. Exp. Eye Res 76, 393–396. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt TM, Taniguchi K, and Kofuji P (2008). Intrinsic and extrinsic light responses in melanopsin-expressing ganglion cells during mouse development. J. Neurophysiol 100, 371–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu DC, Zhang D, Demas J, Slutsky EB, Provencio I, Holy TE, and Van Gelder RN (2005). Physiologic Diversity and Development of Intrinsically Photosensitive Retinal Ganglion Cells. Neuron 48, 987–999. [DOI] [PubMed] [Google Scholar]
- 9.Sekaran S, Lupi D, Jones SL, Sheely CJ, Hattar S, Yau KW, Lucas RJ, Foster RG, and Hankins MW (2005). Melanopsin-dependent photoreception provides earliest light detection in the mammalian retina. Curr. Biol 15, 1099–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt TM, Chen S-K, and Hattar S (2011). Intrinsically photosensitive retinal ganglion cells: many subtypes, diverse functions. Trends Neurosci. 34, 572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quattrochi LE, Stabio ME, Kim I, Ilardi MC, Michelle Fogerson P, Leyrer ML, and Berson DM (2019). The M6 cell: A small-field bistratified photosensitive retinal ganglion cell. J. Comp. Neurol 527, 297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quattrochi LE, Stabio ME, Kim I, Ilardi MC, Fogerson PM, Leyrer ML, and Berson DM (2018). The M6 cell: A small-field bistratified photosensitive retinal ganglion cell. J. Comp. Neurol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stabio ME, Sabbah S, Quattrochi LE, Ilardi MC, Fogerson PM, Leyrer ML, Renna JM, Kim MT, Kim I, Schiel M, et al. (2017). The M5 Cell: A Color-Opponent Intrinsically Photosensitive Retinal Ganglion Cell. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estevez ME, Fogerson PM, Ilardi MC, Borghuis BG, Chan E, Weng S, Auferkorte ON, Demb JB, and Berson DM (2012). Form and Function of the M4 Cell, an Intrinsically Photosensitive Retinal Ganglion Cell Type Contributing to Geniculocortical Vision. J. Neurosci 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt TM, Alam NM, Chen S, Kofuji P, Li W, Prusky GT, and Hattar S (2014). A Role for Melanopsin in Alpha Retinal Ganglion Cells and Contrast Detection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt TM, and Kofuji P (2011). Structure and function of bistratified intrinsically photosensitive retinal ganglion cells in the mouse. J. Comp. Neurol 519, 1492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt TM, and Kofuji P (2009). Functional and morphological differences among intrinsically photosensitive retinal ganglion cells. J. Neurosci 29, 476–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas JA, and Schmidt TM (2019). Cellular properties of intrinsically photosensitive retinal ganglion cells during postnatal development. Neural Dev. 14, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arroyo DA, Kirkby LA, and Feller MB (2016). Retinal Waves Modulate an Intraretinal Circuit of Intrinsically Photosensitive Retinal Ganglion Cells. J. Neurosci 36, 6892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirkby LA, and Feller MB (2013). Intrinsically photosensitive ganglion cells contribute to plasticity in retinal wave circuits. Proc. Natl. Acad. Sci 110, 12090–12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereda AE, Curti S, Hoge G, Cachope R, Flores CE, and Rash JE (2013). Gap junction-mediated electrical transmission: Regulatory mechanisms and plasticity. Biochim. Biophys. Acta - Biomembr 1828, 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Brien J, and Bloomfield SA (2018). Plasticity of Retinal Gap Junctions: Roles in Synaptic Physiology and Disease. Annu. Rev. Vis. Sci 4, 79–100. [DOI] [PubMed] [Google Scholar]
- 23.Bloomfield SA, and Völgyi B (2009). The diverse functional roles and regulation of neuronal gap junctions in the retina. Nat. Rev. Neurosci 10, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spitschan M, Aguirre GK, Brainard DH, and Sweeney AM (2016). Variation of outdoor illumination as a function of solar elevation and light pollution. Sci. Rep 6, 26756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tischbirek C, Birkner A, Jia H, Sakmann B, and Konnerth A (2015). Deep two-photon brain imaging with a red-shifted fluorometric Ca 2+ indicator. Proc. Natl. Acad. Sci 112, 11377–11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen AE, Storchi R, Martial FP, Petersen RS, Montemurro MA, Brown TM, and Lucas RJ (2014). Melanopsin-Driven Light Adaptation in Mouse Vision. Curr. Biol 24, 2481–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berson DM, Castrucci AM, and Provencio I (2010). Morphology and mosaics of melanopsin-expressing retinal ganglion cell types in mice. J. Comp. Neurol 518, 2405–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baden T, Berens P, Franke K, Román Rosón M, Bethge M, and Euler T (2016). The functional diversity of retinal ganglion cells in the mouse. Nature 529, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt TM, Alam NM, Chen S, Kofuji P, Li W, Prusky GT, and Hattar S (2014). A Role for Melanopsin in Alpha Retinal Ganglion Cells and Contrast Detection. Neuron 82, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reifler AN, Chervenak AP, Dolikian ME, Benenati BA, Li BY, Wachter RD, Lynch AM, Demertzis ZD, Meyers BS, Abufarha FS, et al. (2015). All Spiking, Sustained ON Displaced Amacrine Cells Receive Gap-Junction Input from Melanopsin Ganglion Cells. Curr. Biol, 1–11. [DOI] [PubMed] [Google Scholar]
- 31.Sabbah S, Berg D, Papendorp C, Briggman KL, and Berson DM (2017). A Cre Mouse Line for Probing Irradiance- and Direction-Encoding Retinal Networks. eneuro 4, ENEURO.0065-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veruki ML, and Hartveit E (2009). Meclofenamic Acid Blocks Electrical Synapses of Retinal AII Amacrine and on -Cone Bipolar Cells. J. Neurophysiol 101, 2339–2347. [DOI] [PubMed] [Google Scholar]
- 33.Pan F, Mills SL, and Massey SC (2007). Screening of gap junction antagonists on dye coupling in the rabbit retina. Vis. Neurosci 24, 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo SP, Schwartz GW, and Rieke F (2016). Nonlinear Spatiotemporal Integration by Electrical and Chemical Synapses in the Retina. Neuron 90, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bramley JR, Wiles EM, Sollars PJ, and Pickard GE (2011). Carbenoxolone blocks the light-evoked rise in intracellular calcium in isolated melanopsin ganglion cell photoreceptors. PLoS One 6, e22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Z, Yue WWS, Chen L, Sheng Y, and Yau K-W (2018). Cyclic-Nucleotide- and HCN-Channel-Mediated Phototransduction in Intrinsically Photosensitive Retinal Ganglion Cells. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonoda T, Lee K, Birnbaumer L, and Schmidt Correspondence TM (2018). Melanopsin Phototransduction Is Repurposed by ipRGC Subtypes to Shape the Function of Distinct Visual Circuits. Neuron 99, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hampson EC, Vaney DI, and Weiler R (1992). Dopaminergic modulation of gap junction permeability between amacrine cells in mammalian retina. J. Neurosci 12, 4911–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arroyo, David A; Kirkby LA (2016). Retinal waves modulate an intra-retinal microcircuit of intrinsically photosensitive retinal ganglion cells. J. Neurosci [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Hook MJ, Wong KY, and Berson DM (2012). Dopaminergic modulation of ganglion-cell photoreceptors in rat. Eur. J. Neurosci 35, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng Y-R, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222–1237.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, Sun L, Robson P, and Trakhtenberg EF (2018). Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun 9, 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vlasits AL, Euler T, and Franke K (2019). Function first: classifying cell types and circuits of the retina. Curr. Opin. Neurobiol 56, 8–15. [DOI] [PubMed] [Google Scholar]
- 44.Chichilnisky EJ, and Kalmar RS (2002). Functional asymmetries in ON and OFF ganglion cells of primate retina. J. Neurosci 22, 2737–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tufford AR, Onyak JR, Sondereker KB, Lucas JA, Earley AM, Mattar P, Hattar S, Schmidt TM, Renna JM, and Cayouette M (2018). Melanopsin Retinal Ganglion Cells Regulate Cone Photoreceptor Lamination in the Mouse Retina. Cell Rep. 23, 2416–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sexton TJ, Bleckert A, Turner MH, and Van Gelder RN (2015). Type I intrinsically photosensitive retinal ganglion cells of early post-natal development correspond to the M4 subtype. Neural Dev. 10, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt TM, Taniguchi K, and Kofuji P (2008). Intrinsic and Extrinsic Light Responses in Melanopsin-Expressing Ganglion Cells During Mouse Development. J. Neurophysiol 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X, Stafford BK, Godin AL, King WM, and Wong KY (2014). Photoresponse diversity among the five types of intrinsically photosensitive retinal ganglion cells. J. Physiol 592, 1619–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trenholm S, McLaughlin AJ, Schwab DJ, Turner MH, Smith RG, Rieke F, and Awatramani GB (2014). Nonlinear dendritic integration of electrical and chemical synaptic inputs drives fine-scale correlations. Nat. Neurosci 17, 1759–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller LP de S, Do MTH, Yau K-W, He S, and Baldridge WH (2010). Tracer coupling of intrinsically photosensitive retinal ganglion cells to amacrine cells in the mouse retina. J. Comp. Neurol 518, 4813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reifler AN, Chervenak AP, Dolikian ME, Benenati BA, Li BY, Wachter RD, Lynch AM, Demertzis ZD, Meyers BS, Abufarha FS, et al. (2015). All spiking, sustained ON displaced amacrine cells receive gap-junction input from melanopsin ganglion cells. Curr. Biol 25, 2763–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao HW, Ren X, Peterson BB, Marshak DW, Yau KW, Gamlin PD, and Dacey DM (2016). Melanopsin-expressing ganglion cells on macaque and human retinas form two morphologically distinct populations. J. Comp. Neurol 524, 2845–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ecker JL, Dumitrescu ON, Wong KY, Alam NM, Chen S-K, LeGates T, Renna JM, Prusky GT, Berson DM, and Hattar S (2010). Melanopsin-expressing retinal ganglion-cell photoreceptors: cellular diversity and role in pattern vision. Neuron 67, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blankenship AG, Ford KJ, Johnson J, Seal RP, Edwards RH, Copenhagen DR, and Feller MB (2009). Synaptic and Extrasynaptic Factors Governing Glutamatergic Retinal Waves. Neuron 62, 230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stosiek C, Garaschuk O, Holthoff K, and Konnerth A (2003). In vivo two-photon calcium imaging of neuronal networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pologruto TA, Sabatini BL, and Svoboda K (2003). ScanImage: Flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe D, Inokawa H, Hashimoto K, Suzuki N, Kano M, Shigemoto R, Hirano T, Toyama K, Kaneko S, Yokoi M, et al. (1998). Ablation of Cerebellar Golgi Cells Disrupts Synaptic Integration Involving GABA Inhibition and NMDA Receptor Activation in Motor Coordination. Cell 95, 17–27. [DOI] [PubMed] [Google Scholar]
- 59.Sjostrand K, Clemmensen LH, Einarsson G, Larsen R, and Ersboll B (2018). SpaSM: A Matlab Toolbox for Sparse Statistical Modeling. J. Stat. Softw 84. [Google Scholar]
- 60.Padmanabhan K, and Urban NN (2010). Intrinsic biophysical diversity decorrelates neuronal firing while increasing information content. Nat. Neurosci 13, 1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Blockade of gap junctions reduces light sensitivity in the neonatal retina. Related to Figure 3 and Figure S7.
Scalebar is 50 μm. Original video acquired at 1.5 Hz. Playback is at 15 frames per second. Red arrowheads indicate ipRGCs that retained their light response after bath application of 50 μm MFA.
Video S2: Blockade of type-1 dopamine receptors enhances light sensitivity in the neonatal retina. Related to Figure 6 and Figure S7.
Scalebar is 50 μm. Original video acquired at 1.5 Hz. Playback is at 15 frames per second. Red arrowheads indicate cells that gained light responses after bath application of 8 μm SCH23390.
Data Availability Statement
The calcium imaging dataset is available on Mendeley Data at: https://data.mendeley.com/datasets/ddsj9pjhf6. The dataset is organized as a spreadsheet that contains a row for each cell with columns for fluorescence traces for each imaging trial, summary statistics computed from the fluorescence traces, functional cluster assignments, morphological criteria, and experimental metadata.
The code for extracting traces for calcium imaging analysis and unsupervised clustering is available on GitHub at https://github.com/FellerLabCodeShare/ipRGC-development-project. Other datasets generated during and/or analyzed during the current study and all custom scripts and functions generated or used during the current study are available from the Lead Contact (mfeller@berkeley.edu) on request.