Abstract
Background
Integrins play a crucial role in the regulation process of cell proliferation, migration, differentiation, tumor invasion and metastasis. ITGA11, ITGB4 and ITGB8 are three encoding genes of integrins family. Accumulative evidences have proved that abnormal expression of ITGA11, ITGB4 and ITGB8 are a common phenomenon in different malignances. However, their expression patterns and prognostic roles for patients with non-small cell lung cancer (NSCLC) have not been completely illustrated.
Methods
We investigated the expression patterns and prognostic values of ITGA11, ITGB4 and ITGB8 in patients with NSCLC through using a series of databases and various datasets, including ONCOMINE, GEPIA, HPA, TCGA and GEO datasets.
Results
We found that the expression levels of ITGA11 and ITGB4 were significantly upregulated in both LUAD and LUSC, while ITGB8 was obviously upregulated in LUSC. Additionally, higher expression level of ITGB4 revealed a worse OS in LUAD.
Conclusion
Our findings suggested that ITGA11 and ITGB4 might have the potential ability to act as diagnostic biomarkers for both LUAD and LUSC, while ITGB8 might serve as diagnostic biomarker for LUSC. Furthermore, ITGB4 could serve as a potential prognostic biomarker for LUAD.
Keywords: ITGA11, ITGB4, ITGB8, Non-small cell lung cancer, Expression and prognostic analysis
Introduction
Lung cancer is the most frequent malignancy and the leading cause of cancer-related death all over the world. Five-year survival rate for lung cancer patients ranges from 4% to 17% depending on disease stage and regional differences (Hirsch et al., 2017). Non-small cell lung cancer (NSCLC) is the most common pathological type of lung cancer and responsible for 85% to 90% of all lung cancer (Osmani et al., 2018). Owing to the problems in early diagnosis, patients with NSCLC are often diagnosed at advanced stage, which contributes a lot to the dismal prognosis (Ellis & Vandermeer, 2011; Jan et al., 2019). Thus, there is an urgent need to discover new diagnostic and prognostic biomarkers for NSCLC.
Integrins function as bridges between the extracellular matrix (ECM) and the cytoskeleton and work as radars to detect changes in the cellular microenvironment, which enables cells to react according the external milieu (Bianconi, Unseld & Prager, 2016; Ginsberg, 2014). They play a crucial role in the regulation process of cell proliferation, migration, differentiation, tumor invasion and metastasis (Slack-Davis & Parsons, 2004). Integrins family include 24 different transmembrane, multifunctional heterodimers and are composed of an α and a β subunit (Brakebusch et al., 2002). There are 18 different α subunits and eight different β subunits in human body (Hynes, 1992). Recently, the effects of integrins in tumor progression have been receiving a great deal of attention.
ITGA11 encodes integrin subunit α11, which dimerizes with β1 subunit and forms as a cell surface collagen receptor involved in the process of cell migration and collagen reorganization (Tiger et al., 2001). Integrin α11 was overexpressed in the stroma of most head and neck squamous cell carcinomas (HNSCC) and correlated positively with alpha smooth muscle actin expression (Parajuli et al., 2017). In addition, ITGA11 was overexpressed by cancer-associated fibroblast (CAFs) in Pancreatic Ductal Adenocarcinoma (PDAC) stroma and may serve as an interesting stromal therapeutic target (Schnittert et al., 2019). Integrin subunit β4, also known as a laminin-5 receptor, is a protein encoded by ITGB4 (Wang et al., 2012). Inhibition of ITGB4 in glioma cells would decrease the self-renewal abilities of glioma stem cells and suppress the malignant behaviors of glioma cells in vitro and in vivo (Ma et al., 2019). Moreover, higher ITGB4 expression level was detected in tumor than adjacent non-tumor tissues in patients with hepatocellular carcinoma (HCC). Silencing of ITGB4 could repress cell proliferation, colony forming ability and cell invasiveness (Li et al., 2017). Integrin β8, paired with αv subunit, is encoded by ITGB8. It has been reported that ITGB8 is upregulated in laryngeal squamous cell carcinoma (Ni et al., 2012). Additionally, the expression level of ITGB8 can be regulated by the tumor-promoting receptor tyrosine kinase-EphB4, while knockdown of ITGB8 may suppress migration and invasion in prostate cancer cell lines (Mertens-Walker et al., 2015). These studies have shown that ITGA11, ITGB4 and ITGB8 might be candidate biomarkers and therapeutic targets with great potential.
Recent years, there have been developed multifarious platforms, databases as well as various datasets on the web that allow cancer researchers to make in-depth bioinformatic analysis in cancer with multi omics data. Several prognostic biomarkers with great potential for NSCLC have also been identified. For instance, it has been reported that STMN1 expression was correlated with poor OS in patients with Squamous Cell Lung Carcinoma (LUSC) and might serve as a prognostic biomarker (Bao et al., 2017). Using bioinformatics methods, Xie et al. (2019) have found that KRT8 expression might be an independent prognostic biomarker for poor OS and PFS in Lung Adenocarcinoma (LUAD). Sun et al. (2019) have identified five genes that could predict metastasis in NSCLC and might serve as potential targets. As far as we know, bioinformatics analysis has not been applied to explore the roles of ITGA11, ITGB4 and ITGB8 in NSCLC. Therefore, we conducted this study to analyze the expression patterns and prognostic values of these three genes in NSCLC based on online databases, platforms and various datasets.
Materials and Methods
ONCOMINE analysis
The expression levels of ITGA11, ITGB4 and ITGB8 and genes co-expressed with ITGA11, ITGB4 and ITGB8 were analyzed in ONCOMINE database (https://www.oncomine.org) (Rhodes et al., 2007; Rhodes et al., 2004). The cut-off of p value and fold change were defined as 0.01 and 2, respectively (Huang et al., 2019).
GEPIA (Gene Expression Profiling Interactive Analysis) analysis
GEPIA (http://gepia.cancer-pku.cn/) is an interactive web application for gene expression analysis based on 9736 tumors and 8587 normal samples from the TCGA (The Cancer Genome Atlas) and the GTEx (Genotype-Tissue Expression) databases (Tang et al., 2017). The GEPIA database was used to compare mRNA levels of ITGA11, ITGB4 and ITGB8 between TCGA and GTEx databases. Meanwhile, the association among ITGA11, ITGB4 and ITGB8 in NSCLC were also analyzed in GEPIA.
Bioinformatics analysis of data using The Cancer Genome Atlas lung cancer datasets
The level 3 data of TCGA-LUAD and TCGA-LUSC were obtained from UCSC Xena platform (https://xenabrowser.net/datapages/) (Goldman et al., 2015) and RTCGA package (https://rtcga.github.io/RTCGA). The LUAD and LUSC gene expression RNAseq datasets included 524 tumor tissues and 499 tumor tissues, respectively. 502 of the LUAD patients and 492 of the 499 LUSC patients had complete survival data. The differences in overall survival (OS) of LUAD and LUSC patients with high and low expression of ITGA11, ITGB4 and ITGB8 were assessed by Kaplan–Meier curves. Meanwhile, the association between tumor stage and the expression levels of ITGA11, ITGB4 and ITGB8 were also analyzed. Clinicopathological parameters, including age at diagnosis, gender, vital status, tumor stage, smoking history and OS time, were extracted for univariate and multivariate cox regression analysis.
Gene Expression Omnibus (GEO) microarray datasets analysis
To validate the expression profiles of ITGA11, ITGB4 and ITGB8 in NSCLC, we collected a total of 21 datasets including tumor and non-tumor tissues of NSCLC in GEO database (https://www.ncbi.nlm.nih.gov/geo/). We analyzed the mRNA levels of ITGA11, ITGB4 and ITGB8 between tumor and non-tumor controls for each GEO dataset. In addition, we performed a meta-analysis based on the enrolled GEO microarray datasets.
Immunohistochemistry analysis
The protein expression of ITGA11, ITGB4 and ITGB8 in normal lung and tumor tissues were examined using the Human Protein Atlas (HPA) (https://www.proteinatlas.org/) (Uhlen et al., 2015; Uhlen et al., 2017).
Statistical analysis
Statistical analysis was performed on R software (3.6.1) (https://www.r-project.org/) and an integrated development environment RStudio (1.2.1335) (https://rstudio.com/). The mRNA expression of ITGA11, ITGB4 and ITGB8 between NSCLC tissues and normal controls were compared using Student’s t-test. Data visualization was performed using an R package called “ggstatsplot” (https://CRAN.R-project.org/package=ggstatsplot). Kaplan–Meier curves of OS were performed in TCGA-LUAD and TCGA-LUSC raw data by setting median expression of ITGA11, ITGB4 and ITGB8 as cut-off. Statistical differences were assessed by the log-rank test. Univariate and multivariate survival analyses were performed using cox regression model, risk factors (p < 0.2) analyzed by univariate analysis were selected for multivariate analysis.
For GEO datasets analysis, mean (M) and standard deviation (SD) were calculated for each NSCLC tumor and normal control group. In addition, an R package called “meta” was used in R to perform a comprehensive meta-analysis (Schwarzer, 2007). The Q test and I2 statistic were calculated to assess the heterogeneity among the enrolled studies. If p < 0.05 or I2>50%, a random effects model would be selected. Sensitivity analysis was conducted to explore whether a specific study played a crucial influence in significant heterogeneity. Finally, the publication bias was examined through funnel plots and Egger’s test (Egger et al., 1997). Once there was a publication bias, the “fill and trim” method would be selected to adjust for the bias (Duval & Tweedie, 2000). p < 0.05 deemed statistically significant.
Results
The expression levels of ITGA11,ITGB4andITGB8 in patients with non-small cell lung cancer
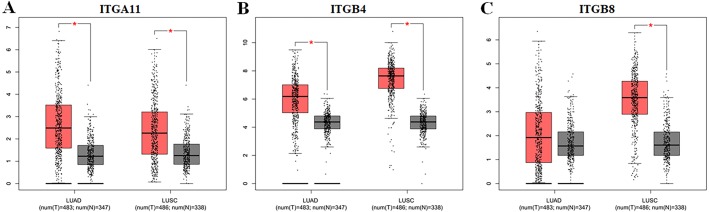
Using ONCOMINE database, we investigated the transcription levels of ITGA11, ITGB4 and ITGB8 in lung cancer vs. normal samples. ONCOMINE analysis revealed that the mRNA expression of ITGA11, ITGB4 and ITGB8 were obviously overexpressed in NSCLC tissues in ten datasets (Fig. 1). These datasets were summarized in Table 1. The GEPIA analysis results also suggested that the expression levels of ITGA11 and ITGB4 were significantly higher in both LUAD and LUSC than that in normal tissues, while the expression level of ITGB8 was only significantly upregulated in LUSC tissues (Fig. 2). Furthermore, we analyzed ITGA11, ITGB4 and ITGB8 mRNA expression level in both lung cancer and normal tissues using the TCGA-LUAD and TCGA-LUSC original data. The results revealed that the expression levels of ITGA11, ITGB4 and ITGB8 were all significantly upregulated in tumor tissues compared with normal tissues (Fig. S1).
Figure 1. The transcription levels of ITGA11, ITGB4 and ITGB8 in different cancers compared with normal tissues in the ONCOMINE dabase.
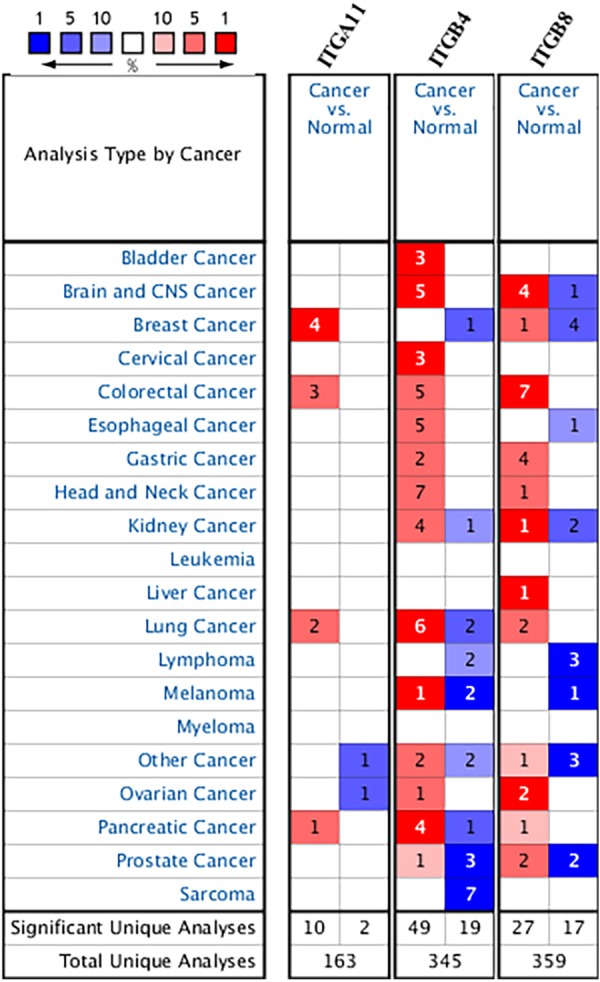
Cell color is determined by the best gene rank percentile for the analysis within the cell.
Table 1. The transcription levels of ITGA11, ITGB4 and ITGB8 between lung cancer and normal samples in ONCOMINE database.
| Gene ID | Types of lung cancer vs. normal | Fold change | P value | t-Test | References |
|---|---|---|---|---|---|
| ITGA11 | Lung Adenocarcinoma vs. Normal | 2.047 | 6.79E–16 | 10.685 | Selamat et al. (2012) |
| Lung Adenocarcinoma vs. Normal | 2.968 | 7.47E–09 | 7.945 | Okayama et al (2012) | |
| ITGB4 | Squamous Cell Lung Carcinoma vs. Normal | 2.867 | 1.32E–05 | 8.706 | Wachi et al. (2005) |
| Squamous Cell Lung Carcinoma vs. Normal | 3.505 | 5.33E–06 | 6.406 | Garber et al. (2001) | |
| Squamous Cell Lung Carcinoma vs. Normal | 2.637 | 4.64E–10 | 7.458 | Talbot et al. (2005) | |
| Squamous Cell Lung Carcinoma vs. Normal | 6.818 | 5.21E–04 | 3.57 | Bhattacharjee et al. (2001) | |
| Lung Adenocarcinoma vs. Normal | 2.99 | 1.17E–14 | 9.575 | Selamat et al. (2012) | |
| Squamous Cell Lung Carcinoma vs. Normal | 3.591 | 8.92E–10 | 8.599 | Hou et al. (2010) | |
| ITGB8 | Squamous Cell Lung Carcinoma vs. Normal | 2.455 | 1.95E–05 | 5.627 | Garber et al. (2001) |
| Squamous Cell Lung Carcinoma vs. Normal | 2.876 | 1.26E–07 | 6.484 | Hou et al. (2010) |
Figure 2. The expression levels of ITGA11 (A), ITGB4 (B) and ITGB8 (C) between NSCLC tissues and normal tissues in GEPIA.
*Indicate that the results are statistically significant.
To further explore the protein expression of ITGA11, ITGB4 and ITGB8 in NSCLC, we analyzed the IHC images using the Human Protein Atlas (HPA) database. As shown in Fig. 3, the protein expression of ITGA11 and ITGB4 were upregulated in both LUAD and LUSC cancer tissues compared with normal lung tissues (Figs. 3A–3C and 3D–3F). In comparison, the protein expression of ITGB8 was obviously upregulated in LUSC with medium staining, but not in LUAD (Figs. 3G–3I).
Figure 3. Immunohistochemistry analysis for ITGA11, ITGB4 and ITGB8 in NSCLC (HPA database).
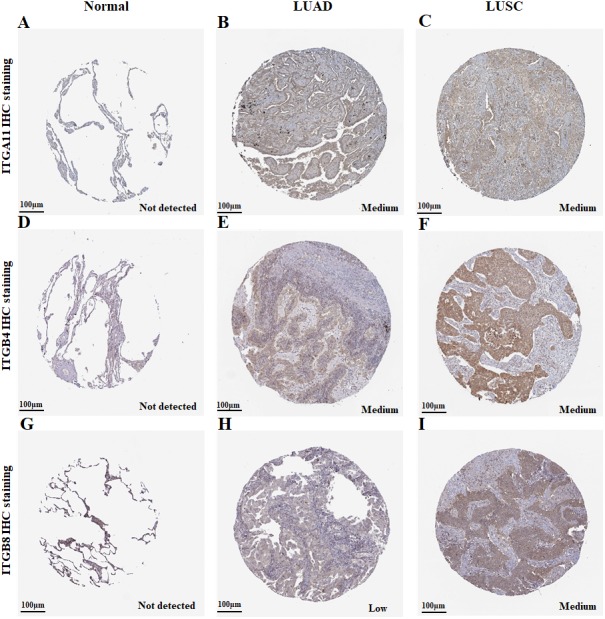
(A–F) The protein expression of ITGA11 and ITGB4 were significantly higher in both LUAD and LUSC tissues compared with the normal lung, respectively. (G–I) The protein expression level of ITGB8 was significantly higher in LUSC tissues compared with the normal lung.
Confirmation of the expression profiles of ITGA11,ITGB4andITGB8 in non-small cell lung cancer using GEO datasets
We also performed a data-mining analysis to investigate the differences in the expression levels of ITGA11, ITGB4 and ITGB8 between tumor and normal tissues in NSCLC using GEO datasets. The main characteristics of the enrolled GEO studies were described in Table S1. The results were shown in Fig. 4 and Figs. S2–S4. As illustrated in Fig. 4A and Fig. S2D, the expression level of ITGB4 was significantly increased in tissues from patients with LUAD (SMD: 0.94; 95% CI [0.65–1.24]; p < 0.01) as well as LUSC (SMD:1.37; 95% CI [0.71–2.04]; p < 0.01) compared to the normal tissues. The heterogeneity was apparent for LUAD (I 2= 80%; p < 0.01) and LUSC (I 2= 89%; p < 0.01). The following sensitivity analysis demonstrated that no study was found to have a vital influence in the enrolled studies (Fig. 4B and Figs. S3D). In addition, we didn’t find evidence of publication bias based on the funnel plot and the Egger’s test (Fig. 4C, p = 0.7759). However, the Figs. S4D indicated publication bias (Egger’s test, p = 0.04729). Therefore, we used the fill and trim method to adjust for the bias. The adjusted random effects model result showed that ITGB4 was also significantly upregulated in LUSC tissues (SMD: 0.77; 95% CI [0.03–1.52]; p = 0.04).
Figure 4. Meta-analysis of ITGB4 expression in LUAD tissues compared with normal controls based on GEO datasets.
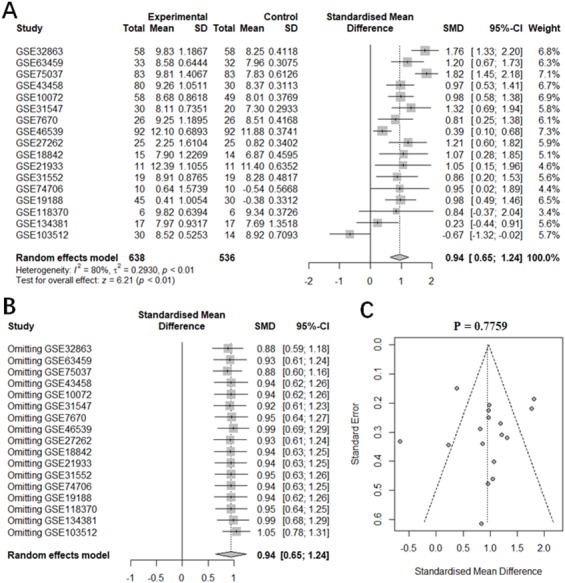
(A) Forest plot of SMD comparing ITGB4 expression in LUAD tissues with normal controls from the enrolled GEO datasets. (B) Sensitivity analysis of the enrolled GEO datasets. (C) The evaluation of the publication bias of the enrolled GEO datasets (Egger’s test, p = 0.7759).
The analysis results of ITGA11 and ITGB8 mRNA levels in LUAD and LUSC were the same as the above results (Figs. S2–S4). The separate analyses of the expression levels of ITGA11, ITGB4 and ITGB8 in LUAD and LUSC tissues compared with normal tissues for each GEO dataset were presented in the Figs. S5 and S6.
The prognostic values of ITGA11,ITGB4andITGB8 in non-small cell lung cancer
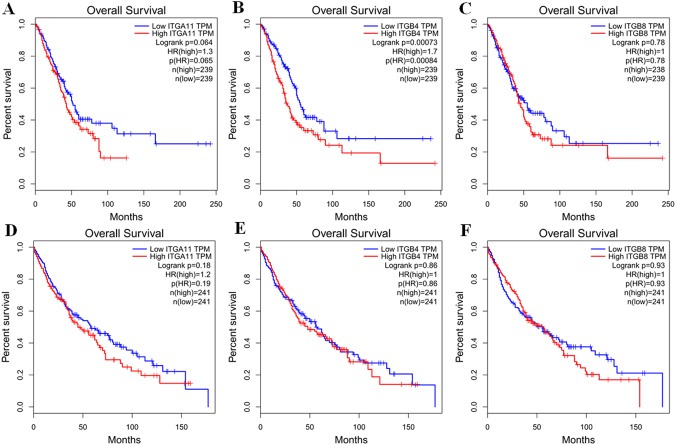
By using GEPIA, we investigated the prognostic values of ITGA11, ITGB4 and ITGB8 in NSCLC. The survival curves revealed that high expression level of ITGB4 could indicate a poor OS in LUAD (p <0.001; Fig. 5B), while ITGA11 and ITGB8 were not related with OS in LUAD (p = 0.064 and p = 0.78, respectively, Figs. 5A and 5C). In comparison, there were no obvious associations between the expression levels of ITAG11, ITGB4 and ITGB8 and LUSC (Figs. 5D–5F). Moreover, using the TCGA original data, we performed survival analysis to validate these associations. The results were consistent with GEPIA analysis (Fig. S7).
Figure 5. Kaplan–Meier survival curves of overall survival (OS) in LUAD and LUSC (GEPIA database).
Survival curves of OS based on the high and low expression of ITGA11, ITGB4 and ITGB8 in LUAD (A–C) and LUSC (D–F), respectively.
Next, we performed cox regression analysis to further assess and validate the prognostic values of ITGA11, ITGB4 and ITGB8 in NSCLC based on TCGA original data. The univariate cox analysis indicated that high ITGB4 expression and advanced stages were significantly correlated with worse OS in LUAD (Table 2). Meanwhile, multivariate cox analysis confirmed that high ITGB4 expression was an independent prognostic biomarker for patients with LUAD (HR: 1.417; 95%CI [1.042–1.926]; p = 0.026; Table 2). In addition, no significant results were found with other genes in the OS of LUAD and LUSC (Table 2). These results were consistent with that analyzed by GEPIA. Furthermore, we investigated the correlation between tumor stage and the expression levels of ITGA11, ITGB4 and ITGB8 (Fig. S8). The results showed that there was a significant correlation between tumor stage and mRNA expression of ITGB8 in LUSC (Fig. S8F).
Table 2. Univariate and multivariate cox analysis of OS in LUAD and LUSC.
Smoking history: 1. lifelong non-smoker; 2. current smoker; 3. current reformed smoker (for >15 years); 4. Current reformed smoker (for ≤ 15 years); 5. current reformed smoker (duration not specified).
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| pvalue | HR | 95%CI | pvalue | HR | 95%CI | |
| LUAD-OS | ||||||
| Gender Male vs. Female | 0.745 | 1.050 | 0.784-1.405 | |||
| Age >65 vs. ≤65 |
0.229 | 1.198 | 0.892-1.610 | |||
| Smoking history 2∕3∕4∕5 vs. 1 | 0.530 | 0.875 | 0.578-1.325 | |||
| Clinical stage III/IV vs. I/II | 0 | 2.466 | 1.786–3.404 | 0 | 2.329 | 1.682–3.226 |
| ITGA11 expression High vs. Low |
0.076 | 1.306 | 0.973–1.753 | 0.361 | 1.153 | 0.849–1.566 |
| ITGB4 expression High vs. Low |
0.002 | 1.575 | 1.175–2.112 | 0.026 | 1.417 | 1.042–1.926 |
| ITGB8 expression High vs. Low |
0.925 | 0.986 | 0.737–1.320 | |||
| LUSC-OS | ||||||
| Gender Male vs. Female | 0.179 | 1.251 | 0.902–1.736 | 0.177 | 1.253 | 0.903–1.739 |
| Age >65 vs. ≤65 |
0.124 | 1.253 | 0.940–1.670 | 0.049 | 1.343 | 1.001–1.803 |
| Smoking history 2∕3∕4∕5 vs. 1 | 0.430 | 0.698 | 0.286–1.704 | |||
| Clinical stage III/IV vs. I/II | 0.002 | 1.655 | 1.199–2.284 | 0.002 | 1.665 | 1.204–2.301 |
| ITGA11 expression High vs. Low |
0.385 | 1.128 | 0.860–1.479 | |||
| ITGB4 expression High vs. Low |
0.388 | 1.127 | 0.859–1.479 | |||
| ITGB8 expression High vs. Low |
0.875 | 0.978 | 0.746–1.283 | |||
Co-expression and correlation analyses of ITGA11,ITGB4andITGB8 in non-small cell lung cancer
The co-expression analysis was conducted using ONCOMINE database. Based on Hou Lung dataset (Hou et al., 2010), we analyzed genes that were co-expressed with ITGA11, the result showed that ITGA11 was co-expressed with COL10A1, THBS2, SULF1, CTRHC1, GREM1, C5orf46, COL11A1, NOX4 (Fig. S9A). The Bild Lung dataset indicated that ITGB4 was co-expressed with LAD1, SFN, FXYD3, KRT19, DSG2, JUP, DSP, PERP (Bild et al., 2006) (Fig. S9B). Based on Yamagata Lung dataset (Yamagata et al., 2003), we analyzed genes that were co-expressed with ITGB8, the result showed that ITGB8 was co-expressed with ERC2, PDE6D, C17orf99, SNRNP27, C1orf61, GATA1, PPP2R2B, CCK, CRYBA1, APBA3, CYP3A4, UROS (Fig. S9C).
By using GEPIA, we investigated the association among ITGA11, ITGB4 and ITGB8 in NSCLC based on Pearson correlation analysis. The results indicated that there was no correlation between ITGA11 and ITGB4 (R = − 0.018; p > 0.05) (Fig. S10A). Also, there was scarcely any correlation between ITGA11 and ITGB8 (R = 0.069; p < 0.05) (Fig. S10B). In addition, a weak positive correlation was found between ITGB8 and ITGB4 (R = 0.32; p < 0.05) (Fig. S10C).
Discussion
Numerous studies have suggested that ITGA11, ITGB4 and ITGB8 are involved in migration, epithelial-mesenchymal transition, invasion, and metastasis in different cancers (Gan et al., 2018; Huang et al., 2017; Kitajiri et al., 2002; Li et al., 2017). The aberrant expression of ITGA11, ITGB4, and ITGB8 have been reported in many cancers (Grossman et al., 2000; Mertens-Walker et al., 2015; Parajuli et al., 2017; Tagliabue et al., 1998). Regrettably, the expression profiles and prognostic roles of ITGA11, ITGB4 and ITGB8 in NSCLC are still not clear. Thus, we conducted this study to explore the expression patterns and prognostic values of ITGA11, ITGB4 and ITGB8 in NSCLC.
It has been reported that ITGA11 could serve as an important stromal factor in NSCLC, which can enhance tumorigenicity of human non-small cell lung cancer cells by regulating IGF2 expression in fibroblasts (Zhu et al., 2007). Moreover, in carcinoma-associated fibroblasts (CAFs), ITGA11 signaling pathway may play an important role in carcinoma-associated fibroblasts (CAFs), which means Integrin α11β1 can promote tumor growth and metastatic potential of NSCLC cells by regulating cancer stromal stiffness (Navab et al., 2016). These results suggested that ITGA11 might play an important role for NSCLC. In our study, ONCOMINE analysis showed that mRNA expression level of ITGA11 was highly expressed in Lung Adenocarcinoma compared with that in normal controls. GEPIA revealed that the expression level of ITGA11 was obviously higher in both LUAD and LUSC than that in normal tissues. In addition, we also downloaded TCGA original data, GEO datasets, and protein expression data from HPA to validate ITGA11 expression profile, the results were consistent with the GEPIA analysis results. These results indicated that ITGA11 might be a diagnostic biomarker for patients with LUAD and LUSC. Furthermore, we investigated the association between the expression level of ITGA11 and OS in LUAD and LUSC using GEPIA and cox regression analysis. However, the results showed ITGA11 expression had no prognostic role in terms of OS in LUAD and LUSC.
ITGB4 was found to have a strong positive correlation with tumor size (p = 0.01) and tumor nuclear grade (p < 0.01) in early breast cancer (Diaz et al., 2005). Furthermore, it is reported that ITGB4 could promote the invasion and metastasis of tumor cells through a series of processes (Stewart & O’Connor, 2015). These results imply us that ITGB4 might also play a crucial role in NSCLC. In our report, ONCOMINE and GEPIA analysis revealed that the expression level of ITGB4 was significantly upregulated in LUAD and LUSC. Additionally, we confirmed this expression feature by analysis TCGA original data and GEO datasets. The protein level was also consistent with the mRNA expression level. Taken together, these results implied that ITGB4 expression could act as a diagnostic biomarker for patients with LUAD and LUSC. Moreover, the survival curve showed that high ITGB4 expression was strong correlated with inferior OS in LUAD. The following univariate cox and multivariate cox regression analysis confirmed that high ITGB4 expression level was an independent prognostic biomarker for poor OS in LUAD.
It has been reported that ITGB8 could mediate the activation of latent TGF- β, which subsequently derives the epithelial-to-mesenchymal (EMT) transition of some cancers and contributes to cancer cell migration and growth (Mu et al., 2002; Pozzi & Zent, 2011). Furthermore, ITGB8 was significantly upregulated in ovarian cancer tissues compared with that in normal ovary tissues (He et al., 2018). Moreover, It has been reported that ITGB8 silencing could suppress the metastatic potential of human lung cancer cell lines A549 and PC (Xu & Wu, 2012). These studies suggested that ITGB8 might play an important role in NSCLC. In our study, we found that the mRNA expression level of ITGB8 was highly overexpressed in LUSC both in ONCOMINE and GEPIA analysis. This expression feature was successfully validated by analyzing the TCGA original data and GEO datasets. These results suggested that ITGB8 might act as a diagnostic biomarker in LUSC. It was worth mentioning that there was no significant correlation in ITGB8 expression level between LUAD and normal tissues by GEPIA analysis. However, the expression feature was not showed when we analyzed the TCGA original data and GEO datasets. This may due to the lack of normal controls in TCGA datasets and the differences in enrolled participants in GEO datasets. Future large-scale studies are required to assess and validate this expression pattern. In addition, we explored the association between the expression level of ITGB8 and OS in LUAD and LUSC using GEPIA and cox regression analysis. the results showed ITGB8 expression had no prognostic role in terms of OS in LUAD and LUSC. Furthermore, we found that there was a strong correlation between ITGB8 expression level and tumor stage in LUSC.
The potential limitations of our study need to be noted. First, the biological mechanisms of these three candidate markers in LUAD and LUSC are still unknown. Second, although this study had a comprehensive analysis based on several databases such as TCGA and GEO, traditional in-house experimental studies including enough specimens are required to further validate our findings.
Conclusions
In summary, we systematically analyzed the expression patterns and prognostic values of ITGA11, ITGB4 and ITGB8 in patients with LUAD and LUSC by conducting a bioinformatics analysis based on several web platforms and various datasets. Our results indicated that ITGA11 and ITGB4 might act as diagnostic biomarkers for both LUAD and LUSC, while ITGB8 may serve as diagnostic biomarker for LUSC. Furthermore, ITGB4 might serve as a potential prognostic biomarker for LUAD. We hope our findings will enrich the knowledge of diagnostic and therapy designs for patients with NSCLC.
Supplemental Information
(A–B) The forest plots of overall analysis of ITGA11 and ITGB8 between LUAD patients and normal controls, respectively. (C–E) The forest plots of overall analysis of ITGA11, ITGB4 and ITGB8 between LUSC patients and normal controls, respectively.
(A–B) Sensitivity analysis for the enrolled GEO datasets in analyzing ITGA11 and ITGB8 expression between LUAD patients and normal controls, respectively. (C–E) Sensitivity analysis for the enrolled GEO datasets in analyzing ITGA11, ITGB4 and ITGB8 expression between LUSC patients and normal controls, respectively.
(A–B) Funnel plots and Egger’s test for the enrolled GEO datasets in analyzing ITGA11 and ITGB8 expression between LUAD tissues and normal controls, respectively. (C–E) Funnel plots and Egger’s test for the enrolled GEO datasets in analyzing ITGA11, ITGB4 and ITGB8 expression between LUSC tissues and normal controls, respectively.
(A–C) The expression levels of ITGA11, ITGB4 and ITGB8 between LUAD and normal tissues for each GEO dataset, respectively.
(A–C) The expression levels of ITGA11, ITGB4 and ITGB8 between LUSC and normal tissues for each GEO dataset, respectively.
Survival curves of OS based on the high and low expression of ITGA11, ITGB4 and ITGB8 in LUAD (A–C) and LUSC (D–F), respectively.
The association between the expression levels of ITGA11, ITGB4 and ITGB8 and tumor stages in LUAD (A–C) and LUSC (D–F), respectively.
Acknowledgments
We’d like to thank Dr. Yi Zheng for her kind encouragement during the study period.
Funding Statement
This work was supported by the Natural Science Foundation of Beijing, China (Grant No. 7182132). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Pancheng Wu and Yanyu Wang conceived and designed the experiments, performed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Yijun Wu, Ziqi Jia and Yang Song analyzed the data, prepared figures and/or tables, approved the final draft.
Naixin Liang conceived and designed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The TCGA-LUAD and TCGA-LUSC raw data is available at UCSC Zena (available at https://xenabrowser.net/datapages/).
NCBI GEO datasets: GSE32863, GSE63459, GSE75037, GSE43458, GSE10072, GSE31547, GSE7670, GSE46539, GSE27262, GSE18842, GSE21933, GSE31552, GSE74706, GSE19188, GSE118370, GSE134381, GSE103512, GSE2088, GSE12428, GSE33479 and GSE30219.
The raw code is available in a Supplementary Files.
References
- Bao et al. (2017).Bao P, Yokobori T, Altan B, Iijima M, Azuma Y, Onozato R, Yajima T, Watanabe A, Mogi A, Shimizu K, Nagashima T, Ohtaki Y, Obayashi K, Nakazawa S, Bai T, Kawabata-Iwakawa R, Asao T, Kaira K, Nishiyama M, Kuwano H. High STMN1 expression is associated with cancer progression and chemo-resistance in lung squamous cell carcinoma. Annals of Surgical Oncology. 2017;24:4017–4024. doi: 10.1245/s10434-017-6083-0. [DOI] [PubMed] [Google Scholar]
- Bianconi, Unseld & Prager (2016).Bianconi D, Unseld M, Prager GW. Integrins in the spotlight of cancer. International Journal of Molecular Sciences. 2016;17(12):2037. doi: 10.3390/ijms17122037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild et al. (2006).Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson Jr JA, Marks JR, Dressman HK, West M, Nevins JR. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee et al. (2001).Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakebusch et al. (2002).Brakebusch C, Bouvard D, Stanchi F, Sakai T, Fassler R. Integrins in invasive growth. Journal of Clinical Investigation. 2002;109:999–1006. doi: 10.1172/JCI15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz et al. (2005).Diaz LK, Cristofanilli M, Zhou X, Welch KL, Smith TL, Yang Y, Sneige N, Sahin AA, Gilcrease MZ. Beta4 integrin subunit gene expression correlates with tumor size and nuclear grade in early breast cancer. Modern Pathology. 2005;18:1165–1175. doi: 10.1038/modpathol.3800411. [DOI] [PubMed] [Google Scholar]
- Duval & Tweedie (2000).Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341X.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- Egger et al. (1997).Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis & Vandermeer (2011).Ellis PM, Vandermeer R. Delays in the diagnosis of lung cancer. Journal of Thoracic Disease. 2011;3:183–188. doi: 10.3978/j.issn.2072-1439.2011.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan et al. (2018).Gan L, Meng J, Xu M, Liu M, Qi Y, Tan C, Wang Y, Zhang P, Weng W, Sheng W, Huang M, Wang Z. Extracellular matrix protein 1 promotes cell metastasis and glucose metabolism by inducing integrin beta4/FAK/SOX2/HIF-1alpha signaling pathway in gastric cancer. Oncogene. 2018;37:744–755. doi: 10.1038/onc.2017.363. [DOI] [PubMed] [Google Scholar]
- Garber et al. (2001).Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg (2014).Ginsberg MH. Integrin activation. BMB Reports. 2014;47:655–659. doi: 10.5483/BMBRep.2014.47.12.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman et al. (2015).Goldman M, Craft B, Swatloski T, Cline M, Morozova O, Diekhans M, Haussler D, Zhu J. The UCSC cancer genomics browser: update 2015. Nucleic Acids Research. 2015;43:D812–D817. doi: 10.1093/nar/gku1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman et al. (2000).Grossman HB, Lee C, Bromberg J, Liebert M. Expression of the alpha6beta4 integrin provides prognostic information in bladder cancer. Oncology Reports. 2000;7:13–16. [PubMed] [Google Scholar]
- He et al. (2018).He J, Liu Y, Zhang L, Zhang H. Integrin subunit beta 8 (ITGB8) upregulation is an independent predictor of unfavorable survival of high-grade serous ovarian carcinoma patients. Medical Science Monitor. 2018;24:8933–8940. doi: 10.12659/MSM.911518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch et al. (2017).Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran Jr WJ, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- Hou et al. (2010).Hou J, Aerts J, Den Hamer B, Van Ijcken W, Den Bakker M, Riegman P, Van der Leest C, Van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLOS ONE. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang et al. (2019).Huang C, Liu J, Xiong B, Yonemura Y, Yang X. Expression and prognosis analyses of forkhead box A (FOXA) family in human lung cancer. Gene. 2019;685:202–210. doi: 10.1016/j.gene.2018.11.022. [DOI] [PubMed] [Google Scholar]
- Huang et al. (2017).Huang L, Cai JL, Huang PZ, Kang L, Huang MJ, Wang L, Wang JP. miR19b-3p promotes the growth and metastasis of colorectal cancer via directly targeting ITGB8. American Journal of Cancer Research. 2017;7:1996–2008. [PMC free article] [PubMed] [Google Scholar]
- Hynes (1992).Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-S. [DOI] [PubMed] [Google Scholar]
- Jan et al. (2019).Jan YH, Lai TC, Yang CJ, Huang MS, Hsiao M. A co-expressed gene status of adenylate kinase 1/4 reveals prognostic gene signature associated with prognosis and sensitivity to EGFR targeted therapy in lung adenocarcinoma. Scientific Reports. 2019;9:12329. doi: 10.1038/s41598-019-48243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajiri et al. (2002).Kitajiri S, Hosaka N, Hiraumi H, Hirose T, Ikehara S. Increased expression of integrin beta-4 in papillary thyroid carcinoma with gross lymph node metastasis. Pathology International. 2002;52:438–441. doi: 10.1046/j.1440-1827.2002.01379.x. [DOI] [PubMed] [Google Scholar]
- Li et al. (2017).Li XL, Liu L, Li DD, He YP, Guo LH, Sun LP, Liu LN, Xu HX, Zhang XP. Integrin beta4 promotes cell invasion and epithelial-mesenchymal transition through the modulation of Slug expression in hepatocellular carcinoma. Scientific Reports. 2017;7:40464. doi: 10.1038/srep40464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2019).Ma B, Zhang L, Zou Y, He R, Wu Q, Han C, Zhang B. Reciprocal regulation of integrin beta4 and KLF4 promotes gliomagenesis through maintaining cancer stem cell traits. Journal of Experimental & Clinical Cancer Research. 2019;38:23. doi: 10.1186/s13046-019-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens-Walker et al. (2015).Mertens-Walker I, Fernandini BC, Maharaj MS, Rockstroh A, Nelson CC, Herington AC, Stephenson SA. The tumour-promoting receptor tyrosine kinase, EphB4, regulates expression of integrin-beta8 in prostate cancer cells. BMC Cancer. 2015;15:164. doi: 10.1186/s12885-015-1164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu et al. (2002).Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H, Sheppard D, Broaddus VC, Nishimura SL. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. Journal of Cell Biology. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navab et al. (2016).Navab R, Strumpf D, To C, Pasko E, Kim KS, Park CJ, Hai J, Liu J, Jonkman J, Barczyk M, Bandarchi B, Wang YH, Venkat K, Ibrahimov E, Pham NA, Ng C, Radulovich N, Zhu CQ, Pintilie M, Wang D, Lu A, Jurisica I, Walker GC, Gullberg D, Tsao MS. Integrin alpha11beta1 regulates cancer stromal stiffness and promotes tumorigenicity and metastasis in non-small cell lung cancer. Oncogene. 2016;35:1899–1908. doi: 10.1038/onc.2015.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni et al. (2012).Ni RS, Shen X, Qian X, Yu C, Wu H, Gao X. Detection of differentially expressed genes and association with clinicopathological features in laryngeal squamous cell carcinoma. Oncology Letters. 2012;4:1354–1360. doi: 10.3892/ol.2012.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama et al (2012).Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Research. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- Osmani et al. (2018).Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Seminars in Cancer Biology. 2018;52:103–109. doi: 10.1016/j.semcancer.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajuli et al. (2017).Parajuli H, Teh MT, Abrahamsen S, Christoffersen I, Neppelberg E, Lybak S, Osman T, Johannessen AC, Gullberg D, Skarstein K, Costea DE. Integrin alpha11 is overexpressed by tumour stroma of head and neck squamous cell carcinoma and correlates positively with alpha smooth muscle actin expression. Journal of Oral Pathology and Medicine. 2017;46:267–275. doi: 10.1111/jop.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi & Zent (2011).Pozzi A, Zent R. TGF-beta sequestration by mesangial cell integrin alphavbeta8: a novel mechanism of glomerular endothelial cell regulation. American Journal of Pathology. 2011;178:485–489. doi: 10.1016/j.ajpath.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes et al. (2007).Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18, 000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes et al. (2004).Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittert et al. (2019).Schnittert J, Bansal R, Mardhian DF, Van Baarlen J, Ostman A, Prakash J. Integrin alpha11 in pancreatic stellate cells regulates tumor stroma interaction in pancreatic cancer. FASEB Journal. 2019;33(5):6609–6621. doi: 10.1096/fj.201802336R. [DOI] [PubMed] [Google Scholar]
- Schwarzer (2007).Schwarzer G. meta: an R package for meta-analysis. R News. 2007;7:40–45. [Google Scholar]
- Selamat et al. (2012).Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, Lam S, Gazdar AF, Laird-Offringa IA. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Research. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack-Davis & Parsons (2004).Slack-Davis JK, Parsons JT. Emerging views of integrin signaling: implications for prostate cancer. Journal of Cellular Biochemistry. 2004;91:41–46. doi: 10.1002/jcb.10665. [DOI] [PubMed] [Google Scholar]
- Stewart & O’Connor (2015).Stewart RL, O’Connor KL. Clinical significance of the integrin alpha6beta4 in human malignancies. Laboratory Investigation. 2015;95:976–986. doi: 10.1038/labinvest.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun et al. (2019).Sun R, Meng X, Wang W, Liu B, Lv X, Yuan J, Zeng L, Chen Y, Yuan B, Yang S. Five genes may predict metastasis in non-small cell lung cancer using bioinformatics analysis. Oncology Letters. 2019;18:1723–1732. doi: 10.3892/ol.2019.10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabue et al. (1998).Tagliabue E, Ghirelli C, Squicciarini P, Aiello P, Colnaghi MI, Menard S. Prognostic value of alpha 6 beta 4 integrin expression in breast carcinomas is affected by laminin production from tumor cells. Clinical Cancer Research. 1998;4:407–410. [PubMed] [Google Scholar]
- Talbot et al. (2005).Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, P Oc, Socci ND, Ngai I, Carlson D, Ghossein R, Viale A, Park BJ, Rusch VW, Singh B. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Genome Research. 2005;65:3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- Tang et al. (2017).Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiger et al. (2001).Tiger CF, Fougerousse F, Grundstrom G, Velling T, Gullberg D. alpha11beta1 integrin is a receptor for interstitial collagens involved in cell migration and collagen reorganization on mesenchymal nonmuscle cells. Developmental Biology. 2001;237:116–129. doi: 10.1006/dbio.2001.0363. [DOI] [PubMed] [Google Scholar]
- Uhlen et al. (2015).Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, Von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, Von Heijne G, Nielsen J, Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Uhlen et al. (2017).Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, Von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnstrom H, Glimelius B, Sjoblom T, Edqvist PH, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2012).Wang L, Dong Z, Zhang Y, Miao J. The roles of integrin beta4 in vascular endothelial cells. Journal of Cellular Physiology. 2012;227:474–478. doi: 10.1002/jcp.22769. [DOI] [PubMed] [Google Scholar]
- Wachi et al. (2005).Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie et al. (2019).Xie L, Dang Y, Guo J, Sun X, Xie T, Zhang L, Yan Z, Amin H, Guo X. High KRT8 expression independently predicts poor prognosis for lung adenocarcinoma patients. Genes. 2019;10(1):36. doi: 10.3390/genes10010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu & Wu (2012).Xu Z, Wu R. Alteration in metastasis potential and gene expression in human lung cancer cell lines by ITGB8 silencing. Anatomical Record. 2012;295:1446–1454. doi: 10.1002/ar.22521. [DOI] [PubMed] [Google Scholar]
- Yamagata et al. (2003).Yamagata N, Shyr Y, Yanagisawa K, Edgerton M, Dang TP, Gonzalez A, Nadaf S, Larsen P, Roberts JR, Nesbitt JC, Jensen R, Levy S, Moore JH, Minna JD, Carbone DP. A training-testing approach to the molecular classification of resected non-small cell lung cancer. Clinical Cancer Research. 2003;9:4695–4704. [PubMed] [Google Scholar]
- Zhu et al. (2007).Zhu CQ, Popova SN, Brown ER, Barsyte-Lovejoy D, Navab R, Shih W, Li M, Lu M, Jurisica I, Penn LZ, Gullberg D, Tsao MS. Integrin alpha 11 regulates IGF2 expression in fibroblasts to enhance tumorigenicity of human non-small-cell lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11754–11759. doi: 10.1073/pnas.0703040104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–B) The forest plots of overall analysis of ITGA11 and ITGB8 between LUAD patients and normal controls, respectively. (C–E) The forest plots of overall analysis of ITGA11, ITGB4 and ITGB8 between LUSC patients and normal controls, respectively.
(A–B) Sensitivity analysis for the enrolled GEO datasets in analyzing ITGA11 and ITGB8 expression between LUAD patients and normal controls, respectively. (C–E) Sensitivity analysis for the enrolled GEO datasets in analyzing ITGA11, ITGB4 and ITGB8 expression between LUSC patients and normal controls, respectively.
(A–B) Funnel plots and Egger’s test for the enrolled GEO datasets in analyzing ITGA11 and ITGB8 expression between LUAD tissues and normal controls, respectively. (C–E) Funnel plots and Egger’s test for the enrolled GEO datasets in analyzing ITGA11, ITGB4 and ITGB8 expression between LUSC tissues and normal controls, respectively.
(A–C) The expression levels of ITGA11, ITGB4 and ITGB8 between LUAD and normal tissues for each GEO dataset, respectively.
(A–C) The expression levels of ITGA11, ITGB4 and ITGB8 between LUSC and normal tissues for each GEO dataset, respectively.
Survival curves of OS based on the high and low expression of ITGA11, ITGB4 and ITGB8 in LUAD (A–C) and LUSC (D–F), respectively.
The association between the expression levels of ITGA11, ITGB4 and ITGB8 and tumor stages in LUAD (A–C) and LUSC (D–F), respectively.
Data Availability Statement
The following information was supplied regarding data availability:
The TCGA-LUAD and TCGA-LUSC raw data is available at UCSC Zena (available at https://xenabrowser.net/datapages/).
NCBI GEO datasets: GSE32863, GSE63459, GSE75037, GSE43458, GSE10072, GSE31547, GSE7670, GSE46539, GSE27262, GSE18842, GSE21933, GSE31552, GSE74706, GSE19188, GSE118370, GSE134381, GSE103512, GSE2088, GSE12428, GSE33479 and GSE30219.
The raw code is available in a Supplementary Files.