Abstract
Objective
To evaluate a new tool to aid interpretation of copy number variants (CNVs) in individuals with neurodevelopmental disabilities.
Methods
Critical exon indexing (CEI) was used to identify genes with critical exons (CEGs) from clinically reported CNVs, which may contribute to neurodevelopmental disorders (NDDs). The 742 pathogenic CNVs and 1,363 variants of unknown significance (VUS) identified by chromosomal microarray analysis in 5,487 individuals with NDDs were subjected to CEI to identify CEGs. CEGs identified in a subsequent random series of VUS were evaluated for relevance to CNV interpretation.
Results
CEI identified a total of 2,492 unique CEGs in pathogenic CNVs and 953 in VUS compared with 259 CEGs in 6,965 CNVs from 873 controls. These differences are highly significant (p < 0.00001) whether compared as frequency, average, or normalized by CNV size. Twenty-one percent of VUS CEGs were not represented in Online Mendelian Inheritance in Man, highlighting limitations of existing resources for identifying potentially impactful genes within CNVs. CEGs were highly correlated with other indices and known pathways of relevance. Separately, 136 random VUS reports were reevaluated, and 76% of CEGs had not been commented on. In multiple cases, further investigation yielded additional relevant literature aiding interpretation. As one specific example, we discuss GTF2I as a CEG, which likely alters interpretation of several reported duplication VUS in the Williams-Beuren region.
Conclusions
Application of CEI to CNVs in individuals with NDDs can identify genes of potential clinical relevance, aid laboratories in effectively searching the clinical literature, and support the clinical reporting of poorly annotated VUS.
Neurodevelopmental disorders (NDDs) including developmental delay (DD), intellectual disability (ID), and autism spectrum disorder (ASD) are often caused by copy number variants (CNVs) in the genome.1–6 Chromosomal microarray analysis (CMA) for the detection of CNVs is the first-tier clinical diagnostic test for the evaluation of NDDs.2–6 CMA frequently identifies variants of uncertain significance (VUS), which limit clinical utility in such cases. Interpretation of VUS is relevant to clinical care of these children and important to their families.7,8
Clinical application of an ultra-high-resolution whole-genome chromosomal microarray optimized for the detection of genetic changes associated with a variety of NDDs provides an increased diagnostic yield, particularly due to careful attention to VUS of potential clinical relevance, constituting up to 67% of reported CNVs.9 These VUS could not be definitively linked to patients' neurodevelopmental traits despite extensive analysis of the extant literature using standard approaches, which include analysis of relevant gene content.10
The recently described11–14 identification of “critical” exons with high levels of brain expression and low mutation burden recognizes genes (e.g., OTUD7A)14 that might be NDD candidate genes. This was adapted to create a categorical classification tool that focuses analysis on specific genes among many contained in CNVs, which are likely to contribute to interpretation. This tool retrospectively identified many genes that have relevant clinical literature, but had not been specifically detailed in the clinical reports previously issued. Prospective application of this tool may aid in the classification of VUS and affect diagnosis and management of a wide range of NDDs.
Methods
Patient data
CMA results were reviewed from 1,602 cases with clinically relevant findings from a previously described retrospective series of 5,487 consecutive patients with indications for testing for broader NDDs that includes ASD, DD, ID, multiple congenital anomaly (MCA), and seizures or combinations of these.9 Medical records and indications for testing provided by referring providers were used to collect phenotype information. Subsequent to this series' analysis for critical exon genes (CEGs) prevalence, a separate group of 136 randomly selected reports of VUS was assessed to determine the impact critical exon indexing (CEI) might have on searching for the relevant literature and revised clinical reporting.
Standard protocol approvals, registrations, and patient consents
Written informed consent was obtained for all participants' data in this study. Deidentified data were collected under Western Institutional Review Board Protocol #20162032.
Chromosomal microarray and reporting
Between September 2012 and December 2015, 5,487 genomic DNAs were extracted with either a buccal swab or blood Gentra Puregene kit (Qiagen, Redwood City, CA), fragmented, labeled, and hybridized to a previously described microarray optimized for NDDs9 (FSDX; Lineagen, Salt Lake City, UT) as specified by the manufacturer (Affymetrix).15 Washed arrays were scanned and raw data files analyzed as CYCHP files using Chromosome Analysis Suite version 2.0.1 (Affymetrix, Santa Clara, CA) with reference genome from the Genome Reference Consortium: human build 37/human genome 19 (GRCh37/hg19). American Board of Medical Genetics and Genomics-certified cytogeneticists used established interpretation standards.10 In brief, CNVs were classified pathogenic if multiple peer-reviewed publications, representing at least 2 families indicate clinical features result from sensitivity of the region or gene(s) to dosage.10 If there was preliminary but insufficient evidence for pathogenicity, CNVs were classified as variants of unknown significance (VUS). CNVs are classified as benign and not reported if the region had been well characterized as a common variant in the general population by multiple peer-reviewed publications or by curated databases (e.g., the Database of Genomic Variants16).
Critical exon indexing
Identification of CEGs by CEI was adapted from previously described methods11,12 The mutation burden of rare (<0.05)11 missense and loss-of-function mutations for each exon was calculated from the 1000 Genomes Project (internationalgenome.org). The BrainSpan Atlas provided spatiotemporal exon level RNA-seq expression from 388 tissue samples from 16 brain regions of prenatal, early childhood, and adult brains from 32 postmortem donors.17 Our cutoffs, which were modified for high stringency clinical translation, considered an exon critical if expressed at >90th percentile throughout all 3 developmental stages, and has a mutation burden <10th percentile (normalized by exon size). “Critical exon” enrichment between CNVs classified as pathogenic, VUS, or normal was tested with the Fisher exact test, Welch t test, or χ2 analysis depending on the nature of the variables.
Online Mendelian Inheritance in Man (OMIM, omim.org) and PubMed (ncbi.nlm.nih.gov/pubmed/) were used to search for published data and disorder associations for each CEG.
Ingenuity Pathway Analysis (IPA; Qiagen, Redwood City, CA)18 was used to determine the association of CEGs within CNVs with known pathways based on literature-curated ontologies for genes, their functions, and their interactions. The number of CEG genes overlapping the list of curated genes identified in each pathway by IPA compared with CEGs not in a given pathway was tested statistically using a 1-tailed χ2 test for each pathway.
Data availability
The authors take full responsibility for conduct of the research, its data, analyses, and interpretation. We have full access to the data and rights to publish any and all data, separate and apart from sponsor guidance.
Results
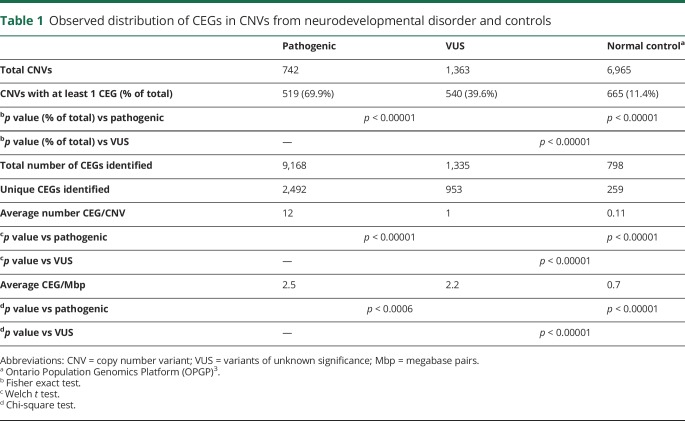
CMA results on 5,487 individuals with NDD (1,558 females and 3,929 males) on a custom microarray in a Clinical Laboratories Improvement Act certified laboratory from September 2012 to December 2015 were previously reported.9 A total of 2,105 rare CNVs (742 pathogenic and 1363 VUS) were clinically reported based on American College of Medical Genetics and Genomics Guidelines9,10 (table 1).
Table 1.
Observed distribution of CEGs in CNVs from neurodevelopmental disorder and controls
Analysis of these CNVs for CEGs yielded 2,492 unique CEGs in pathogenic CNVs and 953 in VUS, including 469 genes found in both pathogenic CNVs and VUS alike. The presence of at least 1 CEG in a CNV was significantly higher in pathogenic CNVs (69.9%) compared with either VUS (39.6%) (p < 0.00001) or a control population13 (11.4%) (p < 0.00001). On average, there were significantly more CEGs per pathogenic variant (12) compared with VUS (1) (p < 0.00001), and both contained significantly more than control's CNVs (0.17) (p < 0.00001) (table 1). Because pathogenic CNVs are typically larger than VUS,9 these data were normalized for CNV size, resulting in slightly more similar CEG/megabase pairs (Mbp) in pathogenic and VUS with 2.5 and 2.2 CEG/Mbp, respectively, but still significantly higher for pathogenic (p < 0.0006) (table 1). Both are significantly greater than the 0.6 CEG/Mbp observed in the control population13 (p < 0.00001) (table 1).
We searched for overlaps between our list of CEGs present in clinically reported CNVs and (1) genes containing de novo variants identified in individuals with ASD19,20 and (2) fragile X (fragile X mental retardation protein [FMRP]) target genes.21 ASD candidate genes were highly represented in the list of CE genes identified in our CNVs (1,023 unique out of 2,228 candidates, p = 0.0039, Fisher exact test). Of the observed CEGs from both pathogenic and VUS, 38% overlap with known FMRP targets (p < 0.00001, Fisher exact test).
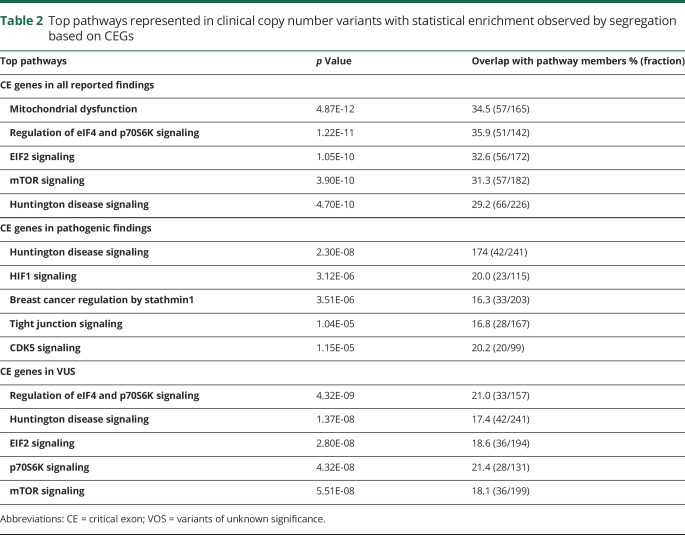
Of the observed CEGs, 18% interact significantly in pathways with gene families that have key neurodevelopmental functions (p values <1.15E-05 to 4.87E-12, 1-tailed χ2). (table 2).
Table 2.
Top pathways represented in clinical copy number variants with statistical enrichment observed by segregation based on CEGs
When the CEGs in our cohort are cross-referenced to the OMIM database (omim.org) commonly used to aid interpretations, 6.3% (158/2,492) of the unique CEGs from pathogenic CNVs and 21.0% (200/953) of those from VUS were not yet represented in OMIM.
CEGs in VUS reports
These population-based observations of enrichment of CEGs in NDDs and their enrichment in relevant pathways led us to evaluate the utility of translating CEI to previous clinical reports of VUS, i.e., CNVs where board-certified analysts using external and internal clinical databases and established clinical guidelines10 failed to find clearly pathogenic genes. We selected a random subset of 136 VUS reports to evaluate the potential impact of CEI on clinical reports and compared how gene content was discussed. Of these reports, 21% of VUS CEGs were not represented in OMIM, highlighting the limitations of existing resources for identifying potentially impactful genes in CNVs. Although some well-known CEGs were mentioned in reports as being potentially causal, in 76% of clinical reports (103/136), at least 1 CEG was not described specifically in the report. Targeted PubMed searches on 49 CEGs not previously commented on in clinical reports yielded 25 articles (51%) relevant for NDDs, which might aid the interpretation of these VUS.
CEGs within subregions of large pathogenic regions
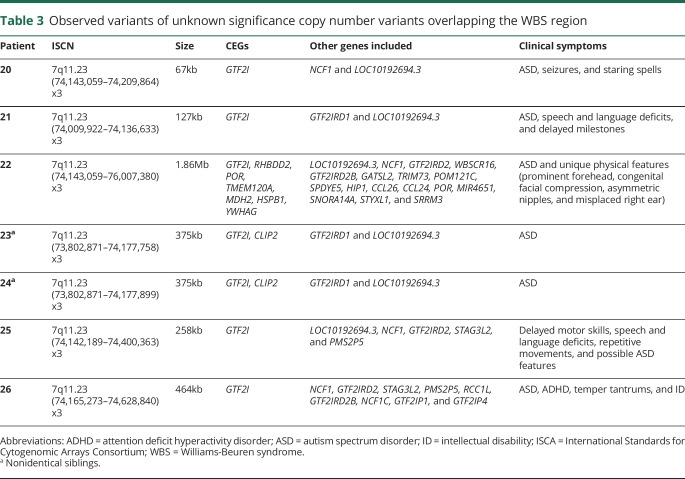
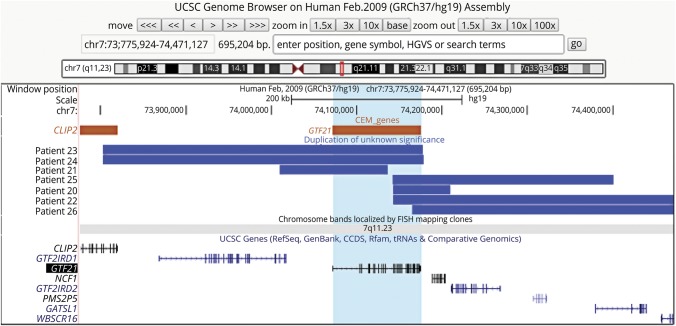
VUS can occur as smaller CNVs overlapping or completely within known pathogenic CNVs. We hypothesized that CEGs within such VUS may indicate a critical region for pathogenicity in both the VUS and the larger known pathogenic CNV. As an example, several cases with small duplications interpreted clinically as VUS overlap the 7q11.23 Williams-Beuren syndrome (WBS) region, but from patients with only ASD clinically (table 3). One duplication VUS (table 3, patient 20) affects part of a single CEG, GTF2I, which encodes a multifunctional transcription factor.22,23 It also slightly overlaps a noncoding gene, LOC10192694.3, and NCF1, a gene expressed primarily in neutrophils. Two brothers (table 3, patients 23 and 24) shared a slightly larger overlapping VUS that completely duplicates GTF2I and partially including another CEG, CLIP2, and a nonCEG, GTF2IRD1. Patient 21 (table 3) has a VUS that affects GTF2I, GTF2IRD1, and LOC10192694.3. Another individual (table 3, patient 22) has a partial duplication of GTF2I that extends 1.9 Mbp proximally and includes several additional genes. Patients 25 and 26 have overlapping VUS of intermediate length. CNV coordinates and testing indications of each patient are shown in table 3 and figure. The only gene contained, wholly or partially, in all of these VUS from individuals with ASD is GTF2I, which has been previously associated with neurobehavioral and cognitive features of WBS.22,23
Table 3.
Observed variants of unknown significance copy number variants overlapping the WBS region
Figure. Map of VUS overlapping CEG GTF2I from individuals with ASD phenotypes.
Discussion
The presence of critical genomic content within CNVs is central to clinical interpretation.10 The recognition that gene expression and conservation at the exon level opens new understanding of candidate genes also showed CEGs correlate significantly with patients with ASD and DD compared with their control siblings.11,12 CEI led to the identification of the OTUD7A gene as a key phenotype regulator for 15q13.3 microdeletion syndrome14 and has been cited in multiple publications across a variety of neurodevelopmental and behavioral disabilities including epilepsies, ASD, cerebral palsy, schizophrenia, and obsessive-compulsive disorder.24–30 The application of CEI by clinical laboratories may assist clarifying pathogenicity of CNVs and expands where gene content is factored into interpretation and discussion of results with referring clinicians and families.
VUS remain a major challenge in clinical interpretation. VUS tend to be ultra-rare, relatively small, and encompass fewer genes than known pathogenic deletions and duplications. Improved CMAs make analytical detection of very small CNVs more robust,9,31,32 increasing the number of VUS requiring analysis. Interpretation is dependent on the coincidence of multiple rare cases and their successful alignment from the literature and databases such as OMIM. Without compelling clinical case experience compared with control individuals, functional evidence, or unambiguous genotype-phenotype correlations, inferences of pathogenicity are difficult and laborious processes for the standard clinical laboratory.
This observational study on the prevalence of CEGs identified by CEI was conducted on CNVs found in consecutive individuals presenting with NDDs and/or MCA by CMA. In total, 2,976 unique CEGs were identified within reported CNVs. It is not particularly surprising that the CEG content of pathogenic CNVs is significantly higher than in VUS or CNVs in controls, given the literature on CEI to date,11,12,14,24–30 (table 1); however, it is of interest that whether looked at in terms of CNVs with at least one CEG (p < 0.00001), average number per CNV (p < 0.00001), or CEG/Mbp basis (p < 0.00001) in VUS is also significantly higher than is controls, too. It is perhaps noteworthy than when adjusted for size differences in VUSs, the CEGs/Mbp are more similar with 2.5 vs 2.2 per Mbp (p = 0.0006) for pathogenic and VUS, respectively, than when compared in terms of overall prevalence (p < 0.0001). This supports possible clinical relevance in some VUS, as does the overlapping presence of 469 specific CEGs (18.8% of 2,492) in both pathogenic and VUS CNVs. We suggest that this supports their importance in the VUS potential for pathogenicity and may therefore aid interpretation.
CEI on a subsequent random set of clinical reports issued by the same clinical laboratory showed that 76% (103/136) failed to mention at least one of these potentially important genes. Of CEGs from VUS, 21% (200/953) were not represented in OMIM at the time but may well be in the future as the clinical literature evolves. Further targeted PubMed searches of VUS CEGs yielded articles relevant for NDDs in many. This gap between clinical reporting of rare events and genes that segregate in terms of frequency between affected and control populations highlights the value of this CEI to assessment of VUS.
Of the observed CEGs, 18% interact in pathways with gene families that have key neurodevelopmental functions (p values: <1.15E-05 to 4.87E-12, χ2 test) (table 2). Of the observed CEGs from both pathogenic and VUS CNVs, 38% overlap with known FMRP targets (p < 0.00001), which are believed to be associated with NDDs, and some are now targets of directed drug development.
Comparison to genes containing de novo variants identified in individuals with ASD19 showed a statistically significant representation of the CEGs in the clinically reported CNVs (p = 0.0039). As the research literature suggests, these overlapping genes could represent causal genes lying within the larger pathogenic CNVs.14,24–30
Clinical genomic literature does not always include discussion of VUS, and clinical reporting is inconsistent within and across laboratories as guidelines10 focus on clinically definitive findings. In the context of genomic testing of affected individuals, the a priori significance of VUS is different than in the predictive setting, e.g., cancer predisposition, and may be relevant to both clinician's management and family perceptions.7,8 Over time, further clinical experience will eventually allow for reclassification of many VUS as either benign or pathogenic; however, the rarity of these sometimes “private” genetic findings delays their clinical utility. As an independent and nonoverlapping resource, CEI can focus the efforts of cytogeneticists and others interpreting test results on specific genes within CNVs rather than the affected region in total or the comprehensive list of the genes affected, and this can aid in the determination of pathogenicity of a VUS.
Another strategy using a model trained on nonpathogenic deletions in the general population has been applied to attempt to estimate the effect size of pathogenic deletions and correlate quantitatively the effects of haploinsufficiency on Intelligence Quotient (IQ). This approach may also help predict the impact of VUS in NDD; however, studies to date have exclusively evaluated deletions and largely focus on size alone33 rather than specific genomic content as prescribed by interpretive guidelines.10 The databases available to clinical laboratories to interpret genomic content, including CNVs, are constantly improving and expanding. Notably, the Exome Aggregation Consortium created a metric based on the probability of loss of function (pLI) of the protein products of genes in that database for similar filtering of candidate disease-causing variants. A pLI ≥ 0.9 is considered “high” and serves as a cutoff to separate genes of sufficient length into loss of function intolerant,34 and we are studying the application of “high pLI” now as a nonredundant resource along with CEI for clinical interpretation.
Finally, we describe a specific example in this series where several clinically reported VUS overlap a single well-known CEG, GTF2I. This gene is located within the Williams-Beuren 7q11.23 microdeletion-duplication syndrome region, which contains nearly 100 other genes.22 However, our VUS cases show a remarkable phenotype-genotype consistency (ASD) distinct from these well-described syndromes. Dosage of the GTF2I gene has been proposed to be responsible for the differing neurobehavioral phenotypes of WBS and the 7q11.23 microduplication syndrome.22,23,35–39 Individuals with partial WBS deletions that do not include GTF2I are reported without ID, but typically have the WBS-associated visual-spatial cognition impairment.39 GTF2I mediates oxytocin reactivity, and individuals with WBS have elevated oxytocin levels, whereas unaffected individuals with different polymorphisms in GTF2I manifest varying oxytocin levels, which correspond with their self-reported levels of social anxiety.36,37 GTF2I encodes a multifunctional transcription factor, a gene class previously associated with NDDs,35–39 and specific single nucleotide polymorphisms are associated with ASD.38 GTF2I is the only gene contained, wholly or partially, in all of these VUS, suggesting that its partial or complete duplication might be responsible for ASD features noted. The additional cases identified here by application of CEI merit reassessment of these CNVs and correlating phenotypes for pathogenicity.
CEI pinpointed clinically relevant genes in VUS and streamlined the identification of citations relevant to the patients' indications for testing. Such relevant literature adds to VUS reports and keys in on potentially critical genes that may define pathogenicity. CEI also has the potential to identify networks and pathways of relevant genes shared within one CNV or across several CNVs in a single individual or in multiple individuals. By narrowing down the list of genes within the breakpoints of CNVs, CEI could aid assessment of “multiple-hit” interactions. This could target potential drug development for some individuals with NDDs who share common causal networks and pathways, which otherwise might not be evident.
Of course, there are limitations and caveats to this approach. CEI as described defines CEGs as those that contain exons with high brain expression or genes with at least 1 splice isoform that is highly expressed in the brain. Genes that do not display this exact expression profile, i.e., ubiquitously expressed in the body but still perform an essential brain development function, would not be identified as a CEG despite potential importance to the etiology of NDDs, e.g., MECP2 (Rett syndrome). These could represent false-negative results; however, the CEI tool is always used in conjunction with other methods, and well-described genes should not in fact be missed in analysis. Similarly, some CNVs common in the general population do also harbor CEGs, such as BTRC and ZNF90, both CEGs, but apparently commonly duplicated or deleted. Whether these CEGs are simply not dose sensitive or whether they only affect NDD etiology when combined with “second hits” (single nucleotide variants in other genes, rare unreported CNVs, or structural variants not detected by CMA) remains to be determined. The cross-referencing of databases should identify these as exceptions and not cause a false-positive misinterpretation.12,13,16
As whole-exome/genome sequencing becomes more commonly used clinically,40 and the analytical capability of these tools to accurately detect CNVs improves41 the challenge of interpreting VUS and apparent “multi-hit” situations grows and should similarly be aided by CEI. Further assessment of this tool is therefore warranted, while cautiously applying its demonstrated strengths in enhancing the clinical interpretation of rare CNVs.
Acknowledgment
The authors thank Dr. Patricia Mowery-Rushton, Dr. Colleen Bilancia, Dr. Ankita Patel, and the entire Lineagen clinical team for helpful discussions. SWS is supported by The Centre for Applied Genomics, the University of Toronto McLaughlin Centre, and the GlaxoSmithKline-CIHR Endowed Chair in Genome Sciences at SickKids and the University of Toronto. This work has been presented in part at the NSGC and ASHG annual meetings in 2017 and 2016.
Glossary
- ASD
autism spectrum disorder
- CEG
critical exon gene
- CEI
critical exon indexing
- CMA
chromosomal microarray analysis
- CNV
copy number variant
- DD
developmental delay
- FMRP
fragile X mental retardation protein
- ID
intellectual disability
- IPA
ingenuity pathway analysis
- Mbp
megabase pairs
- MCA
multiple congenital anomaly
- NDD
neurodevelopmental disorder
- OMIM
Online Mendelian Inheritance in Man
- pLI
probability of loss of function index
- VUS
variants of unknown significance
- WBS
Williams-Beuren syndrome
Appendix. Authors
Study funding
Lineagen, Inc.; S.W. Scherer is supported by The Centre for Applied Genomics, the University of Toronto McLaughlin Centre, and the GlaxoSmithKline-CIHR Endowed Chair in Genome Sciences at SickKids and the University of Toronto.
Disclosure
Disclosures available: Neurology.org/NG.
References
- 1.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet 2004;36:949–951. [DOI] [PubMed] [Google Scholar]
- 2.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010;86:749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaefer GB, Mendelsohn NJ. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med 2013;15:399–407. [DOI] [PubMed] [Google Scholar]
- 4.Volkmar F, Siegel M, Woodbury-Smith M, King B, McCracken J, State M. Practice parameter for the assessment and treatment of children and adolescents with autism spectrum disorder. J Am Acad Child Adolesc Psychiatry 2014;53:237–257. [DOI] [PubMed] [Google Scholar]
- 5.Moeschler JB, Shevell M, Saul RA, et al. Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics 2014;134:e903–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kearney HM, South ST, Wol DJ, Lamb A, Hamosh A, Rao KW. American College of Medical Genetics recommendations for the design and performance expectations for clinical genomic copy number microarrays intended for use in the postnatal setting for detection of constitutional abnormalities. Genet Med 2011;13:676–679. [DOI] [PubMed] [Google Scholar]
- 7.Rei M, Bernhardt BA, Mulchandani S, et al. “What does it mean?”: uncertainties in understanding results of chromosomal microarray testing. Genet Med 2012;14:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jez S, Martin M, South S, Vanzo R, Rothwell E. Variants of unknown significance on chromosomal microarray analysis: parental perspectives. J Community Genet 2015;6:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho KS, Twede H, Vanzo R, et al. Real-world clinical performance of an ultra-high resolution chromosomal microarray optimized for neurodevelopmental disorders. Biomed Res Int 2016;2016:3284534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.South ST, Lee C, Lamb AN, Higgins AW, Kearney HM; Working Group for the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee. ACMG standards and guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013. Genet Med 2013;15:901–909. [DOI] [PubMed] [Google Scholar]
- 11.Uddin M, Tammimies K, Pellecchia G, et al. Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat Genet 2014;46:742–747. [DOI] [PubMed] [Google Scholar]
- 12.Uddin M, Pellecchia G, Thiruvahindrapuram B, et al. Indexing effects of copy number variation on genes involved in developmental delay. Sci Rep 2016;6:28663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uddin M, Thiruvahindrapuram B, Walker S, et al. A high-resolution copy-number variation resource for clinical and population genetics. Genet Med 2015;17:747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uddin M, Unda BK, Kwan V, et al. OTUD7A regulates neurodevelopmental phenotypes in the 15q13.3 microdeletion syndrome. Am J Hum Genet 2018;102:278–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason-Suares H, Kim W, Grimmett L, et al. Density matters: comparison of array platforms for detection of copy-number variation and copy-neutral abnormalities. Genet Med 2013;15:706–712. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald JR, Ziman R, Yuen RK, et al. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2014; 42(Database issue):D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunkin SM, Ng L, Lau C, et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res 2013;41(Database issue):D996–D1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014;30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuen RKC, Merico D, Bookman M, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 2017;20:602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iossifov I, O'Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014;515:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darnell JC, Van Driesche SJ, Zhang C. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011;146:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakurai T, Dorr NP, Takahashi N, McInnes LA, Elder GA, Buxbaum JD, Haploinsufficiency of GTF2I, a gene deleted in Williams syndrome, leads to increases in social interactions. Autism Res 2011;4:28–39. [DOI] [PubMed] [Google Scholar]
- 23.Crespi BJ, Hurd PL. Cognitive-behavioral phenotypes of Williams syndrome are associated with genetic variation in the GTF2I gene, in a healthy population. BMC Neurosci 2014;15:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal D, Ruppert AK, Trucks H, et al. Burden analysis of rare microdeletions suggests a strong impact of neurodevelopmental genes in genetic generalised epilepsies. PLoS Genet 2015;11:e1005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gazzellone MJ, Zhou X, Lionel AC, et al. Copy number variation in Han Chinese individuals with autism spectrum disorder. J Neurodev Disord 2014;6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Zhao D, Dong R, et al. De novo exon 1 deletion of AUTS2 gene in a patient with autism spectrum disorder and developmental delay: a case report and a brief literature review. Am J Med Genet A 2015;167:1381–1385. [DOI] [PubMed] [Google Scholar]
- 27.Lowther C, Speevak M, Armour CM, et al. Molecular characterization of NRXN1 deletions from 19,263 clinical microarray cases identifies exons important for neurodevelopmental disease expression. Genet Med 2017;19:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarrei M, Fehlings DL, Mawjee K, et al. De novo and rare inherited copy-number variations in the hemiplegic form of cerebral palsy. Genet Med 2018;20:172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Li M, Yang Z, et al. Increased co-expression of genes harboring the damaging de novo mutations in Chinese schizophrenic patients during prenatal development. Scientific Rep 2015;5:18209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gazzellone MJ, Zarrei M, Burton CL, et al. Uncovering obsessive-compulsive disorder risk genes in a pediatric cohort by high-resolution analysis of copy number variation. J Neurodev Disord 2016;8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad A, Merico D, Thiruvahindrapuram B, et al. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda) 2012;2:1665–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tucker T, Montpetit A, Chai D, et al. Comparison of genome-wide array genomic hybridization platforms for the detection of copy number variants in idiopathic mental retardation. BMC Med Genomics 2011;4:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huguet G, Schramm C, Douard E, et al. Measuring and estimating the effect sizes of copy number variants on general intelligence in community-based samples. JAMA Psychiatry 2018;75:447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borralleras C, Sahun I, Pérez-Jurado LA, Campuzano V. Intracisternal GTF2I gene therapy ameliorates deficits in cognition and synaptic plasticity of a mouse model of williams-Beuren syndrome. Mol Ther 2015;23:1691–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai L, Carter CS, Ying J, Bellugi U, Pournajafi-Nazarloo H, Korenberg JR. Oxytocin and vasopressin are dysregulated in Williams syndrome, a genetic disorder affecting social behavior. PLoS One 2012;7:e38513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Procyshyn TL, Spence J, Read S, Watson NV, Crespi BJ. The Williams syndrome prosociality gene GTF2I mediates oxytocin reactivity and social anxiety in a healthy population. Biol Lett 2017;13:20170051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malenfant P, Liu X, Hudson ML, et al. Association of GTF2I in the Williams-Beuren syndrome critical region with autism spectrum disorders. J Autism Dev Disord 2012;42:1459–1469. [DOI] [PubMed] [Google Scholar]
- 39.Shirai Y, Li W, Suzuki T. Role of splice variants of GTF2I, a transcription factor localizing at postsynaptic sites, and its relation to neuropsychiatric diseases. Int J Mol Sci 2017;18:E411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srivastava S, Love-Nichols JA, Dies KA, et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med 2019;21:2413–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trost B, Walker S, Wang Z. A comprehensive workflow for read depth-based identification of copy-number variation from whole-genome sequence data. Am J Hum Genet 2018;102:142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors take full responsibility for conduct of the research, its data, analyses, and interpretation. We have full access to the data and rights to publish any and all data, separate and apart from sponsor guidance.