Abstract
Background:
Drinking water disinfection inadvertently leads to the formation of numerous disinfection by-products (DBPs), some of which are cytotoxic, mutagenic, genotoxic, teratogenic, and potential carcinogens both in vitro and in vivo.
Objectives:
We investigated alterations to global gene expression (GE) in nontransformed human small intestine epithelial cells (FHs 74 Int) after exposure to six brominated and two chlorinated DBPs: bromoacetic acid (BAA), bromoacetonitrile (BAN), 2,6-dibromo-p-benzoquinone (DBBQ), bromoacetamide (BAM), tribromoacetaldehyde (TBAL), bromate (), trichloroacetic acid (TCAA), and trichloroacetaldehyde (TCAL).
Methods:
Using whole-genome cDNA microarray technology (Illumina), we examined GE in nontransformed human cells after exposure to DBPs at predetermined equipotent concentrations, identified significant changes in gene expression (), and investigated the relevance of these genes to specific toxicity pathways via gene and pathway enrichment analysis.
Results:
Genes related to activation of oxidative stress–responsive pathways exhibited fewer alterations than expected based on prior work, whereas all DBPs induced notable effects on transcription of genes related to immunity and inflammation.
Discussion:
Our results suggest that alterations to genes associated with immune and inflammatory pathways play an important role in the potential adverse health effects of exposure to DBPs. The interrelationship between these pathways and the production of reactive oxygen species (ROS) may explain the common occurrence of oxidative stress in other studies exploring DBP toxicity. Finally, transcriptional changes and shared induction of toxicity pathways observed for all DBPs caution of additive effects of mixtures and suggest further assessment of adverse health effects of mixtures is warranted. https://doi.org/10.1289/EHP4945
Introduction
Disinfection of drinking water is vital for the protection of public health since it greatly reduces pathogen risks and associated incidences of waterborne diseases (Cutler and Miller 2005) and is considered one of the major public health achievements of the 20th century (Calderon 2000). However, the powerful oxidants used during disinfection (e.g., chlorine or ozone) can react with natural and synthetic organic matter to inadvertently produce a multitude of potentially harmful chemicals, collectively known as disinfection by-products (DBPs) (Richardson and Postigo 2015). Evidence suggests that many DBPs exhibit cytotoxic, genotoxic, mutagenic, teratogenic, neurotoxic, and potentially carcinogenic properties, and may consequently elicit various adverse health effects (Du et al. 2013; Koivusalo et al. 1995; Muellner et al. 2010; Plewa and Wagner 2015; Rahman et al. 2010; Rivera-Núñez and Wright 2013; Villanueva et al. 2004; Wagner and Plewa, 2017; Wright et al. 2017). The presence of toxic DBPs is a concern for legislators and suppliers of drinking water, and identifying the forcing agents for toxicity is a research priority for ensuring responsible water management that protects public health and the environment (Plewa et al. 2017; Li and Mitch 2018).
To date, a limited number of DBPs have been characterized, and only a small fraction of these have been evaluated toxicologically (Wagner and Plewa 2017; Stalter et al. 2016). Brominated DBPs (Br-DBPs) tend to display higher toxicity than their chlorinated analogs (Cl-DBPs) (Escobar-Hoyos et al. 2013; Plewa and Wagner 2015; Yang et al. 2014) and are readily produced through chlorination of bromide-containing source waters (Postigo et al. 2018). This is common in coastal areas suffering from seawater intrusion (Wang et al. 2010), where bromide anions undergo rapid oxidation reactions with hypochlorous acid to produce hypobromous acid (Bougeard et al. 2010; Manasfi et al. 2016; Wang et al. 2010). Br-DBPs are also a dominant by-product in swimming pools using chlorinated seawater and may thus represent a concern for exposure routes other than drinking water (Manasfi et al. 2016). Notwithstanding the apparent differential risks, few studies have comprehensively evaluated or compared mechanistic molecular toxicity of different DBPs. There is consequently a pressing need for research aimed at identifying those by-products posing the greatest threat to humans and the environment and at understanding the molecular mechanisms leading to adverse health effects such as cancer (Hanigan et al. 2017; Plewa and Wagner 2015).
The potential association of DBPs with urinary bladder cancer (Villanueva et al. 2004, 2007) and colorectal cancer (Rahman et al. 2010; Villanueva et al. 2015) is an area of high interest. Both are among the most common types of cancer globally and display increased incidences in developed countries that benefit from higher levels of water disinfection (Siegel et al. 2016; Ploeg et al. 2009). The exact mechanism(s) leading to genotoxic and carcinogenic outcomes are still unclear but are believed to relate in some capacity to the production of reactive oxygen species (ROS) and the subsequent activation of oxidative stress pathways (Pals et al. 2013). Interestingly, other effects, such as alterations to immune function and inflammation, have also been associated with exposure to DBPs (Vlaanderen et al. 2017; Munson et al. 1982). However, despite a rather well-established relationship between inflammation responses and the development and progression of cancer (Coussens and Werb 2002; Westbrook et al. 2009; Ioannidou et al. 2016), there have been limited mechanistic studies in this area in relation to DBPs. This clearly warrants further investigation, since chronic inflammation is known to generate ROS through a variety of mechanisms (Ioannidou et al. 2016; Reuter et al. 2010).
Transcriptomics is a molecular technique that can help elucidate underlying mechanisms of toxicity by quantifying expressional changes of various genes with known biological functions (Cui and Paules 2010). Microarrays are particularly useful as an untargeted, or global, approach to gene expression profiling, yielding information for the entire set of genes expressed in a biological sample at a given time (Joseph 2017). Based on our limited knowledge of how DBP exposure elicits adverse health effects and ultimately cancer, there are significant benefits to be gained from using untargeted transcriptomics to explore chemical–gene interactions caused by DBPs.
The objective of the present study was to build on prior research to address the identified knowledge gaps. We characterized the effects of low concentrations of selected DBPs on global gene expression (GE) in normal nontransformed human enterocytes (FHs 74 Int) and used the generated GE profiles to identify affected toxicity signaling pathways through pathway enrichment analysis.
Methods
Disinfection By-Product Selection
We selected six brominated and two chlorinated DBPs: bromoacetic acid (BAA), bromoacetonitrile (BAN), 2,6-dibromo-p-benzoquinone (DBBQ), bromoacetamide (BAM), tribromoacetaldehyde (TBAL), bromate (), trichloroacetic acid (TCAA), and trichloroacetaldehyde (TCAL). The six Br-DBPs were chosen based on their known or modeled toxicity, representing classes of carbonaceous (C-DBPs) and nitrogen-containing DBPs (N-DBPs) (Plewa et al. 2008). An example of the different toxicological characteristics expressed by representatives BAA and BAN is that both are cytotoxic and genotoxic, but additionally, BAN was shown in Chinese hamster ovary (CHO) cells to disrupt the cell cycle by what was suggested to be an M-phase black that generated aberrant cells with an abnormal number of chromosomes (Komaki et al. 2014; Komaki and Plewa 2017). These DBPs were detected in disinfected drinking waters (Villanueva et al. 2003; Richardson and Postigo 2015; Postigo et al. 2015). Conversely, the two Cl-DBPs were chosen for comparison due to their low overall cytotoxicity. In the case of TCAA, evidence for genotoxicity and mutagenicity is lacking, and there is limited weight of evidence for carcinogenicity, whereas TCAL has confirmed mutagenicity, genotoxicity, and carcinogenicity (reviewed in Richardson et al. 2007). All of the selected DBPs are organic molecules, with the exception of the inorganic oxo-anion bromate.
Reagents
BAA, BAN, BAM, TBAL, sodium bromate, TCAA, TCAL, and neutral red (NR) solution [0.33%, in Dulbecco’s phosphate-buffered saline (DPBS)] were purchased from Sigma-Aldrich, and DBBQ was purchased from Apin Chemicals. All stock solutions were prepared in methanol (MeOH), stored at , and brought to room temperature immediately prior to exposure treatments. DMSO was avoided as a solvent because it can affect gene expression even at the low concentrations often used in toxicological testing (Leusch et al. 2017; Sumida et al. 2011), while evidence suggests MeOH is tolerable at slightly higher concentrations with lower impact on enzymatic activity (Busby et al. 1999) and reporter gene assays (Escher et al. 2012; Leusch et al. 2017).
The cell culture medium was purchased from American Type Culture Collection (ATCC). PBS, epidermal growth factor (EGF), fetal bovine serum (FBS), and nonessential amino acids were purchased from Thermo Fisher Scientific.
Cell Culture
Homo sapiens small intestine normal cells (FHs 74 Int) (CCL-241™; ATCC) were maintained in sterile culture flasks (Corning, catalog no. 431,080) at 100% humidity, 37°C, and 5% . Cells were maintained in Hybri-Care medium (ATCC® 46-X™; ATCC) supplemented with EGF (90%) and 10% FBS and subcultured twice a week upon reaching confluence to maintain exponential growth phase using warm PBS (pH 7.4; Invitrogen) and 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA) solution (Invitrogen).
Cytotoxicity Assay
The cytotoxicity assays were performed using neutral red dye uptake (NRU) as an indicator of cell viability. NRU is one of the most widely applied in vitro cytotoxicity assays with numerous biomedical (Cavanaugh et al. 1990) and environmental applications (Llorente et al. 2012; Sawyer 1995). The assays were carried out using a previously described method for Caco-2 cells (Leusch et al. 2014) with minor modifications for FHs 74 Int cells. Briefly, each of the tested DBPs was prepared as a concentrated stock in methanol (MeOH) up to a concentration of or, in the case of DBBQ, to the limit of solubility (). On day 1 of the assay, plates were seeded at a density of per well () in clear, sterile, flat-bottom 96-well microtiter plates (Greiner Bio-One CELLSTAR®; catalog no. 655-180), using PBS (pH 7.4; Invitrogen), 0.25% (wt/vol) trypsin/EDTA (Invitrogen) solution and growth medium (Hybri-Care medium) supplemented with EGF (90%, Thermo Fisher Scientific) and 10% FBS (Gibco). Eighteen hours later, the growth medium was removed by use of a vacuum aspirator, and wells were washed twice with of warm (37°C) PBS (pH 7.4). The assay medium, spiked with serially diluted DBPs, was added into the test wells to a total volume of per well. After of incubation at 37°C and 5% , cells were again washed with PBS (), of NR solution (0.33%, in DPBS) was added and the plate incubated for . Finally, the NR solution was aspirated from the wells, cells were gently washed with warm PBS ( per well), of NR desorbing fixative (50% EtOH/, 1% acetic acid) was added, and the plate was incubated for 5 min at room temperature. Absorbance was measured at using a FLUOstar Omega® (BMG LABTECH) plate reader.
A cytotoxicity concentration–effect curve was generated using FHs 74 Int cells for each of the DBPs combining the data from all the individual runs (; a minimum of two individual runs on two separate days) (Figure 1). Absolute absorbance values were converted to percent mean absorbance of untreated cell control wells (i.e., percent negative control) by first subtracting the mean background absorbance from the absolute absorbance value of each test well, then dividing the resulting value by the mean absorbance of the negative control, and finally multiplying by 100. Data were normalized in GraphPad Prism for Windows (version 6.05; GraphPad) using the program’s Normalize function to standardize slight fluctuations between each run, and the percentages of negative control values were plotted against the log concentration (M). The median inhibition concentrations () for each of the DBPs were then calculated using Equation 1 in GraphPad Prism 6.05 for Windows, anchoring the bottom constraint to 0% and the top to 100%.
| (1) |
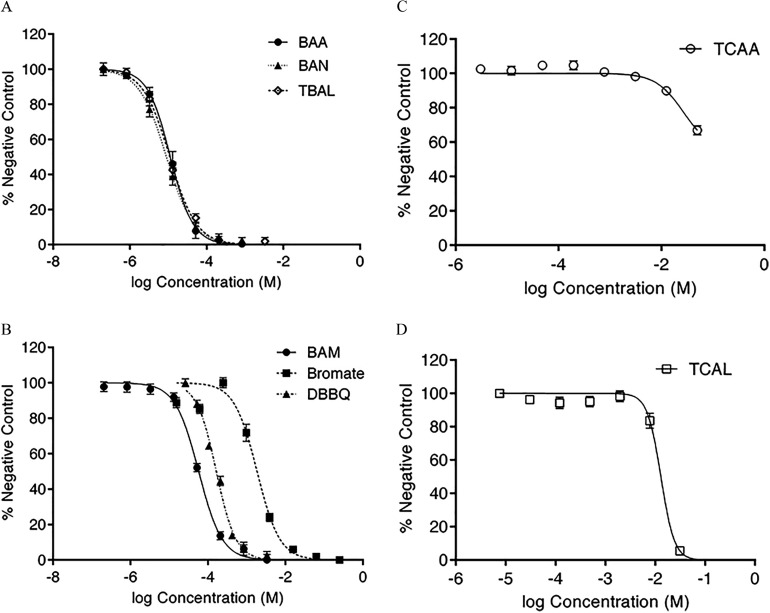
Figure 1.
FHs 74 Int acute cytotoxicity [ neutral red dye uptake (NRU) test; ] concentration–effect curves for: (A) bromoacetic acid (BAA), bromoacetonitrile (BAN), and tribromoacetaldehyde (TBAL); (B) bromoacetamide (BAM), bromate, and dibromobenzoquinone (DBBQ); (C) trichloroacetic acid (TCAA); and (D) trichloroacetaldehyde (TCAL), presented as percent negative control (unexposed cells). Each point is the average of two to three independent runs on separate . Calculated 10% inhibitory concentration () values from these concentration–effect curves are presented in Table 1.
The value was calculated from those parameters by use of Equation 2:
| (2) |
The values for all eight DBPs obtained with FHs 74 Int cells in this study were compared to previously published values in CHO cells (Table 5 in Wagner and Plewa 2017) by correlation analysis [Pearson Product Moment Correlation in SPSS Statistics for Windows (version 22; IBM Corporation)].
Exposure and RNA Preparation
FHs 74 Int cells (passages 3–7) were seeded in 6-well plates at a density of per well prior to treatment with the DBPs. Each well received an concentration (Table 1) of test DBP or vehicle control, in duplicate, and was incubated at 37°C for . The treatment time was selected empirically based on experiments on the induction of genomic DNA damage by DBPs in CHO cells. That data demonstrated that a period allowed for the induction of DNA damage before the effect of DNA repair was observed (Komaki et al. 2009). In addition, the exposure time was established in other studies on the toxic mode of action by DBPs by use of CHO (Dad et al. 2013; Komaki et al. 2014) and FHs 74 Int cells (Pals et al. 2013). Finally, using CHO cells, the treatment period was established as a standard procedure to determine genomic DNA damage across a wide range of DBP chemical classes (Wagner and Plewa 2017).
Table 1.
Equipotent 10% inhibitory concentration () values and numbers of differentially expressed genes (DEGs) identified for the tested disinfection by-product (DBP).
| DBP | Log () (mol/L) () | (mol/L) | No. of DEGs | ||||
|---|---|---|---|---|---|---|---|
| All FCs () | All FCs () | () | |||||
| All | Up | Down | |||||
| BAA | 2,612 | 450 | 417 | 179 | 238 | ||
| BAN | 2,614 | 451 | 406 | 176 | 230 | ||
| DBBQ | 2,614 | 356 | 334 | 156 | 178 | ||
| BAM | 2,610 | 381 | 367 | 164 | 203 | ||
| 2,615 | 398 | 370 | 157 | 213 | |||
| TBAL | 2,620 | 412 | 383 | 170 | 213 | ||
| TCAA | 2,604 | 266 | 201 | 82 | 119 | ||
| TCAL | 2,599 | 269 | 175 | 67 | 108 | ||
Note: values were calculated from the concentration–effect curves presented in Figure 1, based on the cytotoxicity assay in FHs 74 Int cells [ neutral red dye uptake (NRU) test; ). DEGs obtained by rank product analysis (1,500 permutations, ). BAA, bromoacetic acid; BAM, bromoacetamide; BAN, bromoacetonitrile; , bromate; DBBQ, dibromobenzoquinone; SE, standard error; TBAL, tribromoacetaldehyde; TCAA, trichloroacetic acid; TCAL, trichloroacetaldehyde.
The wells were then washed with warm PBS and cells were lysed with QIAzol (Qiagen) ( per well), collected in microcentrifuge tubes, and frozen () overnight. The top aqueous layer was then used for RNA extraction using an RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol with minor modification. In brief, the cell lysates in tubes were brought to room temperature (), homogenized by vortexing for 1 min, and placed on the benchtop for 5 min. Chloroform [, molecular biology (MB) grade, Sigma-Aldrich] was added to each tube, which was then shaken vigorously for and placed on the benchtop for 2–3 min. The lysates were then centrifuged for 15 min at at 4°C to ensure efficient phase separation. A aliquot of the top aqueous layer of each sample was then carefully transferred to a fresh, nuclease-free, microcentrifuge tube, to which of 100% ethanol (MB grade, Sigma-Aldrich) was added and mixed thoroughly by pipetting up and down several times. Each sample () was then immediately loaded onto the RNeasy Mini spin column and centrifuged at for at room temperature. The flow-through was discarded and the process repeated until all of the sample was processed. Next, of Buffer RPE (from the RNeasy Mini Kit) was added to each spin column and centrifuged at for to wash the column, and the flow-through was discarded. Another of Buffer RPE was added to each spin column and centrifuged at for 2 min to dry the spin column membrane. The spin column was placed into a fresh collection tube and centrifuged at full speed for 1 min. Finally, each spin column was transferred into a fresh microcentrifuge tube, of nuclease-free water was pipetted directly onto the spin column membrane, and the column was centrifuged at for 1 min to elute the RNA. This final process was repeated with an additional of nuclease-free water, for a total of of total RNA extract.
The yield and purity of extracted RNA was measured spectrophotometrically using a BioSpectrometer® (Eppendorf South Pacific) equipped with a Traycell microliter measurement cell (Hellma GmbH & Co. KG), and RNA integrity was determined at the Ramaciotti Centre for Genomics using a 2100 Bioanalyzer (Agilent Technologies) prior to hybridization and microarray sample analysis (Table 2).
Table 2.
Summary of RNA yield (), purity (absorbance ratios, A260/A280 and A260/A230), and RNA integrity number (RIN).
| RNA extract ID | RNA concentration () | A260/A280 | A260/A230 | RIN |
|---|---|---|---|---|
| BAA 1 | 69.9 | 2.03 | 2.01 | 9.5 |
| BAA 2 | 47.5 | 1.97 | 2.11 | 9.9 |
| BAN 1 | 66.9 | 1.99 | 2.08 | 9.1 |
| BAN 2 | 64.8 | 1.88 | 1.95 | 10.0 |
| DBBQ 1 | 58.3 | 2.04 | 2.24 | 8.4 |
| DBBQ 2 | 62.7 | 2.00 | 2.10 | 9.6 |
| BAM 1 | 72.7 | 2.00 | 2.20 | 8.5 |
| BAM 2 | 57.4 | 2.06 | 1.96 | 9.9 |
| 1 | 66.9 | 2.00 | 2.25 | 9.6 |
| 2 | 79.2 | 1.95 | 2.07 | 9.7 |
| TBAL 1 | 54.8 | 2.08 | 2.12 | 9.5 |
| TBAL 2 | 55.8 | 1.98 | 1.90 | 9.6 |
| TCAA 1 | 67.6 | 1.95 | 2.04 | 9.2 |
| TCAA 2 | 63.7 | 2.10 | 2.01 | 9.4 |
| TCAL 1 | 62.1 | 1.97 | 2.05 | 9.4 |
| TCAL 2 | 58.9 | 2.07 | 2.03 | 9.4 |
| Negative control 1 | 47.8 | 2.06 | 2.12 | 9.7 |
| Negative control 2 | 63.4 | 1.92 | 2.11 | 9.5 |
Note: BAA, bromoacetic acid; BAM, bromoacetamide; BAN, bromoacetonitrile; , bromate; DBBQ, dibromobenzoquinone; TBAL, tribromoacetaldehyde; TCAA, trichloroacetic acid; TCAL, trichloroacetaldehyde.
Microarray Transcriptomics and Statistical Data Analysis
The RNA extracts were analyzed using HumanHT-12 v4 Expression BeadChip arrays (Illumina). Hybridization and scanning were performed at the Ramaciotti Centre for Genomics with the supplied total RNA extracts. The raw fluorescence data was then transformed using the preprocessing variance stabilization algorithm (Lin et al. 2008), base-2 log transformation, and quantile normalization using the lumi package in the Bioconductor application suite (version 3.2) for R statistical programming language (version 3.5, R Development Core Team) (Du et al. 2008; see Supplemental Material for the transformed microarray expression data). Ultimately, sets of statistically significant differentially expressed genes (DEGs) for each sample–control pair () were identified with the Multiple Experiment Viewer (MeV) suite (version 4.90; The Institute for Genomic Research) for Windows (Saeed et al. 2003) using rank product algorithm (Breitling et al. 2004) set to 1,500 random permutations. DEGs with and fold change (FC) were considered statistically significant and were further used in biological context analysis using pathway enrichment.
Confirmatory quantitative real-time polymerase chain reaction.
For comparison, the expression of the gene heme oxygenase 1 (HMOX1) was analyzed by quantitative real-time polymerase chain reaction (qPCR) in a parallel set of experiments with the same concentrations of DBPs and exposure durations. We selected to perform the confirmatory real-time qPCR on HMOX1, which: a) was detected consistently in a quantitative manner in our microarray experiments for all tested DBPs; b) has an established role in the response to oxidative stress (Poss and Tonegawa 1997); and c) was previously shown to be dysregulated in response to inflammation in mice (Takagi et al. 2018). Briefly, we exposed the FHs 74 Int cells to the selected DBPs and extracted total RNA using the methodology described earlier in this study; we then reverse transcribed of the total RNA using the iScript™ cDNA Synthesis Kit (Bio-Rad) and amplified it on a CFX96 Touch™ Real-Time PCR System (Bio-Rad) using iTaq™ Universal SYBR® Green Supermix (Bio-Rad) per the manufacturer’s instructions under the following conditions: initialization , followed by 44 cycles of denaturation , annealing , and extension . Next, we transformed the resulting raw data of triplicate cycle threshold values into relative expression quantities considering the primer amplification efficiencies (E; ) and normalized them using expression values for the ribosomal protein L27 (RPL27) using the method described in Pfaffl (2004), yielding normalized relative quantities (NRQs). These are shown as the mean values () of a minimum of two repeat experiments (Figure 2 and Table 3). Finally, we determined the statistical significance () of the resulting NRQ values by ordinary one-way analysis of variance with Dunnett’s multiple comparison correction method in GraphPad Prism (version 7.05; GraphPad). For comparison, the qPCR NRQ values are equivalent to the FC values obtained from the microarray analysis (Figure 2).
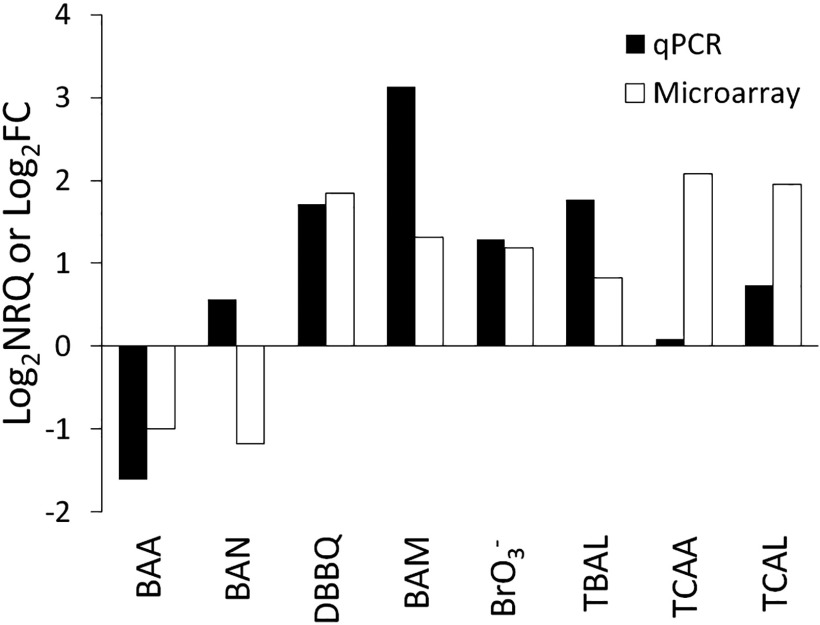
Figure 2.
Gene expression levels of heme oxygenase 1 (HMOX1) following exposure to equipotent concentrations of eight disinfection by-products (DBPs) obtained using quantitative real-time polymerase chain reaction (qPCR) [expressed as relative quantities (); ] and microarray analysis [expressed as fold changes (); ]. Note: BAA, bromoacetic acid; BAM, bromoacetamide; BAN, bromoacetonitrile; , bromate; DBBQ, dibromobenzoquinone; TBAL, tribromoacetaldehyde; TCAA, trichloroacetic acid; TCAL, trichloroacetaldehyde.
Table 3.
Gene expression levels of heme oxygenase 1 (HMOX1) following exposure to equipotent concentrations of eight disinfection by-products (DBPs) obtained using quantitative real-time polymerase chain reaction (qPCR) and microarray.
| Chemical | qPCR | Microarray | ||||
|---|---|---|---|---|---|---|
| NRQ | p-Valuea | FC | p-Valuea | |||
| BAA | 0.33 | 0.0092 | 0.50 | |||
| BAN | 1.47 | 0.56 | 0.0006 | 0.44 | ||
| DBBQ | 3.28 | 1.72 | 3.58 | 1.84 | 0.0002 | |
| BAM | 8.75 | 3.13 | 2.49 | 1.32 | 0.0014 | |
| 2.43 | 1.28 | 0.0001 | 2.27 | 1.19 | 0.0007 | |
| TBAL | 3.41 | 1.77 | 1.76 | 0.82 | 0.0016 | |
| TCAA | 1.06 | 0.09 | ns () | 4.22 | 2.08 | |
| TCAL | 1.67 | 0.74 | 3.87 | 1.95 | ||
Note: BAA, bromoacetic acid; BAM, bromoacetamide; BAN, bromoacetonitrile; , bromate; DBBQ, dibromobenzoquinone; FC, fold change; , fold changes; , relative quantities; NRQ, normalized relative quantity; ns, not significant; TBAL, tribromoacetaldehyde; TCAA, trichloroacetic acid; TCAL, trichloroacetaldehyde.
p-Value for qPCR differential expression based on one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison (); p-value for microarray expression based on rank product analysis () in Multiple Experiment Viewer (MeV) suite (version 4.90) for Windows.
The primer set sequences for HMOX1 were designed using the Primer-BLAST (NCBI) tool (Ye et al. 2012) (forward primer: 5′-ACTCCCTGGAGATGACTCCC-3′; reverse primer: 5′-GGGGGCAGAATCTTGCACTT-3′), and for RPL27, adopted from Ersahin et al. (2014) (forward primer: 5′-ATCGCCAAGAGATCAAAGATAA-3′; reverse primer: 5′-TCTGAAGACATCCTTATTGACG-3′). Both primer sets were synthesized commercially (GeneWorks) and evaluated for amplification efficiency (E) (95.8 and 90.5% for HMOX1 and RPL27, respectively).
Hierarchical Clustering and Biological Context Analysis
Agglomerative hierarchical clustering analysis of the DEG data was performed using unweighted pair-group averages and Pearson’s correlation coefficient (XLSTAT version 2016 for Windows; Addinsoft).
To investigate biological significance of the identified deregulated genes, we enriched the resulting gene sets, represented by the Illumina gene identifiers (; ), using tools available in GeneGo’s MetaCore bioinformatics suite (version 6.33 build 69110, Clarivariate Analytics; see Supplementary Material for MetaCore output file). From the obtained data, we focused on statistically significant toxicity networks (). This allowed us to identify a small number of altered biological processes indicative of the mechanisms of toxicity of the selected DBPs.
Quality Assurance and Quality Control
We performed all the exposure tests with careful consideration of in vitro quality assurance and quality control measures, which included replicate wells for each of the tested concentrations, at least one independent replicate run on a separate day, multiple cell-free wells to correct for baseline variability and to serve as a negative control for DEG determination, multiple wells containing cell culture media only, and solvent control wells. In addition, we employed nontransformed human cells in order to prevent altered gene expression associated with neoplastic cell lines.
Each RNA extract was tested for purity and integrity using the Agilent 2100 electrophoresis bioanalyzer by the Ramaciotti Centre for Genomics, and only those with RNA integrity numbers were subsequently used in the microarray hybridization (Schroeder et al. 2006) (see Table 2 for details).
Results
Cytotoxicity Assay
The 10% inhibition concentration () values of the cytotoxicity assay are summarized in Table 1 (see Figure 1 for concentration–effect curves). The values for each of the tested DBPs were subsequently used in exposure treatments () of the same cell culture for microarray analysis. As values ranged over a factor of 10,000, dosing equimolar concentrations would not have yielded comparable gene expression levels. These DBP concentrations and their resulting cytotoxicity in FHs 74 Int cells were highly correlative with the published median lethal concentration ( values using CHO cell cytotoxicity analyses (; ; ) (Wagner and Plewa 2017).
Global Gene Expression Microarray Analysis
GE analysis using cDNA microarray revealed that only a small subset of genes was affected by treatment with the selected DBPs at concentrations. From the total of 47,231 gene probes corresponding to 23,775 genes annotated to Illumina tags (ILMN_ID) by GeneGo’s MetaCore, less than 10% were identified as differentially expressed (Table 1). A slight dissymmetry toward down-regulation was observed, ranging between 1.1 (DBBQ) and 1.6 (TCAL).
Treatment with BAN resulted in the highest number of DEGs (; ), followed by BAA, TBAL, , BAM, DBBQ, TCAA, and, finally, TCAL (Table 1). Treatment with Br-DBPs resulted in up to 2-fold higher numbers of DEGs () than the chlorine-substituted Cl-DBPs (Table 1). All DEGs with and were analyzed by hierarchical clustering, and subsequent biological context analysis (i.e., gene and pathway enrichment analysis) was performed with DEGs relevant to each individual cluster group, separately.
The change in HMOX1 expression upon exposure to the different DBPs in this study was confirmed by qPCR, and both the qPCR and microarray data were in good agreement for this gene (paired t-test, ; Figure 2).
Hierarchical Clustering and Biological Context Analysis
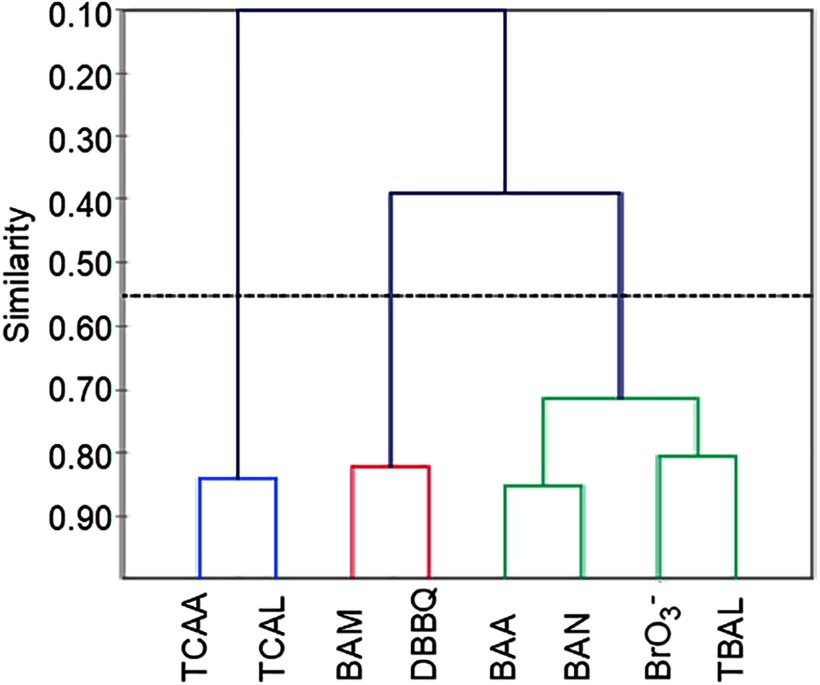
Similarity-based hierarchical clustering identified three main groups (Figure 3): Cluster 1 consisted solely of the two Cl-DBPs (TCAA and TCAL), while Cluster 2 and Cluster 3 incorporated the remaining six Br-DBPs. Cluster 2 contained BAM and DBBQ, and Cluster 3 contained BAA, BAN, , and TBAL (Figure 3).
Figure 3.
Hierarchical clustering of differentially expressed genes (DEGs) based on Pearson similarity coefficient. Cluster 1: TCAA, trichloroacetic acid; TCAL, trichloroacetaldehyde. Cluster 2: BAM, bromoacetamide; DBBQ, dibromobenzoquinone. Cluster 3: BAA, bromoacetic acid; BAN, bromoacetonitrile; , bromate; TBAL, tribromoacetaldehyde.
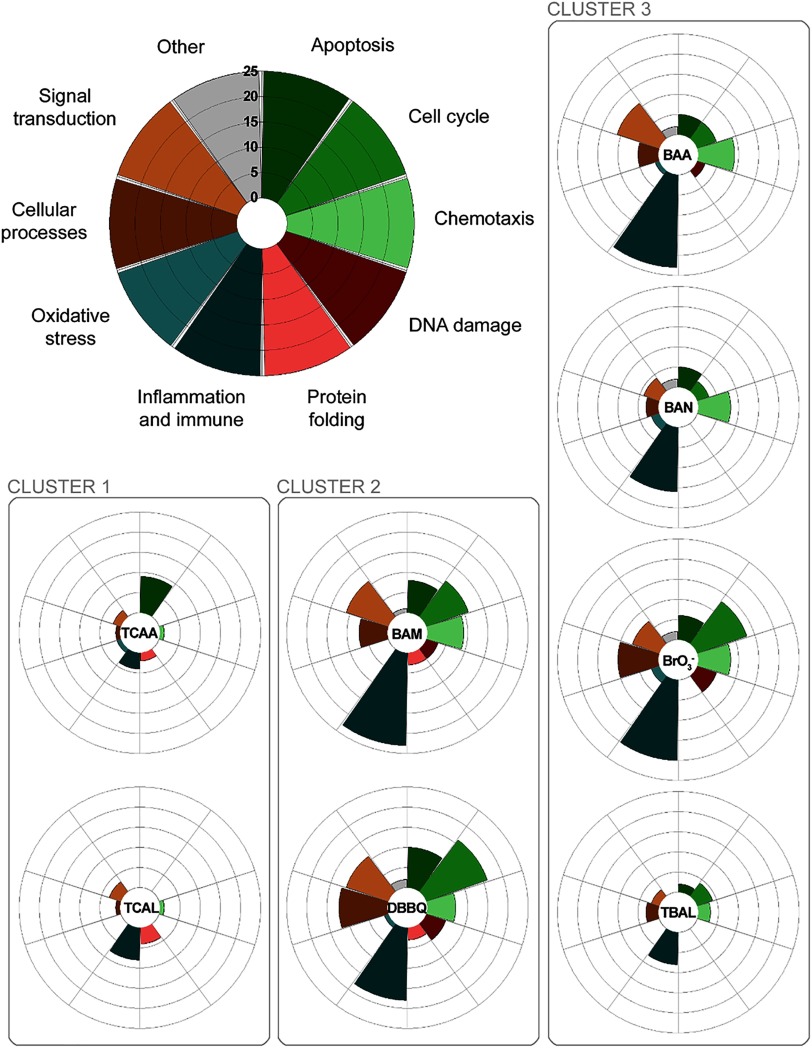
The results of querying GeneGo’s toxicity network libraries, using previously identified DEG signatures, were used to assign the biological context of genes and pathways significantly altered by exposure to the studied DBPs. These were subsequently grouped according to the results of hierarchical clustering analysis (Table 4). This analysis revealed similarities, but also differences, in altered pathways between the three clusters. While there was evidence of effects on oxidative stress pathways in DBPs from all three clusters, the number of altered genes associated with oxidative stress was much lower than other pathways (Figure 4). Most notably, all three groupings exhibited a comparatively large number of altered genes related to inflammation and immune responses (Table 4).
Table 4.
Cellular processes and associated toxicity networks () with up- and down-regulated genes (), fold change (FC) identified using GeneGo’s MetaCore toxicity network enrichment tool.
| Cluster | Affected cellular process | Dominant toxicity network(s) | Upregulated genes | Downregulated genes |
|---|---|---|---|---|
| Cluster 1 | Chemotaxis | MAPK cascades | HMOX1, HSP70 (HSPA1A, HSPA1B), CRK, GRP78 | IRF1 |
| Inflammation/immune response | Antigen presentation/MHC class 1 signaling/MAPK signaling | HMOX1, HSP70, HLA-A, GRP78 | IRF1, BDNF | |
| Protein folding | Unfolded protein response (UPR) via heat shock protein (HSP) 70, HSP90, and p53 | HSP70 (HSPA1A, HSPA1B, HSPA6), HSP40 (DNAJB1), Aha1 (AHSA1), GRP78 | HSP10 (mitochondrial) | |
| Apoptosis | TNFR signaling | APAF-1 | BIRC2, BIRC3 | |
| Oxidative stress response | Nrf2 regulation | HMOX1, TXNRD1, GSTM3 | — | |
| Cluster 2 | Inflammation | IL-1 pro-inflammatory signalling | HMOX1, COX-2, IRAK2, NF-kB, C/EBPbeta, IL-1RI, IL-1b, IL-1a, I-kB (NFKBIA, NFKBIE) | AP-1, endothelin 1 (EDN1) |
| IL-6 receptor anti-inflammatory response | IL-6, IRAK2, CXCL2, CXCL5, GNA13, NF-kB, HRH1, JAK1, I-kB (NFKBIA, NFKBIE), CXCL1 | AP-1, PI3K reg class IA (p85) | ||
| Chemotaxis | MAPK cascades/GRO signaling | HSP70 (HSPA6, HSPA1L, HSPA4L, HSPA1A, HSPA1B), HMOX1, IRAK2, IL-1a, IL-1b, IL-1IR, NF-AT2, JAK1, CRK | AP-1 | |
| Cell cycle dysregulation | Signaling to E2F | c-Abl (ABL1) | H2AX, GADD45a, GADD45b, GADD45g, cyclin D, cyclin E, cyclin A, CDC45L, MCM6, MAP3K, AP-1, FEN1, CDK1 (p34) | |
| APC Regulation of G1/S | I-kB (NFKBIA, NFKBIE), CDC34 | KPNA2, GADD45a, GADD45b, GADD45g, cyclin D, cyclin E, cyclin A, CDC25A, p21, CDK1 (p34) | ||
| Protein folding | UPR via HSP90 | CRYAB, HSP40 (DNAJB1), HSP105 (HSPH1), HSP90AB1, DNAJA1, AHSA1, HSPB8, HSP70 (HSPA1A) | — | |
| Apoptosis | MAPK cascades (MAPK4 & MAPK9) | c-Abl (ABL1), IL-1b, IL-1a, IL-1RI, IRAK2, HMOX1 | GADD45a, GADD45b, AP-1, endothelin 1 (EDN1), CDK1 (p34) | |
| DNA damage response | Inhibition of apoptosis, dysregulation of cell cycle, up-regulation of double-stranded DNA repair | NF-kB | CDK1 (p34), AP-1, PCNA, GADD45a, GADD45b | |
| Cluster 3 | Inflammation/immune response | IL-1 pro-inflammatory signaling/IL-6 signaling | COX-2, HMOX1, IL-1a, IL1-b, NF-kB, IRAK2, C/EBPbeta, IRF1, IL-6, I-kB, IL4R, IL13RA2, JAK1, CXCL2 | ERK1, AP-1, HMOX1, IL13RA1, PI3K reg class IA |
| Chemotaxis | HGF signaling, Cell communication | COX-2, CXCL1, EGFR, IL-8, CXCL5 | AP-1, PI3K reg class IA, calmodulin | |
| Cell cycle dysregulation | Signaling to E2F via cyclin D and cyclin E | BCAR1, MEKK4 (MAP3K4) | GADD45b, GADD45a, AP-1, MCM3, cyclin D, cyclin E, CDC45L, PCNA, TCF | |
| Signal transduction | Signaling via IL-1b and IRF1 | COX-2, IL-1a, IL-1b, HMOX1, NF-kB, IRAK2, IRF1, IFN-α/β receptor, ISG15, CCL5 | HMOX1, AP-1, calmodulin | |
| Proliferation induction | PDGF signaling | COX-2, NF-kB p50/RelB, PA24A, PDGF-C | AP-1, ERK1, calmodulin | |
| Apoptosis | MAPK cascades | IL-1a, IL-1b, IRAK2, MEKK4 (MAP3K4), HMOX1 | GADD45a, GADD45b, AP-1, HMOX1 | |
| Oxidative stress response | HNF4 regulation | COX-2, SOD2, HMOX1, TXNRD1, SMAD3 | AP-1, PRDX5, HMOX1 |
Note: Clusters (outlined in Figure 3) are defined as follows: Cluster 1: TCAA, trichloroacetic acid; TCAL, trichloroacetaldehyde; Cluster 2: BAM, bromoacetamide; DBBQ, dibromobenzoquinone; Cluster 3: BAA, bromoacetic acid; BAN, bromoacetonitrile; , bromate; TBAL, tribromoacetaldehyde. —, no data; IL, interleukin; MAPK, mitogen-activated protein kinase, Nrf2, nuclear factor (erythroid-derived 2)-like.
Figure 4.
Number of significantly affected GeneGo toxicity networks in 10 functional categories (apoptosis, cell cycle, chemotaxis, DNA damage, protein folding, inflammation and immune response, oxidative stress, cellular processes, signal transduction, and others) for each cluster of disinfection by-products (DBPs). The farther away the section expands from the center of each radar plot, the more toxicity networks were affected (on a scale from 0 to 25). Cluster 1: TCAA, trichloroacetic acid; TCAL, trichloroacetaldehyde. Cluster 2: BAM, bromoacetamide; DBBQ, dibromobenzoquinone. Cluster 3: BAA, bromoacetic acid; BAN, bromoacetonitrile; , bromate; TBAL, tribromoacetaldehyde.
Discussion
The most recent working hypothesis is that DBPs primarily cause adverse effects through mechanisms related to the production of ROS, which subsequently result in the induction of oxidative stress pathways (Stalter et al. 2016). Our observation of comparatively few altered genes associated with oxidative stress (Figure 4) is therefore an interesting outcome and potentially very important for understanding DBP toxicity. It was recently proposed that ROS formation is not the sole mechanism of DBP toxicity per se (Procházka et al. 2015), since ROS can arise as a physiological response resulting from other forms of cellular dysfunction caused by chemical insult (Reuter et al. 2010). Indeed, while it is generally agreed that cells respond to DBPs through pathways sharing ROS-mediated mechanisms (Stalter et al. 2016; Procházka et al. 2015; Luo et al. 2017), it is becoming clearer that the specific genes associated with oxidative stress involve additional layers of complexity (Pals et al. 2013). One key piece of the puzzle therefore involves identifying the functional basis of ROS production elicited by DBP exposure (Pals et al. 2013). Untargeted transcriptomic analysis offers a powerful means to broadly identify DBP-responsive genes and thus reveal other deregulations that might be associated with ROS production and the manifestation of oxidative stress responses. Results of the present study offer compelling evidence suggesting a role of pro- and anti-inflammatory response pathways, which we hypothesize forms a significant aspect of the documented cellular injury in DBP-exposed cells.
We observed, in a nontransformed human cell line, FHs 74 Int, the induction of several genes associated with oxidative stress pathways in DBPs from all three clusters, which is consistent with the findings of preceding literature using the same cell line (Pals et al. 2013; Attene-Ramos et al. 2010). However, in all cases, the number of DEGs and pathways associated with inflammation and immune responses were by far the most prominent (Figure 4). Clear differences were observed between the less toxic Cl-DBPs (Cluster 1) and more toxic Br-DBPs (Clusters 2 and 3). This may suggest a more complex and mechanistically distinct set of early cellular responses associated with Br-DBPs and supports existing evidence of a lower toxicity risk for their chlorinated analogs (Procházka et al. 2015; Plewa and Wagner 2015; Wagner and Plewa 2017). For Br-DBPs in particular, we hypothesize an increased risk of genotoxicity associated with considerable activation of inflammatory responses, for example, characterized by the production of pro-inflammatory (e.g., interleukin 1; IL-1) and anti-inflammatory (e.g., IL-6) cytokines. It is plausible that such responses represent a key mechanism initiating various downstream pathways (van der Veen et al. 2016), including the subsequent generation of ROS (Pals et al. 2013, 2017). Based on concurrent up-regulation of nuclear factor kappa B (Nf-kB) and mitogen-activated protein kinase (MAPK) pathways, we speculate that this could involve downstream activation of Toll-like receptors (TLRs) (van der Veen et al. 2016; Gilbert et al. 2004). While it is unlikely that Br-DBPs interact directly with such cellular surface receptors, TLR signaling pathways can be activated by production of pro-inflammatory cytokines (Ceribelli 2016), potentially resulting in chronic inflammation and increased production of ROS (Lucas and Maes 2013). The process whereby activation of TLR pathways contributes to tumorigenesis has been relatively well characterized, albeit not in relation to DBP exposure (Rakoff-Nahoum and Medzhitov 2009; Wang et al. 2014). As indicated, this is merely speculation based on the observed results in relation to existing literature surrounding TLR signaling pathways, and thus, further research is needed to explore this hypothesis and further reveal the mechanistic basis of genotoxicity and potential carcinogenicity related to DBP exposure.
There is a growing realization that inflammatory response pathways are important contributors and regulators of a diverse range of adverse toxicity outcomes (Angrish et al. 2016; Villeneuve et al. 2018). Earlier research has established a strong association between inflammatory networks and ROS production and revealed a high level of interconnectivity that could perpetuate the oxidative damage associated with inflammation responses (Reuter et al. 2010). Observed activation of anti-inflammatory response pathways alongside the pro-inflammatory response is most likely a result of a feedback mechanism, which, in the absence of additional toxic insult or injury, may ultimately lead to homeostasis and recovery (Medzhitov 2010). However, chronic inflammation and oxidative stress may pose an enhanced risk of activating various associated downstream pathways, due to what has been termed a “vicious cycle” of adaptive responsiveness (Federico et al. 2007). To clarify, inflammatory responses induce the production of ROS, and the resulting ROS lead to further production of intermediaries that, in turn, induce additional inflammation (Reuter et al. 2010). Oxidative stress–related xenobiotic-induced ROS production may therefore help explain the various other affected pathways, for example, cell proliferation and regulation of apoptosis (Burdon 1995).
By increasing cellular level of oxidants, many xenobiotics alter gene expression via activation of cellular signaling pathways, including adenylyl cyclase pathway, calcium-dependent signaling pathways, and transcription factors (TFs) such as nuclear factor (erythroid-derived 2)-like (Nrf2), activator protein 1 (AP-1) and NF-kB. Other pathways reportedly influenced by ROS-mediated oxidative stress include altered expression of MAPKs, for example, extracellular signal-regulated kinases (ERKs), c-Jun N-terminal kinases (JNKs), and p38 kinases (Amstad et al. 1992; Angel and Karin 1991; Brown et al. 1998; Gius et al. 1999). Indeed, these various pathways are highly consistent with the DBP-responsive genes (most notably for Br-DBPs in Clusters 2 and 3) identified using microarray analysis of FHs 74 Int cells in the present study. Importantly, many of these pathways could be activated either indirectly or directly as a consequence of alterations to genes associated with inflammation and immune responses.
The use of nontransformed human epithelial cells and untargeted transcriptomics highlighted this connection where many other studies have not, probably due to the greater scope for cellular transcriptional responses compared with more targeted transformed cell lines (Zhang et al. 1997; Hoheisel 2006; Chang et al. 2013). A recent study using a transformed human uroepithelial cell line (SV-HUC-1) observed increased expression of several Nrf-2 TF-mediated oxidative stress–response genes, including PTGS2 and HMOX1 (Li et al. 2018), as did our previous work with Caco-2 cells (Pals et al. 2013; Procházka et al. 2015).
The present study evaluated individual DBPs to explore differences in gene expression and subsequently compare the mechanistic basis of toxicity. One limitation of this study is the lack of PCR confirmation of genes of interest other than HMOX1, which we were not able to include due to budgetary constraints. While we do delve into individual DEGs in the discussion, our conclusions are based on analyses of whole pathways, integrative of multiple DEGs, thus providing a degree of resilience against potential occasional inaccuracies in microarray gene expression data. Still, confirmation by qPCR of individual genes affected by exposure to the DBPs highlighted in this study would be warranted in the future.
Future research is now needed to investigate the potential augmented risk associated with the presence of DBPs as complex mixtures (Teuschler and Simmons 2003; Massalha et al. 2018; Dong et al. 2017; Plewa et al. 2017). Information regarding chemical mode of action, such as that provided by the present study, is critical for determining whether there is a likelihood of enhanced toxicity due to mixture effects (Qin et al. 2011). Despite the identification of three distinct clusters and notable differences between the toxicity and gene expression profiles of Br- and Cl-DBPs, there were similarities in the transcriptional responses of FHs 74 Int cells to all compounds (Figure 4). It is therefore likely that enhanced toxicity might occur from exposure to a mixture compared to individual DBPs (Groten 2000; Yeatts et al. 2010). The observed differences between Cl-DBPs and Br-DBPs may suggest unique mechanisms of action for these compounds. However, it is also possible that the lower toxicity of Cl-DBPs allows cells to compensate through protein repair mechanisms (e.g., heat shock proteins), whereas the greater toxicity of Br-DBPs overloaded such compensatory mechanisms, resulting in subsequent activation of other more damaging signaling pathways, including inflammatory responses.
Conclusions
Gene expression–based toxicogenomic analysis can be a sensitive and robust tool for comparative assessment of biological activity of chemical compounds. However, several crucial factors must be considered to obtain meaningful data. For example, the provision of concentrations from cytotoxicity data employing specific and constant exposure times to DBPs at predetermined equipotent concentrations was an important step aimed at reducing the possibility of transcriptome alterations associated with dead or dying cells. Additionally, where many in vitro bioassays use tumor cell lines because of their rapid growth and ease of maintenance, nontransformed human cell lines have the added advantage of further avoiding erroneous gene expression profiles associated with neoplastic cell lines. Finally, the FHs 74 Int cell line offers the additional advantage of being very well suited for evaluating effects on immunomodulatory and inflammatory response pathways. With these strengths, our results offer considerable evidence that ROS-mediated oxidative stress pathways may be associated with inflammatory response pathways, which could contribute to a cycle of toxic insult. Considering the well-documented relationship between inflammation and cancer progression, further research exploring this relationship is warranted. Effects of individual DBPs are needed to unravel mechanistic information, but there is clear potential for complex mixtures to occur, and the toxicity of relevant DBP mixtures must be investigated in the future.
Supplementary Material
Acknowledgments
We thank Dr. Vinod Gopalan for technical assistance and Dr. Peta Neale for her assistance and thoughtful advice on various elements of the data analysis and manuscript preparation.
Footnotes
Supplemental Material is available online (https://doi.org/10.1289/EHP4945).
The authors declare they have no actual or potential competing financial interests.
Note to readers with disabilities: EHP strives to ensure that all journal content is accessible to all readers. However, some figures and Supplemental Material published in EHP articles may not conform to 508 standards due to the complexity of the information being presented. If you need assistance accessing journal content, please contact ehponline@niehs.nih.gov. Our staff will work with you to assess and meet your accessibility needs within 3 working days.
References
- Amstad PA, Krupitza G, Cerutti PA. 1992. Mechanism of c-fos induction by active oxygen. Cancer Res 52(14):3952–3960, PMID: 1617671. [PubMed] [Google Scholar]
- Angel P, Karin M. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta 1072(2–3):129–157, PMID: 1751545, 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- Angrish MM, Pleil JD, Stiegel MA, Madden MC, Moser VC, Herr DW. 2016. Taxonomic applicability of inflammatory cytokines in adverse outcome pathway (AOP) development. J Toxicol Environ Health A 79(4):184–196, PMID: 26914248, 10.1080/15287394.2016.1138923. [DOI] [PubMed] [Google Scholar]
- Attene-Ramos MS, Wagner ED, Plewa MJ. 2010. Comparative human cell toxicogenomic analysis of monohaloacetic acid drinking water disinfection byproducts. Environ Sci Technol 44(19):7206–7212, PMID: 20540539, 10.1021/es1000193. [DOI] [PubMed] [Google Scholar]
- Bougeard CM, Goslan EH, Jefferson B, Parsons SA. 2010. Comparison of the disinfection by-product formation potential of treated waters exposed to chlorine and monochloramine. Water Res 44(3):729–740, PMID: 19910014, 10.1016/j.watres.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573(1–3):83–92, PMID: 15327980, 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Brown JR, Nigh E, Lee RJ, Ye H, Thompson MA, Saudou F, et al. 1998. Fos family members induce cell cycle entry by activating cyclin D1. Mol Cell Biol 18(9):5609–5619, PMID: 9710644, 10.1128/mcb.18.9.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon RH. 1995. Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Radic Biol Med 18(4):775–794, PMID: 7750801, 10.1016/0891-5849(94)00198-s. [DOI] [PubMed] [Google Scholar]
- Busby WF Jr, Ackermann JM, Crespi CL. 1999. Effect of methanol, ethanol, dimethyl sulfoxide, and acetonitrile on in vitro activities of CDNA-expressed human cytochrome p-450. Drug Met Disp 27:246–249, PMID: 9929510. [PubMed] [Google Scholar]
- Calderon RL. 2000. The epidemiology of chemical contaminants of drinking water. Food Chem Toxicol 38(suppl 1):S13–20, PMID: 10717366, 10.1016/s0278-6915(99)00133-7. [DOI] [PubMed] [Google Scholar]
- Cavanaugh PF Jr, Moskwa PS, Donish WH, Pera PJ, Richardson D, Andrese AP. 1990. A semi-automated neutral red based chemosensitivity assay for drug screening. Invest New Drugs 8(4):347–354, PMID: 2084068, 10.1007/bf00198590. [DOI] [PubMed] [Google Scholar]
- Ceribelli A. 2016. Environment and autoimmunity: facts and gaps. In: Environmental Influences on the Immune System. Esser C, ed. Vienna, Austria: Springer-Verlag, 305–320. [Google Scholar]
- Chang CW, Chen CR, Huang CY, Shu WY, Chiang CS, Hong JH, et al. 2013. Comparative transcriptome profiling of an SV40-transformed human fibroblast (MRC5CVI) and its untransformed counterpart (MRC-5) in response to UVB irradiation. PLoS One 8(9):e73311, PMID: 24019915, 10.1371/journal.pone.0073311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens L, Werb Z. 2002. Inflammation and cancer. Nature 420(6917):860–867, PMID: 12490959, 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Paules RS. 2010. Use of transcriptomics in understanding mechanisms of drug-induced toxicity. Pharmacogenomics 11(4):573–585, PMID: 20350139, 10.2217/pgs.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler D, Miller G. 2005. The role of public health improvements in health advances: the twentieth-century United States. Demography 42(1):1–22, PMID: 15782893, 10.1353/dem.2005.0002. [DOI] [PubMed] [Google Scholar]
- Dad A, Jeong CH, Pals JA, Wagner ED, Plewa MJ. 2013. Pyruvate remediation of cell stress and genotoxicity induced by haloacetic acid drinking water disinfection by-products. Environ Mol Mutagen 54(8):629–637, PMID: 23893730, 10.1002/em.21795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Masalha N, Plewa MJ, Nguyen TH. 2017. Toxicity of wastewater with elevated bromide and iodide after chlorination, chloramination, or ozonation disinfection. Environ Sci Technol 51(16):9297–9304, PMID: 28691804, 10.1021/acs.est.7b02345. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. 2008. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24(13):1547–1548, PMID: 18467348, 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Du H, Li J, Moe B, McGuigan CF, Shen S, Li XF. 2013. Cytotoxicity and oxidative damage induced by halobenzoquinones to T24 bladder cancer cells. Environ Sci Technol 47(6):2823–2830, PMID: 23368424, 10.1021/es303762p. [DOI] [PubMed] [Google Scholar]
- Ersahin T, Carkacioglu L, Can T, Konu O, Atalay V, Cetin-Atalay R. 2014. Identification of novel reference genes based on mesh categories. PLoS One 9(3):e93341, PMID: 24682035, 10.1371/journal.pone.0093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher BI, Dutt M, Maylin E, Tang JYM, Toze S, Wolf CR, et al. 2012. Water quality assessment using the AREc32 reporter gene assay indicative of the oxidative stress response pathway. J Environ Monit 14(11):2877–2885, PMID: 23032559, 10.1039/c2em30506b. [DOI] [PubMed] [Google Scholar]
- Escobar-Hoyos LF, Hoyos-Giraldo LS, Londoño-Velasco E, Reyes-Carvajal I, Saavedra-Trujillo D, Carvajal-Varona S, et al. 2013. Genotoxic and clastogenic effects to monohaloacetic acid drinking water disinfection by-products in primary human lymphocytes. Water Res 47(10):3282–3290, PMID: 23602619, 10.1016/j.watres.2013.02.052. [DOI] [PubMed] [Google Scholar]
- Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. 2007. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer 121(11):2381–2386, PMID: 17893868, 10.1002/ijc.23192. [DOI] [PubMed] [Google Scholar]
- Gilbert KM, Whitlow AB, Pumford NR. 2004. Environmental contaminant and disinfection by-product trichloroacetaldehyde stimulates T cells in vitro. Int Immunopharmacol 4(1):25–36, PMID: 14975357, 10.1016/j.intimp.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Gius D, B A, Shah S, Curry HA. 1999. Intracellular oxidation/reduction status in the regulation of transcription factors NF-kB and AP-1. Toxicol Lett 106(2–3):93–106, PMID: 10403653, 10.1016/S0378-4274(99)00024-7. [DOI] [PubMed] [Google Scholar]
- Groten JP. 2000. Mixtures and interactions. Food Chem Toxicol 38(suppl 1):S65–S71, PMID: 10717373, 10.1016/s0278-6915(99)00135-0. [DOI] [PubMed] [Google Scholar]
- Hanigan D, Truong L, Simonich M, Tanguay R, Westerhoff P. 2017. Zebrafish embryo toxicity of 15 chlorinated, brominated, and iodinated disinfection by-products. J Environ Sci (China) 58:302–310, PMID: 28774621, 10.1016/j.jes.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Hoheisel JD. 2006. Microarray technology: beyond transcript profiling and genotype analysis. Nat Rev Genet 7(3):200–210, PMID: 16485019, 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- Ioannidou A, Goulielmaki E, Garinis GA. 2016. DNA damage: from chronic inflammation to age-related deterioration. Front Genet 7:187, PMID: 27826317, 10.3389/fgene.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph P. 2017. Transcriptomics in toxicology. Food Chem Toxicol 109(Pt 1):650–662, PMID: 28720289, 10.1016/j.fct.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivusalo M, Vartiainen T, Hakulinen T, Pukkala E, Jaakkola JJ. 1995. Drinking water mutagenicity and leukemia, lymphomas, and cancers of the liver, pancreas, and soft tissue. Arch Environ Health 50(4):269–276, PMID: 7677425, 10.1080/00039896.1995.9935953. [DOI] [PubMed] [Google Scholar]
- Komaki Y, Mariñas BJ, Plewa MJ. 2014. Toxicity of drinking water disinfection by-products: cell cycle alterations induced by monohaloacetonitriles. Environ Sci Technol 48(19):11662–11669, PMID: 25185076, 10.1021/es5032344. [DOI] [PubMed] [Google Scholar]
- Komaki Y, Pals J, Wagner ED, Mariñas BJ, Plewa MJ. 2009. Mammalian cell DNA damage and repair kinetics of monohaloacetic acid drinking water disinfection by-products. Environ Sci Technol 43(21):8437–8442, PMID: 19924981, 10.1021/es901852z. [DOI] [PubMed] [Google Scholar]
- Komaki Y, Plewa MJ. 2017. Investigation of nuclear enzyme topoisomerase as a putative molecular target of monohaloacetonitrile disinfection by-products. J Environ Sci (China) 58:231–238, PMID: 28774614, 10.1016/j.jes.2017.04.024. [DOI] [PubMed] [Google Scholar]
- Leusch FDL, Khan SJ, Laingam S, Prochazka E, Froscio S, Trinh T, et al. 2014. Assessment of the application of bioanalytical tools as surrogate measure of chemical contaminants in recycled water. Water Res 49:300–315, PMID: 24355290, 10.1016/j.watres.2013.11.030. [DOI] [PubMed] [Google Scholar]
- Leusch FDL, Neale PA, Hebert A, Scheurer M, Schriks MCM. 2017. Analysis of the sensitivity of in vitro bioassays for androgenic, progestagenic, glucocorticoid, thyroid and estrogenic activity: suitability for drinking and environmental waters. Environ Int 99:120–130, PMID: 28017361, 10.1016/j.envint.2016.12.014. [DOI] [PubMed] [Google Scholar]
- Li XF, Mitch WA. 2018. Drinking water disinfection byproducts (DBPs) and human health effects: multidisciplinary challenges and opportunities. Environ Sci Technol 52(4):1681–1689, PMID: 29283253, 10.1021/acs.est.7b05440. [DOI] [PubMed] [Google Scholar]
- Li J, Moe B, Liu Y, Li XF. 2018. Halobenzoquinone-induced alteration of gene expression associated with oxidative stress signaling pathways. Environ Sci Technol 52(11):6576–6584, PMID: 29737854, 10.1021/acs.est.7b06428. [DOI] [PubMed] [Google Scholar]
- Lin SM, Du P, Huber W, Kibbe WA. 2008. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res 36(2):e11, PMID: 18178591, 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente MT, Parra JM, Sánchez-Fortún S, Castaño A. 2012. Cytotoxicity and genotoxicity of sewage treatment plant effluents in rainbow trout cells (RTG-2). Water Res 46(19):6351–6358, PMID: 23022116, 10.1016/j.watres.2012.08.039. [DOI] [PubMed] [Google Scholar]
- Lucas K, Maes M. 2013. Roll of the Toll like receptor (TLR) radical cycle in chronic inflammation: possible treatments targeting the TLR4 pathway. Mol Neurobiol 48(1):190–204, PMID: 23436141, 10.1007/s12035-013-8425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Zhai L, Yang H, Xu L, Liu J, Liang H, et al. 2017. Dichloroacetonitrile induces cytotoxicity through oxidative stress-mediated and p53-dependent apoptosis pathway in LO2 cells. Toxicol Mech Methods 27(8):575–581, PMID: 28573904, 10.1080/15376516.2017.1337257. [DOI] [PubMed] [Google Scholar]
- Manasfi T, De Méo M, Coulomb B, Di Giorgio C, Boudenne JL. 2016. Identification of disinfection by-products in freshwater and seawater swimming pools and evaluation of genotoxicity. Environ Int 88:94–102, PMID: 26735347, 10.1016/j.envint.2015.12.028. [DOI] [PubMed] [Google Scholar]
- Massalha N, Dong S, Plewa MJ, Borisover M, Nguyen TH. 2018. Spectroscopic indicators for cytotoxicity of chlorinated and ozonated effluents from wastewater stabilization ponds and activated sludge. Environ Sci Technol 52(5):3167–3174, PMID: 29359929, 10.1021/acs.est.7b05510. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. 2010. Inflammation 2010: new adventures of an old flame. Cell 140(6):771–776, PMID: 20303867, 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Muellner MG, Attene-Ramos MS, Hudson ME, Wagner ED, Plewa MJ. 2010. Human cell toxicogenomic analysis of bromoacetic acid: a regulated drinking water disinfection by-product. Environ Mol Mutagen 51(3):205–214, PMID: 19753638, 10.1002/em.20530. [DOI] [PubMed] [Google Scholar]
- Munson AE, Sain LE, Sanders VM, Kauffmann BM, White KL Jr, Page DG, et al. 1982. Toxicology of organic drinking water contaminants: trichloromethane, bromodichloromethane, dibromochloromethane and tribromomethane. Environ. Health Perspect 46:117–126, PMID: 7151752, 10.2307/3429428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pals J, Attene-Ramos M, Xia M, Wagner E, Plewa M. 2013. Human cell toxicogenomic analysis linking reactive oxygen species to the toxicity of monohaloacetic acid drinking water disinfection byproducts. Environ Sci Technol 47(21):12514–12523, PMID: 24050308, 10.1021/es403171b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pals JA, Wagner ED, Plewa MJ, Xia M, Attene-Ramos MS. 2017. Monohalogenated acetamide-induced cellular stress and genotoxicity are related to electrophilic softness and thiol/thiolate reactivity. J Environ Sci (China) 58:224–230, PMID: 28774613, 10.1016/j.jes.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2004. Quantification strategies in real-time PCR. In: A-Z of Quantitative PCR. Bustin SA, ed. La Jolla, CA: International University Line (IUL), 87–112. [Google Scholar]
- Plewa MJ, Wagner ED, Muellner MG, Hsu KM, Richardson SD. 2008. Comparative mammalian cell toxicity of N-DBPs and C-DBPs. In: Disinfection By-Products in Drinking Water, vol. 995 Karanfil T, Krasner SW, Westerhoff P, Xie Y, eds. Washington, DC: American Chemical Society, 36–50, 10.1021/bk-2008-0995.ch003. [DOI] [Google Scholar]
- Plewa MJ, Wagner ED. 2015. Charting a new path to resolve the adverse health effects of DBPs. In: Recent Advances in Disinfection By-Products, vol. 1190 Karanfil T, Mitch B, Westerhoff P, Xie Y, eds. Washington, DC: American Chemical Society, 3–23, 10.1021/bk-2015-1190.ch001. [DOI] [Google Scholar]
- Plewa MJ, Wagner ED, Richardson SD. 2017. TIC-Tox: a preliminary discussion on identifying the forcing agents of DBP-mediated toxicity of disinfected water. J Environ Sci (China) 58:208–216, PMID: 28774611, 10.1016/j.jes.2017.04.014. [DOI] [PubMed] [Google Scholar]
- Ploeg M, Aben KK, Kiemeney LA. 2009. The present and future burden of urinary bladder cancer in the world. World J Urol 27(3):289–293, PMID: 19219610, 10.1007/s00345-009-0383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Tonegawa S. 1997. Reduced stress defense in heme oxygenase 1-deficient cells. Proc Natl Acad Sci USA 94(20):10925–10930, PMID: 9380736, 10.1073/pnas.94.20.10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo C, Emiliano P, Barceló D, Valero F. 2018. Chemical characterization and relative toxicity assessment of disinfection byproduct mixtures in a large drinking water supply network. J Hazard Mater 359:166–173, PMID: 30025226, 10.1016/j.jhazmat.2018.07.022. [DOI] [PubMed] [Google Scholar]
- Postigo C, Jeong CH, Richardson SD, Wagner ED, Plewa MJ, Simmons JE, et al. 2015. Analysis, occurrence and toxicity of haloacetaldehydes in drinking waters: Iodoacetaldehyde as an emerging disinfection byproduct. Occurrence, Formation, Health Effects, and Control of Disinfection By-Products, Karanfil T, Mitch W, Westerhoff P, Xie Y, Eds. Washington, D.C.: Am Chem Soc, Vol. 1190, pp. 25–43, 10.1021/bk-2015-1190.ch002. [DOI] [Google Scholar]
- Procházka E, Escher BI, Plewa MJ, Leusch FD. 2015. In vitro cytotoxicity and adaptive stress responses to selected haloacetic acid and halobenzoquinone water disinfection byproducts. Chem Res Toxicol 28(10):2059–2068, PMID: 26327680, 10.1021/acs.chemrestox.5b00283. [DOI] [PubMed] [Google Scholar]
- Qin LT, Liu SS, Zhang J, Xiao QF. 2011. A novel model integrated concentration addition with independent action for the prediction of toxicity of multi-component mixture. Toxicology 280(3):164–172, PMID: 21182889, 10.1016/j.tox.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Rahman M, Driscoll T, Cowie C, Armstrong B. 2010. Disinfection by-products in drinking water and colorectal cancer: a meta-analysis. Int J Epidemiol 39(3):733–745, PMID: 20139236, 10.1093/ije/dyp371. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Medzhitov R. 2009. Toll-like receptors and cancer. Nat Rev Cancer 9(1):57–63, PMID: 19052556, 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. 2010. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med 49(11):1603–1616, PMID: 20840865, 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SD, Plewa MJ, Wagner ED, Schoeny R, DeMarini DM. 2007. Occurrence, genotoxicity, and carcinogenicity of regulated and emerging disinfection by-products in drinking water: a review and roadmap for research. Mutat Res 636(1–3):178–242, PMID: 17980649, 10.1016/j.mrrev.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Richardson SD, Postigo C. 2015. Formation of DBPs: state of the science. In: Recent Advances in Disinfection By-Products, vol. 1190 Karanfil T, Mitch WA, Westerhoff P, Xie Y, eds. Washington, DC: American Chemical Society, 189–214, 10.1021/bk-2015-1190.ch011. [DOI] [Google Scholar]
- Rivera-Núñez Z, Wright JM. 2013. Association of brominated trihalomethane and haloacetic acid exposure with fetal growth and preterm delivery in Massachusetts. J Occup Environ Med 55(10):1125–1134, PMID: 24064786, 10.1097/JOM.0b013e3182a4ffe4. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34(2):374–378, PMID: 12613259, 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sawyer TW. 1995. Practical applications of neuronal tissue culture in in vitro toxicology. Clin Exp Pharmacol Physiol 22(4):295–296, PMID: 7671445, 10.1111/j.1440-1681.1995.tb02000.x. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7:3–3, PMID: 16448564, 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2016. Cancer statistics, 2016. CA Cancer J Clin 66(1):7–30, PMID: 26742998, 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Stalter D, O'Malley E, von Gunten U, Escher BI. 2016. Fingerprinting the reactive toxicity pathways of 50 drinking water disinfection by-products. Water Res 91:19–30, PMID: 26773486, 10.1016/j.watres.2015.12.047. [DOI] [PubMed] [Google Scholar]
- Sumida K, Igarashi Y, Toritsuka N, Matsushita T, Abe-Tomizawa K, Aoki M, et al. 2011. Effects of DMSO on gene expression in human and rat hepatocytes. Hum Exp Toxicol 30(10):1701–1709, PMID: 21339255, 10.1177/0960327111399325. [DOI] [PubMed] [Google Scholar]
- Takagi T, Naito Y, Mizushima K, Hirai Y, Harusato A, Okayama T, et al. 2018. Heme oxygenase-1 prevents murine intestinal inflammation. J Clin Biochem Nutr 63(3):169–174, PMID: 30487665, 10.3164/jcbn.17-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuschler LK, Simmons JE. 2003. Approaching DBP toxicity as a mixture problem. J. Am. Water Works Assoc 95(6):131–138, 10.1002/j.1551-8833.2003.tb10393.x. [DOI] [Google Scholar]
- Van der Veen JW, Paskel RF, Smits NAM, Hodemaekers H, van Loveren H, Ezendam J. 2016. The involvement of the Toll-like receptor signalling and Nrf2-Keap1 pathways in the in vitro regulation of IL-8 and HMOX1 for skin sensitization. J Immunotoxicol 13(1):1–6, PMID: 25377948, 10.3109/1547691X.2014.975897. [DOI] [PubMed] [Google Scholar]
- Villanueva CM, Cantor KP, Cordier S, Jaakkola JJ, King WD, Lynch CF, et al. 2004. Disinfection byproducts and bladder cancer: a pooled analysis. Epidemiology 15(3):357–367, PMID: 15097021, 10.1097/01.ede.0000121380.02594.fc. [DOI] [PubMed] [Google Scholar]
- Villanueva CM, Cantor KP, Grimalt JO, Malats N, Silverman D, Tardon A, et al. 2007. Bladder cancer and exposure to water disinfection by-products through ingestion, bathing, showering, and swimming in pools. Am J Epidemiol 165(2):148–156, PMID: 17079692, 10.1093/aje/kwj364. [DOI] [PubMed] [Google Scholar]
- Villanueva CM, Cordier S, Font-Ribera L, Salas LA, Levallois P. 2015. Overview of disinfection by-products and associated health effects. Curr Environ Health Rep 2(1):107–115, PMID: 26231245, 10.1007/s40572-014-0032-x. [DOI] [PubMed] [Google Scholar]
- Villanueva CM, Kogevinas M, Grimalt JO. 2003. Haloacetic acids and trihalomethanes in finished drinking waters from heterogeneous sources. Water Res 37(4):953–958, PMID: 12531279, 10.1016/S0043-1354(02)00411-6. [DOI] [PubMed] [Google Scholar]
- Villeneuve DL, Landesmann B, Allavena P, Ashley N, Bal-Price A, Corsini E, et al. 2018. Representing the process of inflammation as key events in adverse outcome pathways. Toxicol Sci 163(2):346–352, PMID: 29850905, 10.1093/toxsci/kfy047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaanderen J, van Veldhoven K, Font-Ribera L, Villanueva CM, Chadeau-Hyam M, Portengen L, et al. 2017. Acute changes in serum immune markers due to swimming in a chlorinated pool. Environ Int 105:1–11, PMID: 28478232, 10.1016/j.envint.2017.04.009. [DOI] [PubMed] [Google Scholar]
- Wagner ED, Plewa MJ. 2017. CHO cell cytotoxicity and genotoxicity analyses of disinfection by-products: an updated review. J Environ Sci (China) 58:64–76, PMID: 28774627, 10.1016/j.jes.2017.04.021. [DOI] [PubMed] [Google Scholar]
- Wang JQ, Jeelall YS, Ferguson LL, Horikawa K. 2014. Toll-like receptors and cancer: MYD88 mutation and inflammation. Front Immun 5:367, PMID: 25132836, 10.3389/fimmu.2014.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu D, Zhao Z, Cui F, Zhu Q, Liu T. 2010. Factors influencing the formation of chlorination brominated trihalomethanes in drinking water. J Zhejiang Univ Sci A 11(2):143–150, 10.1631/jzus.A0900343. [DOI] [Google Scholar]
- Westbrook AM, Wei B, Braun J, Schiestl RH. 2009. Intestinal mucosal inflammation leads to systemic genotoxicity in mice. Cancer Res 69(11):4827–4834, PMID: 19487293, 10.1158/0008-5472.CAN-08-4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JM, Evans A, Kaufman JA, Rivera-Núñez Z, Narotsky MG. 2017. Disinfection by-product exposures and the risk of specific cardiac birth defects. Environ Health Perspect 125(2):269–277, PMID: 27518881, 10.1289/EHP103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Komaki Y, Kimura SY, Hu HY, Wagner ED, Mariñas BJ, et al. 2014. Toxic impact of bromide and iodide on drinking water disinfected with chlorine or chloramines. Environ Sci Technol 48(20):12362–12369, PMID: 25222908, 10.1021/es503621e. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-blast: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134, PMID: 22708584, 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatts SD, Gennings C, Wagner ED, Simmons JE, Plewa MJ. 2010. Detecting departure from additivity along a fixed-ratio mixture ray with a piecewise model for dose and interaction thresholds. J Agric Biol Environ Stat 15(4):510–522, PMID: 21359103, 10.1007/s13253-010-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, et al. 1997. Gene expression profiles in normal and cancer cells. Science 276(5316):1268–1272, PMID: 9157888, 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.