Abstract
The cytokine interleukin-1β (IL-1β) is a key mediator of anti-microbial immunity as well as autoimmune inflammation. Production of IL-1β requires transcription by innate immune receptor signaling and maturational cleavage by inflammasomes. Whether this mechanism applies to IL-1β production seen in T cell-driven autoimmune diseases remains unclear. Here, we describe an inflammasome-independent pathway of IL-1β production that was triggered upon cognate interactions between effector CD4+ T cells and mononuclear phagocytes (MPs). The cytokine TNF produced by activated CD4+ T cells engaged its receptor TNFR on MPs, leading to pro-IL-1β synthesis. Membrane-bound FasL, expressed by CD4+ T cells, activated death receptor Fas signaling in MPs resulting in caspase-8-dependent pro-IL-1β cleavage. The T cell-instructed IL-1β resulted in systemic inflammation, while absence of TNFR or Fas signaling protected mice from CD4+ T cell-driven autoimmunity. The TNFR-Fas-caspase-8-dependent pathway provides a mechanistic explanation for IL-1β production and its consequences in CD4+ T cell-driven autoimmune pathology.
The cytokine IL-1β mediates host immunity through its ability to influence both innate and adaptive immune responses. It promotes innate immunity by inducing the acute phase response and recruiting inflammatory cells1,2. In the adaptive immune system, IL-1β enhances T cell priming and differentiation, and more importantly, acts as a licensing cytokine to enable the function of memory CD4+ T cells3. However, aberrant production of IL-1β in the absence of pathogenic insult can result in immunopathology associated with several auto-immune and auto-inflammatory diseases4. Autoinflammatory diseases occur due to abnormal activation of macrophages or monocytes in the absence of any conventional microbial or danger signal5. On the other hand, autoimmune diseases are caused by a break in immunological tolerance resulting in the activation of B cell or T cell in response to self-antigens6.
Genome-wide association studies (GWAS) have uncovered heritable traits of autoinflammatory diseases that often result in dysregulated production of IL-1β7. IL-1β−driven autoinflammatory diseases include familial Mediterranean fever, periodic fever syndrome and pyogenic and granulomatous disorders7, which are characterized by an increase in acute phase proteins and systemic amyloidosis. A unifying mechanism of inflammation in these diseases is the dysregulated activation of the inflammasome, due to gain-of-function mutations leading to overproduction of IL-1β. In addition to detrimental systemic effects, IL-1β can cause severe pathology in the tissues. Because of the pivotal role of IL-1β in these diseases, blocking IL-1β activity through various approaches has delivered promising results.
Autoimmune diseases such as type 1 diabetes, pericarditis, rheumatoid arthritis and psoriasis are also responsive to neutralization of IL-1β 8. The autoimmune flares in patients are often associated with presence of cytokine-secreting T cells9. Genetic mouse models have shown that these autoimmune diseases are primarily caused by the dysregulated activation of autoreactive T cells10. IL-1β can promote T cell-mediated autoimmunity by enhancing T cell function, as well as inhibiting suppression mediated by regulatory T cells (Treg cells) 3,11. While targeting of IL-1β has shown promise in clinical trials, the exact mechanism for the production of IL-1β in T cell-mediated autoimmunity is not known. The inflammasome has an established role in autoinflammatory diseases, but its role in IL-1β-dependent T cell-driven autoimmune inflammation remains obsure12. GWAS have failed to report significant genetic association between inflammasome proteins and T cell-dependent autoimmunity. Additionally, disease progression in mouse models of rheumatoid arthritis (RA) is independent of the inflammasome components NLRP3 and caspase-1 (casp-1)13. Similarly, casp-1 deficiency does not mitigate diabetes in NOD mice14.
Due to its highly inflammatory nature, IL-1β is produced under strict regulation in a two-step mechanism. The transcription and translation of pro-IL-1β, which is dependent on the activation of the transcription factor NF-κB 15 is induced by the activation of pattern recognition receptors (PRRs) such as the Toll-like receptors (TLRs). Because pro-IL-1β is not biologically active, it requires the proteolytic cleavage of pro-IL-1β into its bioactive form. Activation of the inflammasomes by damage-associated molecules or microbial virulence factors induces the casp-1-dependent processing of pro-IL-1β7.
Here, we investigated how bioactive IL-1β was produced during T cell-driven autoimmune diseases in the absence of overt infection or injury. We describe a mechanism of IL-1β production that is independent of signaling through PRRs and inflammasome activation. We found that during cognate interaction, effector-memory CD4+ T cells instructed antigen-presenting myeloid cells to produce mature IL-1β. This T cell-induced IL-1β was dependent on the expression of the cytokine TNF and the membrane-bound protein FasL by the activated T cells during their interaction with the macrophages or DCs (hereafter, mononuclear phagocytes, MPs). Signaling through the TNF receptor (TNFR) was required for the synthesis of pro-IL-1β in MPs. The interaction with activated T cells also triggered signaling through the surface receptor for FasL, Fas, in MPs, which resulted in casp-8-dependent maturation of pro-IL-1β. This TNFR-Fas pathway of IL-1β production was responsible for the induction of inflammation and pathology during experimental autoimmune encephalomyelitis (EAE), a T cell-mediated autoimmune disease, suggesting this pathway was likely responsible for the production of IL-1β during T cell-driven autoimmune pathology.
Results
T cell-interacting BMDCs produce IL-1β
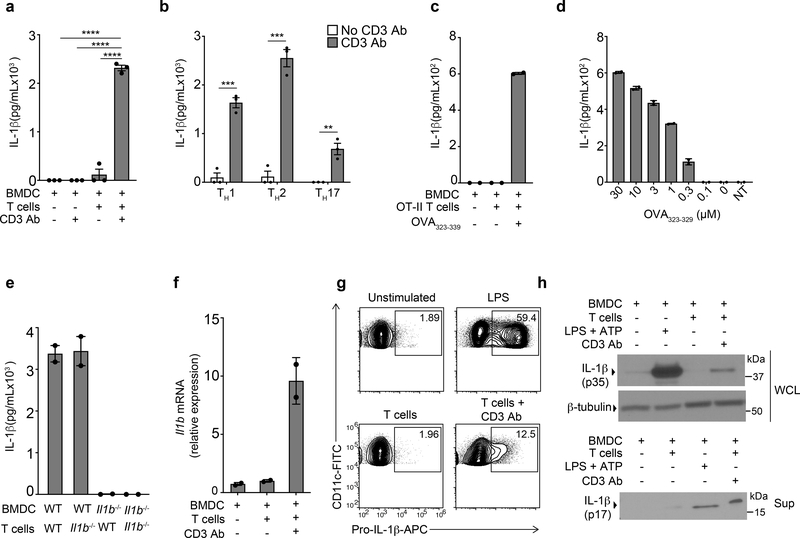
T cell-intrinsic signaling through IL-1R is critical for optimal cytokine production by effector and memory CD4+ T cells following their reactivation by splenic CD11c+ DCs3. We therefore tested whether cognate interactions between DCs and effector CD4+ T cells could elicit the production of IL-1β independently of signaling through PRRs. We generated bone marrow-derived DCs (BMDCs) derived by culturing wild-type bone marrow cells with GM-CSF-containing media for 5 days, and differentiated naïve wild-type CD4+ T cells into TH0 lineage cells through 5 days of culture with plate-bound CD3 Ab and CD28 Ab in the presence of IL-2. BMDCs were co-cultured with rested TH0 CD4+ T cells and their interaction was induced with soluble CD3 Ab. In order to prevent consumption of IL-1β by T cells, we added IL-1R antagonist (IL-1Ra), which acts as a potent biological inhibitor of IL-1β. The CD3 Ab-mediated interaction between BMDCs and effector CD4+ T cells induced the secretion of IL-1β (Fig. 1a). Furthermore, interaction of BMDCs with CD4+ T cells differentiated into TH1, TH2 or TH17 cells using in vitro cytokine polarization also led to secretion of IL-1β (Fig. 1b), indicating a broadly-conserved pathway of IL-1β production across multiple T cell lineages. Because all CD4+ T cell lineages instructed the production of IL-1β in BMDCs, we used differentiated CD4+ TH0 cells, hereafter referred to as effector CD4+ T cells unless otherwise noted, for all further experiments. Next, we examined if production of IL-1β was dependent on the presentation of cognate peptide by BMDCs. In co-cultures of OVA peptide-restricted OT-II TCR transgenic effector CD4+ T cells and wild-type BMDCs, production of IL-1β was detected only in the presence of the cognate OVA323–339 peptide (Fig. 1c). Additionally, the amount of IL-1β detected in the supernatants directly correlated with the concentration of OVA323–339 provided in the cultures (Fig. 1d), indicating that the avidity of the MHC-TCR interaction was a major determinant of the quantity of IL-1β secreted. CD4+ T cells have been reported to produce IL-1β16. However, we found that use of Il1b−/− BMDCs, but not Il1b−/− T cells, led to a complete loss of T cell-instructed IL-1β production (Fig. 1e), suggesting that BMDCs were the only source of IL-1β in this co-culture system. Co-culture of Il1b−/− CD4+ T cells with wild-type BMDCs induced the Il1b transcript 3 h post-stimulation (Fig. 1f), and intracellular pro-IL-1β could be detected by flow cytometry in CD11c+ BMDCs 6 h post-stimulation (Fig. 1g and Extended Data Fig. 1a). Total and cleaved bioactive IL-1β were detected by immunoblotting analysis of the co-culture whole cell lysate and supernatant, respectively, after 18 h of stimulation (Fig. 1h). These data indicated that the interaction between BMDCs and effector CD4+ T cells triggered the transcriptional induction of pro-IL-1β and its bioactive cleavage in BMDCs.
Figure 1. Cognate interaction between BMDCs and effector CD4 T cells leads to inflammasome independent production of bioactive IL-1β by BMDCs.
(a) lL-1β, as quantified by ELISA, in the supernatants of wild-type (WT) effector CD4+ cells (TH0 cells) co-cultured with WT BMDCs in the presence or absence of CD3 Ab for 18 h. Error bars indicate SEM for n=3 independent experiments. (b) Production of IL-1β measure by ELISA in supernatants of effector CD4+ T cells polarized to TH1, TH2 and TH17 lineages cultured with WT BMDCs in the presence of CD3 Ab for 18 h or left untreated. Error bars indicate SEM for n=3 independent experiments. (c-d) IL-1β in the supernatants following stimulation of TH17-polarized OT-II T cells for 18 h with fixed (100μM) (c) or titrating (d) concentrations of OT-II323–339 peptide presented by WT BMDCs. NT, no T cells in culture. Error bars indicate SEM from n=2 technical replicates. Data are representative of 3 independent experiments. (e) IL-1β measured by ELISA after 18 h of culture of effector WT or Il1b−/− CD4+ TH0 cells with WT or Il1b−/− BMDCs in the presence of CD3 Ab. Error bars indicate SEM from two (n=2) independent experiments. (f) qPCR of Il1b mRNA in lysates of WT BMDCs co-cultured with Il1b−/− TH0 cells and treated with CD3 Ab for 3h. Data are normalized to 18s rRNA. Error bars indicate SEM from n=2 technical replicates. Data are representative of three independent experiments. (g) Expression of intracellular pro-IL-1β measured by flow cytometry in WT live,CD90-CD11c+ BMDCs stimulated with LPS (100ng/mL) or cultured with TH0 cells in the presence of CD3 Ab for 6h. Data are representative of three independent experiments. (h) Immunoblot analysis of pro-IL-1β (p35) or cleaved IL-1β (p17) in the cell lysates or the supernatants, respectively, of WT BMDCs co-cultured with WT TH0 cells treated with CD3 Ab for 18 h. Data are representative of three independent experiments. (a, b) Statistical analysis was performed by paired, one-tailed Student’s t-test. Individual p values: **p<0.01, ***p<0.001, ****p<0.0001.
TNFR signaling in BMDCs leads to pro-IL-1β synthesis
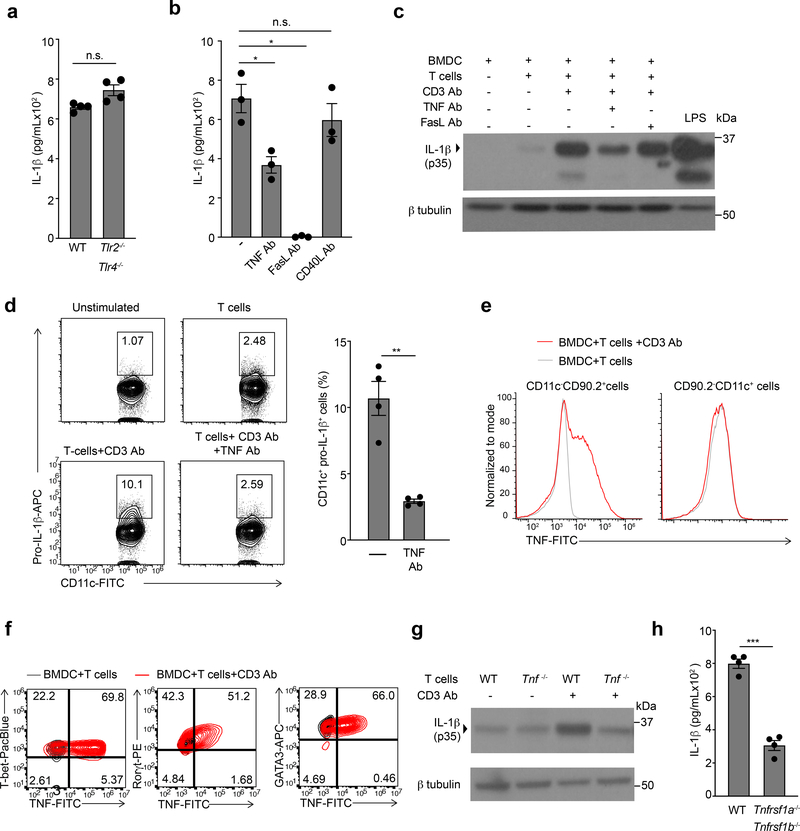
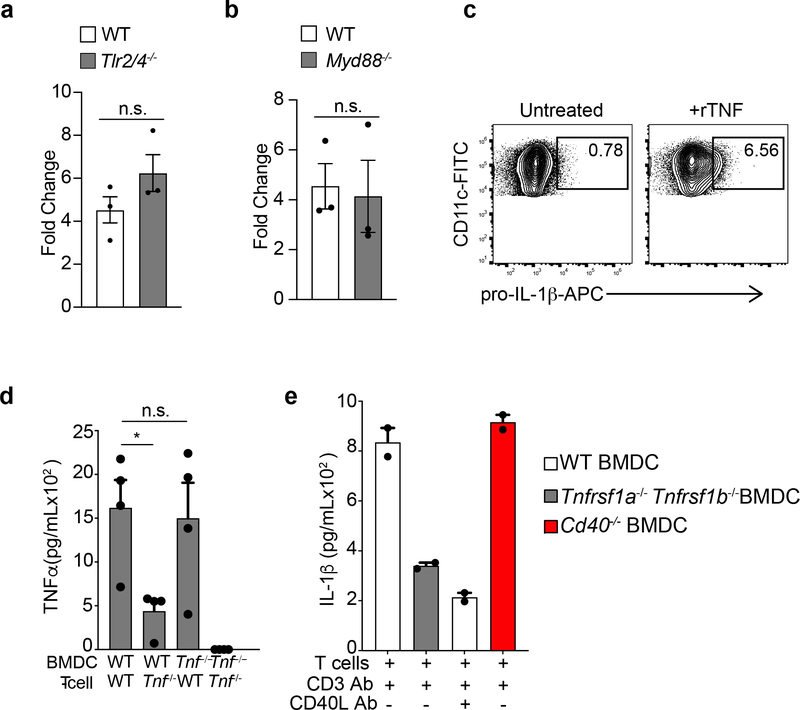
Next, we characterized the signaling events which enabled the BMDC intrinsic production of T cell-induced IL-1β. Inflammasome activation is a major mechanism for the production of IL-1β, in which TLR signaling is responsible for the transcriptional upregulation of pro-IL-1β17. Induction of intracellular pro-IL-1β and secretion of mature IL-1β in Tlr2−/−Tlr4−/− or Myd88−/− BMDCs co-cultured with effector CD4+ T cells, however, was similar to that of wild-type BMDCs (Fig 2a and Extended Data Fig. 2a and 2b), ruling out endotoxin contamination in these cultures. Because transcription of pro-IL-1β is mediated by NF-κB15 and AP-118, which can be activated downstream of TNF superfamily receptors, we examined the role of TNF receptors in the T cell-induced production of pro-IL-1β in BMDCs. Antibody-mediated neutralization of TNF and FasL, but not CD40L, significantly reduced the production of IL-1β compared to PBS (Fig. 2b). Pro-IL-1β was reduced by TNF, but not FasL, neutralization as assessed by immunoblotting (Fig. 2c), suggesting that Fas-FasL interaction was critical for the production of mature IL-1β, but not the transcriptional induction of pro-IL-1β. Consistent with this finding, neutralization of TNF in wild-type BMDCs-effector CD4+ T cell co-cultures reduced the amount of intracellular pro-IL-1β detected by flow cytometry, as compared to PBS (Fig. 2d). Moreover, stimulation with recombinant TNF drove the synthesis of pro-IL-1β in wild-type BMDCs (Extended Data Fig. 2c).
Figure 2. T cell derived TNFα is critical for induction of pro-IL-1β in BMDCs.
(a) lL-1β, as quantified by ELISA, in the supernatants of WT TH0 cells co-cultured with WT or Tlr2/4−/− BMDCs in the presence of CD3 Ab for 6 h. Error bars indicate SEM for n=4 independent experiments. (b) IL-1β measured by ELISA following 6 h of culture with WT TH0 cells and WT BMDCs in the presence of CD3 Ab and neutralizing TNF Ab (20μg/mL), FasL Ab (10μg/mL), or CD40L Ab (20μg/mL). Error bars indicate SEM for n=3 independent experiments. (c) Immunoblot analysis of pro-IL-1β (p35) in the lysates of WT BMDCs stimulated with LPS (100ng/mL) or cultured with TH0 cells in the presence of CD3 Ab and neutralizing antibodies. Data are representative of two independent experiments. (d) Expression of intracellular pro-IL-1β measured by flow cytometry in WT live, CD90-CD11c+ BMDCs cultured with TH0 cells in the presence of CD3 Ab and neutralizing TNF Ab (20μg/mL) for 6h. Flow plots are representative of four independent experiments. Error bars indicate SEM for n=4 independent experiments. (e) Expression of intracellular TNF measured by flow cytometry in WT TH0 cells (left; live,CD11c-CD90.2+) and WT BMDCs (right; live,CD90.2-CD11c+) that were co-cultured for 3 h in the presence of CD3 Ab and brefeldin A. Data are representative of two independent experiments. (f) Expression of intracellular TNF measured by flow cytometry in effector CD4+ T cells (live,CD11c-CD90.2+) polarized to TH1, TH2 and TH17 lineages cultured with WT BMDCs in the presence of CD3 Ab and brefeldin A for 3h. Cells were considered to be transcription factor positive based on isotype control antibody staining. Data are representative of three independent experiments. (g) Immunoblot analysis of pro-IL-1β (p35) in the lysates of WT or Tnf−/− TH0 cells cultured WT BMDCs in the presence or absence of CD3 Ab for 6h. Data are representative of two independent experiments. (h) lL-1β was quantified by ELISA in the supernatants of WT TH0 cells co-cultured with WT or Tnfrsf1a−/−Tnfrsf1b−/− BMDCs in the presence of CD3 Ab for 6 h. Error bars indicate SEM for n=4 independent experiments. (a, b, d, h) Statistical analysis was performed by paired, one-tailed Student’s t-test. *p<0.05, **p<0.01, ***p<0.001, n.s.=not significant.
Although TNF is largely of myeloid origin during PRR-driven inflammation19, we found that reactivating Tnf−/− effector CD4+ T cells with wild-type BMDCs resulted in significantly less secreted TNF compared to wild-type effector CD4+ T cells co-cultured with wild-type BMDCs (Fig. 2e and Extended Data Fig. 2d), indicating that activated T cells were the predominant producers of TNF during interaction with BMDCs. While TNF is primarily known to be an effector cytokine for TH17 cells20, primed TH1 and TH2 cells also rapidly upregulated TNF upon interaction with BMDCs (Fig. 2f), suggesting that multiple activated CD4+ T cell lineages can signal through the TNFR on BMDCs. Indeed Tnf−/− effector CD4+ T cells induced appreciably diminished pro-IL-1β in wild-type BMDCs compared to wild-type effector CD4+ T cells (Fig 2g). Moreover, Tnfrsf1a−/−Tnfrsf1b−/− BMDCs secreted significantly less T cell-induced IL-1β compared to wild-type BMDCs (Fig. 2h), indicating that TNFR signaling in BMDCs was required for optimal production of IL-1β. Because IL-1β production was not completely abrogated in Tnfrsf1a−/−Tnfrsf1b−/− BMDCs, we examined if there was also a role for CD40 signaling in this context. Blocking CD40L using neutralizing antibody in Tnfrsf1a−/−Tnfrsf1b−/− BMDCs further reduced the production of IL-1β (Extended Data Fig. 2e). These data indicate that TNFR signaling lead to induction of IL-1β in BMDCs during their interaction with CD4+T cells, while other TNF superfamily proteins, such as CD40, could contribute to this induction in the absence of TNFR signaling.
T cell-instructed IL-1β is independent of casp-1
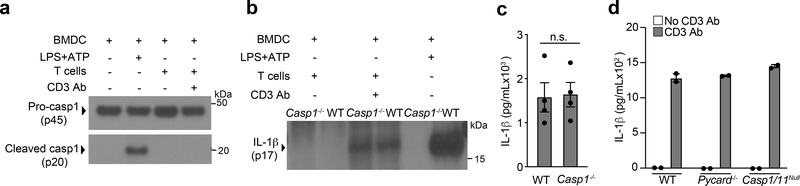
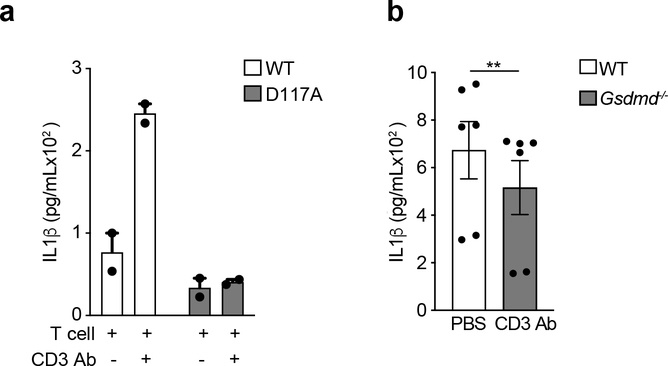
Casp-1, the effector protease of all inflammasomes21, is largely responsible for the production of bioactive IL-1β through its cleavage of pro-IL-1β at the aspartate residue in position 117 (D117)12. Expression of an D117A IL-1β mutant in Il1b−/− BMDCs co-cultured with effector CD4+ T cells led to significantly less secretion of IL-1β compared to expression of wild-type IL-1β (Extended Data Fig. 3a), indicating the D117 casp-1 cleavage site of pro-IL-1β was critical for the T cell-induced production of IL-1β in BMDCs. Despite this, we did not detect active casp-1 in T cell-interacting BMDCs after 18 h of co-culture (Fig. 3a). Furthermore, Casp1−/− BMDCs showed no reduction in T cell-induced IL-1β production compared to wild-type BMDCs (Fig. 3b, c). Pycard−/− and Casp4−/− BMDCs, lacking ASC and casp-11 respectively, also had normal production of IL-1β (Fig. 3d), indicating T cell-induced IL-1β production was independent of canonical as well as non-canonical inflammasomes16,22. During inflammasome activation, gasdermin-D drives pyroptosis and the secretion of mature IL-1β23. Gsdmd−/− BMDCs showed a partial reduction in T cell-induced IL-1β compared to wild-type BMDCs (Extended Data Fig. 3b). These observations indicate that T cell-induced IL-1β in BMDCs was independent of casp-1 and casp-11, and likely to be entirely independent of inflammasome activation.
Figure 3. T cell instructed IL-1β production is independent of Caspase-1.
(a) Immunoblot analysis of pro-casp1 (p45) and cleaved casp1 (p20) in the lysates of WT BMDCs stimulated with LPS+ATP or cultured with WT TH0 cells in the presence of CD3 Ab for 18 h. Data are representative of three independent experiments. (b) Immunoblot analysis of cleaved IL-1β (p17) or (c) lL-1β, as quantified by ELISA, in the supernatant of WT or Casp1−/− BMDCs cultured with WT TH0 cells in the presence of CD3 Ab for 18 h. (b) Data are representative of three independent experiments. (c) Error bars indicate SEM for n=3 independent experiments. Statistical analysis was performed by paired, one-tailed Student’s t-test. n.s.=not significant. (d) IL-1β measured by ELISA following 18 h of culture with WT TH0 cells and WT, Casp1/11−/−, or Pycard−/− BMDCs in the presence of CD3 Ab. Data are representative of two independent experiments. Error bars indicate SEM for n=2 technical replicates.
Fas-casp-8 mediates maturation of T cell-induced IL-1β
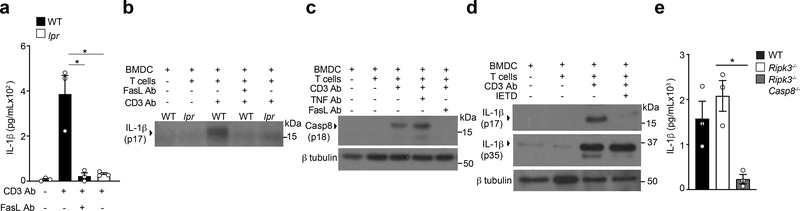
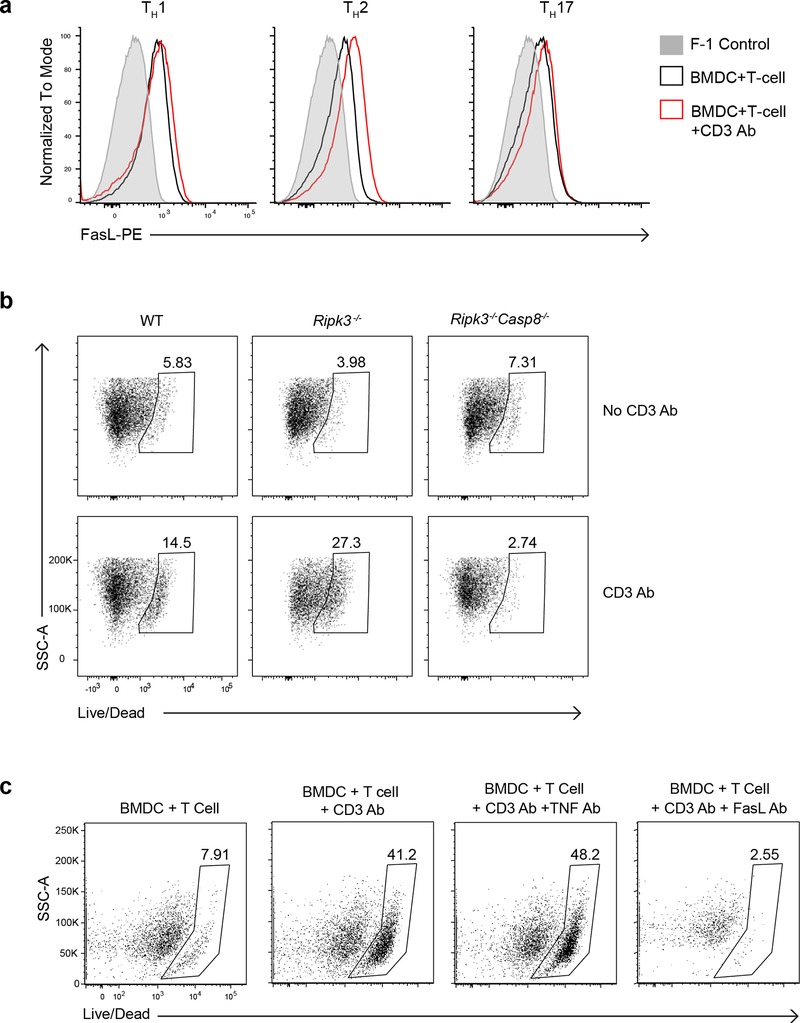
Effector CD4+ T cells constitutively expressed FasL, which was further upregulated upon CD3 Ab mediated interaction with BMDCs (Extended Data Fig. 4a). As such, we investigated whether Fas signaling induced cleavage of pro-IL-1β in BMDCs. BMDCs derived from lpr mice, which encodes a spontaneous homozygous loss-of-function mutation in the Fas gene, did not secrete cleaved IL-1β upon interaction with wild-type effector CD4+ T cells (Fig. 4a,b), indicating that Fas signaling was required for production of T cell-induced IL-1β in BMDCs. Fas signaling has been reported to trigger casp-8-dependent cleavage of pro-IL-1β in macrophages 24. Interaction of BMDCs with effector CD4+ T cells led to maturational cleavage of casp-8 in the co-culture whole cell lysate (Fig. 4c). TNFR signaling has been reported to cause casp-8 activation25. However, antibody mediated neutralization of FasL, but not TNF, during interaction of BMDCs with effector CD4+ T cells led to reduced amounts of cleaved casp-8 in the co-culture whole cell lysate (Fig. 4c). Addition of the casp-8 inhibitor IETD in the BMDCs-effector CD4+ T cells co-cultures led to loss of mature IL-1β in the supernatant (Fig. 4d), indicating that casp-8 was the effector protease involved in the maturation of T cell-induced pro-IL-1β in BMDCs. Casp-8 has been reported to induce NF-κB-dependent genes, including pro-IL-1β, after activation through TLRs26,27. However, inhibition of casp-8 did not affect the production of pro-IL-1β in BMDCs following their interaction with effector CD4+ T cells (Fig. 4d). Furthermore, Ripk3−/−Casp8−/− BMDCs had a complete loss of secreted T cell-induced IL-1β (Fig. 4e). Because casp-8 is a critical mediator of apoptosis induced by cell-extrinsic signals23, we investigated if T cell-interacting BMDCs were undergoing cell death. Ripk3−/−Casp8−/− BMDCs were completely resistant to T cell interaction-induced cell death (Extended Data Fig. 4b). Furthermore, casp-8 dependent BMDC death was mediated by Fas, but not TNFR signaling (Extended Data Fig. 4c). These observations established a role for casp-8 downstream of Fas signaling in the production of mature IL-1β in T cell-interacting BMDCs.
Figure 4. Fas-FasL interaction between effector CD4 T cells and BMDCs leads to caspase-8 dependent cleavage of pro-IL-1β.
(a) lL-1β, as quantified by ELISA, or (b) detected by immunoblot analysis of cleaved IL-1β (p17) in the supernatants of WT TH0 cells co-cultured with WT or lpr BMDCs in the presence of CD3 Ab and neutralizing FasL Ab (10μg/mL) for 18 h. (a) Error bars indicate SEM for n=3 independent experiments. (b) Data are representative of two independent experiments. (c) Immunoblot analysis of cleaved casp-8 (p18) in the lysates of WT BMDCs cultured with WT TH0 cells in the presence of CD3 Ab and neutralizing FasL Ab (10μg/mL) or TNF Ab (20μg/mL) for 12 h. Data are representative of two independent experiments. (d) Immunoblot analysis of pro-IL-1β (p35) or cleaved IL-1β (p17) in the cell lysates or the supernatants, respectively, of WT BMDCs co-cultured with WT TH0 cells in the presence of CD3 Ab and IETD (10μM) for 18 h. Data are representative of two independent experiments. (e) lL-1β was quantified by ELISA in the supernatants of WT TH0 cells co-cultured with WT, Rip3−/−, or Rip3−/−Casp8−/− BMDCs in the presence of CD3 Ab for 18 h. Error bars indicate SEM for n=3 independent experiments. Statistical analysis was performed by paired, one-tailed Student’s t-test. *p<0.05.
TNFR-Fas-dependent IL-1β is induced in myeloid cells
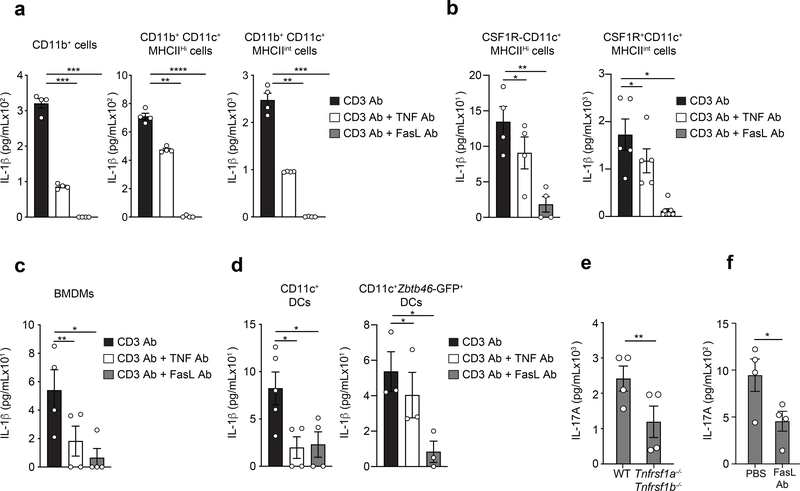
The GM-CSF-derived BMDCs used in this study are a heterogenous mixture of macrophage-like and DC-like cell populations43. To examine the importance of the TNFR-Fas pathway in specific cell types of this heterogenous mixture, we sorted CD11c+CD11b+MHCIIint and CD11c+CD11b+MHChi BMDCs cells from total BMDCs (Extended Data Fig. 5a). Both subsets secreted IL-1β in a manner dependent on TNF and FasL after cognate interaction with effector CD4+ T cells (Fig. 5a). CD11c+ BMDCs are comprised of CSF1R+ GM-Macs and CSF1R- GM-DCs28. CSF1R+ GM-Macs produce IL-1β in response to inflammasome ligands, while CSF1R- GM-DCs do not produce IL-1β or undergo inflammasome activation29. To assess whether these subsets differed in their ability to produce inflammasome-independent T cell-induced IL-1β we sorted them based on the expression of CSF1R (Extended Data Fig. 5b). Purified CSF1R+ GM-Macs and CSF1R- GM-DCs were co-cultured with effector CD4+ T cells and CD3 Ab. We detected secreted IL-1β in both cultures after 18 h (Fig. 5b). The production of T cell-induced IL-1β in GM-Macs and GM-DCs was blocked when cells were co-cultured with neutralizing antibodies against TNF or FasL (Fig. 5b). Moreover, bone marrow derived macrophages (BMDM) differentiated in the presence of MCSF-containing L929 supernatant for 5 days also produced TNFR-Fas-dependent IL-1β following their interaction with effector CD4+ T cells (Fig. 5c). These data suggested that despite the differences between macrophages and DCs to undergo inflammasome activation, both of these myeloid cell populations could use the TNFR-Fas-dependent pathway for the production of IL-1β.
Figure 5. Diverse myeloid cells populations utilize TNFR-Fas pathway for T cell induced IL-1β production.
lL-1β was quantified by ELISA in the supernatants of WT TH0 cells co-cultured with (a) CD11b+ BMDCs, CD11b+CD11c+MHCIIHi BMDCs, CD11b+CD11c+MHCIIint BMDCs, (b) CSF1R-CD11c+MHCIIHi GM-DCs, or CSF1R+CD11c+MHCIIint GM-Macs in the presence of CD3 Ab and neutralizing TNF Ab (20μg/mL) or FasL Ab (10μg/mL) for 18 h. lL-1β was quantified by ELISA in the supernatants of WT TH0 cells co-cultured with (c) BMDMs, or (d) CD11c+ cDCs FACS sorted from WT spleen (left) or CD11c+Zbtb46-GFP+Ly6C- splenic cDCs FACS sorted from Zbtb46-GFP reporter mice spleen (right) as in (a). Error bars indicate SEM for n=3–4 independent experiments. (e) IL-17A was quantified by ELISA in the supernatants of WT CD44hiCD62Llo effector CD4 T cells re-stimulated with CD3 Ab using WT or Tnfrsf1a−/−Tnfrsf1b−/− CD11c+ splenic cDCs or (f) WT CD11c+ splenic cDCs in the presence of neutralizing FasL Ab (10μg/mL) for 48 h. Error bars indicate SEM from n=4 independent experiments. Statistical analysis was performed by paired, one-tailed Student’s t-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Blocking IL-1R signaling during splenic DC-T cell interaction abrogates the production of effector cytokines by T cells3. To test if conventional DCs (cDCs) produced T cell-induced IL-1β, we magnetically sorted CD11c+ DCs from the spleens of wild-type mice and co-cultured them with effector CD4+ T cells. We detected IL-1β in the culture supernatants after 18 h of CD3 Ab-induced interaction (Fig. 5d). The amount of IL-1β was reduced when cells were cultured with neutralizing antibodies to TNF and FasL (Fig. 5d), indicating that splenic CD11c+ cDCs produced T cell-instructed IL-1β in a TNFR-Fas dependent manner. Based on flow cytometry analysis, we could not detect CD11c-CSF1R+ cells among the splenic CD11c+ cells (Extended Data Fig. 5c), indicating that the production of IL-1β was not due to contamination by macrophages. Zbtb46-GFP mice, in which GFP is expressed concomitantly with the cDC-specific transcription factor ZBTB46, were further used to isolate a pure cDC population and exclude monocyte-derived CD11c+GFP-Ly6C+ DCs 30. Pure cDCs (CD11c+Zbtb46-GFP+Ly6C-) (Extended Data Fig. 5d), were again cultured with effector CD4+ T cells and IL-1β was detected in the supernatant following 18 h of culture with CD3 Ab (Fig. 5d). Production of IL-1β was reduced when cells were co-cultured with neutralizing antibodies for TNF or FasL (Fig. 5d).
Because IL-1β is critical for the licensing of IL-17A production during effector memory CD4+ T cell3 reactivation, we tested whether TNFR and Fas signaling were necessary for IL-17A production. Compared to wild-type CD11c+ cDCs, Tnfrsf1a−/−Tnfrsf1b−/− CD11c+ cDCs had diminished capacity to trigger the production of IL-17A in CD62LloCD44hiCD4+ effector memory T cells isolated from the spleen and peripheral LNs, after 18 h of co-culture (Fig. 5e). Blocking FasL using neutralizing antibody during the reactivation of wild-type CD62LloCD44hiCD4+ effector memory T cell by wild-type splenic CD11c+ cDCs also reduced the production of IL-17A when compared to PBS control (Fig. 5f). These observations indicated that the TNFR-Fas-dependent mechanism of IL-1β production was conserved in various macrophage and DC populations and enabled optimal CD4+ T cell effector function.
T cell-induced IL-1β causes systemic inflammation
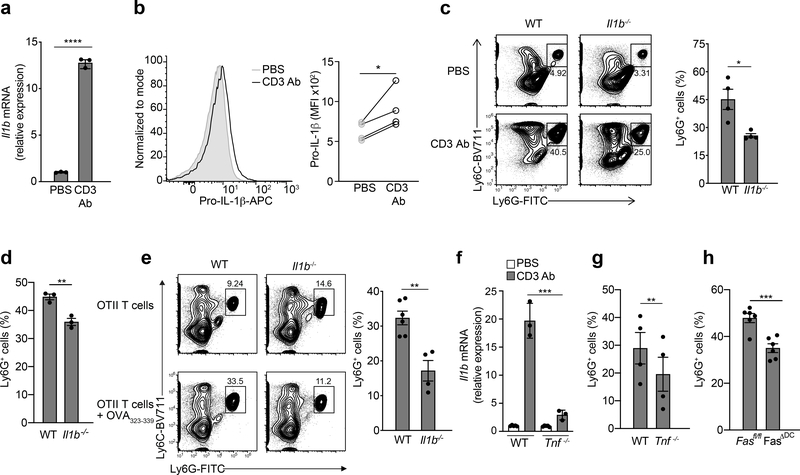
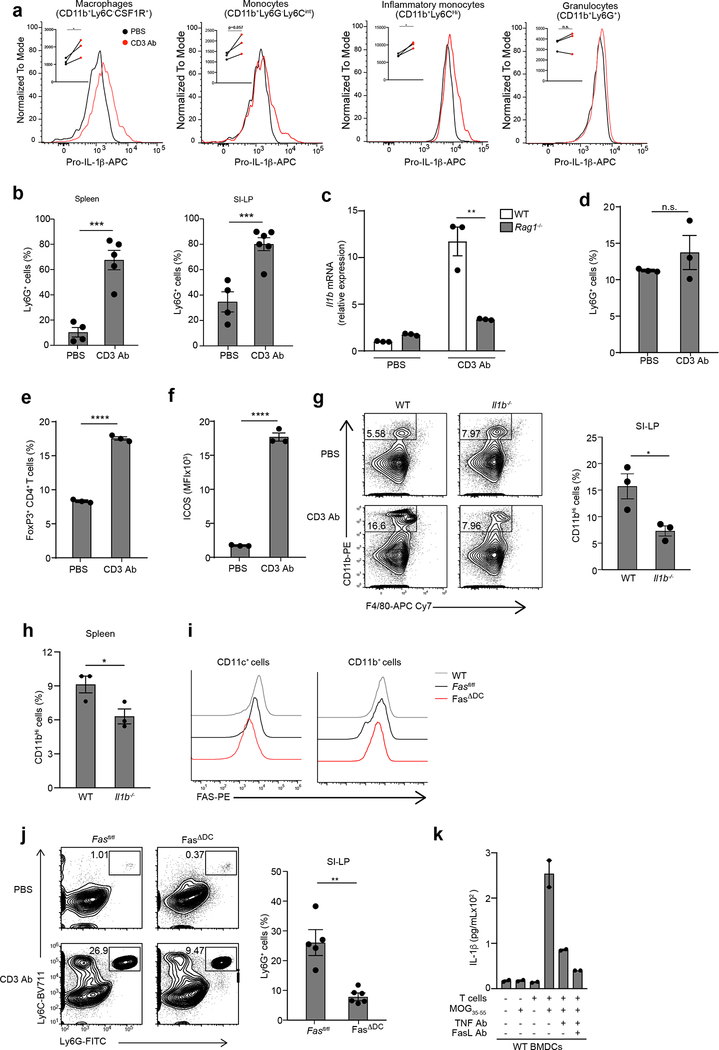
Although cytokines made by self-reactive T cells contribute to autoimmune inflammation, innate immune activation can precipitate autoimmunity31and infiltration of neutrophils and inflammatory monocytes into the affected tissues is a feature of pathology for these diseases. Because the majority of mouse models for T cell-driven autoimmunity rely on initial activation of PRRs, usually through stimulation with Mycobacterium tuberculosis to break tolerance32, we used PRR-independent approaches to mimic cognate antigen presenting cell (APC)-T cell interactions that are likely to occur during autoimmune flares. First, we administered CD3 Ab intraperitoneally in wild-type mice to induce systemic TCR activation and widespread reactivation of T cells mediated by myeloid cells. Transcriptional upregulation of Il1b in total splenocytes was detected 4 h after CD3 Ab administration (Fig. 6a). Upregulation of pro-IL-1β protein was detected by flow cytometry in CD11c+ cDCs, CD11b+Ly6C-CSF1R+ macrophages, CD11b+Ly6G-Ly6Cint monocytes and CD11b+Ly6Chi inflammatory monocytes isolated from the spleen 3–4 h after administration of CD3 Ab (Fig. 6b and Extended Data Fig. 6a). No significant induction of pro-IL-1β was observed in spleen CD11b+Ly6G+ granulocytes (Extended Data Fig. 6a). Significant recruitment of Ly6G+ neutrophils to the spleen and to the small intestinal lamina propria (SI-LP) was detected within 18 h of CD3 Ab administration (Extended Data Fig 6b). Transcriptional upregulation of Il1b or neutrophil infiltration was not detected in Rag1−/− mice injected with CD3 Ab (Extended Data Fig. 6c,d), indicating that IL-1β induction in this system was dependent on adaptive immune cells. CD3 Ab treatment did not lead to reduction in FoxP3+ CD4+ T cells in the spleen of wild-type mice (Extended Data Fig. 6e), indicating the CD3 Ab-induced inflammation was not due to the antibody-mediated depletion of Treg cells. Despite the increased proportion of Treg cells in the spleen (Extended Data Fig. 6e), CD4+ T cells had increased ICOS expression (Extended Data Fig. 6f), indicating their activated status after CD3 Ab treatment. Next, we administered CD3 Ab intraperitoneally to wild-type or Il1b−/− mice and found significantly less CD11b+ monocytes and Ly6G+ neutrophils in the spleen and SI-LP of Il1b−/− mice compared to wild-type controls 18 h post-CD3 Ab (Fig. 6c,d and Extended Data 6g,h), indicating that systemic inflammation was dependent on production of IL-1β.
Figure 6. CD4 T cells engage TNFR and Fas signaling pathways to induce IL-1β mediated systemic inflammation.
(a) qPCR of Il1b mRNA in lysates of splenocytes collected from WT mice 3–4 h post CD3 Ab injection (50μg, i.v.). Data are normalized to Hprt1. Error bars indicate SEM from n=3 technical replicates. Data are representative of three independent experiments. (b) Expression of intracellular pro-IL-1β measured by flow cytometry in WT live,CD11c+ splenocytes from (a). Data are representative of 4 independent experiments. Statistical analysis was performed by paired one-tailed Student’s t-test. *p<0.05. (c) Neutrophil infiltration in the SI-LP or (d) spleen as measured by flow cytometry in WT or Il1b−/− mice 18 h post CD3 Ab injection (20μg, i.p.) Error bars indicate SEM from n=3–4 independent experiments. (e) Neutrophil infiltration in the spleen as measured by flow cytometry 12 h post OVA323–339 peptide injection (50μg, i.v.) into WT or Il1b−/− mice that previously received (1 d prior) OT-II TH17 cells (5×106, i.v.). Error bars indicate SEM from n=4 independent experiments. (f) qPCR of Il1b mRNA in lysates of splenocytes collected from WT or Tnf−/− mice 3–4 h post CD3 Ab injection (50μg, i.v.). Data are normalized to Hprt1. Error bars indicate SEM from n=3 technical replicates. Data are representative of two independent experiments. (g) Neutrophil infiltration in the spleen as measured by flow cytometry in WT and Tnf−/− mice 3 h post CD3 Ab injection or (h) Fasfl/fl and Fasfl/fl x CD11c-cre (FasΔDC) mice 18 h post CD3 Ab injection (20μg, i.p.). All Ly6G+ cells were previously gated on live,CD11b+F480- cells. Error bars indicate SEM from n=4–6 independent experiments. (a, c, d, e, g) Statistical analysis was performed by unpaired, one-tailed Student’s t-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
In the second approach, OT-II TCR Tg T cells were differentiated in vitro into TH17 cells using polarizing cytokines, adoptively transferred into wild-type recipients and reactivated in vivo with OVA323–339 administered intravenously 24 h post-OT-II TH17 transfer. Recruitment of neutrophils to the spleens of wild-type, but not Il1b−/− recipient mice, was observed 12 h post-OVA323–339 injection (Fig. 6e). Induction of pro-IL-1β transcripts in splenocytes (Fig. 6f) and accumulation of neutrophils in the spleen (Fig. 6g) was significantly reduced in Tnf−/− mice compared to wild-type controls 4 h post-CD3 Ab administration. Mice with conditional deletion of Fas in CD11c+ cells (Extended Data Fig. 6i) also had significantly reduced neutrophil infiltration in the SI-LP and spleen 18 h post CD3 Ab treatment compared to their Fasfl/fl littermates (Fig. 6h and Extended Data Fig. 6j). As such, the activation of T cells in vivo resulted in TNF-Fas-dependent production of IL-1β and systemic inflammation.
TNFR-Fas are required for IL-1β-mediated autoimmunity
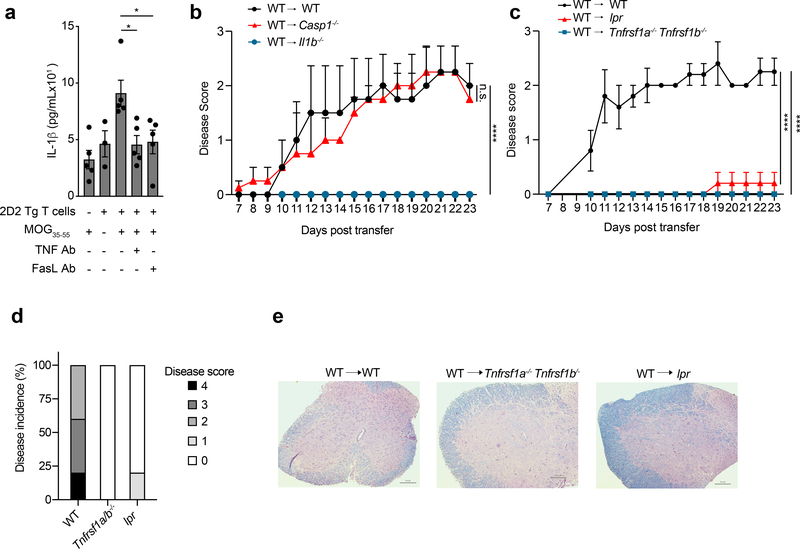
Self-reactive CD4+ T cells are key players in several IL-1β-mediated autoimmune diseases, such as multiple sclerosis, RA and type 1 diabetes33–35. IL-1R signaling is known to be critical for autoimmune inflammation in EAE, however the mechanism leading to production IL-1β in this system has remained unclear 36,37. To test if self-reactive CD4+ T cells could elicit IL-1β production in EAE, we cultured 2D2 TCR Tg CD4+ T cells, which are specific for the myelin oligodendrocyte glycoprotein MOG35–55 peptide, with wild-type BMDCs in the presence of MOG35–55 peptide. Production of IL-1β was detected after 18 h in the co-culture supernatant (Fig. 7a), and was reduced when cells were cultured with neutralizing Ab for TNF and FasL (Fig. 7a).
Figure 7. TNFR and Fas signaling pathways, not Caspase-1, is responsible for IL-1β mediated autoimmune inflammation.
(a) lL-1β was quantified by ELISA in the supernatants of TH17 polarized 2D2 TCR Tg cells cultured with WT BMDCs and MOG35–55 (30μM) in the presence of neutralizing TNF Ab (20μg/mL) or FasL Ab (10μg /mL) for 18 h. Error bars indicate SEM from n=5 independent experiments. Statistical analysis was performed by paired, one-tailed Student’s t-test. *p<0.05. (b) Mean EAE clinical disease scores of WT, Il1b−/−, and Casp1−/− recipients after adoptive transfer of WT CD4+ T cells primed in vivo with MOG35–55. Error bars indicate SEM from n=4 individual mice. Statistical analysis was performed by two-way repeated-measures ANOVA test, ****p<0.0001, n.s.=not significant. DF=16, F=0.0304 (WT vs Casp1−/−), F=4.764 (WT vs Il1b−/−) (c) Mean EAE clinical disease scores of WT, Tnfrsf1a−/−Tnfrsf1b−/−, or lpr recipients after adoptive transfer of WT CD4+ T cells primed in vivo with MOG35–55. Error bars indicate SEM from n=5 individual mice. Statistical analysis was performed by two-way repeated-measures ANOVA test, ****p<0.0001, DF=14, F=7.136 (WT vs Tnfrsf1a−/−Tnfrsf1b−/−), F=5.417 (WT vs Lpr). (d) Percentage of mice with given disease scores in each recipient group from (c). (e) Luxol fast blue staining of spinal cords from given genotypes, 28 days after CD4+ T cell transfer, was performed to assess demyelination. Scale bar is 100μm. Data are representative of two independent experiments.
To investigate the mechanism of T cell-induced sterile autoimmune inflammation, we used a passive model of EAE, in which MOG-specific CD4+ T cells obtained from MOG-immunized wild-type mice were transferred into naive mice. Prior to transfer into naïve mice, the MOG-specific CD4+ T cells were expanded ex vivo and co-cultured with BMDCs to test their ability to induce TNFR-Fas-dependent IL-1β in the BMDCs (Extended Data Fig. 6k). The MOG-specific CD4+ T cells were then transferred intravenously into naïve wild-type or Il1b−/− recipient mice. Unlike the wild-type mice, the Il1b−/− recipient mice were completely resistant to induction of passive EAE by the activated MOG-specific CD4+ T cells, as assessed by the induction of progressive paralysis for up to 23 days after transfer (Fig. 7b). Similarly, Tnfrsf1a−/−Tnfrsf1b−/− and lpr recipient mice were protected from MOG-specific CD4+ T cell-induced neurological autoimmunity (Fig. 7c,d), and had significantly reduced demyelination of the spinal cord (Fig. 7e) when compared to wild-type recipients. Of note, unmanipulated lpr mice develop a lymphoproliferative disorder as they age, but this does not affect the EAE disease score, which relies on progressive paralysis38. After transfer of MOG-specific CD4+ T cells, Casp1−/− recipient mice developed disease comparable to wild-type recipients (Fig. 7b), indicating that casp-1 was dispensable for T cell induced auto-immune inflammation. Together, these data provide evidence that autoreactive T cells engaged TNFR and Fas on antigen presenting myeloid cells to induce IL-1β and autoimmune inflammation (Extended Data Fig. 7).
Discussion
We showed here that effector CD4+ T cells induced IL-1β transcription and cleavage in interacting MPs, including macrophages and DCs, in a TNFR-Fas-dependent manner. We found that effector CD4+ T cells of all lineages expressed TNF and FasL that engaged TNFR and Fas on multiple MP subsets, leading to production of IL-1β in these cells in a casp-8-dependent manner. The CD4+ T cell-induced IL-1β production was completely independent of PRR activation and did not depend on either canonical or non-canonical inflammasome activation. In a mouse model of passive EAE, T cell-instructed IL-1β was critical for auto-immune pathology. Our studies suggest that the pathway of IL-1β production described here is responsible for inflammation and pathology associated with T cell-driven auto-immune diseases.
While secretion of T cell-induced IL-1β was independent of any PRR activation, its production paralleled that of the inflammasome pathway, where two independent signals are required for the synthesis and subsequent proteolytic cleavage of pro-IL-1β. The distinction between these two mechanisms of IL-1β production can be further appreciated with regard to their physiological ramifications. The TLR-NLR inflammasome pathway is primarily employed by monocytes and macrophages, but not DCs, to induce IL-1β necessary for clearance of virulent pathogens. In contrast, the T cell-instructed IL-1β, which can be produced by both macrophages and DCs, appears to be responsible for auto-immune inflammation in the absence of overt pathogenic insult. Because DCs do not undergo inflammasome activation29, the TNFR-Fas-casp-8 pathway for production of IL-1β could be the primary mechanism used by DCs to aid CD4+ T cell function, while macrophages appear to employ both pathways for production of IL-1β, depending on the nature of the stimuli.
In addition to TNFR, other TNFR-family members, such as CD40 can also play a role in pro-IL-1β synthesis. The existence of diverse receptors for T cell-induced pro-IL-1β is parallel to the ability of different innate recognition receptors to upregulate pro-IL-1β. Fas signaling has been reported to induce the transcription of pro-IL-1β in TLR9-dependent autoimmune inflammation39. We did not find evidence for a role for Fas in the synthesis of pro-IL-1β, suggesting that Fas could contribute to the synthesis of IL-1β only in the context of canonical PRR ligands.
Analogous to the inflammasome pathway that culminates in pyroptosis, we observed that MPs underwent cell death upon interaction with T cells. The Fas-casp-8 pathway that was critical for production of T cell-induced IL-1β was also responsible for MP cell death. While we found a partial role for gasdermin-D in the secretion of T cell-instructed IL-1β, and casp-8 has been shown to cleave gasdermin-D40, it remains to be examined if gasdermin-D participates in casp-8-dependent cell death. Cell death is not essential for inflammasome-dependent IL-1β secretion41, therefore whether the casp-8-dependent cell death observed here is coupled to secretion of IL-1β or is merely a byproduct of casp-8 activation needs further investigation.
Aberrant inflammasome activation caused by gain-of-function mutations in NLRP3 and pyrin drive IL-1β-dependent auto-inflammatory conditions that are also initiated independently of TLRs and other PRRs12. However, the role of the inflammasome in T cell-driven autoimmune diseases is neither discernible nor established. While a role for casp-1 in EAE induction has been reported42, it is likely due to the ability of Mycobacterium tuberculosis in the MOG emulsion to trigger TLR32- and inflammasome-dependent IL-1β. We found that inflammasome was dispensable for T cell-dependent neuroinflammation, although neuroinflammation was completely dependent on IL-1β.
The ability of T cells to induce IL-1β in MPs engaging the T cells can be a double-edged sword for the health of the host. On one hand, it could license the function of memory CD4+ T cells, which is critical for anti-microbial immunity, but it could also cause systemic inflammation, as seen in many autoimmune scenarios. It is possible that the TNFR-Fas pathway of IL-1β production evolved primarily to support T cell function during interaction with cDCs. However, the interaction of auto-reactive T cells with macrophages, which might happen in tissues, likely produces higher quantities of IL-1β, which would further contribute to pathology. Because the amount of IL-1β produced by MPs was directly proportional to the concentration of the cognate peptide present, it is unlikely that bystander or low-avidity interaction between MPs and T cells would trigger the production of IL-1β. At the same time, self-reactive CD4+ T cells that have escaped thymic selection could engage this pathway during their reactivation, thereby leading to autoimmunity.
Single nucleotide polymorphisms have been reported in the Tnfrsf1a, Faslg and Fas loci in strong association with IL-1β-mediated autoimmune diseases43–47. Moreover, several lines of evidence support the involvement of TNFR and Fas signaling in autoimmune inflammation, yet their exact roles remain obscure48–50. Our data provides a mechanism for the role of TNFR and Fas signaling in IL-1β-driven autoimmunity, and suggests that in T cell-mediated autoimmunity, IL-1β is produced through T cell-instruction rather than activation of the inflammasome.
Methods
Mice
C57BL/6 wild-type control mice were obtained from the UT Southwestern Mouse Breeding Core Facility. IL-1β−/− mice were generated by David Chaplin, UA at Birmingham and provided to us by Fayyaz S. Sutterwala at Cedars Sinai. Rip3−/− and Rip3−/−Casp8−/− mice were provided by Andrew Oberst at the University of Washington. ASC KO mice were a gift from Genentech. Gsdmd−/− mice were provided by Jonathan Kagan at Boston Children’s Hospital. Caspase-1 KO were provided by Russell Vance at University of California, Berkeley. B6.MRL-Faslpr/J (lpr), B6.129S-Tnftm1Gkl/J (Tnfa−/−), B6.129S-Tnfrsf1atm1Imx Tnfrsf1btm1Imx/J (Tnfrsf1a/b−/−), Rag1tm1Mom, Casp1/11null, 2D2 TCR transgenic mice and Zbtb46gfp
were obtained from Jackson Laboratories. All mice were bred and housed in a specific pathogen-free facility at UT Southwestern Medical Center and Cincinnati Children’s Hospital Medical Center. For isolation of steady state CD4 memory T cells, mice were housed in a conventional facility for 2–4 weeks before tissue isolation. All mouse experiments were done as per protocols approved by Institutional Animal Care and Use Committee (IACUC) at UT Southwestern Medical Center and Cincinnati Children’s Hospital Medical Center.
Reagents
IL-1Ra (R&D Systems; 480-RM, 50ng/mL), rTNFα (Peprotech; 315–01Am, 20ng/mL), αCD3e (Biolegend; 100331 for in vitro assays and 100340 for in vivo assays), αTNF (Biolegend; 506332, 20μg/mL), αFasL (Biolegend; 106608, 10μg/mL), αCD40L (Biolegend; 310812, 10 μg/mL), IL-1β ELISA (R&D Systems; DY-401), TNFα ELISA(Biolegend; 510802 for capture and 506312 for detection), Naïve T cell Mojosort (Biolegend; 480040), total CD4+ T cell Mojosort (Biolegend; 480006), Zombie yellow (Biolegend; 423103), DAPI (Biolegend; 422801), IETD (R&D Systems; FMK007, 10μM), rGMCSF (Biolegend; 576306), Anti-biotin beads (Miltenyi Biotec; 130–090-485), OVA323–339 (Invivogen; vac-isq), LPS (Sigma, 100ng/mL), ATP (Invivogen; tlrl-atpl, 5nM), IL17a ELISA (Biolegend; 506902 for capture and 507002 for detection), EAE immunization kit (Hooke Laboratories, EK-2110) mouse MOG35–55 (CSbiologicals, CS0681)
Antibodies for flow cytometry
Anti-mouse Pro-IL-1β APC (eBioscience, 17–7114-80; 1:500), Anti-mouse CD11b BV785 (Biolegend, 101243; 1:400), Anti-mouse CD11c FITC (Biolegend; 117306, 1:400), Anti-mouse CD115 PE (Biolegend; 135505, 1:400), Anti-mouse Ly6G FITC (Biolegend, 127605; 1:400), Ly6C BV711 (Biolegend, 12803; 1:400), Anti-mouse TNFα FITC (Biolegend, 506304, 1:1000), Anti-mouse F4/80 APC-eflour 780 (Invitrogen, 47–4801-80; 1:400), Anti-mouse CD90 Pacific blue (Biolegend, 105324; 1:400), Anti-mouse FasL PE (Biolegend, 106605; 1:100), Anti-mouse CD62L biotin (Biolegend, 104404; 1:200), Anti-mouse CD11c biotin (Biolegend,117304; 1:500), Rorgt PE (ebioscience, 12–6988-82, 1:100), Tbet Pacific Blue (Biolegend, 644807, 1:100), GATA3 APC (Biolegend, 653806, 1:20), ICOS APC (Biolegend, 313509, 1:400), Foxp3 AF488 (Biolegend, 126405, 1:100). Additional information can be found in the Life science reporting summary.
Cell culture
Complete RPMI media (RPMI1640, Hyclone) supplemented with L-glutamine, penicillin-streptomycin, sodium pyruvate, β-mercaptoethanol (Sigma) was used throughout the experiments.
In vitro differentiation of naïve CD4+ T cells
Cell culture treated plates were coated with αCD3 (5μg mL−1) and αCD28 (5μg mL−1) for 3–4 h at 37°C. CD4+CD62Lhi naïve CD4+ T cells were isolated from splenocytes using a Mojosort naïve CD4 T cell isolation kit according to manufacturer’s protocol. Purified naïve T cells were plated in antibody-coated plates with appropriate polarizing conditions for 5 days. T cell were cultured in complete RPMI supplemented with 10% FCS. Cytokine cocktails for in vitro polarization: Th1 – IL-12 (10ng mL−1, peprotech), IL-2 (50U mL−1, peprotech), αIL-4 (10μg mL−1, Biolegend); Th17 – IL-6 (20ng mL−1, peprotech), hTGFβ (5ng mL−1, peprotech), αIL-4 (10μg mL−1, Biolegend), αIFNγ (10μg mL−1, Biolegend), IL-23 (20ng mL−1, Biolegend) and IL-1β (20ng mL−1, peprotech), Th2- IL-4 (4ng mL−1, peprotech), IL-2 (50U mL−1, Biolegend), and αIFNγ (10μg mL−1, Biolegend), Th0 – IL-2 (50U mL−1). For co-culture experiments T cells were rested in 10% RPMI supplemented with IL-2 (10U mL−1) for 36hrs before co-culture.
In vitro differentiation of bone marrow derived dendritic cells and retroviral transduction
Mouse progenitors were isolated from bone marrow (femurs and tibias). Following RBC lysis, cells were plated at 0.75×106 mL−1 in BMDC media (5% FCS containing complete RPMI + 1% rGMCSF (Biolegend, 100ng mL−1). Media was replaced on day 2 and day 4 and cells were harvested for experiments on day 5 by gently flushing each well. For sorting of BMDC subpopulations, total BMDC cells were stained with fluorescent antibodies and FACS sorted as shown (Extended Data Fig. 5a) For retroviral transduction, following RBC lysis cells were plated at 10×106 mL−1 in 2mL of retroviral supernatant containing 8μL mL−1 polybrene. Cells were spinfected at 2500 rpm, 32°C, for 90 minutes, then 3mL of BMDC media was added to each well and cells were incubated overnight. The next morning, ~70% of the media was removed from the wells and spinfection was repeated with fresh retroviral supernatant. On day 5 cells were harvested for experiments.
In vitro differentiation of bone marrow derived macrophages
Mouse progenitors were isolated from bone marrow (femurs and tibias). Following RBC lysis, cells were plated in BMDM media in TC treated culture dish (5% FCS containing complete RPMI + 30% L929 supernatant). On day one, non-adherent cells were transferred to a non-treated petri dish. After 5–7 days of culture in BMDM media, macrophages were obtained from the dish by EDTA treatment.
Isolation of CD44hiCD62Llo cells from spleen and lymph nodes
Single cell suspension was obtained from spleen and peripheral lymph nodes. 4–8 mice were pooled for each experiment. CD4+ T cells were isolated using mouse CD4 T cells isolation kit (Biolegend) following manufacturer’s instructions. Cells were then labeled with anti-CD62L biotin antibody followed by anti-biotin microbeads (Miltenyi). CD62Llo cells were isolated using negative selection on AutoMacs. Approximately 95% cells were CD4+CD44hiCD62Llo.
Purification of various myeloid cells populations
Splenocytes or total BMDCs were labelled with anti-CD11c biotin antibody followed by anti-biotin microbeads (Miltenyi). CD11c+ cells were magnetically sorted using positive selection on AutoMacs. CD11c+ enriched cells were then sorted based on various markers as described in the Extended Data Fig. 5 using Moflo cell sorter. Sorting strategy and post-sort purities are shown in Extended Data Fig. 5.
Co-culture of DC and T cells for induction of IL-1β
1 million DCs were cultured with 4 million T cells in 12 well plate. DC-T cell interaction was triggered by adding either αCD3ε (αCD3, 200ng mL−1) or OVA323–339 as described in the legends. In experiments measuring secreted IL-1β, IL-1R antagonist (50ng mL−1) was added 1hr prior to DC stimulation to block IL-1β consumption. Co-cultures were also pretreated with neutralizing antibodies and inhibitor wherever described. Co-culture experiments were performed in complete RPMI supplemented with 10% FCS. For Western blot analysis of the culture supernatant cells were cultured in 1% FCS containing complete RPMI.
T cell re-stimulation (in vivo)
6–8 weeks old mice were treated with 20μg αCD3 or PBS by i.p. injection. Lamina propria cells and splenocytes were isolated at given time points after stimulation followed by surface and intracellular staining.
Quantitative real-time PCR
RNA was isolated using Qiagen RNA extraction kit using manufacturer’s protocol. cDNA was synthesized using Random primers and MMLV reverse transcriptase (Invitrogen; 2805–013). The QuantStudio 7 Flex Real-Time PCR System (ThermoFisher Scientific) was used to measure SyBR green (ThermoFisher Scientific) incorporation. All data is normalized to 18s. qPCR primers sequences are as follows: Il1b: Fwd-5’TGTGCTCTGCTTGTGAGGTGCTG 3’, Rev 5’CCCTGCAGCTGGAGAGTGTGGA3’ | Hprt1: Fwd-5’ CAGTCCCAGCGTCGTGATTA-3’, Rev-5’ TGGCCTCCCATCTCCTTCAT-3’ | 18S: Fwd-5’ GTAACCCGTTGAACCCCATT, Rev-5’ CCATCCAATCGGTAGTAGCG
Immunoblot analysis
Cells were lysed in 1X RIPA Buffer and protein was quantified using Pierce™ BCA Protein Assay Kit. Cell lysates where boiled in 1X Laemmli buffer at 95°C for 10 mins. Cell lysates were separated by SDS–PAGE and transferred onto PVDF membranes. Blots were incubated with anti-IL1β [1:1000] (R&D AF-401-SP), anti-caspase8 [1:1000] (Enzo ALX-804–447-C100), anti-caspase1 [1:1000] (Genentech), anti-βtubulin [1:5000] (CST 2146S). As secondary antibodies, anti-rabbit-IgG-HRP (Biorad) [1:5000], anti-mouse-IgG-HRP and anti-goat-IgG-HRP [1:10000] (Jackson ImmunoResearch Laboratory) were used. Anti-β-actin (C4, Santa Cruz, 1:5000) was used as control. Western blot was develop using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher) and ECL signal was recorded on X-Ray Films using a developer (Kodak).
Surface and intracellular staining and flow cytometry
Cells were stained with relevant antibodies for 30 min on ice and washed. For intracellular staining, Foxp3 staining buffer set (eBioscience; 00–5523-00) was used according to manufacturer’s protocol. The stained cells were analyzed with BDLSRII or Novocyte (ACEA biosciences). For cytokine receptor staining, control refers to fluorescence minus one control. Data were analyzed with FlowJo 10 Software.
Isolation of lamina propria lymphocytes
Small intestines were flushed with cold PBS and carefully cut longitudinally. 1–2cm pieces of the tissue were digested twice with 2mM EDTA buffer followed by 3 rounds of enzymatic digestion with Collagenase IV (10 μg/mL, Sigma) and DNase I (500 μg/mL, Sigma). Cell suspensions obtained after digestions were loaded on a 40–70% percoll gradient as described before.
Passive EAE induction
9–10 weeks old WT female mice were immunized with MOG35–55 emulsion obtained from the Hooke lab EAE immunization kit as per manufacturer’s protocol. Mice were also injected with 80ng Pertussis toxin intraperitoneally on day 0 and day 1. Total splenocytes were harvested 11–14 days after immunization and cultured in vitro with MOG (20μg/mL), anti-IFNγ (10μg/mL) and IL-23 (10ng/mL). After 3 days of reactivation, total CD4+ T cells isolated using Mojosort Kit (Biolegend). 5–10×106 CD4+ T cells were transferred intravenously into given genotypes of mice that were sub-lethally irradiated (400 rad) the day before transfer. Recipients also received 80ng Pertussis toxin intraperitoneally on day 0 and day 1 of transfer. Mice were monitored daily and disease severity was scored as follows: 0 = no clinical signs of paralysis, 1 = tail paralysis, 2 = tail paralysis and hind limb weakness, 3 = Complete hind limb weakness, 4 = forelimb paralysis or moribund.
Histology
On day 28 after adoptive transfer, mice were sacrificed and perfused with ice cold PBS. Spinal cords were collected and fixed with 10% neutral-buffered formalin for 36h. Fixed tissues submitted to Cincinnati Children’s Hospital Medical Center research pathology core for tissue embedding and Luxol fast blue stain. Slides were imaged at 10X using a Nikon Eclipse Tι.
Quantification and statistical analysis
Based on previous and preliminary studies in our lab, we predicted that the reported samples sizes would be sufficient to ensure adequate power. Statistical analyses were performed in Prism (Graph pad) using unpaired or paired Student’s t test as indicated in the figure legends. Data are presented as means +/− SEM. Significance was considered at *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. n.s. = not significant.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon request
Extended Data
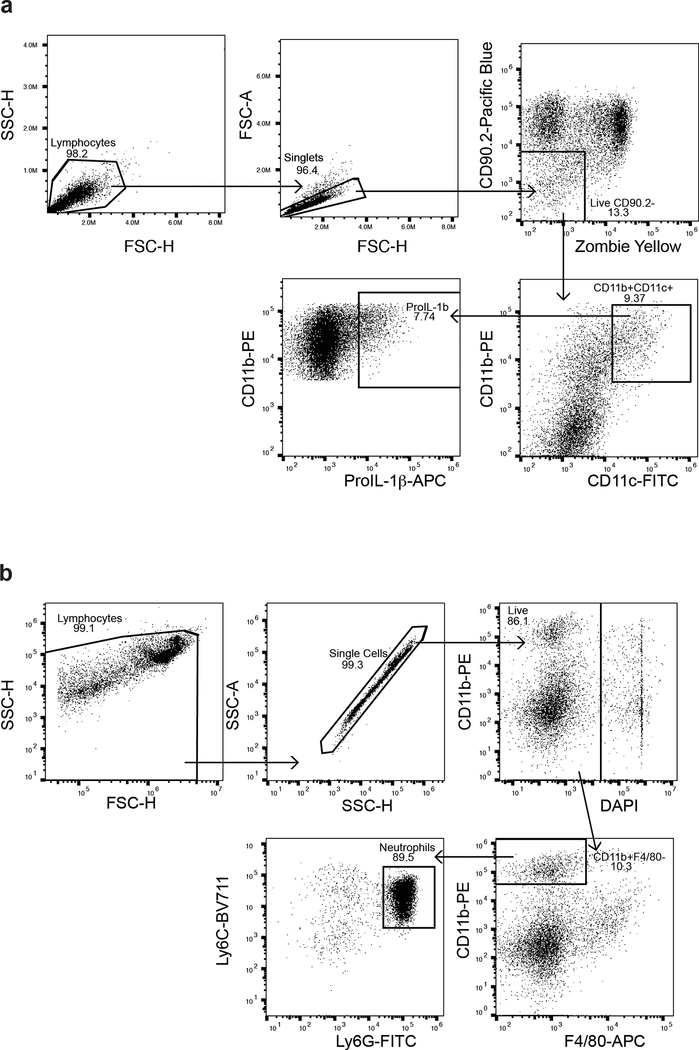
Extended Data Fig. 1: Representative Gating strategy for flow cytometric analysis.
(a) Gating strategy for flow cytometric analysis of intracellular pro-IL-1β in CD11c+ DCs following BMDC-T cell co-culture. (b) Gating strategy for flow cytometric analysis of CD11b+ monocytes and neutrophils frequency in various tissues.
Extended Data Fig. 2: Induction of pro-IL-1β in BMDCs is independent of TLR activation but dependent on TNF.
(a and b) Expression of intracellular pro-IL-1β measured by flow cytometry in WT, Tlr2−/−Tlr4−/−, or Myd88−/− BMDCs (live,CD90-CD11c+) cultured with TH0 cells in the presence of CD3 Ab for 6 h. Fold change indicates proportion of pro-IL-1β+ BMDCs, compared to PBS controls. Error bars indicate SEM three (n=3) independent experiments. (c) Expression of intracellular pro-IL-1β measured by flow cytometry in WT BMDCs (live, CD11c+) stimulated in vitro with recombinant TNF (20ng/mL) for 6 h. Data are representative of two independent experiments. (d) TNFα, as quantified by ELISA, in the supernatants of WT or Tnf−/− TH0 cells cultured with WT or Tnf−/− BMDCs in the presence of CD3 Ab for 6 h. Error bars indicate SEM from n=4 independent experiments. (e) lL-1β was quantified by ELISA in the supernatants of WT TH0 cells cultured with BMDCs of the indicated genotypes in the presence of CD3 Ab and neutralizing CD40L Ab (10μg/mL) for 6 h. Error bars indicate SEM from n=2 technical replicates. Data are representative of two independent experiments. (a, b, d) Statistical analysis was performed by paired, two-tailed Student’s t-test. *p<0.05.
Extended Data Fig. 3: T cell-induced IL-1β production is only partially impaired in gasdermin-D deficient BMDCs.
(a) lL-1β was quantified by ELISA in the supernatants of WT TH0 cells cultured with Il1b−/− BMDCs transduced with retrovirus expressing WT or D117A pro-IL-1β in the presence of CD3 Ab for 18 h. Error bars indicate SEM from n=2 technical replicates. Data are representative of two independent experiments. (b) lL-1β was quantified by ELISA in the supernatants of WT TH0 cells cultured with WT or Gsdmd−/− BMDCs in the presence of CD3 Ab for 12 h. Error bars indicate SEM from n=6 independent experiments. Statistical analysis was performed by paired, two-tailed Student’s t-test. **p<0.01.
Extended Data Fig. 4: T cells induce Fas-caspase-8 dependent death of interacting BMDCs.
(a) Expression of surface FasL measured by flow cytometry in in vitro polarized TH1, TH2 and TH17 cells (live, CD90.2+) cultured with WT BMDCs in the presence of CD3 Ab for 1 h. Data are representative of two independent experiments. (b and c) Cell death as assayed by Zombie Yellow viability dye was measured by flow cytometry in WT, Ripk3−/−, or Ripk3−/−Casp8−/− live,CD90.2+CD11c+ BMDCs cultured with WT TH0 cells in the presence or absence of CD3 Ab and neutralizing TNF Ab (20μg/mL) or FasL Ab (10μg/mL) for 6 h. Data are representative of two independent experiments.
Extended Data Fig. 5: Gating strategy and post sort purity of various myeloid cell populations using in Figure 5.
(a and b) Pre-sort and post-sort purity of FACS sorted BMDC subsets: CD11b+, CD11b+CD11c+MHCIIhi, or CD11b+CD11c+MHCIIint BMDCs (b) and CD11c+MHCIIintCSF1R+ (GM-Macs) or CD11c+MHCIIhiCSF1R- (GM-DCs). Data are representative of 4 independent experiments. (c) Post-sort purity of CD11c+ splenic DCs showing lack of CSF1R+ cell contamination. Data are representative of 4 independent experiments. (d) Pre-sort and post-sort purity of CD11c+Zbtb46-GFP+Ly6C- splenic cDCs that were FACS sorted from pre-sorted total CD11c+ Zbtb46-GFP splenocytes to obtain a pure cDC population. Data are representative of three independent experiments.
Extended Data Fig. 6: Anti-CD3 stimulation of T cells in vivo leads to IL-1β dependent inflammatory cell recruitment dictated by Fas expression on CD11c+ cells.
(a) Expression of intracellular pro-IL-1β measured by flow cytometry in CD11b+Ly6C-CSF1R+ macrophages, CD11b+Ly6G-Ly6Cint monocytes, CD11b+Ly6Chi inflammatory monocytes, and CD11b+Ly6G+ granulocytes quantified from the spleens of WT mice 3–4 h post CD3 Ab injection (50μg, i.v.). n=3 independent experiments are quantified in the inset. Statistical analysis was performed by paired, one-tailed Student’s t-test. (b) Neutrophil infiltration as measured by flow cytometry in the spleen (left panel) or SI-LP (right panel) of WT mice 18 h post CD3 Ab injection (20μg, i.p.). Error bars indicate SEM from n=4 independent experiments. (c) qPCR of Il1b mRNA in lysates of splenocytes collected from WT or Rag1−/− mice 4 h post CD3 Ab injection (50μg, i.v.). Data are normalized to Hprt. Error bars indicate SEM from n=3 technical replicates. Data are representative of two independent experiments. (d) Neutrophil infiltration in the spleen of Rag1−/− mice as measured by flow cytometry 3–4 h post CD3 Ab injection (50μg, i.v.). Error bars indicate SEM from n=3 independent experiments. (e) Expression of Foxp3 and (f) ICOS measured by flow cytometry in splenic live, CD4+ T cells from WT mice, 18 h post CD3 Ab injection (20μg, i.p.). Error bars indicate SEM from n=3 independent experiments. (g) Infiltration of CD11b+ cells as measured by flow cytometry in the SI-LP or (h) spleen of WT and Il1b−/− mice, 18 h post CD3 Ab injection (20μg, i.p.). Error bars indicate SEM from n=3 independent experiments. (i) Expression of cell surface Fas measured by flow cytometry in CD11b+ or CD11c+ BMDCs from given genotypes. Data are representative of two independent experiments. (j) Neutrophil infiltration (live, CD11b+F480-) as measured by flow cytometry in the SI-LP of Fasfl/fl or Fasfl/fl x CD11c-cre (FasΔDC) mice, 18 h post CD3 Ab injection (20μg, i.p.). Error bars indicate SEM from n=5 independent experiments. Splenocytes were taken from WT mice immunized with MOG35–55, and stimulated for 4 d in vitro with MOG in the presence of IL-1β, IL-23, and anti-IFNγ. (k) lL-1β was quantified by ELISA in the supernatants of WT BMDCs cultured with CD4+ T cells isolated from the stimulated splenocyte culture in the presence of MOG35–55 and neutralizing antibodies TNF Ab (20μg/mL) or FasL Ab (10μg/mL) for 24 h. Error bars indicate SEM from n=2 independent experiments. (b-h,j) Statistical analysis was performed by unpaired, one-tailed Student’s t-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, n.s.=not significant.
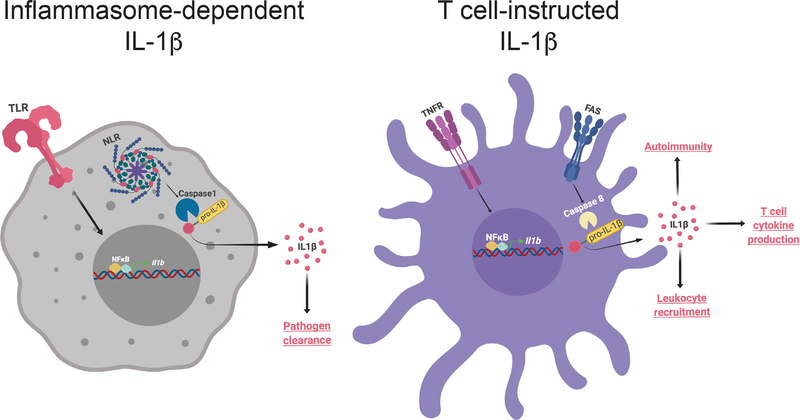
Extended Data Fig. 7: Illustration of “T cell-instructed” IL-1β production by MPs and its comparison to inflammasome induced IL-1β production by macrophages.
(Left) During inflammasome activation in monocytes and macrophages, TLR and NLR-caspase-1 activation leads to synthesis and cleavage of pro-IL-1β, respectively. Robust production of IL-1β as a result of inflammasome activation is critical for pathogen clearance. (Right) In contrast, “T cell-instructed” IL-1β production by antigen presenting MPs utilizes TNFR signaling for pro-IL1β synthesis while the cleavage signal is provided by Fas-caspase-8 axis. The IL-1β produced upon such T cell instruction drives cytokine production by effector CD4 T cells, systemic leukocyte recruitment, and autoimmunity. Figure was created using Biorender.
Acknowledgements
We thank all the members of the Pasare lab and R. Bagirzadeh for helpful discussions. Thanks to A. Ma for sharing WT and D117A Il1b constructs. Thanks to M. Jordan for sharing Zbtb46-GFP reporter mice. This work was supported by grants from the National Institutes of Health (AI113125 and AI123176) to C.P. M.M.M was supported by National Science Foundation Graduate Research Fellowship under Grant No. 2017220107.
Footnotes
Competing interest statement
The authors have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
References
- 1.Zheng H et al. Resistance to fever induction and impaired acute-phase response in interleukin-1 beta-deficient mice. Immunity 3, 9–19 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Miller LS et al. MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity 24, 79–91, doi: 10.1016/j.immuni.2005.11.011 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Jain A, Song R, Wakeland EK & Pasare C T cell-intrinsic IL-1R signaling licenses effector cytokine production by memory CD4 T cells. Nat Commun 9, 3185, doi: 10.1038/s41467-018-05489-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinarello CA Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 117, 3720–3732, doi: 10.1182/blood-2010-07-273417 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broderick L, De Nardo D, Franklin BS, Hoffman HM & Latz E The inflammasomes and autoinflammatory syndromes. Annu Rev Pathol 10, 395–424, doi: 10.1146/annurev-pathol-012414-040431 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Goodnow CC, Sprent J, Fazekas de St Groth B & Vinuesa CG Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 435, 590–597, doi: 10.1038/nature03724 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Masters SL, Simon A, Aksentijevich I & Kastner DL Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*). Annu Rev Immunol 27, 621–668, doi: 10.1146/annurev.immunol.25.022106.141627 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopalco G et al. Interleukin-1 as a common denominator from autoinflammatory to autoimmune disorders: premises, perils, and perspectives. Mediators Inflamm 2015, 194864, doi: 10.1155/2015/194864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raphael I, Nalawade S, Eagar TN & Forsthuber TG T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 74, 5–17, doi: 10.1016/j.cyto.2014.09.011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bluestone JA, Bour-Jordan H, Cheng M & Anderson M T cells in the control of organ-specific autoimmunity. J Clin Invest 125, 2250–2260, doi: 10.1172/JCI78089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schenten D et al. Signaling through the Adaptor Molecule MyD88 in CD4(+) T Cells Is Required to Overcome Suppression by Regulatory T Cells. Immunity 40, 78–90, doi: 10.1016/j.immuni.2013.10.023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw PJ, McDermott MF & Kanneganti TD Inflammasomes and autoimmunity. Trends Mol Med 17, 57–64, doi: 10.1016/j.molmed.2010.11.001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ippagunta SK et al. Inflammasome-independent role of apoptosis-associated speck-like protein containing a CARD (ASC) in T cell priming is critical for collagen-induced arthritis. J Biol Chem 285, 12454–12462, doi: 10.1074/jbc.M109.093252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schott WH et al. Caspase-1 is not required for type 1 diabetes in the NOD mouse. Diabetes 53, 99–104 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Cogswell JP et al. NF-kappa B regulates IL-1 beta transcription through a consensus NF-kappa B binding site and a nonconsensus CRE-like site. J Immunol 153, 712–723 (1994). [PubMed] [Google Scholar]
- 16.Martin BN et al. T cell-intrinsic ASC critically promotes T(H)17-mediated experimental autoimmune encephalomyelitis. Nat Immunol 17, 583–592, doi: 10.1038/ni.3389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinon F, Burns K & Tschopp J The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10, 417–426 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Baldassare JJ, Bi Y & Bellone CJ The role of p38 mitogen-activated protein kinase in IL-1 beta transcription. J Immunol 162, 5367–5373 (1999). [PubMed] [Google Scholar]
- 19.Geppert TD, Whitehurst CE, Thompson P & Beutler B Lipopolysaccharide signals activation of tumor necrosis factor biosynthesis through the ras/raf-1/MEK/MAPK pathway. Mol Med 1, 93–103 (1994). [PMC free article] [PubMed] [Google Scholar]
- 20.Li CR, Mueller EE & Bradley LM Islet antigen-specific Th17 cells can induce TNF-alpha-dependent autoimmune diabetes. J Immunol 192, 1425–1432, doi: 10.4049/jimmunol.1301742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franchi L, Eigenbrod T, Munoz-Planillo R & Nunez G The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol 10, 241–247, doi: 10.1038/ni.1703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayagaki N et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671, doi: 10.1038/nature15541 (2015). [DOI] [PubMed] [Google Scholar]
- 23.He WT et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res 25, 1285–1298, doi: 10.1038/cr.2015.139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bossaller L et al. Cutting edge: FAS (CD95) mediates noncanonical IL-1beta and IL-18 maturation via caspase-8 in an RIP3-independent manner. J Immunol 189, 5508–5512, doi: 10.4049/jimmunol.1202121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Du F & Wang X TNF-alpha induces two distinct caspase-8 activation pathways. Cell 133, 693–703, doi: 10.1016/j.cell.2008.03.036 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Lemmers B et al. Essential role for caspase-8 in Toll-like receptors and NFkappaB signaling. J Biol Chem 282, 7416–7423, doi: 10.1074/jbc.M606721200 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Philip NH et al. Activity of Uncleaved Caspase-8 Controls Anti-bacterial Immune Defense and TLR-Induced Cytokine Production Independent of Cell Death. PLoS Pathog 12, e1005910, doi: 10.1371/journal.ppat.1005910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helft J et al. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity 42, 1197–1211, doi: 10.1016/j.immuni.2015.05.018 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Erlich Z et al. Macrophages, rather than DCs, are responsible for inflammasome activity in the GM-CSF BMDC model. Nat Immunol 20, 397–406, doi: 10.1038/s41590-019-0313-5 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Satpathy AT et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med 209, 1135–1152, doi: 10.1084/jem.20120030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bachmann MF & Kopf M On the role of the innate immunity in autoimmune disease. J Exp Med 193, F47–50 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janeway CA Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54 Pt 1, 1–13 (1989). [DOI] [PubMed] [Google Scholar]
- 33.VanderBorght A, Geusens P, Raus J & Stinissen P The autoimmune pathogenesis of rheumatoid arthritis: role of autoreactive T cells and new immunotherapies. Semin Arthritis Rheum 31, 160–175, doi: 10.1053/sarh.2001.27736 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Fletcher JM, Lalor SJ, Sweeney CM, Tubridy N & Mills KH T cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Clin Exp Immunol 162, 1–11, doi: 10.1111/j.1365-2249.2010.04143.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pugliese A Autoreactive T cells in type 1 diabetes. J Clin Invest 127, 2881–2891, doi: 10.1172/JCI94549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mannie MD, Dinarello CA & Paterson PY Interleukin 1 and myelin basic protein synergistically augment adoptive transfer activity of lymphocytes mediating experimental autoimmune encephalomyelitis in Lewis rats. J Immunol 138, 4229–4235 (1987). [PubMed] [Google Scholar]
- 37.Lin CC & Edelson BT New Insights into the Role of IL-1beta in Experimental Autoimmune Encephalomyelitis and Multiple Sclerosis. J Immunol 198, 4553–4560, doi: 10.4049/jimmunol.1700263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adachi M et al. Enhanced and accelerated lymphoproliferation in Fas-null mice. Proc Natl Acad Sci U S A 93, 2131–2136, doi: 10.1073/pnas.93.5.2131 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mande P et al. Fas ligand promotes an inducible TLR-dependent model of cutaneous lupus-like inflammation. J Clin Invest 128, 2966–2978, doi: 10.1172/JCI98219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orning P et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069, doi: 10.1126/science.aau2818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conos SA, Lawlor KE, Vaux DL, Vince JE & Lindqvist LM Cell death is not essential for caspase-1-mediated interleukin-1beta activation and secretion. Cell Death Differ 23, 1827–1838, doi: 10.1038/cdd.2016.69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furlan R et al. Caspase-1 regulates the inflammatory process leading to autoimmune demyelination. J Immunol 163, 2403–2409 (1999). [PubMed] [Google Scholar]
- 43.De Jager PL et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 41, 776–782, doi: 10.1038/ng.401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinks A et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet 45, 664–669, doi: 10.1038/ng.2614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.International Multiple Sclerosis Genetics, C. et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 45, 1353–1360, doi: 10.1038/ng.2770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin Y et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat Genet 48, 1418–1424, doi: 10.1038/ng.3680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsoi LC et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat Commun 8, 15382, doi: 10.1038/ncomms15382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itoh N et al. Requirement of Fas for the development of autoimmune diabetes in nonobese diabetic mice. J Exp Med 186, 613–618 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kruglov AA, Lampropoulou V, Fillatreau S & Nedospasov SA Pathogenic and protective functions of TNF in neuroinflammation are defined by its expression in T lymphocytes and myeloid cells. J Immunol 187, 5660–5670, doi: 10.4049/jimmunol.1100663 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Varanasi V, Avanesyan L, Schumann DM & Chervonsky AV Cytotoxic mechanisms employed by mouse T cells to destroy pancreatic beta-cells. Diabetes 61, 2862–2870, doi: 10.2337/db11-1784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request