Abstract
Efficient HIV-1 transduction depends on a number of cellular co-factors. Cellular double-strand DNA break (DSB) repair proteins have been proposed, by ourselves and others, to be required for efficient HIV-1 transduction. Expression and/or activity of these DNA repair proteins can be induced by the introduction of DSBs into the host cell genome. HIV-1 transduction was up-regulated by treatment with DSB-inducing agents in both drug-arrested cells and differentiated neuronal cells. The presented data support the hypothesis that DSB repair proteins are involved in the early steps of the retroviral life-cycle.
Keywords: gene therapy, HIV, vector, DNA damage
Introduction
Gene therapy depends on successful transfer of a desired gene into a patient’s cells. A vast number of gene therapy trials have employed retroviral vectors to this purpose. These are versatile tools that can successfully transduce a gene into a host cell’s genome, in a process called retroviral DNA integration. Efficiency of retroviral transduction is a rate-limiting step of gene therapy and is, in many cases, quite low and insufficient to achieve the therapeutic objectives (Tomanin & Scarpa 2004).
Integration of viral DNA into host DNA is an essential step in the replication cycle of retroviruses, retroviral vectors, and retrotransposons (Coffin et al. 1997; Skalka 1999; Flint 2004). The first two steps in integration, denoted as processing and joining, are catalyzed by the retroviral protein integrase. In the first step, nucleotides (usually 2) are removed from the 3´-ends of the viral DNA, and in the second step, these newly created ends are joined to staggered phosphates in the complementary strands of the host cell DNA. The host cell DNA essentially suffers a double-strand break whose ends are held together by single-strand links to viral DNA sequences. Opposite short gaps in the complementary strands are generated by the staggered joining reaction. The gaps are filled and chromatin structure is reconstituted in the process of post-integration repair. Host cell proteins were usually assumed to be required for this step of the retroviral life-cycle. Cumulative evidence suggests that post-integration repair requires proteins that are involved in cellular double-strand DNA break (DSB) repair (see below).
Post-integration repair is absolutely required for successful retroviral transduction and occurs quite efficiently in cells expressing high levels of normal DSB repair proteins. In contrast, post-integration repair fails in cells expressing mutant forms or low intracellular amounts of these proteins (Daniel et al. 1999; Lau et al. 2004; Skalka & Katz 2005; Smith & Daniel 2006; Smith et al. 2008). Thus, efficiency of post-integration repair appears to be one of the factors which determines the efficiency of retroviral transduction. Intracellular levels and activities of DNA repair proteins increase when cells are exposed to DNA damage (Negroni et al. 2008). Because of this, we hypothesized that treatment with low, subtoxic concentrations of DSB-causing agents may increase the efficiency of retroviral transduction. This hypothesis was partly tested in dividing cells where the efficiency of stable integration increased following treatment with these agents (Groschel & Bushman 2005). This was proposed to be due to these agents arresting growth thereby permitting more time for integration to be completed (Groschel & Bushman 2005). However, we note that most cells in the body are non-dividing at the time of retroviral transduction in gene therapy applications, and therefore it needs to be tested to see if this approach works in non-dividing cells. In addition, if the proposed mechanism (i.e. induction of growth arrest) should apply, treatment with DSB-causing agents should not work in nondividing cells, since they are arrested regardless of the presence or absence of DSB-causing agents. In this study, we demonstrate that DSB-causing agents significantly upregulate the efficiency of stable retroviral transduction and integration in both growth-arrested fibroblasts and differentiated neurons. Our results suggest that it is feasible to upregulate transduction using this approach in gene therapy applications. Additionally, our data demonstrates that the DNA damage-caused growth arrest is not a necessary component of the mechanism underlying this effect.
Materials and methods
Cells
293T/17 and rat B35 cells were purchased from ATCC (Manassas, VA) and carried in DMEM medium with 10% (v/v) fetal bovine serum (FBS) and 1% (w/v) penicillin/streptomycin. Primary A-T fibroblasts (deficient in the ATM protein - GM02052) and matched controls (GM01661) were purchased from the Coriell Cell Repository (Camden, NJ). Fibroblasts were maintained in RPMI-1640 medium 10% (v/v) FBS, 5×10−6M 2-mercaptoethanol, non-essential amino acids, and 1% penicillin/streptomycin.
HIV-1-based vectors
The VSV G-pseudotyped HIV-1 based vector was prepared as described previously (Naldini et al. 1996; Daniel et al. 2004), and carried an EGFP reporter gene.
Infections
Following growth arrest (see below), cells were infected overnight in the presence 10 ug DEAE-dextran per ml, at multiplicity of infection (m.o.i.) of 0.1. Real-time PCR assays (see below) were performed two days post-infection. Virus was added to cells together with DSB-causing agents. Both virus and the DSB-causing agents were kept on cells until they were harvested for real-time PCR assays.
Induction of growth arrest
To induce G2/M arrest, 293T/17 cells were treated with 1 ug nocodazole/ml for 24 h. To induce G1/S arrest, cells were treated with 1 ug aphidicolin/ml for 24 h. To differentiate B35 cells, cells were treated for five days with 1 mM dibutyryl-cAMP, (Biomol, Plymouth Meeting, PA). Following induction of growth arrest, DSB-causing agents and/or virus were added. Nocodazole, aphidicolin or dibutyryl-cAMP were kept on cells throughout the experiment.
Cytotoxicity assay
Cells were plated at 103 cells (B35/dibutyryl-cAMP) or 3×104 cells (fibroblasts/aphidicolin) or 5×104 cells (fibroblasts/nocodazole) per well of a 96-well plate and growth arrested as described above. Bleomycin or etoposide were then added to the indicated final concentrations. Cytotoxicity of DSB-causing agents was measured using the Cell Proliferation Kit II (XTT) (Roche Diagnostics, Indianapolis, IN), following the company protocol, at 3 days after the addition of bleomycin or etoposide.
Real-time PCR assays
To detect and quantify fully integrated proviral DNA, a two-step nested Alu-PCR technique was conducted. Cells were infected with HIV-1-based vector (with an EGFP reporter, see above). Two days post-infection genomic DNA was extracted (QIAmp DNA Mini Kit, Qiagen) and DNA from 8×103 cells was used for Alu-PCR. The first round of Alu-PCR employed a primer targeting the cellular Alu sequence 5’-GCC TCC CAA AGT GCT GGG ATT ACA G-3’ as well as the primer targeting the HIV-1 gag region, 5’-TTT TGG CGT ACT CAC CAG TCG-3’. In addition, all samples were run also in the absence of the Alu primer during this first round of PCR in order to confirm that our final readout was indeed integrated DNA. In this initial amplification step 150 ng genomic DNA was used as a template. Samples were subjected to 30 PCR cycles of 95°C for 30 s, 60°C for 45 s, and 72°C for 5 min and, after the final round, samples were kept at 72°C for 10 min. Products of the first round were used in the second, real-time PCR round with a FAM/Iowa Black FQ dual-labeled probe 5’-(FAM)-CAG TGG CGC CCG AAC AGG GA-(TAMRA)-3’ (Integrated DNA Technologies), an LTR forward primer: 5’-TGT GTG CCC GTC TGT TGT GT-3’; and a GAG reverse primer 5’-CCT GCG TCG AGA GAG CTC-3’. Real-time PCR reactions were performed using a LightCycler 1.5 with software 3.5.3 (Roche). Reaction mixtures contained QuantiFast Probe 2× mix (Qiagen, Valencia, CA), 100 nM probe, and 200 nM primers. The standard cycling conditions were 95C – 3 min followed by 50 cycles at 95°C for 3 s and 60°C for 30 s. Samples were run in duplicate. We note that B35 (neuronal progenitor) cells are of rat origin. The rat genome does not have Alu sequences, but they possess B1 elements, which have high homology to Alu. Thus, we were able to employ the Alu primer to detect integration in these cells as well.
To detect the overall amount of viral DNA in cells (both unintegrated and integrated), real-time PCR was performed under the conditions described for the second round of Alu-PCR, except the genomic DNA was used as input, instead of the first round PCR product as in the case of Alu-PCR experiments.
Results
Toxicity of selected DSB-causing agents
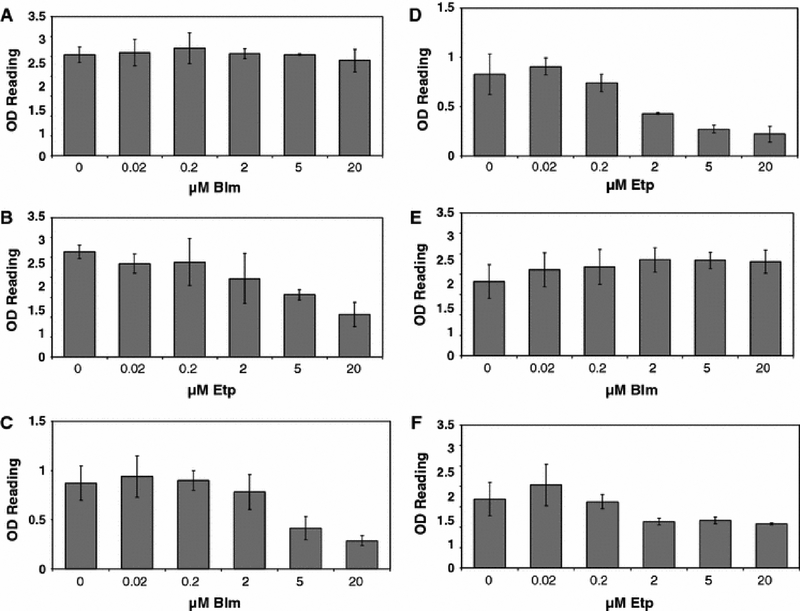
There are several ways to induce DSBs in cells. A commonly used approach is to expose cells to ionizing radiation. However, for potential gene therapy applications it appears to be more feasible to use chemicals that induce DSBs rather than ionizing radiation as a DSB causing agent. In this study, we employed two well-characterized agents. First, we used etoposide, which is a topoisomerase II poison and bocks the ligation step by this enzyme, thus causing the break (Li & Liu 2001). Second, we employed belomycin sulfate (bleomycin). This agent induces DSBs by the production of superoxide and hydroxide free radicals, which cleave DNA (Saunders & Schultz 1972; Bennett & Reich 1979; Povirk et al. 1979). Given the different mechanism of action, it is expected that these agents may exhibit different levels of toxicity in different cell types. We examined the effects of etoposide and bleomycin on the viability of growth-arrested primary fibroblasts and neural B35 cells, which were induced to terminally differentiate. As shown in Fig. 1A, bleomycin had no toxicity on nocodazole (G2/M)-arrested primary fibroblasts up to a very high concentration of 20 uM, whereas etoposide was somewhat more toxic, notably so starting at around 2–5 uM (Fig. 1B). In contrast, both agents exhibited higher toxicity on aphidicolin (G1S)-arrested cells (Fig. 1C and D). The OD reading for bleomycin treated samples was reduced to 50% of the control sample reading at around 5 uM, while the reading of etoposide treated samples was reduced by half at 2 uM. Neuronal cells showed a pattern similar to that of nocodazole-arrested fibroblasts. Bleomycin was not toxic up to 20 uM, whereas etoposide was somewhat toxic starting at 2 uM (Fig. 1E and F).
Fig. 1.
Cytotoxicity of bleomycin and etoposide. A. Fibroblasts (GM01661) were growth arrested with nocodazole, and exposed to bleomycin at the indicated concentrations. Cell viability was measured using the XTT assay. B. Nocodazole-treated fibroblasts exposed to etoposide at the indicated concentrations. C. Aphidicolin-treated fibroblasts exposed to bleomycin concentrations as indicated. D. Aphidicolin-treated fibroblasts exposed to etoposide at the indicated concentrations. E. Neuronal cells exposed to bleomycin. F. Neuronal cells exposed to etoposide. Error bars indicate standard deviations.
Effects of bleomycin and etoposide on transduction efficiency in nocodazole-arrested primary fibroblasts
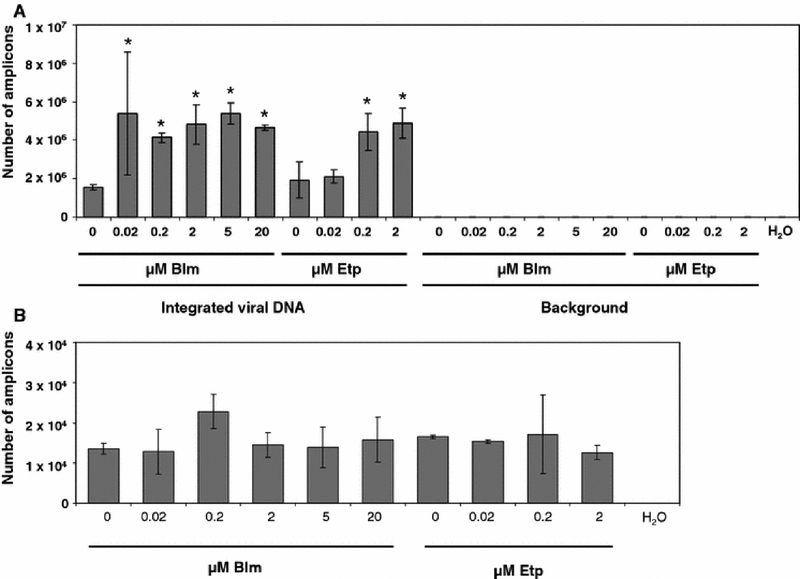
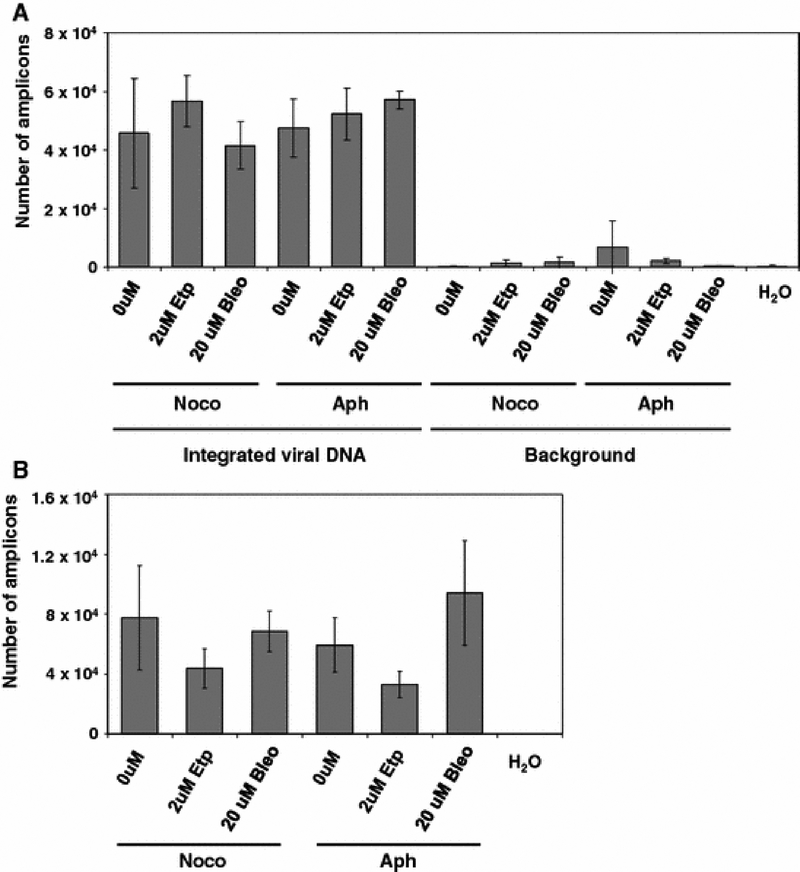
Primary fibroblasts were arrested in the G2/M phase using nocodazole. Following growth arrest we infected these cells with an HIV-1-based vector which can infect nondividing cells (Weinberg et al. 1991; Lewis et al. 1992). We then measured the efficiency of stable integration using Alu-PCR. In addition, we measured the total viral DNA synthesis in these cells. As shown in Fig. 2A, bleomycin treatment increased the number of transduced cells 2.7-fold, when cells were treated with 0.2 uM bleomycin, which is does not affect cell viability (Fig. 1). Further increase in bleomycin concentration did not notably enhance transduction any further. A different pattern was observed in etoposide treated samples. As shown in Fig. 2A, this agent also increased transduction. Transduction was upregulated by 2.5-fold in samples treated with 2 uM etoposide. These results suggest that treatment with DSB-causing agents upregulates the efficiency of stable transduction of nondividing cells. To determine if steps of the retroviral life-cycle preceding integration were affected, we performed a real-time PCR assay to detect the total viral DNA synthesis in these cells. As shown in Fig. 2B, neither bleomycin nor etoposide affected this step of retroviral life-cycle. We conclude that these DSB-causing agents appear to exert their effect at the integration or post-integration repair step of the retroviral life-cycle.
Fig. 2.
Effects of DSB-causing agents on the transduction of nocodazole-arrested fibroblasts. A. Effect of bleomycin or etoposide on stable integration in nocodazole-arrested fibroblasts. Integration was measured with a nested Alu-PCR assay (see Methods), data is shown as the number of amplicons quantitated in the real-time PCR (second round) reaction. Background was measured from samples subjected only to the gag primer in the first round of PCR. * - indicates samples that were transduced at significantly higher rates than control, untreated samples. B. Effects of DSB-causing agents on total viral DNA synthesis. Error bars indicate standard deviations.
Effects of etoposide and bleomycin on transduction efficiency in aphidicolin-arrested primary fibroblasts
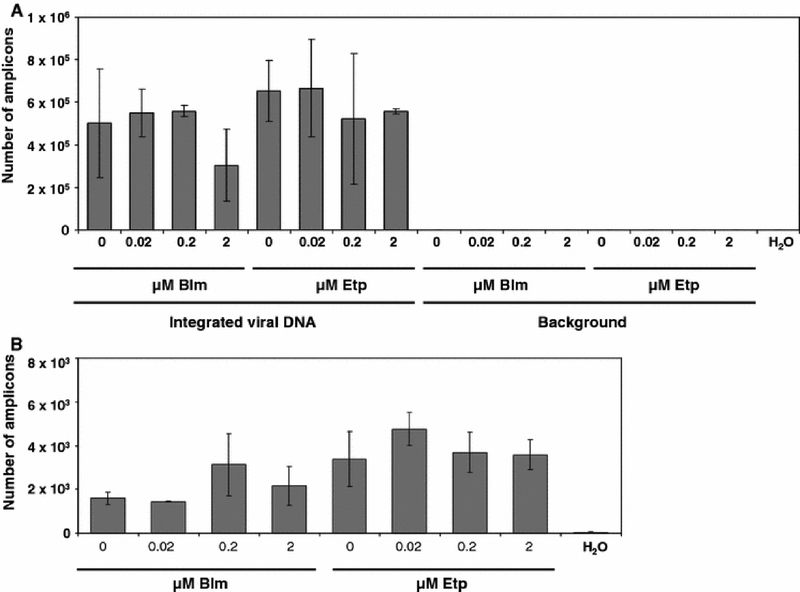
Primary fibroblasts were arrested with aphidicolin in the G1/S phase since treatment with aphidicolin results in a collapse of replication forks (Tibbetts et al. 2000). We again treated primary fibroblasts with aphidicolin and infected them with the HIV-1-based vector. Transduction efficiency was evaluated as described above. We did not observe a significant effect of these agents on the transduction efficiency of these cells. (Fig. 3A). Similarly, there was no effect on total viral DNA synthesis (Fig. 3B). We conclude that DSB-causing agents do not affect the transduction efficiency of aphidicolin-arrested cells.
Fig. 3.
Effects of DSB-causing agents on the transduction of aphidicolin-arrested fibroblasts. A. Effect of bleomycin or etoposide on stable integration in nocodazole-arrested fibroblasts. Integration was measured with a nested Alu-PCR assay, data is shown as the number of amplicons quantitated in the real-time PCR (second round) reaction. Background was measured from samples subjected only to the gag primer in the first round of PCR. B. Effects of DSB-causing agents on total viral DNA synthesis. Error bars indicate standard deviations.
Effects of bleomycin and etoposide on transduction efficiency in differentiated neurons
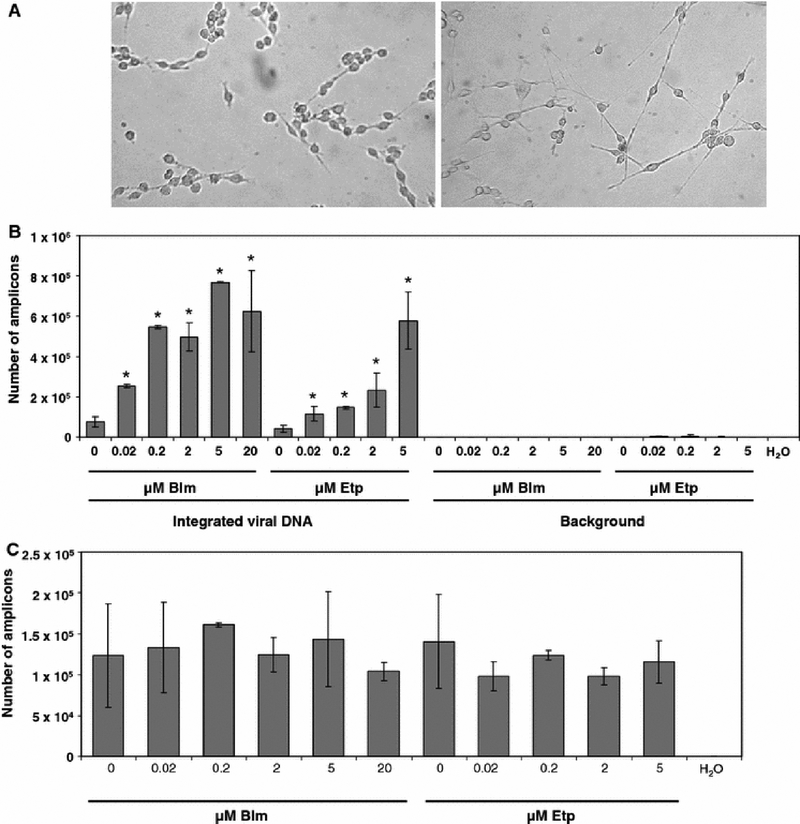
Many genetic diseases are associated with cognitive and other problems of the central nervous system. Therefore, neurons are an attractive target for gene therapy (Manfredsson & Mandel 2010). However, transduction efficiency in neurons is one of the major issues in gene therapy of neuronal diseases (Lundberg et al. 2008). To determine if DSB-causing agents affect HIV-1 transduction of neurons, we differentiated neural progenitors using a cAMP analogue [(Otey et al. 2003), Fig. 4A]. We then treated these cells with bleomycin and etoposide. As shown in Fig. 4B, bleomycin increased the transduction efficiency by approximately 10 fold. Etoposide had a similar effect, but as observed for nocodazole-arrested fibroblasts, the same degree of upregulation was achieved at higher concentrations than those of bleomycin (Fig. 4B). Finally, we did not observe any effects on the total viral DNA synthesis due to DSB-causing agents in these cells (Fig. 4C).
Fig. 4.
Effects of DSB-causing agents on the transduction of differentiated neuronal cells. A. photography of undifferentiated (left) and differentiated (right) B35 cells. B. Effect of bleomycin or etoposide on stable integration in differentiated neuronal B35 cells. * - indicates samples that were transduced at significantly higher rates than control, untreated samples. C. Effects of DSB-causing agents on total viral DNA synthesis. Error bars indicate standard deviations.
Effects of ATM deficiency on etoposide- and bleomycin-mediated upregulation of retroviral transduction
The presented data indicate that DSB-causing agents upregulate transduction of at least some nondividing cells. One of the major proteins, which was recently shown to play a role in post-integration repair, is the ATM protein (Daniel et al. 2001; Daniel & Pomerantz 2005; Lau et al. 2005; Smith et al. 2008). Mutations in ATM cause a general defect in DSB repair (Durocher & Jackson 2001; Shiloh 2001). To determine if the effect of DSB-causing agents on retroviral transduction could be caused by the upregulation of ATM, we have examined effects of bleomycin and etoposide on the transduction of ATM-deficient primary fibroblasts (GM02052). These cells are homozygous for the 103C>T transition in exon 5 of the ATM gene, which creates a premature stop codon at amino acid 35 (Gilad et al. 1996). We have compared the efficiency of transduction of these cells in the presence and absence of bleomycin or etoposide. We have observed that neither bleomycin nor etoposide upregulate transduction of nocodazole-arrested ATM-deficient cells (Fig. 5A), in contrast to ATM-proficient fibroblasts. As expected, these agents did not affect transduction of aphidicolin-arrested cells (Fig. 5A). As observed with normal fibroblasts, these DSB-causing agents did not affect the level of total viral DNA synthesis in these cells (Fig. 5B). We conclude that ATM is required for the observed upregulation of HIV-1-based vector transduction in nondividing cells.
Fig. 5.
Effects of DSB-causing agents on transduction of growth arrested ATM-deficient fibroblasts (GM02052 cells). A. Effects of bleomycin and etoposide on stable integration in nocodazole-arrested fibroblasts. B. Effects of DSB-causing agents on total viral DNA synthesis. Error bars indicate standard deviations.
Discussion
Efficiency of gene transfer is a major factor that determines the success or failure of gene therapy. A vast number of gene therapy trials use retroviral vectors. Hence, the efficiency of retroviral transduction is of crucial importance to gene therapy trials which use these vectors. In this study, we demonstrate that gene transduction by retroviral vectors, which are HIV-1-based, can be enhanced by subtoxic doses of DSB-causing chemical agents. We have observed this phenomenon in both nocodazole-arrested fibroblasts and differentiated neurons. Perhaps surprisingly, we did not see a similar upregulation in aphidicolin-arrested cells. However, as noted above, aphidicolin causes the collapse of replication forks. This is known to be recognized by cells as DNA damage, and the DNA damage response to collapsed replication forks involves proteins that are also involved in DSB repair (Sancar et al. 2004). Thus, it seems likely that the activity and intracellular levels of these proteins are already upregulated in aphidicolin-arrested cells, and further treatment with DSB-causing agents does not provide an additional stimulus, and consequently does not upregulate transduction.
According to our data, the increase in transduction by DSB inducing agents appears to have occurred at the level of integration or post-integration repair. We attempted to determine if the ATM protein, which was shown to play a role in post-integration repair, is the mediator of the effects of the DSB-causing agents on retroviral transduction. Our results indicate that these effects are indeed dependent on the ATM pathway, and the upregulation of transduction thus likely occurs at the post-integration repair step of the retroviral life-cycle.
In addition to ATM, it is possible that other DSB repair pathways are involved. Our future experiments will focus on identification of the pathways that mediate these effects. If known, we can then develop more specific stimulants of retroviral transduction, which may then be used in clinical trials.
As noted above, DSB-causing agents have been shown to upregulate transduction of dividing cells (Groschel & Bushman 2005). In addition, it had been hypothesized that this is due to the fact that these agents can induce growth arrest, allowing more time for the completion of early steps of the retroviral life-cycle (Groschel & Bushman 2005). Our results demonstrate that these agents increase transduction even in nondividing cells, such as neurons, and are therefore not consistent with the aforementioned hypothesis. Rather, the presented data are consistent with the hypothesis that DSB repair is involved in the post-integration repair step of the retroviral life-cycle. These DSB-causing agents upregulate activity and/or the expression of DSB repair proteins thereby enhancing the efficiency of post-integration repair, which is required to fully complete the integration process.
References
- Bennett JM, Reich SD (1979). Bleomycin. Ann Intern Med. 90, 945–948. [DOI] [PubMed] [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE (1997). Retroviruses. Plainview, NY: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Daniel R, Greger JG, Katz RA, Taganov KD, Wu X, Kappes JC, Skalka AM (2004). Evidence that stable retroviral transduction and cell survival following DNA integration depend on components of the nonhomologous end joining repair pathway. J Virol. 78, 8573–8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R, Katz RA, Merkel G, Hittle JC, Yen TJ, Skalka AM (2001). Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficiency of stable transduction by retroviruses. Mol Cell Biol. 21, 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R, Katz RA, Skalka AM (1999). A role for DNA-PK in retroviral DNA integration. Science. 284, 644–647. [DOI] [PubMed] [Google Scholar]
- Daniel R, Pomerantz RJ (2005). ATM: HIV-1’s Achilles heel? Nat Cell Biol. 7, 452–453. [DOI] [PubMed] [Google Scholar]
- Durocher D, Jackson SP (2001). DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol. 13, 225–231. [DOI] [PubMed] [Google Scholar]
- Flint SE LW; Racaniello VR; Skalka AM (2004). Principles of Virology. Washington, DC: ASM Press. [Google Scholar]
- Gilad S, Bar-Shira A, Harnik R, Shkedy D, Ziv Y, Khosravi R, Brown K, Vanagaite L, Xu G, Frydman M, Lavin MF, Hill D, Tagle DA, Shiloh Y (1996). Ataxia-telangiectasia: founder effect among north African Jews. Hum Mol Genet. 5, 2033–2037. [DOI] [PubMed] [Google Scholar]
- Groschel B, Bushman F (2005). Cell cycle arrest in G2/M promotes early steps of infection by human immunodeficiency virus. J Virol. 79, 5695–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Kanaar R, Jackson SP, O’Connor MJ (2004). Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 23, 3421–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Swinbank KM, Ahmed PS, Taylor DL, Jackson SP, Smith GC, O’Connor MJ (2005). Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat Cell Biol. 7, 493–500. [DOI] [PubMed] [Google Scholar]
- Lewis P, Hensel M, Emerman M (1992). Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11, 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TK, Liu LF (2001). Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol. 41, 53–77. [DOI] [PubMed] [Google Scholar]
- Lundberg C, Bjorklund T, Carlsson T, Jakobsson J, Hantraye P, Deglon N, Kirik D (2008). Applications of lentiviral vectors for biology and gene therapy of neurological disorders. Curr Gene Ther. 8, 461–473. [DOI] [PubMed] [Google Scholar]
- Manfredsson FP, Mandel RJ (2010). Development of gene therapy for neurological disorders. Discov Med. 9, 204–211. [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D (1996). In vivo gene delivery and stable transduction of nondividing cells by alentiviral vector. Science. 272, 263–267. [DOI] [PubMed] [Google Scholar]
- Negroni A, Stronati L, Grollino MG, Barattini P, Gumiero D, Danesi DT (2008). Radioresistance in a tumour cell line correlates with radiation inducible Ku 70/80 end-binding activity. Int J Radiat Biol. 84, 265–276. [DOI] [PubMed] [Google Scholar]
- Otey CA, Boukhelifa M, Maness P (2003). B35 neuroblastoma cells: an easily transfected, cultured cell model of central nervous system neurons. Methods Cell Biol. 71, 287–304. [DOI] [PubMed] [Google Scholar]
- Povirk LF, Hogan M, Dattagupta N (1979). Binding of bleomycin to DNA: intercalation of the bithiazole rings. Biochemistry. 18, 96–101. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 73, 39–85. [DOI] [PubMed] [Google Scholar]
- Saunders PP, Schultz GA (1972). Mechanism of action of bleomycin. I. Bacterial growth studies. Biochem Pharmacol. 21, 1657–1666. [DOI] [PubMed] [Google Scholar]
- Shiloh Y (2001). ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev. 11, 71–77. [DOI] [PubMed] [Google Scholar]
- Skalka A (1999). Retroviral DNA Integration. 52. [Google Scholar]
- Skalka AM, Katz RA (2005). Retroviral DNA integration and the DNA damage response. Cell Death Differ. 12 Suppl 1, 971–978. [DOI] [PubMed] [Google Scholar]
- Smith JA, Daniel R (2006). Following the path of the virus: the exploitation of host DNA repair mechanisms by retroviruses. ACS Chem Biol. 1, 217–226. [DOI] [PubMed] [Google Scholar]
- Smith JA, Wang FX, Zhang H, Wu KJ, Williams KJ, Daniel R (2008). Evidence that the Nijmegen breakage syndrome protein, an early sensor of double-strand DNA breaks (DSB), is involved in HIV-1 post-integration repair by recruiting the ataxia telangiectasia-mutated kinase in a process similar to, but distinct from, cellular DSB repair. Virol J. 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT (2000). Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 14, 2989–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanin R, Scarpa M (2004). Why do we need new gene therapy viral vectors? Characteristics, limitations and future perspectives of viral vector transduction. Curr Gene Ther. 4, 357–372. [DOI] [PubMed] [Google Scholar]
- Weinberg JB, Matthews TJ, Cullen BR, Malim MH (1991). Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J Exp Med. 174, 1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]