Abstract
Antigen presentation is the key first step in the establishment of an antigen-specific T cell response. Among professional antigen presenting cells (APCs), dendritic cells (DCs) are the major population responsible for the priming of both CD4+ and CD8+ naïve T cells. This priming requires physical interaction between the DC and the T cell, during which signals are exchanged that determine both the magnitude and the quality of the ensuing response. The nature of these signals varies widely depending on the nature of the antigen, the anatomical site in which they take place, and the phenotype of the antigen-presenting DC, making the study of the dynamics, microanatomical distribution and phenotypic variation of DCs a key part of our understanding of adaptive immunity. Here, we provide a brief survey of how our view of T cell activation by DCs has evolved over recent years as intravital multiphoton microscopy and other emerging technologies have expanded our ability to study these events in vivo.
Introduction
Dendritic cells (DCs) are the immune system’s prototypical antigen-presenting cell (APC), responsible for the vast majority of the priming of naïve CD4+ and CD8+ T cells under most conditions [1–3]. As such, DCs are uniquely positioned to influence the magnitude and quality of T cell responses. While steady-state DCs are classically thought to promote T cell tolerance to self-antigens [4], DCs that have been activated (e.g., with Toll-like receptor ligands) promote effector T cell responses, which vary quantitatively and qualitatively according to the exact nature of the activating stimulus [5]. DCs are therefore key players in the balance between tolerance and the various types of cellular and humoral immunity [2–5]. T cell activation by DCs involves physical contact between the DC and the T cell. During these interactions, DCs present antigen to T cells in the context of major histocompatibility (MHC) molecules, with additional input delivered in the form of costimulatory surface ligands and cytokines [5]. Thus, DCT cell interactions have become a subject of study in their own right [6–17]. In the present review, we discuss how technological advances, primarily in microscopy, have improved our ability to monitor such DC-T cell interactions, highlighting selected findings emerging from these studies.
Conventional (c)DCs in lymph-nodes can be subsetted according to two major axes. First, DCs can be classified according to the route they take from bone marrow to the LN: resident DCs arise from blood borne precursors that enter LN through HEVs [18]; migratory DCs initially travel from blood to non-lymphoid tissues, subsequently migrating to the tissue-draining LN through lymphatics carrying antigen sampled while in the tissue [19]. Migratory DCs are characterized by high expression of MHCII class II and intermediate expression of the integrin CD11c, while the reverse is true for resident DCs (although there is evidence that this dichotomy may be influenced by the DCs activation state) [20,21]. Both migratory and resident DCs can be further subdivided, along a second axis based on phenotype, into cDC1 and cDC2 [2,3]. cDC1 express the chemokine receptor XCR1, and the E-cadherin receptor CD103 (when of migratory origin) or CD8 (when resident) [2]. cDC2 are characterized by expression of the integrin CD11b [2,3]. A number of studies have shown that cDC1 are superior to cDC2 in their capacity to cross-present antigen to CD8+ T cells [2,3,22,23]. cDC2, on the other hand are more potent in direct presentation of antigen to CD4+ T cells [2,3,24–27].
Studying T cell-DC interactions by intravital imaging
Mainstream immunological techniques such as flow cytometry, cell co-culture systems, and gene expression profiling have been powerful in defining how the phenotypes of different DC populations correspond to different roles in initiating and maintaining cellular immune responses. Classic histological approaches add a spatial dimension to these functional and phenotypic analyses, so that the general compartmentalization of DC functions within the LN can be discerned. A common feature of classic techniques, however, is that they do provide information on fine spatiotemporal aspects antigen presentation—including the dynamics of cellular migration and cell-cell interactions—beyond the limited resolution provided by analyzing different mice at arbitrary time intervals. As a remedy to some of these problems, intravital multiphoton laser scanning microscopy (MPLSM) has emerged in the last two decades as the tool of choice for studying antigen presentation in situ (Fig. 1A). The use of brief, concentrated pulses of near-infrared laser light restricts the excitation of shorter wavelength fluorophores to the focal plane, allowing deeper tissue penetration with less photodamage [28–30]. The practical consequence of this property is that cells can be imaged in real time while performing their functions, deep inside intact LN of live mice, providing key insight into cellular dynamics of antigen presentation [7–17].
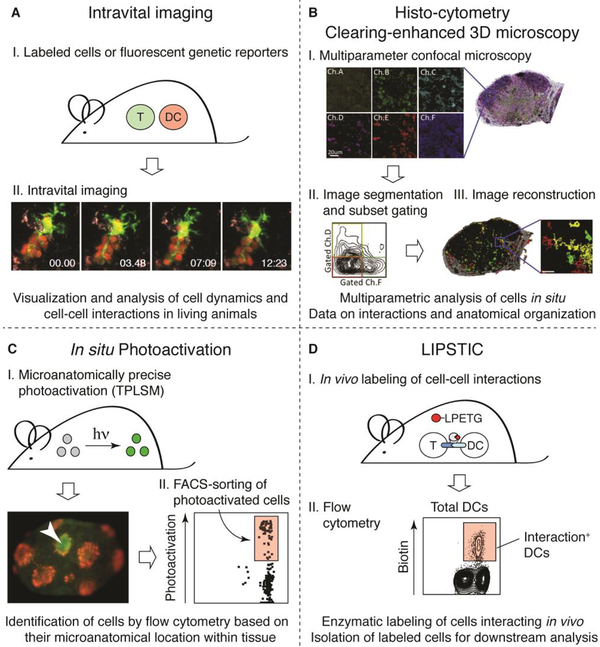
Figure 1. Studying the spatiotemporal dynamics of antigen presentation.
(A) Intravital imaging. (I) Interacting cells are labeled with different fluorescent dyes or by expression of different fluorescent reporter proteins. (II) A two-photon microscope is used to visualize DC-T cell interactions in LN or other locations in explanted tissues or more often within living mice. A cluster of CD8+ T cells (red) undergoing stable interactions with a single antigen-pulsed DC (yellow) is shown (image by G. Victora and K. Swee). (B) Histo-cytometry. Histo-cytometry combines multicolor immunophenotyping with automated image analysis, providing detailed information on the microanatomical location and phenotypic identity of cells in a tissue. (I) A series of confocal images of tissue sections is taken. (II) Images are segmented into cells and, analogously to flow cytometry, channels are compensated for fluorophore spillover. (III) Image analysis allows for quantitative visualization of phenotypically distinct immune cell populations. Images by M. Gerner and R. Germain, adapted with permission from [42]. (C) In situ photoactivation. (I) A microanatomical region of interest is photoactivated within a tissue using MPLSM. An example of photoactivation of cells within a single germinal centers TPLSM is shown (red, follicular dendritic cells; green and arrowhead, photoactivated cells). Image by J. Jacobsen. (II) The tissue is then isolated and dissociated, and photoactivated cells can be easily identified based on their fluorescence in the photoactivation channel by flow cytometry. Cells can be then be used for downstream analyses such as RNA-seq. (D) LIPSTIC. (I) A receptor-ligand pair of interest is genetically tagged with the transpeptidase sortase A (SrtA) or with the SrtA target, 5 N-terminal glycines (G5). The peptide substrate biotin (red)-LPETG is administered by injection, and DCs that interacted with T cells can be isolated based on presence of the biotin-LPETG tag using flow-cytometry. (II) Flow-cytometry contour plot showing biotin-positive cells after in vivo LIPSTIC labeling of DC-T cell interactions via the CD40-CD40L axis.
Introduction of intravital imaging quickly provided a wealth of insight into the T cell priming process that would not be available otherwise. For example, DCs are highly efficient at screening large swaths of the T cell repertoire prior to engaging with specific T cells in long-lasting interactions [7,14,31]. One DC can simultaneously engage several (often as many as 10) antigen-specific T cells, with the amount of antigen present on the DC determining both the number of T cells per DC and the duration of interactions [7]. Intravital imaging also delineated three major phases in T cell priming by DCs: in phase 1 (up to 8 hours after entering the LN), motile naïve T cells scan multiple DCs for antigen; in phase 2 (8 to 24 hours), sessile cognate T cell-DC pairs form, which can last for several hours; finally, in phase 3 (24 to 48 hours), activated T cells disengage from DCs, resume their motile behavior, and start proliferating [10]. Largely equivalent findings were reported for CD4+ T cells after vaccination [13]. By targeting antigen to endogenous DCs using a fusion of ovalbumin to an antibody against the DC surface lectin DEC-205 (anti-DEC-OVA) under inflammatory and steady-state conditions, it was later determined that long-term interactions between antigen-specific CD4 T cells and DC is a shared feature of tolerogenic and immunogenic T cell priming [15].
More recent studies have used intravital imaging to probe the role of DCs in priming CD8+ and CD4+ T cell responses during viral infection. In early stages of vaccinia virus infection, for example, CD4+ and CD8+ T cell priming occurs in distinct locations in the LN, on anatomically segregated and phenotypically distinct DC subsets. Depletion of XCR1+ DCs during this early time point did not affect initial priming of CD8+ T cells. During later stages of the infection, however, both CD4+ and CD8+ T cells formed clusters around XCR1+ DCs. Late depletion of either XCR1+ DCs or CD4+ T cells in this model impaired effector and memory CD8+ T cells responses, indicating that CD4+ T cell help to CD8+ T cells is delivered later in the response, and primarily through licensing of XCR1+ DCs [32]. A second group reported similar findings using herpes simplex virus as an infection model. By painting skin with the fluorescent dye tetramethylrhodamine (TRITC), the authors demonstrated that during early time point after infection (18–20 hours) TRITC+ migratory DCs travel through the paracortex and form clusters with antigen specific CD4+ T cells in the proximity to medullary regions of inguinal LN. At later time points (40–42 hours) both CD4+ and CD8+ T cells clustered together, but this time around XCR1+ TRITC− (resident) cDCs. [33]. The common model emerging from both models is that CD11b+ cDC1s initially activate CD4+ T cells, which later license XCR1+ cDC2s to effectively prime CD8+ T cells. Thus, XCR1+ DCs serve as the platform for delivery of CD4+ T cell help to CD8+ T cells.
Intravital microscopy has also been used to visualize DC-T cell interactions in nonlymphoid tissues, most prominently within the tumor microenvironment. For example, using a transgenic mouse line expressing mammary tumor virus-polyoma middle T antigen (MMTV-PyMT), Krummel and colleagues showed that DCs directly contact tumor cells in vivo, and also interact with tumor antigen-specific T cells [34]. The same group later showed that CD103+ migratory cDC1s present in the tumor environment can engage in long-lasting interactions with transferred antigen-specific CD8+ T cells, and are required for effective adoptive CTL therapy [35]. Subsequently, multiple studies have confirmed CD103+ DCs as the major population responsible for cross-presenting tumor antigens to CD8+ T cells both in the draining LN and within the tumor itself [36,37]. A very recent study combining intravital microscopy with single-cell RNA sequencing described a novel form of crosstalk between tumor-infiltrating DCs and T cells: in response to anti-PD-1 therapy, CD8+ T cell produces IFN-γ, which directly stimulates tumor-infiltrating DCs produce IL-12. IL-12 in turn acts on CD8+ T cells, enhancing their ability to kill tumor targets [38].
While the earliest studies of DC-T cell interaction in vivo used adoptive transfer of dye-labeled cells into non-fluorescent mice [6,10], several genetically-encoded fluorescent reporter strains are now available to visualize endogenous DCs in their native context, using different promoters that highlight distinct DC populations more or less specifically. These include the CD11c-YFP transgene [11], Zbtb46GFP [39], Xcr1Venus [40], and the photoactivatable Xcr1KikGR[41]. Each of these models have advantages and limitations. For instance, CD11c-YFP is expressed by all DC subtypes, but also by macrophages and B and T cells [2,11]. Zbtb46GFP is much more specific to cDCs, but does not segregate between cDC1 or cDC2, nor between resident and migratory cDC subsets [39]. Xcr1Venus is specific for the XCR1+ cDC1 subset but cannot distinguish migratory and resident DCs. The elegant Xcr1KikGR model addresses the resident vs. migratory issue by using light to photoconvert the reporter protein Kikume Green-Red (KikGR). This allows migratory DCs to be marked by illumination of their tissue of origin (e.g. skin), and subsequently identified after they have migrated to the draining LN [41]. However, no such system is yet available for cDC2. Thus, while several reporter systems exist, even the broad categories of cDC populations are yet to be completely resolved by intravital microscopy. As we will see in the next section, a number of techniques that combine various degrees of spatiotemporal resolution with improved phenotyping capabilities have been developed, and promise to bridge the gap between dynamic behavior, microanatomical localization, and cellular phenotype.
Linking cellular dynamics, microanatomy, and DC phenotype
A drawback of existing immunological techniques is their limited ability to cross-talk with each other. For instance, while flow cytometry provides rich information on DC phenotype, it does so at the expense of obliterating all spatial context; conversely, traditional histology, while maintaining anatomical information, is not quantitative enough to clearly resolve cellular subsets. Likewise, intravital microscopy can demonstrate and quantify interactions between different cell types, but cannot be used to isolate interacting cells for phenotypic analysis. In recent years, several solutions to this problem have emerged, and techniques now exist that are capable of linking positional and phenotypic information to an unprecedented extent.
Deep phenotyping of tissue sections by histo-cytometry.
Histo-cytometry combines flow cytometry-like multicolor immunostaining of histological sections or whole-mount cleared tissues with downstream automated image analysis to provide detailed information on the micoanatomical positioning and phenotype of cells within a tissue. Using computational algorithms, images of tissues are segmented into individual cells, generating a quantitative map of the phenotypic diversity, anatomical distribution, and colocalization patterns of multiple cell subsets within a tissue [42,43] (Fig. 1B). To study T cell-DC interactions, T cells undergoing priming are identified by transfer of labeled or transgenic populations, and the identity of the DCs physically contacting these T cells (information that would be lost in flow cytometry) is then determined by the markers expressed by the engaged DCs [44]. Using histo-cytometry, Germain, Gerner and colleagues have provided detailed descriptions of the spatial distribution of migratory and resident cDC1 and cDC2 subsets during the priming of T cell responses in skin-draining LN [44–46]. Resident CD11b+ cDC2s are located within the lymphatic zone of the LN, whereas migratory cDC2s are positioned in interfollicular regions [42]; in contrast, resident CD8+ and migratory CD103+ cDC1s reside in the paracortical T cell zone [42]. These studies also showed that migratory cDC2s promote clustering of highly suppressive T regulatory (Treg) cells in the outer T cell zone and within interfollicular regions, there they block the activation of self-reactive T helper cells in a TCR dependent manner [46]. Among other findings, studies using histo-cytometry have shown that (i) a population of resident cDC2s populates lymphatic sinuses and samples the antigen directly from the lymph [44]; (ii) MHC-I dependent antigen presentation to CD8 T cells is more sensitive to antigen availability due to the positioning of resident cDC1 deeper within the LN [45]; and (iii) resident CD8+ DCs directly capture Plasmodium sporozoites that migrate from their site of deposition in the skin, triggering CD8+ T cell responses to the pathogen [47]. Variations on the theme of histo-cytometry using DNA barcodes [48], mass spectrometry on tissue sections [49], and in situ hybridization [50–52] are also being developed at a fast pace, and could add further phenotypic depth to this type of analysis.
Labeling microanatomical niches using in situ photoactivation.
A second, conceptually simple, solution to merging microanatomical and phenotypic information is in situ photoactivation (ISPA) [53,54] (Fig. 1C). In ISPA, a photoactivatable protein is expressed in most or all cells of a transgenic mouse using a ubiquitous promoter (e.g., our UBC-PAGFP mouse line). LN or other tissues from these mice can be photoactivated with microanatomical precision using MPLSM, either as explants or in live mice. Tissues are then dissociated, and cells within the photoactivated area can be identified by virtue of their fluorescence in the photoactivation channel. These cells can be further phenotyped using flow cytometry markers, or sorted and subjected to gene expression profiling. This technique was first developed to obtain gene expression profiles of B cells occupying the light and dark zones of germinal centers [53], and has been extensively used in this context [55–59]. More recently, ISPA has been coupled to single-cell RNA-seq (scRNA-seq) approaches to determine the cellular composition of distinct cellular niches (referred to as NICHE-seq [60]). While ISPA has not yet (to our knowledge) been applied to DC-T cell interactions, broader photolabeling approaches (where the entire tissue of origin is illuminated and DC migration from tissue to dLN is monitored) have [41,61–63]. We therefore expect that ISPA, especially if combined with scRNA-seq, may prove useful as a means of identifying the full transcrirptional programs of T cells and DCs occupying the same microanatomical niches, complementing insights obtained histo-cytometric analysis of T cell-DC superposition.
In vivo labeling of cell-cell interactions.
Whereas both intravital microscopy and histo-cytometry can be used to identify cells in direct physical apposition, a limitation common to both techniques is their inability to retrieve cells for further downstream analysis. To circumvent this, we recently developed a novel technique we call LIPSTIC (Labeling Immune Partnerships by SorTagging Intercellular Contacts) which allows us to enzymatically label cells undergoing a given ligand-receptor interaction within live mice in such a manner that these cells are later identifiable by flow cytometry [64] (Fig. 1D). In LIPSTIC, a ligand and receptor are genetically fused to the S. aureus sortase A transpeptidase (SrtA) and to a five-glycine acceptor tag, respectively. Upon cell-cell interaction, a labeled SrtA substrate injected into the live mouse is covalently transferred from the SrtA+ donor cell to the G5+ acceptor, enabling the chemical recording of cell interaction history. We employed this approach to monitor CD40-CD40L interaction in vivo between DCs and CD4+ T cells during the different phases of T cell priming by DCs [10], and found different modalities of CD40-CD40L interaction during these stages. While naïve CD4+ T cells initially interact with DCs in an antigen-dependent fashion during phase 2, subsequent phase 3 interactions do not require TCR engagement for delivery of a CD40L-dependent signal [64]. A recent report has described a LIPSTIC-based system that relies on a SrtA variant with increased ability to label non-modified endogenous N-terminal glycines on the acceptor cell. This may provide additional flexibility to the system by not requiring engineering of an acceptor mouse strain [65].
Future directions.
Each of the technologies delineated above contributes an additional dimension to the study of spatial and dynamic aspects of DC-T cell interaction, but also brings along its own caveats and limitations. While intravital imaging provides the most detailed description of the dynamic behavior of cells, it is not suitable for downstream analysis; histo-cytometry and in situ photoactivation add phenotypic information to microanatomically-defined cell populations but do not provide quantitative information on cellular interactions; and LIPSTIC allows easy quantitative measurement of a cell’s cumulative history of interactions, but provides no dynamic or microanatomical information. We expect that future studies will combine these techniques so as to maximize resolution while compensating for each other’s limitations. In the context of the ongoing revolution in single-cell genomics [60,66], these techniques promise to increase enormously the resolution of our understanding of how DCs shape the adaptive immune response in in vivo systems.
Highlights.
Physical interaction with DCs is the first step in T cell priming, and controls the magnitude and quality of the ensuing response
Intravital microscopy has helped elucidate many of the key principles of T cell-DC interaction.
Emerging technologies have expanded our ability to study T cell priming by DCs in situ and in vivo.
Acknowledgements
We thank R. Germain for critical reading of the manuscript and helpful suggestions. This work was supported by NIH Pioneer award DP1AI144248 and Starr Cancer Consortium grant I10–0044 to G.V.; A.C. is a Damon Runyon Postdoctoral Fellow. G.V. is a Searle Scholar and a MacArthur fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
● of special interest
●● of outstanding interest
- 1.Steinman RM: The dendritic cell system and its role in immunogenicity. Annu Rev Immunol 1991, doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 2.Merad M, Sathe P, Helft J, Miller J, Mortha A: The Dendritic Cell Lineage: Ontogeny and Function of Dendritic Cells and Their Subsets in the Steady State and the Inflamed Setting. Annu Rev Immunol 2013, doi: 10.1146/annurevimmunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenbarth SC: Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol 2018, doi: 10.1038/s41577-018-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinman RM, Hawiger D, Nussenzweig MC: Tolerogenic denritic cells. Annu Rev Immunol 2003, doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki A, Medzhitov R: Toll-like receptor control of the adaptive immune responses. Nat Immunol 2004, doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 6.Stoll S, Delon J, Brotz TM, Germain RN: Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science (80- ) 2002, doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- 7.Bousso P, Robey E: Dynamics of CD8+T cell priming by dendritic cells in intact lymph nodes. Nat Immunol 2003, doi: 10.1038/ni928. [DOI] [PubMed] [Google Scholar]
- 8.Skokos D, Shakhar G, Varma R, Waite JC, Cameron TP, Lindquist RL, Schwickert T, Nussenzweig MC, Dustin ML: Peptide-MHC potency governs dynamic interactions between T cells and dendritic cells in lymph nodes. Nat Immunol 2007, doi: 10.1038/ni1490. [DOI] [PubMed] [Google Scholar]
- 9.Bousso P: T-cell activation by dendritic cells in the lymph node: Lessons from the movies. Nat Rev Immunol 2008, doi: 10.1038/nri2379. [DOI] [PubMed] [Google Scholar]
- 10.Mempel TR, Henrickson SE, Von Andrian UH: T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 2004, doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 11.Lindquist RL, Shakhar G, Dudziak D, Wardemann H, Eisenreich T, Dustin ML, Nussenzweig MC: Visualizing dendritic cell networks in vivo. Nat Immunol 2004, doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 12.Hugues S, Fetler L, Bonifaz L, Helft J, Amblard F, Amigorena S: Distinct T cell dynamics in lymph nodes during the induction of tolerance and immunity. Nat Immunol 2004, doi: 10.1038/ni1134. [DOI] [PubMed] [Google Scholar]
- 13.Miller MJ, Safrina O, Parker I, Cahalan MD: Imaging the Single Cell Dynamics of CD4+ T Cell Activation by Dendritic Cells in Lymph Nodes. J Exp Med 2004, doi: 10.1084/jem.20041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MJ, Hejazi AS, Wei SH, Cahalan MD, Parker I: T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc Natl Acad Sci 2004, doi: 10.1073/pnas.0306407101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shakhar G, Lindquist RL, Skokos D, Dudziak D, Huang JH, Nussenzweig MC, Dustin ML: Stable T cell-dendritic cell interactions precede the development of both tolerance and immunity in vivo. Nat Immunol 2005, doi: 10.1038/ni1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN: Chemokines enhance immunity by guiding naive CD8+T cells to sites of CD4+T cell-dendritic cell interaction. Nature 2006, doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 17.Tadokoro CE, Shakhar G, Shen S, Ding Y, Lino AC, Maraver A, Lafaille JJ, Dustin ML: Regulatory T cells inhibit stable contacts between CD4+ T cells and dendritic cells in vivo. J Exp Med 2006, doi: 10.1084/jem.20050783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu FF, Randolph GJ, Rudensky AY, Nussenzweig M: In Vivo Analysis of Dendritic Cell Development and Homeostasis. Science (80- ) 2009, doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randolph GJ, Angeli V, Swartz MA: Dendritic-cell trafficking to lymph nodes through lymphatic vessels. Nat Rev Immunol 2005, doi: 10.1038/nri1670. [DOI] [PubMed] [Google Scholar]
- 20.Merad M, Ginhoux F, Collin M: Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat Rev Immunol 2008, doi: 10.1038/nri2455. [DOI] [PubMed] [Google Scholar]
- 21.Singh-Jasuja H, Thiolat A, Ribon M, Boissier MC, Bessis N, Rammensee HG, Decker P: The mouse dendritic cell marker CD11c is down-regulated upon cell activation through Toll-like receptor triggering. Immunobiology 2013, doi: 10.1016/j.imbio.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 22.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. : Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science (80- ) 2008, doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.den Haan JMM, Lehar SM, Bevan MJ: Cd8+ but Not Cd8− Dendritic Cells Cross-Prime Cytotoxic T Cells in Vivo. J Exp Med 2000, doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatto D, Wood K, Caminschi I, Murphy-Durland D, Schofield P, Christ D, Karupiah G, Brink R: The chemotactic receptor EBI2 regulates the homeostasis, localization and immunological function of splenic dendritic cells. Nat Immunol 2013, doi: 10.1038/ni.2555. [DOI] [PubMed] [Google Scholar]
- 25.Calabro S, Liu D, Gallman A, Nascimento MSL, Yu Z, Zhang T ting, Chen P, Zhang B, Xu L, Gowthaman U, et al. : Differential Intrasplenic Migration of Dendritic Cell Subsets Tailors Adaptive Immunity. Cell Rep 2016, doi: 10.1016/j.celrep.2016.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Nish SA, Jiang R, Hou L, Licona-Limón P, Weinstein JS, Zhao H, Medzhitov R: Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity 2013, doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumamoto Y, Linehan M, Weinstein JS, Laidlaw BJ, Craft JE, Iwasaki A: CD301b+ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity 2013, doi: 10.1016/j.immuni.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pittet MJ, Garris CS, Arlauckas SP, Weissleder R: Recording the wild lives of immune cells. Sci Immunol 2018, doi: 10.1126/sciimmunol.aaq0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zipfel WR, Williams RM, Webb WW: Nonlinear magic: Multiphoton microscopy in the biosciences. Nat Biotechnol 2003, doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 30.Pittet MJ, Weissleder R: Intravital imaging. Cell 2011, doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller MJ, Wei SH, Cahalan MD, Parker I: Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proc Natl Acad Sci 2003, doi: 10.1073/pnas.2628040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eickhoff S, Brewitz A, Gerner MY, Klauschen F, Komander K, Hemmi H, Garbi N, Kaisho T, Germain RN, Kastenmüller W: Robust Anti-viral Immunity Requires Multiple Distinct T Cell-Dendritic Cell Interactions. Cell 2015, doi: 10.1016/j.cell.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hor JL, Whitney PG, Zaid A, Brooks AG, Heath WR, Mueller SN: Spatiotemporally Distinct Interactions with Dendritic Cell Subsets Facilitates CD4+and CD8+T Cell Activation to Localized Viral Infection. Immunity 2015, doi: 10.1016/j.immuni.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Engelhardt JJ, Boldajipour B, Beemiller P, Pandurangi P, Sorensen C, Werb Z, Egeblad M, Krummel MF: Marginating Dendritic Cells of the Tumor Microenvironment Cross-Present Tumor Antigens and Stably Engage Tumor-Specific T Cells. Cancer Cell 2012, doi: 10.1016/j.ccr.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, Barczak A, Rosenblum MD, Daud A, Barber DL, et al. : Dissecting the Tumor Myeloid Compartment Reveals Rare Activating Antigen-Presenting Cells Critical for T Cell Immunity. Cancer Cell 2014, doi: 10.1016/j.ccell.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 36.Roberts EW, Broz ML, Binnewies M, Headley MB, Nelson AE, Wolf DM, Kaisho T, Bogunovic D, Bhardwaj N, Krummel MF: Critical Role for CD103+/CD141+ Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell 2016, doi: 10.1016/j.ccell.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, Casanova-Acebes M, Khudoynazarova M, Agudo J, Tung N, et al. : Expansion and Activation of CD103+Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity 2016, doi: 10.1016/j.immuni.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C, Engblom C, Pfirschke C, Siwicki M, Gungabeesoon J, et al. : Successful Anti-PD-1 Cancer Immunotherapy Requires T Cell-Dendritic Cell Crosstalk Involving the Cytokines IFN-γ and IL-12. Immunity 2018, doi: 10.1016/j.immuni.2018.09.024.● Using intravital imaging and single-cell RNA-seq, Garris and colleagues demonstrate that PD1 blockade induces Ifn-γ production by T cells. Ifn-γ acts on DCs and triggers them to produce IL-12, which boosts the capacity of CD8+ T cells to kill tumor targets.
- 39.Satpathy AT, KC W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, Murphy TL, Murphy KM: Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med 2012, doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki C, Sugiyama M, Ohta T, Hemmi H, Hamada E, Sasaki I, Fukuda Y, Yano T, Nobuoka M, Hirashima T, et al. : Critical Roles of a Dendritic Cell Subset Expressing a Chemokine Receptor, XCR1. J Immunol 2013, doi: 10.4049/jimmunol.1202798. [DOI] [PubMed] [Google Scholar]
- 41.Kitano M, Yamazaki C, Takumi A, Ikeno T, Hemmi H, Takahashi N, Shimizu K, Fraser SE, Hoshino K, Kaisho T, et al. : Imaging of the cross-presenting dendritic cell subsets in the skin-draining lymph node. Proc Natl Acad Sci 2016, doi: 10.1073/pnas.1513607113.●● Kitano and colleagues use a new mouse strain with photoactivatable cDC1 to show that migratory XCR1+ DCs traveling from skin to the draining LN are potent drivers of CD8+ T cell responses.
- 42.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN: Histo-cytometry: A method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 2012, doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W, Germain RN, Gerner MY: Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce 3D). Proc Natl Acad Sci 2017, doi: 10.1073/pnas.1708981114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerner MY, Torabi-Parizi P, Germain RN: Strategically Localized Dendritic Cells Promote Rapid T Cell Responses to Lymph-Borne Particulate Antigens. Immunity 2015, doi: 10.1016/j.immuni.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 45.Gerner MY, Casey KA, Kastenmuller W, Germain RN: Dendritic cell and antigen dispersal landscapes regulate T cell immunity. J Exp Med 2017, doi: 10.1084/jem.20170335.●● Using histo-cytometry, Gerner and colleagues demonstrate asymmetric distribution of cDC1 and cDC2 within LN, with cDC2 located in the periphery and cDC1 in deeper regions of the LN. Draining antigen forms a steep gradient within LN, which affects priming of T cells by cDC1 and cDC2, with cDC1 being more sensitive to the gradient due to their location in regions of limited antigen availability.
- 46.Liu Z, Gerner MY, Van Panhuys N, Levine AG, Rudensky AY, Germain RN: Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 2015, doi: 10.1038/nature16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radtke AJ, Kastenmüller W, Espinosa DA, Gerner MY, Tse SW, Sinnis P, Germain RN, Zavala FP, Cockburn IA: Lymph-Node Resident CD8α+Dendritic Cells Capture Antigens from Migratory Malaria Sporozoites and Induce CD8+T Cell Responses. PLoS Pathog 2015, doi: 10.1371/journal.ppat.1004637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S, Nolan GP: Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 2018, doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, Yang SR, Kurian A, Van Valen D, West R, et al. : A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018, doi: 10.1016/j.cell.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. : Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science (80- ) 2018, doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, et al. : Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc 2015, doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X: Spatially resolved, highly multiplexed RNA profiling in single cells. Science (80- ) 2015, doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC: Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 2010, doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacobsen JT, Victora GD: Microanatomical labeling of germinal center structures for flow cytometry using photoactivation. In Methods in Molecular Biology. 2017. [DOI] [PubMed] [Google Scholar]
- 55.Shulman Z, Gitlin AD, Targ S, Jankovic M, Pasqual G, Nussenzweig MC, Victora GD: T follicular helper cell dynamics in germinal centers. Science (80- ) 2013, doi: 10.1126/science.1241680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tas JMJ, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill JC, Reynaud CA, Browne EP, et al. : Visualizing antibody affinity maturation in germinal centers. Science (80- ) 2016, doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ersching J, Efeyan A, Mesin L, Jacobsen JT, Pasqual G, Grabiner BC, Dominguez-Sola D, Sabatini DM, Victora GD: Germinal Center Selection and Affinity Maturation Require Dynamic Regulation of mTORC1 Kinase. Immunity 2017, doi: 10.1016/j.immuni.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gitlin AD, Shulman Z, Nussenzweig MC: Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature 2014, doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suan D, Nguyen A, Moran I, Bourne K, Hermes JR, Arshi M, Hampton HR, Tomura M, Miwa Y, Kelleher AD, et al. : T Follicular Helper Cells Have Distinct Modes of Migration and Molecular Signatures in Naive and Memory Immune Responses. Immunity 2015, doi: 10.1016/j.immuni.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 60.Medaglia C, Giladi A, Stoler-Barak L, De Giovanni M, Salame TM, Biram A, David E, Li H, Iannacone M, Shulman Z, et al. : Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science (80- ) 2017, doi: 10.1126/science.aao4277.● This study combines in situ photoactivation with single-cell RNA-seq to provide an unbiased view of cellular heterogeneity within defined microanatomical niches.
- 61.Torcellan T, Hampton HR, Bailey J, Tomura M, Brink R, Chtanova T: In vivo photolabeling of tumor-infiltrating cells reveals highly regulated egress of T-cell subsets from tumors. Proc Natl Acad Sci 2017, doi: 10.1073/pnas.1618446114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomura M, Yoshida N, Tanaka J, Karasawa S, Miwa Y, Miyawaki A, Kanagawa O: Monitoring cellular movement in vivo with photoconvertible fluorescence protein “Kaede” transgenic mice. Proc Natl Acad Sci 2008, doi: 10.1073/pnas.0802278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morton AM, Sefik E, Upadhyay R, Weissleder R, Benoist C, Mathis D: Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc Natl Acad Sci 2014, doi: 10.1073/pnas.1405634111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasqual G, Chudnovskiy A, Tas JMJ, Agudelo M, Schweitzer LD, Cui A, Hacohen N, Victora GD: Monitoring T cell-dendritic cell interactions in vivo by intercellular enzymatic labelling. Nature 2018, doi: 10.1038/nature25442.●● In this paper, we introduce LIPSTIC, an approach to track APC-T cell interaction history in vivo using trans-synaptic enzymatic labeling. This method allows us to determine the identity of DCs interacting physically with T cells via the CD40-CD40L pathway and track how interactions evolve over time.
- 65.Ge Y, Chen L, Liu S, Zhao J, Zhang H, Chen PR: Enzyme-Mediated Intercellular Proximity Labeling for Detecting Cell-Cell Interactions. J Am Chem Soc 2019, doi: 10.1021/jacs.8b10286. [DOI] [PubMed] [Google Scholar]
- 66.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. : Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013, doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]