Abstract
Diphthamide is a unique posttranslational modification on translation elongation factor 2 (EF2) in archaea and eukaryotes. Biosynthesis of diphthamide was proposed to involve four steps. The first step is a C-C bond forming reaction catalyzed by unique radical S-adenosylmethionine (SAM) enzymes. Classical radical SAM enzymes use SAM and [4Fe-4S] clusters to generate a 5′-deoxyadenynal radical and catalyze numerous reactions. Radical SAM enzymes in diphthamide biosynthesis cleave a different C-S bond in SAM to generate a 3-amino-3-carboxypropyl radical and modify a histidine residue of substrate protein EF2. Here, we describe our investigations on these unique radical SAM enzymes, including the preparation, characterization and activity assays we have developed.
1. Introduction
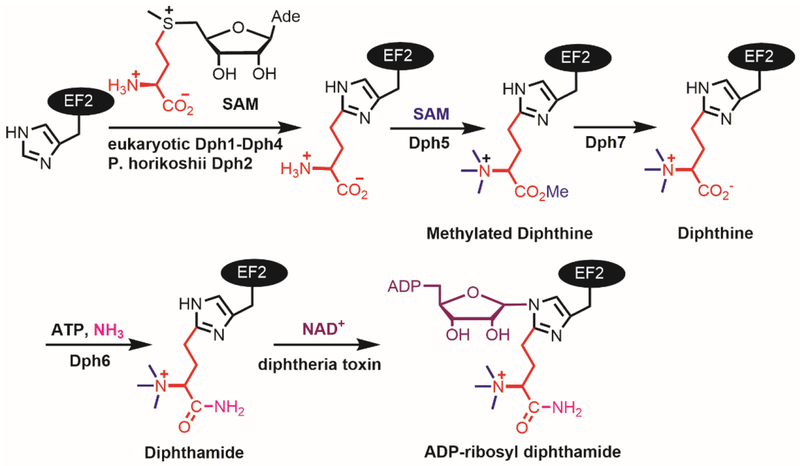
Diphthamide is a unique posttranslational modification on translation elongation factor 2 (EF2) in archaea and eukaryotes (Robinson, Henriksen, & Maxwell, 1974; Van Ness, Howard, & Bodley, 1980a, 1980b). Diphtheria toxin and Pseudomonos exotoxin A recognize diphthamide and catalyze the transfer of an adenosine diphosphate ribose (ADP-ribose) from nicotinamide adenine dinucleotide (NAD) to one of the nitrogen atoms in the imidazole ring of the histidine residue of EF2 (Fig.1)(Collier, 2001). The ADP-ribosylation inhibits the function of EF2 and thus protein synthesis, leading to cell death (Honjo, Nishizuka, Hayaishi, & Kato, 1968).
Figure 1.
Diphthamide biosynthesis pathway.
The biosynthesis of diphthamide was proposed to involve at least four steps and seven proteins (Schaffrath, Abdel-Fattah, Klassen, & Stark, 2014). The first step is the transfer of the 3-amino-3-carboxypropyl group from S-adenosyl-L-methionine (SAM) to the histidine residue of EF2, forming a C–C bond. The enzymes that performs this reaction are Dph2 (diphthamide biosynthesis protein 2) homodimer in archaea, like Pyrococcus horikoshii (PhDph2) (Zhang, Zhu, Torelli, Lee, Dzikovski, Koralewski et al., 2010) or a Dph1-Dph2 heterodimer in eukaryotes (Dong, Su, Dzikovski, Dando, Zhu, Du et al., 2014). These enzymes form a unique family of radical SAM (RS) enzymes. Classical RS enzymes, a family of enzymes with more than 113,000 members found in nature (Akiva, Brown, Almonacid, Barber, Custer, Hicks et al., 2014), all contain a [4Fe-4S] cluster and bind SAM. In these enzymes, the reduced state of the [4Fe-4S] cluster provides one electron to cleave the C5′,Ade-S bond of SAM, generating a 5′-deoxyadenosyl (5′-dA) radical that then initiates downstream reactions (Broderick, Duffus, Duschene, & Shepard, 2014). In contrast, the PhDph2 homodimer or Dph1-Dph2 heterodimer binds [4Fe-4S] clusters, cleaves the Cγ,Met-S bond of SAM and generates a 3-amino-3-carboxypropyl (ACP) radical. The formation of the ACP radical is supported by the PhDph2-catalyzed generation of 2-aminobutyric acid and homocysteine sulfinic acid in the absence of the substrate protein, PhEF2 (Zhang et al., 2010). Further support for a radical mechanism was provided by the reaction with a SAM analogue, SAMCA (Dong, Horitani, Dzikovski, Pandelia, Krebs, Freed et al., 2016).
In this chapter, we detail the preparation, characterization, and activity assays of the RS enzymes involved in diphthamide biosynthesis, PhDph2 and yeast Dph1-Dph2. We also describe the preparation of the related proteins in the first step of diphthamide biosynthesis, such as substrate protein PhEF2 and yeast EF2, electron donor Dph3 and Dph3 reductase Cbr1 in yeast.
2. Protein expression, purification and characterization
2.1. PhDph2 and PhEF2
The first step of diphthamide biosynthesis is the cleavage of the ACP group from SAM and formation of a new C-C bond (H. Lin, 2011). From genetic studies, this step requires four proteins in eukaryotes, Dph1, Dph2, Dph3, and Dph4 (Liu, Milne, Kuremsky, Fink, & Leppla, 2004). Eukaryotic Dph1 and Dph2 share about 20% sequence identity. Iterative BLAST searches (Altschul, Gish, Miller, Myers, & Lipman, 1990) starting with Saccharomyces cerevisiae Dph1 or Dph2 generate both proteins from other eukaryotic species. In contrast, BLAST searches identify only one protein, Dph2, in archaeal species (Zhang et al., 2010). Eukaryotic Dph3 and Dph4 have no orthologues in archaea. To better understand the first step of diphthamide biosynthesis, we initially studied the relatively simpler archaeal system Pyrococcus horikoshii, which is a thermophile and its proteins typically have better stability.
Procedure for PhDph2 expression
Amplify Phdph2 gene from P. horikoshii genomic DNA by PCR and insert into pENTR/TEV/D-TOPO entry vector (Invitrogen), followed by recombination with pDESTF1 destination vector to create expression clones with an N-terminal His6 tag. This vector also contains an ampicillin (Amp) resistance marker for in vivo selection.
Transform the generated plasmid into the E. coli expression strain BL21 (DE3) with pRARE2, which encodes rarely used tRNAs in E. coli and contains a chloramphenicol (Cam) resistance gene. The transformed cells were selected on lysogeny broth (LB) agar plates containing 0.1 g/L Amp and 0.02 g/L Cam.
Pick a single colony to inoculate 20 mL of LB containing 0.1 g/L Amp and 0.02 g/L Cam at 37°C and 200 rpm overnight.
Inoculate the baffled shaker flasks containing 2 L of LB supplemented with 0.02 g/L Cam and 0.1 g/L Amp with the 20 mL starter culture.
Grow cells at 37 °C and 200 rpm. Cool down the cells to 18 °C by ice water when the cultures reach an optical density (OD) of 0.6 measured at 600 nm. Add FeCl3, Fe(NH4)2(SO4)2 and L-cysteine to final concentrations of 50 μM, 50 μM, and 400 μM, respectively.
Induce the expression with 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG), at which point seal the culture flasks to limit the amount of oxygen in the system. Incubate the cells in a shaker at 18 °C and 200 rpm for 20 h.
Harvest the cells by centrifugation at 20,000 g (Beckman Coulter Avanti J-E) for 30 min and flash freeze the cell paste in liquid nitrogen before storage at −80 °C.
Procedure for PhDph2 anaerobic purification
As in classical RS enzymes, the [4Fe-4S] clusters in PhDph2 and Dph1-Dph2 are also oxygen-labile. Therefore, purification of these proteins, as well as the related assays for the activity test needs to be performed in an anaerobic environment. A vinyl anaerobic chamber (Coy LAB PRODUCTS) with an auto vacuum airlock is used. Details about the anaerobic chamber and usage were well described in a previous chapter by Booker’s lab (Lanz, Grove, Gogonea, Lee, Krebs, & Booker, 2012). A slightly different method is used here for preparing anaerobic buffers.
Autoclave the lysis buffer (20 mM Tris-HCl buffer pH 8.0, 500 mM NaCl, 10 mM MgCl2, 5 mM imidazole) and desalting buffer (200 mM Tris-HCl at pH 7.4 containing 150 mM NaCl, and 5% glycerol) at 125 °C for 25 min. Cap the buffer bottles while cooling down. After cooled down to room temperature, transfer the buffers to round flasks and further degas by Schlenk line for 2 h with magnetic stirring. Protect the buffers with N2 and transfer them into the anaerobic chamber.
Transfer the frozen cell paste from the equivalent of 4 L of bacterial culture into a Coy anaerobic chamber and resuspend in 80 mL degassed lysis buffer. Add 1 mM dithiothreitol (DTT), 1 g/L lysozyme (VWR) and 500 units benzonase (Sigma-Aldrich) in the buffer and incubate at 26 °C for 30 min.
Split the solution into two 50 mL tubes. Seal the tubes in the chamber with black tape, transfer them outside of the chamber, and freeze them in liquid nitrogen. Move the tubes back into the chamber and thaw. Repeat this freeze-thaw step again to more efficiently lysis the cells.
Transfer the lysate to 50 mL centrifuge tubes. Seal the tubes, transfer out of the chamber, centrifuge for 30 min at 20,000 g, and then transfer back into the chamber.
Pre-equilibrate 3 mL of Ni-NTA Agarose resin (Qiagen) with the lysis buffer inside the chamber. Incubate the supernatant from centrifuge tubes with the resin for 30 min. Load the resin onto a polypropylene column and wash with 20 mL of lysis buffer.
Wash the resin again with 20 mL of 30 mM imidazole in lysis buffer. Elute PhDph2 (dark brown color) from the column with elution buffers (200 mM imidazole in the lysis buffer).
Take small aliquots out of the chamber and analyze by SDS-PAGE. Combine the fractions inside the chamber based on the SDS-PAGE analysis. Buffer-exchange combined PhDph2 to the desalting buffer with 1 mM DTT using a Bio-Rad 10-DG desalting column inside the chamber.
Concentrate the purified PhDph2 protein using Amicon Ultra 0.5 mL centrifugal filter devices (Millipore) inside the chamber. Aliquot protein into 0.5 mL microcentrifuge tubes and put these tubes into a 50 mL tube. Seal the tube for storage at −80°C. Determine protein concentration by Bradford assay.
Procedure for PhEF2 expression and purification
Clone Phef2 gene following the same protocol as that of Phdph2. Transform the plasmid into E. coli expression strain BL21 (DE3) with a pRARE2 plasmid.
Grow the cells in 2 L of LB medium supplemented with 0.035 g/L Cam and 0.1 g/L Amp at 37 °C. Induce protein expression with 0.1 mM IPTG when the cultures reach an OD of 1.0 measured at 600 nm. Grow the cells for 3 h at 37 °C before harvesting by centrifugation at 20,000 g for 30 min.
Resuspend the cell pellet in 50 mL of buffer containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl and 0.5 mM phenylmethylsulfonyl fluoride.
Lyse cells by an EmulsiFlex-C3 cell disruptor (Avestin, Inc. Canada). Remove cell debris by centrifugation at 12,000 g for 30 min.
Purify the protein from the supernatant on a BioLogic DuoFlow 10 System (Bio-Rad, Hercules, CA) with a HisTrap HP column (GE Healthcare, Piscataway, NJ) with a linear gradient from 30 mM imidazole to 500 mM imidazole in 30 min.
Collect the fractions and dialyze against 200 mM Tris-HCl at pH 7.4 containing 150 mM NaCl, 1 mM DTT and 5% glycerol. Concentrate the purified protein and determine concentration by Bradford assay.
2.2. Saccharomyces cerevisiae Dph1-Dph2
Yeast Dph1 or Dph2 cannot be singly expressed in E. coli as insoluble proteins. They need to be co-expressed to obtain the soluble Dph1-Dph2 heterodimer. Dph1 and dph2 genes were initially put in the co-expression vector pETduet-1. The expression level was not very good. Interestingly, when the whole dph1-dph2 gene piece was cut from pETduet-1 and put into pET28b vector, we obtained higher Dph1-Dph2 expression. To increase the Fe-S cluster loading, Dph1-Dph2 was co-expressed with the ISC (iron-sulfur cluster) proteins, a set of bacterial proteins that assemble and transfer Fe-S clusters to client proteins.(Frazzon & Dean, 2003) Consistent with PhDph2, anaerobically purified yeast Dph1-Dph2 complex displayed a dark brown color and showed a typical [4Fe-4S]2+ cluster absorption band at 410 nm. The 410 nm absorption decreased when dithionite was added. Quantification based on the 410 nm absorption, as well as iron and sulfur content assays suggested the presence of 0.6~1 [4Fe-4S]2+ clusters per Dph1-Dph2 complex. The EPR (electron paramagnetic resonance) spectrum of the reduced Dph1-Dph2 complex showed g values of 2.04 and 1.92, consistent with a [4Fe-4S]+ cluster (Dong et al., 2014). Size exclusion chromatography-multiangle light scattering (SEC-MALS) experiment of the Dph1-Dph2 complex showed that the molecular weight is ~110 kDa, suggesting that these two proteins form a heterodimer.
Procedure for yeast Dph1-Dph2 expression
Extract yeast genomic DNA from BY4741 strain using Pierce Yeast DNA Extraction Kit. Amplify yeast dph1 and dph2 genes from yeast genomic DNA by PCR.
First insert dph2 gene with an N-terminal His tag into multiple cloning sites (MCS) 2 of the pETduet-1 vector using NdeI and XohI restriction sites, then insert dph1 gene into MCS1 using EcoRI and SalI restriction sites.
Cut the whole dph1- dph2 gene piece by EcoRI and XohI, insert into a pET28b vector digested by EcoRI and XohI.
Co-transform the resulting plasmid containing dph1- dph2 gene with pDB1282 (Frazzon & Dean, 2003), which contains the Azotobacter vinelandii ISC operon, into BL21 (DE3) pRARE2 strain.
Grow the cells in 2 liters LB medium with 0.1 g/L Amp, 0.02 g/L Cam, and 0.05 g/L kanamycin (Kan) at 37°C and 200 rpm.
At an OD600nm of 0.3, add solid arabinose to each flask at a final concentration of 0.1 % (w/v) to induce ISC proteins expression.
At an OD600nm of 0.6, cooled down the cultures with an ice-water bath and supplement the cultures with FeCl3, Fe (NH4)2(SO4)2, and L-cysteine to final concentrations of 50 μM, 50 μM, and 400 μM, respectively.
Induce Dph1 and Dph2 protein expression by adding 0.1 mM IPTG. Seal the culture flasks balloon to limit the amount of oxygen in the system. Incubate cells in a shaker at 18 °C and 200 rpm for 20 h before harvest.
Procedure for anaerobic purification of yeast Dph1-Dph2 complex
The purification is carried out in a Coy anaerobic chamber. Suspend cell pellet from 2 liter culture in 40 mL lysis buffer (500 mM NaCl, 10 mM MgCl2, 5 mM imidazole, 1 mM DTT, and 200 mM Tris-HCl at pH 7.4).
Add lysozyme (200 mg) and nuclease (4 μL, 25 U/mL of lysis buffer, Thermo) to the cell suspension and incubate at 25 °C for 0.5 h. Seal the tubes, transfer out of the chamber, and freeze them in liquid nitrogen. Move the tubes back into the chamber and thaw at 25 °C. Repeat this freeze-thaw step to obtain better lysis.
Remove the cell debris by centrifugation at 20,000g for 30 min outside of the anaerobic chamber. Transfer the tubes back into the chamber. Incubate the supernatant for 0.5 h with 1.2 mL of Ni-NTA resin pre-equilibrated with the lysis buffer. Load the Ni-NTA resin onto a polypropylene column and wash with 20 mL of lysis buffer, followed by 20 mL of 30 mM imidazole in the lysis buffer.
Elute Dph1-Dph2 from the column with elution buffers (100 mM, 150 mM, and 200 mM imidazole in the lysis buffer, 1.5 mL each). Buffer-exchange the brown-colored elution fractions to 150 mM NaCl, 200 mM Tris-HCl at pH 7.4 and 5% glycerol using a Bio-Rad 10-DG desalting column.
Concentrate the purified proteins using Amicon Ultra 0.5 mL centrifugal filter devices inside the chamber. Determine protein concentration by Bradford assay.
Procedure for the preparation of 57Fe-enriched Dph1-Dph2
To prepare 57Fe enriched Dph1-Dph2, grow the transformed cells in M9 minimal medium supplemented with 0.2% (w/v) glucose, 2 mM MgSO4 and 0.1 mM CaCl2. Dissolve 50 mg of 57Fe powder (Isoflex USA) in 2 M HCl (1.7mL), and adjust the solution pH to 5~6 as a stock solution. Add the 57Fe stock solution and L-cysteine to M9 medium to final concentrations of 100 μM and 400 μM, respectively. Express and purify 57Fe enriched Dph1-Dph2 following the same protocols as those for Dph1-Dph2 describe above.
2.3. Dph1-Dph2 Characterization
2.3.1. Size-Exclusion Chromatography Coupled Multiangle Light Scattering (SEC-MALS) for Dph1-Dph2 heterodimer.
Apply purified Dph1-Dph2 protein (~10μg/μL) to SEC using a Yarra SEC 4000 column (Phenomenex Inc.) equilibrated in MALS buffer (200 mM Tris-HCl pH 7.4, 150 mM NaCl). The SEC was coupled to a static 18-angle light scattering detector (DAWN HELEOS-II) and a refractive index detector (Optilab T-rex) (Wyatt Technology). Collect data every second at a flow rate of 1 mL/min. Analyze the data using the program ASTRA, giving the molar mass and mass distribution (polydispersity) of the sample. Monomeric BSA (Sigma) was used for normalization of the light scattering detectors and data quality control.
2.3.2. UV-Vis spectroscopy of Dph1-Dph2
Prepare 100μL Dph1-Dph2 complex (25 μM) anaerobically in 150 mM NaCl and 200 mM Tris-HCl at pH 7.4 and 5% glycerol.
Transfer the sample to a quartz cell and seal it before taking out of the anaerobic chamber. Detect UV-Vis spectra on a Cary 50 Bio UV-Vis spectrophotometer (Varian), scanning from 200 nm to 800 nm.
Correct the baseline with the buffer used to prepare the samples before measurement.
Put the quartz cell back into the chamber and treat the sample with dithionite for 10 min at a final concentration of 0.5 mM. Seal the sample and detect by UV-Vis spectrophotometer again.
2.3.3. EPR spectroscopy of Dph1-Dph2
Add dithionite to the purified Dph1-Dph2 complex (350 μM) to a final concentration of 1 mM in an anaerobic chamber. Incubate the mixture for 5 min and transfer to the bottom of an EPR tube (715-PQ-250M, Wilmad–LabGlass) using long glass pipette.
Freeze the sample by dip the EPR tube in liquid N2 promptly several times to prevent the tube from cracking. Cap the EPR tube and transfer outside the chamber.
Record EPR spectra on a Bruker ElexSys E500 EPR spectrometer at a frequency of 9.34 GHz at 12 K using an ESR910 liquid-helium cryostat (Oxford Instruments) with the settings as follows: modulation frequency, 100 kHz; modulation amplitude, 8 G; microwave power, 0.63 mW. Calibrate the field sweeps with a BRUKER ER 035 Gauss meter. The microwave frequency can be monitored with a frequency counter. Xepr software can be used for data acquisition and manipulation.
2.4. Preparation of Saccharomyces cerevisiae EF2, Dph3 and Cbr1
2.4.1. Expression and purification of Saccharomyces cerevisiae EF2
Overexpression of Saccharomyces cerevisiae EF2 in E. coli strains was not successful. We then prepared EF2 with a C-terminal His6 tag in yeast cells. After testing different promoter systems, better protein quality was obtained with the vectors having a MET25 promoter. Yeast cells can be grown either in culture flask or fermenter at 30°C, the latter gives much higher cells density and more proteins.
PCR-amplify the Saccharomyces cerevisiae ef2 coding sequence from yeast genomic DNA with an added C-terminal His6 tag. Clone into p423MET25 vector (ATCC, Manassas, VA), which has a His3 selection marker.
Transform the p423MET25-EF2 plasmid into S. cerevisiae BY4741 strain with deletion of dph2 gene (OpenBiosystems, Huntsville, AL) using a Frozen-EZ Yeast Transformation kit (ZYMO RESEARCH CORP).
To express the EF2 protein, grow the transformed cells in 6 L synthetic complete media lacking histidine for 48 h in a fermenter (BioFlo 110, New Brunswick Scientific) at 30°C with agitation.
Harvest cells by centrifugation until the OD600nm is 4~6. Re-suspend the collected cells in 200 mL of lysis buffer, which contains 20 mM Tris-HCl (pH 8.0), 400 mM (NH4)2SO4, 10 mM MgCl2, and 1 mM protease inhibitor phenylmethylsulfonyl fluoride.
Lyse the cells using glass beads (400 μm, OPS DIAGNOSTICS) with bead beater (Biospec). Fill the beater chamber with 200 mL glass beads and 200 mL lysis buffer containing 50~80 g of cells. Install the ice-water jacket and fill it with ice. As a cycle, run the bead beater for 1 min and let it sit for 2 min to cool down. Repeat this cycle four times.
Remove cell debris by centrifugation at 12,000 g for 30 min. Purify the protein from the supernatant on a BioLogic DuoFlow 10 System with a HisTrap HP column with a linear gradient from 30 mM imidazole to 500 mM imidazole in 30 min.
Collect the fractions and dialyze against 200 mM Tris-HCl buffer (pH 7.4) containing 500 mM NaCl and 5% glycerol. Concentrate the purified proteins and determine protein concentration by Bradford assay.
2.4.2. Cloning, expression and purification of Dph3
Yeast dph3 was found to be identical to Kluyveromyces lactis killer toxin zymocin insensitive 11 (kti11) gene from Saccharomyces cerevisiae (Fichtner & Schaffrath, 2002). Dph3 is a small protein less than 10 kDa that contains a CSL zinc finger domain. The CSL zinc finger uses four conserved Cys residues to coordinate Zn2+(Sun, Zhang, Wu, Xu, Li, Zhao et al., 2005), and the last Cys residue is followed by a Ser and a Leu residue. The first solution NMR structure of yeast Dph3 (Sun et al., 2005) shows a single Zn2+ ion bound by four conserved cysteine residues of Dph3. However, the Dph3 protein was purified from E. coli grown on minimal medium M9 supplemented with ZnSO4. We (Dong et al., 2014) and others (Proudfoot, Sanders, Singer, Zhang, Brown, Binkowski et al., 2008) demonstrated that yeast Dph3 binds both zinc and iron when prepared from E. coli cells grown on rich medium (Proudfoot et al., 2008). Crystal structures of Dph3 binds only iron (Kolaj-Robin, McEwen, Cavarelli, & Seraphin, 2015) and both zinc and iron (Glatt, Zabel, Vonkova, Kumar, Netz, Pierik et al., 2015) were reported. Our in vitro reconstitution experiment demonstrated that the iron-bound form of Dph3 helps to reduce Dph1-Dph2.
Amplify yeast dph3 gene from yeast genomic DNA, which was extracted from BY4741 using Pierce Yeast DNA Extraction Kit.
Clone dph3 gene following the same protocol as that of Phdph2. Transform this plasmid to BL21(DE3) pRARE2 strain.
Grow cells in 2 liters LB medium supplemented with 0.035 g/L Cam and 0.1 g/L Amp at 37°C and 200 rpm. At an OD600nm of 0.6, cool down the cultures to 18°C and supplement with (or without) Fe(NH4)2(SO4)2 to a final concentration of 70 μM.
Induce protein expression by adding IPTG to 0.1 mM. Harvest cells after incubation at 18°C for 20 h. Resuspend the cell pellet in 30 mL of buffer containing 20 mM Tris-HCl (pH 7.4), 500 mM NaCl and 0.5 mM phenylmethylsulfonyl fluoride.
Lyse cells by an EmulsiFlex-C3 cell disruptor. Purify Dph3 through Ni-NTA affinity chromatography following the same protocol as that used for the purification of PhEF2.
Collect the eluted imidazole fractions and dialyze against 20 mM Tris-HCl buffer (pH 7.4) containing 500 mM NaCl and 5% glycerol. Concentrate the purified protein using Amicon Ultra-4 centrifugal filter devices.
2.4.3. Dph3 characterization by UV-Vis spectroscopy
Dph3 can be characterized by UV-Vis spectroscopy. The cysteine-coordinated ferric iron exhibits 350, 490 and 570 nm absorptions. Prepare 100 μL of Dph3 (80 μM) in 20 mM Tris-HCl buffer (pH 7.4) containing 500 mM NaCl and 5% glycerol in a quartz cell. Correct the baseline using the same buffer. Obtain the UV-Vis spectrum of Dph3 by scanning from 200 nm to 800 nm. Then add dithionite to the sample to the final concentration of 0.5 mM and obtain the UV-Vis spectrum again.
2.4.4. Cloning, expression, and purification of Cbr1
To reduce Dph1-Dph2 and Dph3 using electrons from NADH, a flavin-dependent enzyme is needed. We discovered that cytochrome b5 reductase 1 (Cbr1) is the physiological reductase of Dph3 (Z. Lin, Dong, Zhang, Lee, & Lin, 2016). Cbr1 is a flavin-containing protein located on the outer membrane of endoplasmic reticulum and mitochondria. The first 27 amino acids is a transmembrane helix. The soluble form of Cbr1 with truncation of the first 27 amino acids is cloned from the DNA of yeast BY4741 strain.
PCR amplify the truncated cbr1 gene from yeast genomic DNA with primers introducing EcoRI and XhoI restriction sites.
Double digest 1.5 μg of pET28a vector and the PCR product in 50 μL solution containing 5 μL Cutsmart buffer, 2.5 μL EcoRI-HF and 2.5 μL XhoI in 37 °C for 3 h.
Insert the truncated cbr1 gene into pET28a vector.
Transform the resulting plasmid into BL21R2 E. coli strain. Pick single colony to inoculate overnight starter culture with 5 mL LB media containing 50 μg/mL Kan and 25 μg/mL Cam in a 37 °C shaker.
Inoculate 2 liter of LB media containing 50 μg/mL Kan and 25 μg/mL Cam with overnight starter culture in a 37 °C shaker until OD600nm reaches 0.6.
Cool down the culture with ice-water bath for 20 min. Induce the expression of truncated Cbr1 with 1.5 mM of IPTG and 50 μM riboflavin for 20 h at 16 °C.
Collect the pellet, resuspend in 30 mL lysis buffer (25 mM Tris-HCl pH 8.0, 150 mL NaCl and 10% glycerol) and lyse with EmulsiFlex-C3 cell disruptor.
Centrifuge the lysate at 20,000 g for 30 min. Incubate the supernatant with 0.5 mL of Ni-NTA resin pre-equilibrated with the lysis buffer for 30 min at 4 °C.
Load the incubated Ni-NTA resin onto a polypropylene column at 4 °C. Wash with 10 mL lysis buffer, followed by 40 mL of 30 mM imidazole in lysis buffer.
Elute protein with elution buffers (50 mM, 75 mM, 100 mM, and 200 mM imidazole in the lysis buffer, 5 mL each). Collect the yellow fractions to run a 12% SDS-PAGE to check protein purity.
Dialyze the pure fraction against 25 mM Tris-HCl pH 8.0 with 150 mL NaCl.
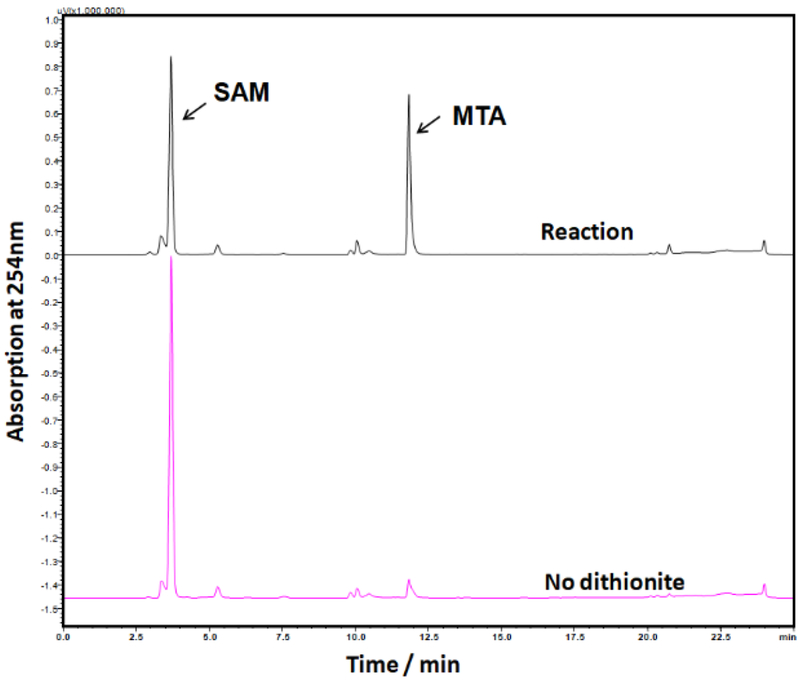
3. In vitro analysis of SAM cleavage by Dph1-Dph2 with high-performance liquid chromatography
Set up reactions under anaerobic condition with 100 μM Dph1-Dph2, 2 mM SAM, 10 mM dithionite, 150 mM NaCl, 1 mM DTT and 200 mM Tris-HCl at pH 7.4 in a final volume of 30 μl. Also carry out negative control reactions without dithionite (Figure. 2).
Incubate the mixture at 30 °C for 1 h.
Quench reactions with 30 μl of 10% trifluoroacetic acid (TFA), followed by centrifugation to separate the precipitated proteins and the supernatant.
Analyze the supernatant by high-performance liquid chromatography (Shimadzu) on a C18 column (Kinetex) monitored at 254 nm, using a linear gradient from 0 to 40% solvent B over 15 min at a flow rate of 0.5 mL /min (solvent A: 0.1% aqueous TFA and solvent B: 0.1% TFA in acetonitrile).
Figure 2.
HPLC assay for Dph1-Dph2 activity. Reduced Dph1-Dph2 is active in the SAM cleavage reaction, generating MTA. SAM is not cleaved without dithionite.
4. In vitro reconstitution of the first step of diphthamide biosynthesis
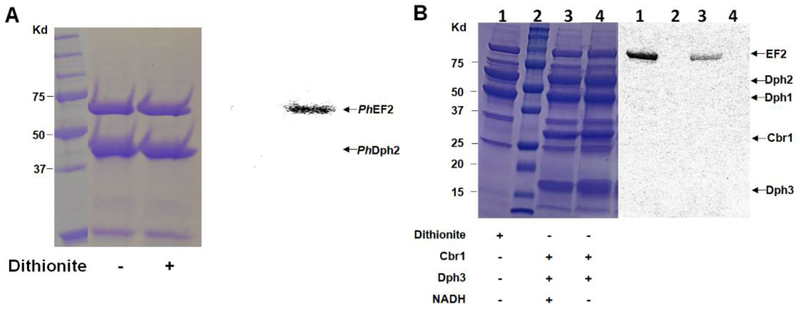
4.1. Anaerobic reconstitution of PhDph2 activity
Set up a reaction mixture containing 20 μM PhDph2, 7 μM PhEF2 and 5 mM dithionite in 150 mM NaCl, 1 mM DTT and 200 mM Tris-HCl at pH 7.4 in an anaerobic chamber. Set up a same reaction but without dithionite added as a control reaction. Seal the reaction vials with rubber stoppers (Sigma-Aldrich Z564680) and then transfer the vials out of the anaerobic chamber.
Inject carboxyl-14C-SAM (18 μM; 55 mCi/mmol, American Radiolabeled Chemicals Inc.) into the reaction vials to start the reaction. Vortex the reaction mixture briefly and incubate at 65 °C for 10 min.
Stop the reactions by adding protein loading dye and heating the vial at 95 °C for 5 min.
Analyze the reaction mixture with 12% SDS–PAGE gel electrophoresis (Figure.3A).
Expose the dried gel to a PhosphorImaging screen (GE Healthcare) and detect the radioactivity using a Typhoon FLA 7000 (GE Healthcare).
Figure 3.
In vitro reconstitution of the first step of diphthamide biosynthesis with 14C-SAM. A) In vitro reconstitution of PhDph2 activity using dithionite. B) In vitro reconstitution of yeast Dph1-3 activity. Lane 2 is protein ladder. All the reactions in lanes 1, 3 and 4 contained EF2 and Dph1-Dph2. The presence of other reagents is indicated below each lane. In both A and B, the left panels show the Coomassie blue-stained gel; the right panels show the autoradiography.
4.2. Anaerobic reconstitution of the first step of diphthamide biosynthesis using yeast Dph1-3
Truncated Cbr1 uses NADH to reduce Dph3 in vitro (Z. Lin et al., 2016). We can use Cbr1 with NADH to reduce Dph3 and reconstitute the first step of diphthamide biosynthesis. By using carboxyl-14C-SAM, we can label yeast EF2 with 14C and the reaction can be monitored with autoradiography using a Typhoon scanner (Figure. 3B).
Transfer at least 100 μL of Dph3, Cbr1, and EF2 separately on ice into three 10 mL round-bottom flasks equipped with magnetic spin bars. Degas these proteins using a Schlenk line before use.
Add proteins and reagents in the following sequence in an anaerobic chamber: 6 μL of desalting buffer (150 mM NaCl, 200 mM Tris-HCl at pH8.0, 5% glycerol), Dph3 (10 μM, final concentration), Cbr1 (5 μM), NADH (200 μM), Dph1-Dph2 (5 μM) and EF2 (2 μM). Add water to make the final volume of the reaction 23 μL. Seal the Eppendorf tubes with rubber stoppers. Transfer all the sealed Eppendorf tubes out of the glove box.
Prepare diluted carboxyl-14C-SAM by adding 4 μL of carboxyl-14C-SAM with a microliter syringe into a tube with 16 μL of desalting buffer (prepared ahead of time in the glove box). Vertex the tube and spin down briefly.
Add 2 μL of diluted carboxyl-14C-SAM into each reaction with a microliter syringe. Vertex and spin down briefly.
Incubate all the reactions at 30 °C for 1 h.
Add 5 μL loading dye to quench the reactions. Spin down all the samples briefly after quenching and boil at 95 °C for 5 min.
Resolve the reaction mixture using 12% SDS-PAGE. Stain the gel with Coomassie staining solution and then de-stain the gel.
Dry the de-stained gel with cellophane (GelAir™, Bio-Rad) support in the gel dryer for 3 h. Expose the dried gel with phosphor screen for 24 h and scan the phosphor screen with a Typhoon scanner
Note:
After quenching, the samples can be stored in −20 °C freezer.
The de-staining step is essential to get rid of the free carboxyl-14C-SAM. Free carboxyl-14C-SAM in the gel will result in higher background signal.
Allow the gel to dry fully to prevent the contamination of the phosphor screen and typhoon scanner.
A positive control with dithionite as reductant and a negative control without reductant should be carried out at the same time.
4.3. In vitro ADP-ribosylation of diphthamide using rhodamine NAD (Rh-NAD).
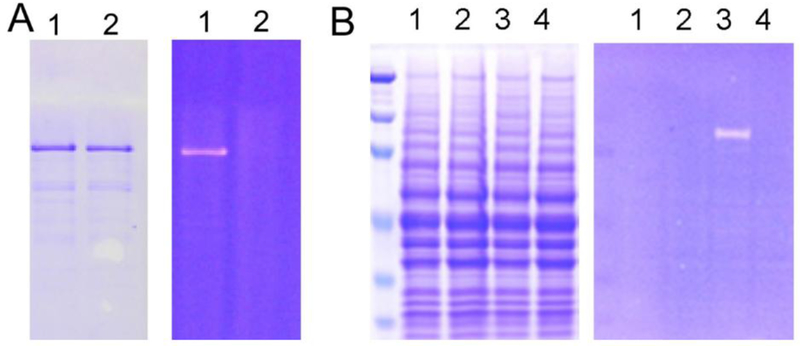
To facilitate the detection of diphthamide formation, we also developed a fluorescent assay to monitor the reaction (Du, Jiang, & Lin, 2009). This assay can be used on both purified EF2 protein and cell lysate. The assay takes advantage of the selective recognition of both diphthamide and diphthine (the intermediate before the amidation step, see Figure 1) by DT. We synthesized a rhodamine-conjugated NAD molecule, Rh-NAD. If diphthamide or diphthine is formed in the reaction mixture, then incubating the reaction mixture with Rh-NAD and DT should allow fluorescent labeling of EF2. The synthesis of Rh-NAD has been reported (Du et al., 2009).
Incubate the purified yeast EF2 and Rh-NAD (50 μM) with DT (100 nM ) at 30 °C in 25 mM Tris-HCl (pH 8.0), 50 mM NaCl, 30 mM DTT and 2 mM ethylenediaminetetraacetic acid (EDTA) for 1 h. Resolve the reaction mixture by SDS-PAGE (Figure 4A). Visualize rhodamine fluorescence signal from protein gel on a Fisher Scientific Ultraviolet Transilluminators or quantify the signal using a Typhoon scanner.
Figure 4.
Rh-NAD labeling diphthamide using diphtheria toxin (DT)-catalyzed ADP-ribosylation of EF-2. (A) Fluorescent labeling of purified eEF-2 with diphthamide modification. Lane 1, with DT; lane 2, without DT. (B) Fluorescent labeling of EF-2 in cell lysate. Lane 1, wild type cell lysate without DT; lane 2, Δdph1 cell lysate without DT; lanes 3, wild type cell lysate with DT; lanes 4, Δdph1 cell lysate with DT. Fig 4B is adapted with permission from (Biochemistry, 2009, 48, 2878). Copyright (2009) American Chemical Society.
The labeling method can be further used on cell lysate from wild-type (wt) yeast strain and Δdph1 strain (Figure 4B). The EF2 from wt strain has diphthamide modification and thus can be labeled by diphtheria toxin, while the EF2 from Δdph1 cells lacks diphthamide modification and thus cannot be labeled.
5. Conclusion
The first step of diphthamide biosynthesis is the transfer of an ACP group from SAM to substrate EF2 by unique radical SAM enzymes. In this chapter, we have discussed the preparation and characterization of these enzymes, PhDph2 homodimer and yeast Dph1-Dph2 heterodimer, as well as the preparation of related proteins in the first step of diphthamide biosynthesis, including substrate protein PhEF2 and yeast EF2, yeast Dph3 and Cbr1. We have highlighted three activity assays for the analysis of diphthamide radical SAM enzymes, HPLC assay to detect the cleavage product MTA, radioactive labeling assay using carboxyl-14C-SAM, and a fluorescent assay using Rh-NAD.
ACKNOLEDGMENTS
This work was supported by grants from the National Institute of Health National Institute of General Medical Sciences (GM 088276).
Abbreviation
- DTT
dithiothreitol
- EF2
elongation factor 2
- EPR
electron paramagnetic resonance
- IPTG
isopropyl-β-D-thiogalactopyranoside
- LB
lysogeny broth
- NAD
nicotinamide adenine dinucleotide
- OD
optical density
- SAM
S-adenosylmethionine
- SEC-MALS
Size-Exclusion Chromatography Coupled Multiangle Light Scattering
- TFA
trifluoroacetic acid
- UV-Vis
Ultraviolet-visible
REFERENCES
- Akiva E, Brown S, Almonacid DE, Barber AE, Custer AF, Hicks MA, … Babbitt PC (2014). The Structure–Function Linkage Database. Nucleic. Acids.Res, 42(D1), D521–D530. doi: 10.1093/nar/gkt1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, & Lipman DJ (1990). Basic local alignment search tool. J. Mol. Biol, 215(3), 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Broderick JB, Duffus BR, Duschene KS, & Shepard EM (2014). Radical S-adenosylmethionine enzymes. Chem. Rev, 114(8), 4229–4317. doi: 10.1021/cr4004709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier RJ (2001). Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon, 39(11), 1793–1803. [DOI] [PubMed] [Google Scholar]
- Dong M, Horitani M, Dzikovski B, Pandelia ME, Krebs C, Freed JH, … Lin H (2016). Organometallic Complex Formed by an Unconventional Radical S-Adenosylmethionine Enzyme. J. Am. Chem. Soc, 138(31), 9755–9758. doi: 10.1021/jacs.6b04155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Su X, Dzikovski B, Dando EE, Zhu X, Du J, … Lin H (2014). Dph3 is an electron donor for Dph1-Dph2 in the first step of eukaryotic diphthamide biosynthesis. J. Am. Chem. Soc, 136(5), 1754–1757. doi: 10.1021/ja4118957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Jiang H, & Lin H (2009). Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry, 48(13), 2878–2890. doi: 10.1021/bi802093g [DOI] [PubMed] [Google Scholar]
- Fichtner L, & Schaffrath R (2002). KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin.Mol. Microbiol, 44(3), 865–875. [DOI] [PubMed] [Google Scholar]
- Frazzon J, & Dean DR (2003). Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol, 7(2), 166–173. [DOI] [PubMed] [Google Scholar]
- Glatt S, Zabel R, Vonkova I, Kumar A, Netz DJ, Pierik AJ, … Muller CW (2015). Structure of the Kti11/Kti13 heterodimer and its double role in modifications of tRNA and eukaryotic elongation factor 2. Structure, 23(1), 149–160. doi: 10.1016/j.str.2014.11.008 [DOI] [PubMed] [Google Scholar]
- Honjo T, Nishizuka Y, Hayaishi O, & Kato I (1968). Diphtheria Toxin-dependent Adenosine Diphosphate Ribosylation of Aminoacyl Transferase II and Inhibition of Protein Synthesis. J. Biol. Chem, 243(12), 3553–3555. [PubMed] [Google Scholar]
- Kolaj-Robin O, McEwen AG, Cavarelli J, & Seraphin B (2015). Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification. FEBS. J, 282(5), 819–833. doi: 10.1111/febs.13199 [DOI] [PubMed] [Google Scholar]
- Lanz ND, Grove TL, Gogonea CB, Lee KH, Krebs C, & Booker SJ (2012). RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins. Methods. Enzymol, 516, 125–152. doi: 10.1016/B978-0-12-394291-3.00030-7 [DOI] [PubMed] [Google Scholar]
- Lin H (2011). S-Adenosylmethionine-dependent alkylation reactions: when are radical reactions used? Bioorg. Chem, 39(5-6), 161–170. doi: 10.1016/j.bioorg.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Dong M, Zhang Y, Lee EA, & Lin H (2016). Cbr1 is a Dph3 reductase required for the tRNA wobble uridine modification. Nat. Chem. Biol, 12(12), 995–997. doi: 10.1038/nchembio.2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Milne GT, Kuremsky JG, Fink GR, & Leppla SH (2004). Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol. Cell. Biol, 24, 9487–9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot M, Sanders SA, Singer A, Zhang R, Brown G, Binkowski A, … Yakunin AF (2008). Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain. J. Mol. Biol, 375(1), 301–315. doi: 10.1016/j.jmb.2007.10.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EA, Henriksen O, & Maxwell ES (1974). Elongation factor 2. Amino acid sequence at the site of adenosine diphosphate ribosylation. J. Biol. Chem, 249(16), 5088–5093. [PubMed] [Google Scholar]
- Schaffrath R, Abdel-Fattah W, Klassen R, & Stark MJ (2014). The diphthamide modification pathway from Saccharomyces cerevisiae--revisited. Mol.Microbiol, 94(6), 1213–1226. doi: 10.1111/mmi.12845 [DOI] [PubMed] [Google Scholar]
- Sun J, Zhang J, Wu F, Xu C, Li S, Zhao W, … Shi Y (2005). Solution Structure of Kti11p from Saccharomyces cerevisiae Reveals a Novel Zinc-Binding Module. Biochemistry, 44, 8801–8809. [DOI] [PubMed] [Google Scholar]
- Van Ness BG, Howard JB, & Bodley JW (1980a). ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J. Biol. Chem, 255(22), 10717–10720. [PubMed] [Google Scholar]
- Van Ness BG, Howard JB, & Bodley JW (1980b). ADP-ribosylation of elongation factor 2 by diphtheria toxin. NMR spectra and proposed structures of ribosyl-diphthamide and its hydrolysis products. J. Biol. Chem, 255(22), 10710–10716. [PubMed] [Google Scholar]
- Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, … Lin H (2010). Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature, 465(7300), 891–896. doi: 10.1038/nature09138 [DOI] [PMC free article] [PubMed] [Google Scholar]