Abstract
Bupropion hydroxylation is a bioactivation and metabolic pathway, and the standard clinical CYP2B6 probe. This investigation determined the influence of CYP2B6 allelic variants on clinical concentrations and metabolism of bupropion enantiomers. Secondary objectives evaluated the influence of CYP2C19 and P450 oxidoreductase variants. Healthy volunteers in specific cohorts (CYP2B6*1/*1, CYP2B6*1/*6, CYP2B6*6/*6, and also CYP2B6*4 carriers) received single-dose oral bupropion. Plasma and urine bupropion and hydroxybupropion was quantified. Subjects were also genotyped for CYP2C19 and P450 oxidoreductase variants. Hydroxylation of both bupropion enantiomers, assessed by plasma hydroxybupropion/bupropion AUC ratios and urine hydroxybupropion formation clearances, was lower in CYP2B6*6/*6 but not CYP2B6*1/*6 compared with CYP2B6*1/*1 genotypes, and numerically greater in CYP2B6*4 carriers. CYP2C19 and P450 oxidoreductase variants did not influence bupropion enantiomers hydroxylation or plasma concentrations. Results show that clinical hydroxylation of both bupropion enantiomers was equivalently influenced by CYP2B6 allelic variation. CYP2B6 polymorphisms affect S-bupropion bioactivation, which may affect therapeutic outcomes.
Keywords: cytochrome P450 2B6, CYP2B6, CYP2C19, P450 oxidoreductase, bupropion, stereochemistry, pharmacogenetics
Introduction
Bupropion is used for treating major depressive disorder, both initial therapy and relapse prevention, either as monotherapy or in antidepressant combinations.1 It is also a first-line switch agent for patients intolerant to or non-responsive to selective serotonin reuptake inhibitors. Bupropion is also used for seasonal affective disorder, smoking cessation,2 obesity,3 and attention deficit hyperactivity disorder in adults and children.4 Bupropion undergoes extensive metabolism, with less than 1% recovered intact in urine, to the three primary metabolites hydroxybupropion (via t-butylhydroxylation), and the isomers threohydrobupropion and erythrohydrobupropion (via keto reduction). Hydroxybupropion exposure is approximately an order of magnitude greater than that of parent drug.
Bupropion is used clinically as a racemate. Metabolism in vitro 5 and disposition in vivo are stereoselective.6–11 Plasma R-bupropion exposures are 2- to 6-fold higher than those of S-bupropion, the apparent oral clearance of S-bupropion exceeds that of R-bupropion, and R,R-hydroxybupropion exposures are 35- to 65-fold higher than S,S hydroxybupropion.7–9
Bupropion hydroxylation is considered to be an important clinical bioactivation pathway, contributing to the antidepressant and smoking-cessation effects of bupropion. Hydroxybupropion is pharmacologically active, inhibiting norepinephrine and dopamine uptake, and antagonizing nicotine and showing antidepressant effects in rodents.12–14 Clinically, smoking-cessation and antidepressant effectiveness of bupropion is attributed to hydroxybupropion.15–17 These activities are attributed to S,S- rather than R,R-hydroxybupropion, despite much lower plasma concentrations.13
Bupropion hydroxylation is also an important catabolic pathway. In vitro it is catalyzed predominantly by CYP2B6,18 and is the standard in vitro probe for assessing CYP2B6 activity and drug interactions. Clinically, racemic bupropion hydroxylation has been used to phenotype CYP2B6 activity, polymorphisms, and drug interactions, and is the recommended standard in vivo CYP2B6 probe.19,20 The CYP2B6 gene is highly polymorphic, and carriers of allelic variants have altered in vivo bupropion metabolism, evidenced by the plasma hydroxybupropion/bupropion AUC ratio, although bupropion clearance is unchanged.16,21–25 Hydroxybupropion may also inhibit P450 to a similar or greater extent than the parent drug 26
Bupropion systemic clearance does not depend on CYP2B6 activity because of the low fraction metabolized to hydroxybupropion. However CYP2C19 is of interest regarding bupropion disposition. CYP2C19 does not catalyze t-butyl hydroxylation,18 but contributes to bupropion metabolism through alternate hydroxylation pathways,27 recently identified as aromatic 4’-hydroxylation.28 It is unclear whether CYP2C19 or CYP2B6 contributes more to clinical bupropion metabolism.29,30 Nevertheless, CYP2C19 may influence parent drug elimination, as carriers of the reduced activity CYP2C19*2 allele had slightly higher bupropion steady-state plasma exposure.31
Polymorphisms of P450 oxidoreductase (POR), many of which have decreased activity (the extent of which is also CYP isoform-specific), including towards CYP2B6,32,33 might also affect bupropion disposition. Recent clinical studies of association between POR variants and CYP2B6 activity have reported increased bupropion metabolism in GG homozygous carriers of the POR g.6593A>G polymorphism.24,25
While certain CYP2B6 allelic variants have altered hydroxylation of racemic bupropion in vitro,34–36 and in vivo,16,21–25 the hydroxylation activity of CYP2B6 variants towards bupropion enantiomers is unknown, as is the influence of CYP2B6 genetic polymorphisms on in vivo bupropion enantiomers hydroxylation and clearance. This investigation determined the influence of CYP2B6 genetics on in vivo bupropion enantiomers metabolism and clearance, specifically the most common and clinically significant variant CYP2B6*6. A secondary objective was to evaluate other, less common allelic variants, when encountered. Healthy volunteers were genotyped, and then CYP2B6 genotype cohorts composed to evaluate bupropion disposition. Another secondary objective was to evaluate the influence of CYP2C19 and POR polymorphisms on bupropion enantiomers clinical metabolism and clearance.
Results
Only 17 CYP2B6*6/*6 subjects could be identified and studied. Others with rare allelic variants were also evaluated, including two CYP2B6*5/*5 homozygotes and one CYP2B6*1/*4 heterozygote and three CYP2B6*4/*6 heterozygotes.
Bupropion disposition and CYP2B6 genotypes
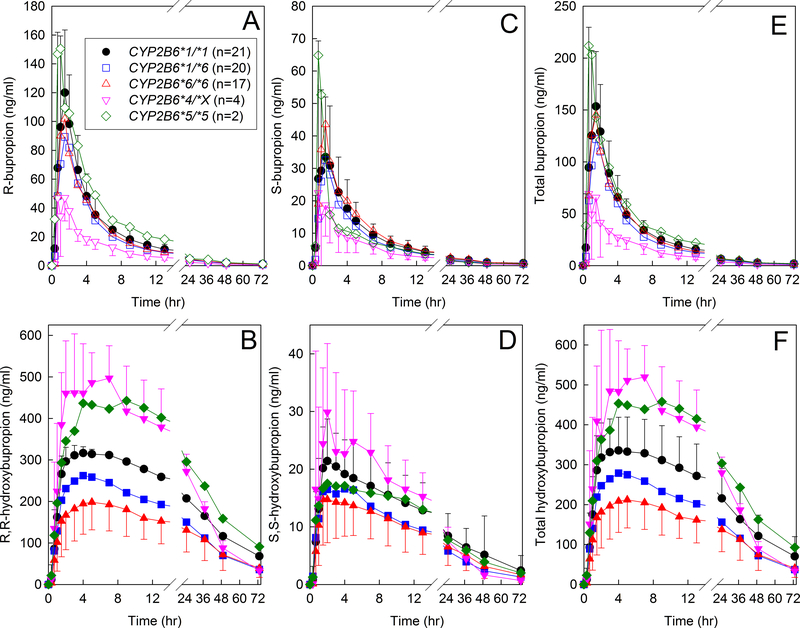
Plasma bupropion enantiomer concentrations are shown in Figure 1 for the three major genotype groups (CYP2B6*1/*1, CYP2B6*1/*6, CYP2B6*6/*6), and also for *4 carriers (CYP2B6*1/*4 and CYP2B6*4/*6, shown together as CYP2B6*4/*X) and for *5 homozygotes. Pharmacokinetic parameters are provided in Table 1. Bupropion disposition was stereoselective, as observed previously.7,8 The overall R-bupropion/S-bupropion plasma AUC0-∞ ratio for all subjects was 2.3 (1.8, 3.1) (median, interquartile range). There were no significant differences between CYP2B6*1/*1 and CYP2B6*6 hetero- or homozygotes in bupropion Cmax, AUC∞ or Cl/F for either enantiomer or the racemate. For the four CYP2B6*4/*X subjects, AUC0-∞ (ng/ml-hr−1) was 322 ±98 and 163 ± 74 for R-and S-bupropion respectively, while that for CYP2B6*1/*1 subjects was 737 ± 214 and 297 ± 123. The R-bupropion/S-bupropion plasma AUC0-∞ ratio in CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6 subjects was 2.5 (1.9, 3.4), 2.5 (2.0, 3.3) and 1.9 (1.8, 2.6) and not significantly different between groups. Urine recovery of R- and S-bupropion (% of dose) across all subjects was 1.0 (0.5, 2.7) and 0.3 (0.1, 0.5). There were no significant differences between the major CYP2B6 genotype groups in bupropion renal clearance (Table 2).
Figure 1.
Influence of CYP2B6 genotype on bupropion disposition. Subjects received 150 mg oral immediate release bupropion. Shown are plasma concentrations of (A) R-bupropion, (B) R,R-hydroxybupropion, (C) S-bupropion, (D) S,S-hydroxybupropion, (E) RS-bupropion, (F) total hydroxybupropion. Each data point is the mean ± SD. Some SD values are omitted for clarity. Genotype cohorts were CYP2B6*1/*1, CYP2B6*1/*6, CYP2B6*6/*6, CYP2B6*4/*X (with results for one CYP2B6*1/*4 and three CYP2B6*4/*6 subjects combined) and CYP2B6*5/*5.
Table 1.
Pharmacokinetic parameters for plasma bupropion and hydroxybupropion in CYP2B6 genotypes
|
CYP2B6*1/*1 (n=21) |
CYP2B6*1/*6 (n=20) |
CYP2B6*6/*6 (n=17) |
All subjects (n=64) |
|
|---|---|---|---|---|
| bupropion | ||||
| Cmax (ng/ml) | 170 (119, 209) | 145 (113, 184) | 149 (123, 195) | 148 (114, 194) |
| AUC0-∞ (hr•ng•ml−1) | 1035 ± 295 | 853 ± 232 | 970 ± 251 | 933 ± 288 |
| CL/F (ml−1•kg−1•min−1) | 37 ± 11 | 41 ±14 | 39 ± 10 | 42 ± 17 |
| Elimination t1/2 (h) | 25 (23, 27) | 22 (18, 28) | 22 (17, 30) | 23 (19,28) |
| R-bupropion | ||||
| Cmax (ng/ml) | 130 (91, 175) | 102 (82, 139) | 106 (84, 130) | 108 (84, 145) |
| AUC0-∞ (ng •h •ml−1) | 737 ± 214 | 610 ± 185 | 652 ± 173 | 657 ± 215 |
| CL/F (ml−1•kg−1•min−1) | 26 (19, 31) | 28 (24, 34) | 29 (24, 37) | 27 (22, 34) |
| Elimination t1/2 (h) | 24 (19, 26) | 22 (19, 28) | 19 (17, 25) | 22 (18, 26) |
| S-bupropion | ||||
| Cmax (ng/ml) | 30 (24, 60) | 35 (26, 49) | 52 (36, 65) | 36 (25, 55) |
| AUC0-∞ (hr•ng•ml−1) | 297 ± 123 | 248 ± 79 | 324 ± 116 | 279 ± 111 |
| CL/F (ml−1•kg−1•min−1) | 64 (45, 86) | 69 (49, 99) | 55 (43, 81) | 67 (49, 86) |
| Elimination t1/2 (h) | 26 (13, 26) | 25 (23, 26) | 26 (25, 27) | 26 (24, 26) |
| hydroxybupropion | ||||
| Cmax (ng/ml) | 370 (268, 419) | 283 (210, 344)* | 234 (157, 282)* | 296 (230, 400) |
| Elimination t1/2 (hr) | 24 (23, 34) | 21 (17, 33) | 28 (22, 30) | 24 (20, 30) |
| AUC0–72 hydroxybupropion/bupropion (hr•ng•ml−1) | 13.5 (9.8, 15.5) | 10.9 (8.4, 13.0) | 9.2 (5.4, 12.7)* | 11.7 (8.4, 15.2) |
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 15.1 (10.4, 18.3) | 12.2 (8.8, 15.1) | 10.2 (6.1, 15.3)* | 13.2 (9.1, 18.0) |
| R,R-hydroxybupropion | ||||
| Cmax (ng/ml) | 336 (252, 401) | 276 (195, 311)* | 220 (147, 261)* | 278 (217, 385) |
| Elimination t1/2 (hr) | 24 (23, 35) | 21 (17, 33) | 28 (23, 30) | 24 (20, 30) |
| AUC0–72 hydroxybupropion/bupropion (hr•ng•ml−1) | 18.0 (13.1, 20.4) | 14.6 (10.4, 20.0) | 14.0 (7.9, 17.7)* | 15.7 (11.5, 20.6) |
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 20.8 (15.1, 24.9) | 15.9 (11.9, 23.5) | 15.7 (8.5, 20.8)* | 27.1 (12.0, 24.8) |
| S,S-hydroxybupropion | ||||
| Cmax (ng/ml) | 21 (17, 29) | 18 (11, 23) | 16 (17, 24) | 19 (14, 26) |
| Elimination t1/2 (h) | 23 ± 6 | 24 ± 9 | 25 ± 8 | 23 ± 8 |
| AUC0–72 hydroxybupropion/bupropion (hr•ng•ml−1) | 2.1 (1.6, 2.6) | 1.6 (1.3, 2.3) | 1.6 (1.0, 2.0)* | 1.9 (1.5, 2.4) |
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 2.1 (1.6, 2.7) | 1.8 (1.3, 2.3) | 1.6 (1.0, 2.0)* | 1.9 (1.5, 2.3) |
Cmax, maximum concentration; Tmax, time to maximum concentration; AUC0–72 area under concentration-time curve from 0 to 72 hr; AUC0--∞, area under concentration-time curve extrapolated to infinity; CL/F, apparent oral clearance; t½, elimination half-life. Results are the mean ± SD or median (25th, 75th quartile)
Significantly different from CYP2B6*1/*1 (p<0.05).
Results are not shown for 3 CYP2B6*4/*6, 1 CYP2B6*1/*4 and 2 CYP2B6*5/*5 subjects
Table 2.
Pharmacokinetic parameters for urine bupropion and hydroxybupropion in CYP2B6 genotypes
|
CYP2B6*1/*1 (n=21) |
CYP2B6*1/*6 (n=19) |
CYP2B6*6/*6 (n=17) |
All subjects (n=64) |
|
|---|---|---|---|---|
| Apparent renal or formation clearance (ml−1•kg−1•min−1) | ||||
| bupropion | 0.23 (0.14, 0.54) | 0.37 (0.14, 0.67) | 0.28 (0.07, 0.62) | 0.34 (0.12, 0.55) |
| R-bupropion | 0.27 (0.14, 0.60) | 0.41 (0.15, 0.85) | 0.36 (0.07, 0.72) | 0.36 (0.14, 0.70) |
| S-bupropion | 0.16 (0.12, 0.28) | 0.25 (0.10, 0.38) | 0.17 (0.07, 0.30) | 0.20 (0.10, 0.32) |
| hydroxybupropion | 1.18 (0.83, 1.93) | 1.29 (0.86, 1.92) | 0.70 (0.32, 1.39)* | 1.27 (0.75, 1.92) |
| R,R-hydroxybupropion | 1.31 (0.77, 2.17) | 1.36 (0.95, 1.95) | 0.67 (0.32, 1.54)* | 1.36 (0.71, 2.05) |
| S,S-hydroxybupropion | 1.15 (0.75, 1.56) | 1.02 (0.70, 1.57) | 0.64 (0.28, 1.12)* | 1.01 (0.67, 1.57) |
| 0–73 hr hydroxybupropion/bupropion molar ratio | ||||
| Hydroxybupropion / bupropion | 4.4 (2.3, 14.6) | 3.5 (1.9, 8.2) | 2.7 (1.9, 5.2) | 3.7 (2.2, 9.1) |
| R,R-hydroxybupropion / R-bupropion | 4.4 (2.1,14.6) | 3.3 (1.7, 8.4) | 2.7 (1.7, 5.2) | 3.5 (1.9, 8.8) |
| S,S-hydroxybupropion / S-bupropion | 5.4 (3.1, 14.1) | 4.3 (2.6, 8.4) | 3.8 (2.3, 6.3) | 5.0 (3.0, 10.8) |
Results are the median (25th, 75th quartile)
Significantly different from CYP2B6*1/*1 (p<0.05).
Hydroxybupropion plasma concentrations were highly stereoselective (Figure 1), as observed previously.7,8 The plasma R,R-hydroxybupropion/S,S-hydroxybupropion AUC0-∞ ratio overall was 24 (17,28). Significant genotypic differences in bupropion metabolism were observed. Bupropion hydroxylation, evaluated by the plasma hydroxybupropion/bupropion AUC ratio, for both enantiomers and the racemate, was significantly lower in CYP2B6*6 homozygotes compared with CYP2B6*1 homozygotes (Table 1, Figure S1). Conversely, bupropion hydroxylation was numerically greater in CYP2B6*4 carriers. For example, median hydroxybupropion/bupropion AUC ratios in CYP2B6*4/X and CYP2B6*1/*1 subjects were 53 (36, 71) and 21 (15, 25) for R,R-hydroxybupropion, 3.4 (2.7, 4.7) and 2.1 (1.6, 2.7) for S,S-hydroxybupropion, and 37 (24, 52) and 15 (10, 18) for total hydroxybupropion. The plasma R,R-hydroxybupropion/S,S-hydroxybupropion AUC0-∞ ratio was not different in the CYP2B6 genotype groups (25 (21, 34), 24 (21, 28) and 18 (16, 26) in CYP2B6*1/*1, CYP2B6*1*/6 and CYP2B6*6*/6 subjects, respectively).
Bupropion hydroxylation was also evaluated using urine excretion data. Overall urine recovery of R,R- and S,S-hydroxybupropion (% of dose) was 5.1 (3.5, 6.6) and 1.6 (1.2, 1.9), respectively. Hydroxybupropion formation clearance for both enantiomers and the racemate was significantly lower in CYP2B6*6/*6 than CYP2B6*1*/1 wild types (Table 2, Figure S2). Conversely, bupropion hydroxylation was numerically greater in CYP2B6*4 carriers. Formation clearances in CYP2B6*4/X and CYP2B6*1/*1 subjects, were 8.0 (4.5, 10.9) and 1.3 (0.8, 2.2) for R,R-hydroxybupropion, 4.4 (2.7, 7.0) and 1.2 (0.8, 1.6) for S,S-hydroxybupropion, and 6.9 (3.8, 9.9) and 1.18 (0.83, 1.93) for total hydroxybupropion, respectively. Bupropion hydroxylation, evaluated by either the plasma hydroxybupropion/bupropion AUC ratio or hydroxybupropion formation clearance, was not different from wild-type in CYP2B6*1/*6 heterozygotes. In contrast to hydroxybupropion formation clearances, the urine hydroxybupropion/bupropion molar ratio was not different in CYP2B6*6/*6 subjects.
Bupropion disposition and CYP2C19 genotypes
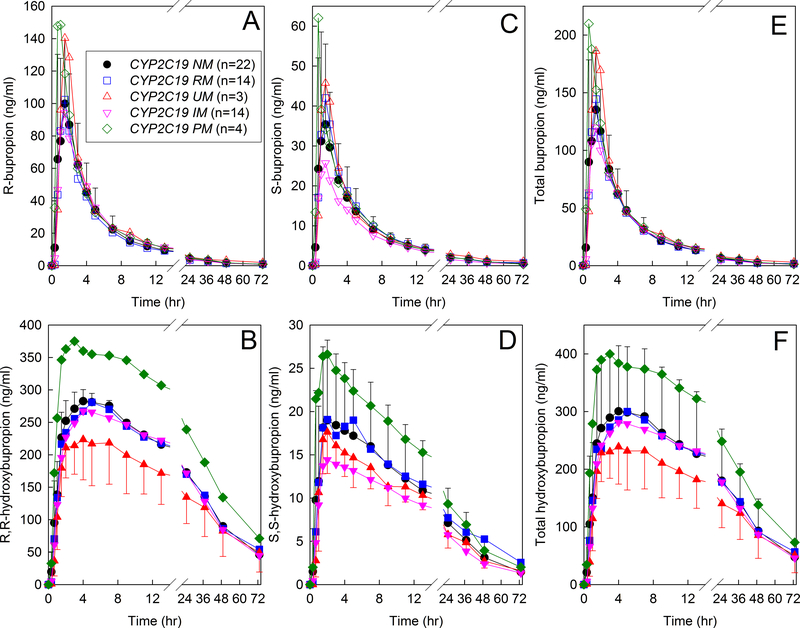
Plasma bupropion and hydroxybupropion enantiomer concentrations are shown for the CYP2C19 phenotypes (Figure 2). The distribution of CYP2C19 phenotypes was a convenience sample, because subjects had been recruited into CYP2B6 genotype cohorts. CYP2C19 genetic data were not available for all subjects. The final cohort was 22 normal, 14 intermediate, 4 poor, 14 rapid, and 3 ultrarapid metabolizers. Neither R-bupropion nor S-bupropion plasma Cmax, AUC0-∞, CL/F, nor elimination t1/2 were significantly different between the CYP2C19 phenotypes, specifically CYP2C19 normal metabolizers (CYP2C19*1/*1) and either intermediate (9 CYP2C19*1/*2, 1 CYP2C19*1/*3, 4 CYP2C19*2/*17) or rapid metabolizers (CYP2C19*1/*17) (Table 3). Results are shown only for the three major phenotype groups for clarity, but were not different in the poor or ultrarapid metabolizers. There were also no significant differences between normal metabolizers and other phenotypes in R,R- or S,S-hydroxybupropion plasma Cmax, elimination t1/2, AUC, hydroxybupropion/bupropion AUC ratios, or urine R,R- or S,S-hydroxybupropion formation clearances (not shown). Results were not different if grouped by allele. Specifically, the AUC0-∞ for CYP2C9*1 (n=22), CYP2C9*2 (n=17), and CYP2C9*17 (n=17) carriers was 620 ± 205, 715 ± 217, and 664 ± 217 hr•ng•ml−1 (P=0.11) for R-bupropion, 275 ± 123, 273 ± 68 and 286 ± 118 hr•ng•ml−1 (P=0.86) for S-bupropion, cand 887 ± 291, 986 ± 244, and 952 ± 32 hr•ng•ml−1 (P=0.33) for racemic bupropion.
Figure 2.
Influence of CYP2C19 genotype on bupropion disposition. Subjects received 150 mg oral immediate release bupropion. Shown are plasma concentrations of (A) R-bupropion, (B) R,R-hydroxybupropion, (C) S-bupropion, (D) S,S-hydroxybupropion, (E) RS-bupropion, (F) total hydroxybupropion. Each data point is the mean ± SD. Some SD values are omitted for clarity. Subjects whose results are shown in Figure 1 were also genotyped for the CYP2C19*2, *3, and *17 alleles, and were grouped into normal, intermediate, poor, rapid, and ultrarapid metabolizer phenotypes. CYP2C19 genetic data were not available for all subjects.
Table 3.
Pharmacokinetic parameters for plasma bupropion and hydroxybupropion in CYP2C19 phenotypes
|
CYP2C19 NM (n=22) |
CYP2C19 IM (n=14) |
CYP2C19 RM (n=14) |
All subjects (n=57) |
|
|---|---|---|---|---|
| Bupropion | ||||
| Cmax (ng/ml) | 134 (108, 187) | 146 (120, 193) | 160 (111, 202) | 148 (114, 193) |
| AUC0-∞ (hr•ng•ml−1) | 887 ± 291 | 964 ± 255 | 912 ± 316 | 943 ± 282 |
| CL/F (ml−1•kg−1•min−1) | 38 (30, 51) | 38 (29, 44) | 37 (34, 45) | 37 (30, 45) |
| Elimination t1/2 (h) | 23 (20, 27) | 21 (18, 25) | 23 (20, 27) | 23 (20,27) |
| R-bupropion | ||||
| Cmax (ng/ml) | 99 (80, 136) | 108 (91, 164) | 127 (84, 148) | 108 (85, 146) |
| AUC0-∞ (ng •h •ml−1) | 620 ± 205 | 716 ± 231 | 629 ± 204 | 668 ± 213 |
| CL/F (ml−1•kg−1•min−1) | 27 (23, 34) | 27 (19, 32) | 28 (23, 34) | 27 (22, 32) |
| Elimination t1/2 (h) | 21 (18, 25) | 21 (17, 26) | 22 (17, 25) | 22 (18, 25) |
| S-bupropion | ||||
| Cmax (ng/ml) | 36 (16, 55) | 28 (20, 50) | 49 (24, 60) | 36 (25, 53) |
| AUC0-∞ (hr•ng•ml−1) | 275 ± 126 | 249 ± 53 | 280 ± 125 | 277 ± 104 |
| CL/F (ml−1•kg−1•min−1) | 69 (44, 90) | 71 (62, 87) | 56 (51, 85) | 65 (49, 86) |
| Elimination t1/2 (h) | 26 (24, 27) | 25 (13, 26) | 25 (19, 27) | 26 (24, 27) |
| Hydroxybupropion | ||||
| Cmax (ng/ml) | 295 (230, 378) | 325 (259, 402) | 238 (184, 352) | 293 (233, 381) |
| Elimination t1/2 (hr) | 23 (18, 32) | 26 (22, 34) | 23 (21, 27) | 24 (20, 30) |
| AUC0–72 hydroxybupropion/bupropion (hr•ng•ml−1) | 11.6 (8.6, 16.7) | 12.9 (10.2, 15.2) | 9.2 (6.9, 14.9) | 11.7 (8.5, 14.8) |
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 12.9 (9.7, 20.0) | 14.2 (11.9, 17.0) | 9.8 (7.1, 16.4) | 13.6 (9.1, 17.9) |
| R,R-hydroxybupropion | ||||
| Cmax (ng/ml) | 278 (219, 348) | 300 (245, 382) | 225 (174, 332) | 277 (220, 363) |
| Elimination t1/2 (hr) | 23 (18, 33) | 27 (22, 35) | 23 (20, 27) | 24 (21, 30) |
| AUC0–72 hydroxybupropion/bupropion (hr•ng•ml−1) | 15.7 (12.8, 23.0) | 16.8 (13.2, 20.7) | 12.6 (9.2, 19.8) | 15.7 (11.5, 20.3) |
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 17.0 (13.2, 27.7) | 18.5 (15.9, 24.0) | 14.0 (9.5, 22.1) | 17.1 (12.8, 24.2) |
| S,S-hydroxybupropion | ||||
| Cmax (ng/ml) | 19 (15, 23) | 21 (15, 24) | 17 (10, 23) | 19 (14, 24) |
| Elimination t1/2 (h) | 21 ± 8 | 24 ± 7 | 23 ± 8 | 23 ± 8 |
| AUC0–72 hydroxybupropion/bupropion (hr•ng•ml−1) | 2.0 (1.5, 2.5) | 1.9 (1.6, 2.3) | 1.5 (1.0, 2.1) | 1.9 (1.5, 2.3) |
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 2.1 (1.5, 2.7) | 2.0 (1.8, 2.3) | 1.5 (0.9, 2.6) | 1.9 (1.5, 2.3) |
Cmax, maximum concentration; Tmax, time to maximum concentration; AUC0–72 area under concentration-time curve from 0 to 72 hr; AUC0--∞, area under concentration-time curve extrapolated to infinity; CL/F, apparent oral clearance; t½, elimination half-life. Results are the mean ± SD or median (25th, 75th quartile).
Results are not shown for 3 UM and 4 PM
Bupropion disposition and POR genotypes
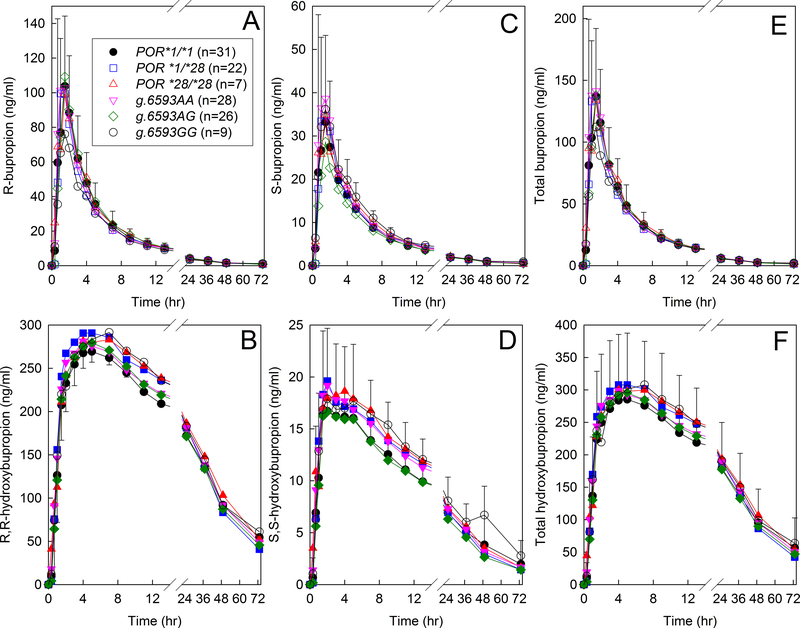
Plasma bupropion and hydroxybupropion enantiomer concentrations are shown in Figure 3 for the POR genotypes. The distribution of POR genotypes was a convenience sample, because subjects had been recruited into CYP2B6 genotype cohorts. POR genetic data were not available for all subjects. Cohorts included 31 POR*1/*1, 22 POR*1/*28, and 7 POR*28/*28 subjects. No POR*5 or POR*8 subjects were identified. Compared with wild-type POR*1/*1, neither the POR*28 heterozygotes or homozygotes had any difference in R- or S-bupropion plasma Cmax, AUC0-∞, CL/F, elimination t1/2, renal clearance, or R,R- or S,S-hydroxybupropion plasma Cmax, elimination t1/2, or AUC (Table 4). There were also no differences in plasma hydroxybupropion/bupropion AUC ratios, or urine R,R- or S,S-hydroxybupropion formation clearances. Subjects were also genotyped for the intronic variant g.6593 A>G, which does not have an assigned allele. Genotype groups were 28 g.6593AA, 26 g.6593AG, and 9 g.6593GG subjects. There were no significant differences in R- or S-bupropion plasma Cmax, AUC0-∞, CL/F, elimination t1/2, renal clearance, or R,R- or S,S-hydroxybupropion plasma Cmax, elimination t1/2, or AUC. There were also no differences in plasma hydroxybupropion/bupropion AUC ratios or urine R,R- or S,S-hydroxybupropion formation clearances between g.6593AA, g.6593AG, or g.6593GG genotypes (Table 4).
Figure 3.
Influence of P450 oxidoreductase (POR) variants on. bupropion disposition. Subjects received 150 mg oral immediate release bupropion. Shown are plasma concentrations of (A) R-bupropion, (B) R,R-hydroxybupropion, (C) S-bupropion, (D) S,S-hydroxybupropion, (E) RS-bupropion, (F) total hydroxybupropion. Each data point is the mean ± SD. Some SD values are omitted for clarity. Subjects whose results are shown in Figure 1 were also genotyped for the POR*5, *8, and *28 alleles, as well as the g.6593 A>G polymorphism. POR genetic data were not available for all subjects.
Table 4.
Pharmacokinetic parameters for hydroxybupropion in POR genotypes
|
POR*1/*1 (n=31) |
POR*1/*28 (n=22) |
POR*28/*28 (n=7) |
|
|---|---|---|---|
| R,R-hydroxybupropion | |||
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 17.3 (11.7, 28.5) | 17.6 (11.9, 23.0) | 16.6 (12.6, 25.8) |
| formation clearance (ml−1•kg−1•min−1) | 1.2 (0.7, 1.9) | 1.7 (1.0, 2.2) | 0.8 (0.7, 2.9) |
| S,S-hydroxybupropion | |||
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 1.9 (1.7, 2.1) | 2.0 (1.5, 2.6) | 1.9 (1.4, 2.5) |
| formation clearance (ml−1•kg−1•min−1) | 1.1 (0.6, 1.6) | 1.0 (0.8, 1.7) | 0.7 (0.7, 1.6) |
| g.6593AA (n=28) | g.6593AG (n=27) | g.6593GG (n=9) | |
| R,R-hydroxybupropion | |||
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 16.9 (12.1, 25.1) | 16.5 (11.6, 24.8) | 20.8 (15.1, 35.9) |
| formation clearance (ml−1•kg−1•min−1) | 1.1 (0.7, 1.9) | 1.4 (0.9, 2.3) | 1.5 (0.9, 2.4) |
| S,S-hydroxybupropion | |||
| AUC0−∞ hydroxybupropion/bupropion (hr•ng•ml−1) | 1.9 (1.4, 2.3) | 2.0 (1.5, 2.6) | 2.1 (1.4, 2.7) |
| formation clearance (ml−1•kg−1•min−1) | 0.8 (0.7, 1.4) | 1.2 (0.7, 1.8) | 0.9 (0.4, 1.4) |
Results are the mean ± SD or median (25th, 75th quartile)
Discussion
The major finding of this investigation is that clinical t-butyl hydroxylation of both bupropion enantiomers was equivalently influenced by CYP2B6 polymorphisms. Specifically, both the plasma R,R-hydroxybupropion/R-bupropion and S,S-hydroxybupropion/S-bupropion AUC ratios, and the R,R- and S,S-hydroxybupropion formation clearances, were significantly lower than wild-type controls in CYP2B6*6/*6, but not CYP2B6*1/*6 genotypes. Racemic hydroxybupropion/bupropion plasma AUC ratios and urine hydroxybupropion formation clearances were also significantly lower than wild-type controls in CYP2B6*6/*6, but not CYP2B6*1/*6 genotypes. Although not statistically analyzed due to the small sample size, plasma bupropion concentrations were lower, hydroxybupropion concentrations were higher, hydroxybupropion/bupropion AUC ratios and urine hydroxybupropion formation clearances were numerically greater in CYP2B6*4 carriers vs CYP2B6*1 controls, for both enantiomers. In contrast to bupropion hydroxylation, R- or S-bupropion disposition was not influenced by CYP2B6 genotype.
CYP2B6 genetic effects on in vivo hydroxylation of both R- and S-bupropion are consistent with prior observation that cDNA-expressed and human liver microsomal CYP2B6.1 protein metabolized both enantiomers, and stereoselectively.5 The in vitro hydroxylation of bupropion enantiomers by expressed CYP2B6 allelic variants has not been reported, however, thus the present in vivo findings cannot be compared with in vitro data. The equivalent influence of CYP2B6 polymorphisms on clinical R- vs S-bupropion hydroxylation is different than the greater influence of CYP2B6 induction on R- vs S-bupropion hydroxylation, based on plasma metabolite/parent ratios.7,8 For example, rifampin induction increased the plasma S,S-hydroxybupropion/S-bupropion AUC ratio 2.3-fold, and the R,R-hydroxybupropion/R-bupropion AUC ratio 1.2-fold, while effects on the formation clearances of R,R- vs S,S-hydroxybupropion were similar. Ritonavir induction increased the plasma S,S-hydroxybupropion/S-bupropion AUC ratio 1.2-fold while the R,R-hydroxybupropion/R-bupropion AUC ratio was unchanged, and R,R- and S,S-hydroxybupropion formation clearances were equally increased. As discussed, below, it is possible that these apparent differences in plasma metabolite/parent AUC ratios may be related more to R,R- and S,S-hydroxybupropion disposition, than to true stereoselective differences in effects of altered CYP2B6 activity, whether due to CYP2B6 polymorphisms or drug interactions.
Enantioselective effects of CYP2B6 polymorphisms can be compared with previous studies using achiral analysis, and which mostly evaluated CYP2B6*6. The present investigation found a gene-dose effect of the *6 allele to reduce hydroxylation of both bupropion enantiomers, based on both plasma (hydroxybupropion/bupropion AUC ratio) and urine (hydroxybupropion formation clearance) data (Tables 1 and 2). Previous plasma racemic hydroxybupropion/bupropion AUC ratios in CYP2B6*1/*1, *1/*6, and *6/6 genotypes were (mean) 8, 6 and 5,21 17, 10 and 9,24 and 16, 9 and 8,25 after single-dose immediate release bupropion. At steady state, ratios were 41, 22, and 16 in the three genotype groups,16 and were less in *6 carriers than wild types (13 vs 17).23 The present investigation also found increased bupropion hydroxylation in CYP2B6*4 carriers, with median hydroxybupropion/bupropion ratios in CYP2B6*1/*1 and CYP2B6*4/X subjects of 15 and 37 for total hydroxybupropion, 21 and 53 for R,R-hydroxybupropion, and 2.1 and 3.4 for S,S-hydroxybupropion. An influence of CYP2B6*4 was also previously observed, with total hydroxybupropion/bupropion ratios of 8 and 14 in CYP2B6*1/*1 and CYP2B6*1/*4 genotypes, respectively.21 Although it has been speculated that CYP2B6 genetic variability may contribute to differences in the R/S-bupropion plasma AUC ratio,29 this did not differ significantly between CYP2B6*1/*1, CYP2B6*1/*6, and CYP2B6*6/*6 subjects.
A secondary finding was that CYP2C19 polymorphisms did not influence bupropion enantiomers disposition. Current understanding of CYP2C19 relevance in bupropion metabolism and elimination is evolving, and less characterized than CYP2B6. The fraction of bupropion metabolized clinically by CYP2C19 was predicted to be only one-third that by CYP2B6.29 CYP2C19 does not catalyze t-butyl hydroxylation to hydroxybupropion in vitro,18 and CYP2C19 genotype did not affect plasma hydroxybupropion/bupropion AUC ratios in vivo.31 CYP2C19 does however catalyze bupropion aromatic 4’-hydroxylation,27,28,30 and secondary 4’-hydroxylation of the primary metabolites erythrohydrobupropion and threohydrobupropion.28,29 Little is known about the influence of CYP2C19 variants on bupropion disposition. Oral bupropion (racemate) systemic clearance was unaffected by CYP2C19 genotype.21 In contrast, a small (1.15-fold) but statistically significantly greater racemic bupropion plasma AUC was reported in CYP2C19*2 loss of function allele carriers, but there was no difference in CYP2C19*17 gain of function carriers.31 The present investigation found no difference in bupropion enantiomers AUC between CYP2C19 normal and either intermediate or rapid metabolizers, or between allele types. These results, albeit with small subject numbers, do not support the conclusion that CYP2C19 polymorphisms influence bupropion plasma concentrations.
Another secondary finding was that POR polymorphisms did not influence bupropion enantiomers disposition. Neither POR*28 homozygotes, nor g.6593GG genotypes, had altered hydroxylation of either bupropion enantiomer, assessed by both plasma hydroxybupropion/bupropion AUC ratios and urine hydroxybupropion formation clearances. Results for POR*28 are congruent with previous observations.24,25 Results for the g.6593A>G polymorphism contrast with previous studies, which found greater bupropion hydroxylation in g.6593GG genotypes.24,25 Reasons for the differences are not apparent, but could be related to ethnic differences in the study populations, as well as small sample sizes.
The intersection of stereochemistry and pharmacogenetics for CYP2B6 substrates is relatively unexplored. Common CYP2B6 substrates, which are chiral and used clinically as racemates include bupropion, methadone, ketamine, cyclophosphamide and ifosfamide, while nevirapine is achiral and efavirenz and artemether are single enantiomers. Ketamine N-demethylation by CYP2B6 is stereoselective, and both enantiomers are metabolized in vitro less by cDNA-expressed CYP2B6.6 than CYP2B6.1 and in liver microsomes from humans carrying the CYP2B6*6 allele.37 Clinical ketamine enantiomers metabolism was found not to differ in CYP2B6*6 carriers compared with CYP2B6*1/*1 genotypes,38 while another study found diminished (racemate) metabolism, although genetic effects could not be distinguished from age effects.39 Methadone in vitro is N-demethylated stereoselectively by CYP2B6, and both enantiomers are metabolized less and more efficiently, respectively, by expressed CYP2B6.6 and CYP2B6.4 compared with CYP2B6.1.40,41 Clinical methadone metabolism and clearance are decreased and increased respectively, in CYP2B6*6/*6 genotypes and CYP2B6*4 carriers.42 In contrast, metabolism of racemic bupropion is less by both CYP2B6.4 and CYP2B6.6,35,43 as is racemic ifosfamide, which is metabolized more efficiently by CYP2B6.7 and CYP2B6.9.44 Ifosfamide metabolism by CYP2B6.1 is enantioselective,45 but stereoselectivity with CYP2B6 variants is unknown. Cyclophosphamide hydroxylation by CYP2B6.6 was greater than CYP2B6.1 in vitro, but this was not apparent clinically.46–48 The influence of CYP2B6 polymorphisms and stereochemistry on human ifosfamide and cyclosphosphamide metabolism is unknown. Available data show that stereoselectivity and genetic influences on disposition of bupropion and methadone are similar, with CYP2B6*6 and CYP2B6*4 associated with decreased and increased metabolism, respectively, and both enantiomers are affected, in vitro and in vivo.
One clinical implication of this investigation attends to bupropion pharmacotherapy. Bupropion hydroxylation is a bioactivation pathway, and hydroxybupropion, specifically S,S-hydroxybupropion, is considered to contribute to bupropion l antidepressant and anti-smoking effects.12,13,15–17 Higher plasma racemic hydroxybupropion concentrations, related to CYP2B6 genetic variation, were associated with better smoking-cessation outcomes,16 and better improvement in depression.17 This investigation shows that genetic differences in CYP2B6 influence S-bupropion metabolism and S,S-hydroxybupropion formation and thus could affect clinical outcomes.15,16
Another clinical implication attends more broadly to CYP2B6 pharmacogenetics. Questions continue regarding the best in vivo CYP2B6 probe. Racemic bupropion, and more specifically racemic bupropion hydroxylation, is the standard in vivo probe for CYP2B6 activity, polymorphisms and drug interactions, and is recommended by regulatory agencies.19,20 Bupropion clearance and plasma AUC do not reflect CYP2B6 activity because hydroxylation is a minor route of elimination. More commonly used is the plasma hydroxybupropion/bupropion AUC ratio, but this is confounded because racemic hydroxybupropion is elimination rather than formation-rate limited. Urine metrics, such as hydroxybupropion formation clearance and the hydroxybupropion/bupropion molar ratio, do reflect CYP2B6 induction,7,8 but there are few other evaluations of these metrics, such as CYP2B6 genetic influences. Urine racemic hydroxybupropion formation clearances differed more than plasma hydroxybupropion/bupropion AUC ratios in response to CYP2B6 induction,7,8 and CYP2B6 genotypes (Tables 1 and 2). Recognition that bupropion hydroxylation is stereoselective,5 led to the clinical evaluation of bupropion enantiomers hydroxylation to probe CYP2B6.7,8 Because S,S-hydroxybupropion was formation rate-limited while R,R-hydroxybupropion and the racemate were elimination-rate limited, S,S-hydroxybupropion formation clearance appeared to be an improved measure of bupropion hydroxylation and CYP2B6 activity. After CYP2B6 induction by rifampin and ritonavir, plasma hydroxybupropion/bupropion AUC ratios were increased more for S,S- than R,R-hydroxybupropion, although the formation clearances of both isomers equivalently reflected CYP2B6 induction.7,8 In the present investigation the hydroxybupropion/bupropion plasma AUC ratios of the isomers differed comparably in CYP2B6*6 homozygotes, as did the S,S- and R,R-hydroxybupropion formation clearances. Based on in vitro data predicting that the fraction of bupropion metabolized to hydroxybupropion was 3-fold greater for R- than S-bupropion, despite more efficient hydroxylation of S- than R-bupropion, it was suggested that R,R-hydroxybupropion formation clearance or R,R-hydroxybupropion/R-bupropion metabolic ratios may be a more sensitive and better probe of CYP2B6 activity than S,S-hydroxybupropion/S-bupropion formation clearance or metabolic ratios.29 This is not consistent with the present and previous data.7,8 It is also unclear whether R,R-hydroxybupropion/R-bupropion formation clearance or metabolic ratios would improve on those of the racemate, since the plasma AUC for R,R-hydroxybupropion constitutes 95% of the total hydroxybupropion AUC. One of the greater unknowns, is the reason why plasma R,R- and S,S-hydroxybupropion concentrations are so different.7,8,29 Alternative CYP2B6 probes have also been suggested, such as 8-hydroxylation of efavirenz,49 whose metabolism in vitro was considered to be a better predictor of clinical drug interactions than bupropion.20
In summary, this investigation shows that CYP2B6 genotype influenced the hydroxylation of both R- and S-bupropion without affecting bupropion plasma concentrations or clearance. CYP2B6*6 homozygotes had decreased hydroxylation, and CYP2B6*4 carriers had increased hydroxylation. CYP2B6 genetic polymorphism had equivalent effects on R- and S-bupropion hydroxylation. Neither the CYP2C19 nor POR genetic variants studied affected bupropion hydroxylation or clearance.
Methods
Study population and protocol
The investigation was registered () and was approved by the Washington University in St. Louis Institutional Review Board, and all subjects provided written informed consent. Subjects were normal healthy volunteers (smokers or nonsmokers) age 18–50 yr in good general health within 30% of ideal body weight (body mass index <33), had no history of hepatic or renal disease, use of prescription or non-prescription medications, herbals or foods known to be metabolized by or affect CYP2B6 activity. Pregnant or nursing females were not eligible. Potential subjects were genotyped for single nucleotide polymorphisms (SNPs) of CYP2B6 516G>T (rs3745274), 785A>G (rs2279343), 983T>C (rs28399499), and 1459C>T (rs3211371) as described previously.42 SNP analysis permitted identification of CYP2B6 *1, *4 (785A>G), *5 (1459C>T), *6 (516G>T, 785A>G), *7 (516G>T, 785A>G, 1459C>T), *9 (516G>T), *16 (785A>G, 983T>C), and *18 (983T>C) alleles. Genotyping results then were used to invite subjects to create target cohorts of twenty subjects each with CYP2B6*1/*1, CYP2B6*1/*6 and CYP2B6*6/*6 genotypes. A 30% difference between groups in hydroxybupropion/bupropion AUC ratio was considered clinically significant. To detect a 30% difference between CYP2B6 genotypes, with 30% variability, ß=0.8, and α=0.05, would require 17 subjects per group. The target was 20 per group. In addition, subjects of other rare CYP2B6 genotypes coincidentally identified were also studied.
Enrolled subjects were also genotyped for CYP2C19 681G>A (rs4244285), 636G>A (rs4986893) and −806C>T (rs12248560); and also for POR 859G>C (rs121912974), Y181D 541T>G (rs72552771), A503V 1508C>T (rs1057868), and intron g.6593 A>G (rs2868177). SNP analysis permitted identification of CYP2C19 *2 (681G>A), *3 (636G>A), and *17 (−806 C>T) alleles. CYP2C19 variants were grouped into normal, intermediate, poor, rapid, and ultrarapid metabolizer phenotypes. SNPs also identified POR *5 (859G>C), *8 (541T>G), and *28 (A503V) alleles, as well as the g.6593 A>G polymorphism.
A total of 63 subjects (33 male, 30 female; 42 Caucasians, 9 African-Americans, 10 Asians, 2 other/unknown), 29 ±8 yr, 74 ± 13 kg, were studied. Detailed demographic data are provided in Table S1.
Subjects were instructed to refrain from: 1) oranges, grapefruit or apples or their juices for 5d before and throughout the study period, 2) non-study medications (including over the counter and/or herbal), for 3d prior to the study day without prior approval, 3) alcohol for 48 hr prior to and during the study day, 4) caffeine-containing beverages on the study day, 5) food/liquids after midnight the day prior to bupropion administration.
The study design was a single-center, open-label, single-session protocol. A peripheral IV catheter was inserted in an arm for blood sampling. Subjects received 150 mg oral racemic bupropion with 200cc water, then a standard breakfast and lunch 3 and 5 hr after bupropion administration, respectively, and free access to food and water thereafter. Venous blood was sampled for 73 hr after bupropion, centrifuged, and plasma stored at −80°C. Continuous 24hr urine samples were collected for 3d and stored at −80°C. Plasma and urine R- and S-bupropion and R,R- and S,S-hydroxybupropion concentrations were measured by HPLC-tandem mass spectrometry as described previously.50 Interday coefficients of variation were 7, 6, and 5% and 7, 5, and 5% for 1, 10 and 100 ng/ml R- and S- bupropion, respectively, and 9, 5, and 6% and 7, 5, and 6% for 5, 50, and 500 ng/ml R,R- and S,S-hydroxybupropion in plasma, and 9, 7, and 6% and 6, 6, and 7% for 10, 100 and 1000 ng/ml R- and S- bupropion, respectively, and 6, 7, and 8% and 7, 7, and 5% for 5, 50, and 500 ng/ml R,R- and S,S-hydroxybupropion in urine.
Data and statistical analysis
Pharmacokinetic data were analyzed using noncompartmental methods (Phoenix, Pharsight Corp, Mountain View, CA), assuming complete absorption, as described previously.7,8 Differences between CYP2B6 genotypes for pharmacokinetic parameters, and major CYP2C19 phenotypes and POR variants were analyzed using one-way analysis of variance followed by the Student-Newman-Keuls test for multiple comparisons (Sigmaplot 12.5, Systat Software, Inc, San Jose, CA). Non-normal data were log transformed for analysis, but reported as the non-transformed results. Statistical significance was assigned at P<0.05. Formal comparison of other CYP2B6 allelic variants to CYP2B6*1/*1 subjects, and comparison of CYP2C19 poor and ultrarapid metabolizer phenotypes to regular metabolizers was not performed due to the small subject numbers studied. Normally distributed data are reported as the mean ± SD, others are reported as the median and interquartile range.
Supplementary Material
Table S1. Subject demographics
Figure S1 Influence of CYP2B6*6 genotype on buprenorphine metabolism
Figure S2 Influence of CYP2B6*6 genotype on buprenorphine metabolism.
STUDY HIGHLIGHTS.
What is the current knowledge on the topic?
Bupropion hydroxylation is a bioactivation pathway, CYP2B6 probe, and is stereoselective. The CYP2B6 gene is highly polymorphic, with some variant allele products catalyzing altered racemic bupropion hydroxylation activity.
What question did this study address?
The influence of CYP2B6 allelic variants, as well as CYP2C19 and P450 oxidoreductase variants, on the clinical plasma concentrations of bupropion enantiomers and bupropion hydroxylation.
What does this study add to our knowledge?
CYP2B6*6/*6 genotypes and CYP2B6*6/*4 carriers had decreased and increased hydroxylation, respectively, of both bupropion enantiomers, based on plasma hydroxybupropion/bupropion AUC ratios and urine hydroxybupropion formation clearance. CYP2C19 and P450 oxidoreductase variants had no effect on plasma concentrations or hydroxylation.
How might this change clinical pharmacology or translational science?
CYP2B6 variants may affect bupropion bioactivation to the active metabolite S,S-hydroxybupropion. Results may influence use of bupropion hydroxylation as a CYP2B6 probe.
Acknowledgements
The authors thank Jennifer Parchomski, RN, and Jane Blood, RN, Department of Anesthesiology, Washington University in St. Louis (St. Louis, MO) for their excellent clinical research assistance, and Chris Sawyer and Richard Head, PhD, of the Genome Technology Access Center and Department of Genetics at Washington University in St. Louis School of Medicine (St. Louis, MO) for genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant P30CA91842 to the Siteman Cancer Center (St. Louis, MO) and by ICTS/CTSA Grant# UL1TR000448 to Washington University in St. Louis (St. Louis, MO) from the National Center for Research Resources (NCRR), a component of the National Institutes of Health.
Funding: Supported by National Institutes of Health grant R01-DA14211 and by FDA grant U01-FD004899 (to Dr. Kharasch), and by National Institutes of Health grant UL1TR000448 to the Washington University in St. Louis Institute of Clinical and Translational Sciences.
Footnotes
Disclosures: The authors declared no competing interests for this work.
References
- 1.Dhillon S, Yang LP, & Curran MP Bupropion: a review of its use in the management of major depressive disorder. Drugs 68,653–89 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Reid RD, Pritchard G, Walker K, Aitken D, Mullen KA, & Pipe AL Managing smoking cessation. CMAJ 188,E484–E92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilding JP Combination therapy for obesity. J Psychopharmacol 31,1503–8 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Verbeeck W, Bekkering GE, Van den Noortgate W, & Kramers C Bupropion for attention deficit hyperactivity disorder (ADHD) in adults. Cochrane Database Syst Rev 10,CD009504 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coles R, & Kharasch ED Stereoselective metabolism of bupropion by CYP2B6 and human liver microsomes. Pharm Res 25,1405–11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H, Loboz KK, Gross AS, & McLachlan AJ Stereoselective analysis of hydroxybupropion and application to drug interaction studies. Chirality 19,163–70 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Kharasch ED, Mitchell D, Coles R, & Blanco R Rapid clinical induction of hepatic cytochrome P4502B6 activity by ritonavir. Antimicrob Agents Chemother 52,1663–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kharasch ED, Mitchell D, & Coles R Stereoselective bupropion hydroxylation as an in vivo phenotypic probe for cytochrome P4502B6 (CYP2B6) activity. J Clin Pharmacol 48,464–74 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Masters AR, Gufford BT, Lu JB, Metzger IF, Jones DR, & Desta Z Chiral plasma pharmacokinetics and urinary excretion of bupropion and metabolites in healthy volunteers. J Pharmacol Exp Ther 358,230–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teitelbaum AM, Flaker AM, & Kharasch ED Development and validation of a high-throughput stereoselective LC-MS/MS assay for bupropion, hydroxybupropion, erythrohydrobupropion, and threohydrobupropion in human plasma. J Chromatogr B 1017–1018,101–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teitelbaum AM, Flaker AM, & Kharasch ED Development, validation and application of a comprehensive stereoselective LC/MS-MS assay for bupropion and oxidative, reductive, and glucuronide metabolites in human urine. J Chromatogr B 1027,239–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bondarev ML, Bondareva TS, Young R, & Glennon RA Behavioral and biochemical investigations of bupropion metabolites. Eur J Pharmacol 474,85–93 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Damaj MI, et al. Enantioselective effects of hydroxy metabolites of bupropion on behavior and on function of monoamine transporters and nicotinic receptors. Mol. Pharmacol. 66,675–82 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Malcolm E, Carroll FI, Blough B, Damaj MI, & Shoaib M Examination of the metabolite hydroxybupropion in the reinforcing and aversive stimulus effects of nicotine in rats. Psychopharmacology (Berl) 232,2763–71 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Lee AM, et al. CYP2B6 genotype alters abstinence rates in a bupropion smoking cessation trial. Biol Psychiatry 62,635–41 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Zhu AZ, et al. CYP2B6 and bupropion’s smoking-cessation pharmacology: the role of hydroxybupropion. Clin Pharmacol Ther 92,771–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laib AK, Brunen S, Pfeifer P, Vincent P, & Hiemke C Serum concentrations of hydroxybupropion for dose optimization of depressed patients treated with bupropion. Ther Drug Monit 36,473–9 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Faucette SR, et al. Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. Drug Metab Dispos 28,1222–30 (2000). [PubMed] [Google Scholar]
- 19.Ilic K, et al. The influence of sex, ethnicity, and CYP2B6 genotype on bupropion metabolism as an index of hepatic CYP2B6 activity in humans. Drug Metab Dispos 41,575–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fahmi OA, et al. Evaluation of CYP2B6 induction and prediction of clinical drug-drug interactions: Considerations from the IQ consortium induction working group-An industry perspective. Drug Metab Dispos 44,1720–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchheiner J, et al. Bupropion and 4-OH-bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6. Pharmacogenetics 13,619–26 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Chung JY, et al. Effects of pregnane X receptor (NR1I2) and CYP2B6 genetic polymorphisms on the induction of bupropion hydroxylation by rifampin. Drug Metab Dispos 39,92–7 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Benowitz NL, Zhu AZ, Tyndale RF, Dempsey D, & Jacob P 3rd. Influence of CYP2B6 genetic variants on plasma and urine concentrations of bupropion and metabolites at steady state. Pharmacogenet Genomics 23,135–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv J, Hu L, Zhuo W, Zhang C, Zhou H, & Fan L Effects of the selected cytochrome P450 oxidoreductase genetic polymorphisms on cytochrome P450 2B6 activity as measured by bupropion hydroxylation. Pharmacogenet Genomics 26,80–7 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Gao LC, et al. The P450 oxidoreductase (POR) rs2868177 and cytochrome P450 (CYP) 2B6*6 polymorphisms contribute to the interindividual variability in human CYP2B6 activity. Eur J Clin Pharmacol 72,1205–13 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Eng H, & Obach RS Use of human plasma samples to identify circulating drug metabolites that inhibit cytochrome P450 enzymes. Drug Metab Dispos 44,1217–28 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Liu HF, Liu L, Nguyen K, Jones EB, & Fretland AJ The in vitro metabolism of bupropion revisited: concentration dependent involvement of cytochrome P450 2C19. Xenobiotica 40,536–46 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Sager JE, Choiniere JR, Chang J, Stephenson-Famy A, Nelson WL, & Isoherranen N Identification and structural characterization of three new metabolites of bupropion in humans. ACS Med Chem Lett 7,791–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sager JE, Price LS, & Isoherranen N Stereoselective metabolism of bupropion to OH-bupropion, threohydrobupropion, erythrohydrobupropion, and 4’-OH-bupropion in vitro. Drug Metab Dispos 44,1709–19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palacharla RC, Nirogi R, Uthukam V, Manoharan A, Ponnamaneni RK, & Kalaikadhiban I Quantitative in vitro phenotyping and prediction of drug interaction potential of CYP2B6 substrates as victims. Xenobiotica,in press (2018). [DOI] [PubMed] [Google Scholar]
- 31.Zhu AZ, Zhou Q, Cox LS, Ahluwalia JS, Benowitz NL, & Tyndale RF Gene variants in CYP2C19 are associated with altered in vivo bupropion pharmacokinetics but not bupropion-assisted smoking cessation outcomes. Drug Metab Dispos 42,1971–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burkhard FZ, Parween S, Udhane SS, Fluck CE, & Pandey AV P450 oxidoreductase deficiency: Analysis of mutations and polymorphisms. J Steroid Biochem Mol Biol 165,38–50 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Pan LQ, Naranmandura H, Zeng S, & Chen SQ Influence of various polymorphic variants of cytochrome P450 oxidoreductase (POR) on drug metabolic activity of CYP3A4 and CYP2B6. PLoS One 7,e38495 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bumpus NN, Sridar C, Kent UM, & Hollenberg PF The naturally occurring cytochrome P450 (P450) 2B6 K262R mutant of P450 2B6 exhibits alterations in substrate metabolism and inactivation. Drug Metab Dispos 33,795–802 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Sridar C, Kenaan C, Amunugama H, Ballou DP, & Hollenberg PF Polymorphic variants of cytochrome P450 2B6 (CYP2B6.4-CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: A charge-reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional cytochrome P450-reductase complex. J Pharmacol Exp Ther 338,803–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radloff R, et al. Novel CYP2B6 enzyme variants in a Rwandese population: Functional characterization and assessment of in silico prediction tools. Hum Mutat 34,725–34 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Li Y, et al. The CYP2B6*6 allele significantly alters the N-demethylation of ketamine enantiomers in vitro. Drug Metab Dispos 41,1264–72 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Rao LK, Flaker AM, Friedel CC, & Kharasch ED Role of cytochrome P4502B6 polymorphisms in ketamine metabolism and clearance. Anesthesiology 125,1103–12 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Li Y, et al. CYP2B6*6 allele and age substantially reduce steady-state ketamine clearance in chronic pain patients: impact on adverse effects. Br J Clin Pharmacol 80,276–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gadel S, Crafford A, Regina K, & Kharasch ED Methadone N-demethylation by the common CYP2B6 allelic variant CYP2B6.6. Drug Metab Dispos 41,709–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gadel S, Friedel C, & Kharasch ED Differences in methadone metabolism by CYP2B6 variants. Drug Metab Dispos 43,994–1001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kharasch ED, Regina KJ, Blood J, & Friedel C Methadone pharmacogenetics: CYP2B6 polymorphisms determine plasma concentrations, clearance, and metabolism. Anesthesiology 123,1142–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu C, Ogburn ET, Guo Y, & Desta Z Effects of the CYP2B6*6 allele on catalytic properties and inhibition of CYP2B6 in vitro: Implication for the mechanism of reduced efavirenz metabolism and other CYP2B6 substrates in vivo. Drug Metab Dispos 40,717–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calinski DM, Zhang H, Ludeman S, Dolan ME, & Hollenberg PF Hydroxylation and N-dechloroethylation of Ifosfamide and deuterated Ifosfamide by the human cytochrome P450s and their commonly occurring polymorphisms. Drug Metab Dispos 43,1084–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu H, Wang JJ, Chan KK, & Philip PA Stereoselectivity in metabolism of ifosfamide by CYP3A4 and CYP2B6. Xenobiotica 36,367–85 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Raccor BS, et al. Potential contribution of cytochrome P450 2B6 to hepatic 4-hydroxycyclophosphamide formation in vitro and in vivo. Drug Metab Dispos 40,54–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veal GJ, et al. Cyclophosphamide pharmacokinetics and pharmacogenetics in children with B-cell non-Hodgkin’s lymphoma. Eur J Cancer 55,56–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shu W, et al. Genetic markers in CYP2C19 and CYP2B6 for prediction of cyclophosphamide’s 4-hydroxylation, efficacy and side effects in Chinese patients with systemic lupus erythematosus. Br J Clin Pharmacol 81,327–40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desta Z, et al. Inhibition of cytochrome P450 2B6 activity by voriconazole profiled using efavirenz disposition in healthy volunteers. Antimicrob Agents Chemother 60,6813–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coles R, & Kharasch ED Stereoselective analysis of bupropion and hydroxybupropion in human plasma and urine by LC/MS/MS. J Chromatogr B 857,67–75 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Subject demographics
Figure S1 Influence of CYP2B6*6 genotype on buprenorphine metabolism
Figure S2 Influence of CYP2B6*6 genotype on buprenorphine metabolism.