Abstract
Background
Gliomas are the most common type of primary tumors in the central nervous system. This study aimed to investigate the biological role of miR-410-3p in glioma and elucidate the potential molecular mechanisms involved.
Methods
The expression levels of miR-410-3p in clinical tissue samples and glioma cell lines were determined using qRT-PCR analysis. The clinical significance of miR-410-3p in glioma was evaluated using Kaplan-Meier survival analysis and Fisher’s exact test. The effects of miR-410-3p on glioma cell proliferation, apoptosis, migration and invasion were investigated using MTT assays, flow cytometry, transwell migration and invasion assays. Besides, corresponding mechanistic studies were carried out.
Results
miR-410-3p was significantly down-regulated in glioma tissues. Besides, Kaplan-Meier analysis demonstrated that patients with low miR-410-3p expression had a shorter overall survival. Decreased miR-410-3p expression was associated with larger tumor size, lower Karnofsky performance score (KPS), and higher World Health Organization (WHO) grade. Over-expression of miR-410-3p suppressed cell proliferation, migration, and invasion, and accelerated apoptosis; whereas depletion of miR-410-3p facilitated cell proliferation, migration, and invasion, and inhibited apoptosis. Mechanistic investigations demonstrated that Ras-related protein 1A (RAP1A) was a direct target of miR-410-3p, and that rescue of RAP1A expression reversed miR-410-3p over-expression-induced inhibitory effects on cell proliferation, migration, and invasion. Notably, miR-410-3p over-expression repressed tumor growth in mouse xenograft models.
Conclusion
Our findings indicate that miR-410-3p functions as a tumor suppressor in glioma by directly targeting RAP1A. Thus, this study may provide some new insights into gliomagenesis and progression.
Keywords: miR-410-3p, RAP1A, poor prognosis, tumorigenesis, glioma
Introduction
Gliomas, accounting for approximately 40% of all intracranial tumors, are the most frequently diagnosed primary tumors in the central nervous system.1–3 About 13,000 glioma-related deaths are estimated to occur annually in the US.4,5 According to the criteria of the World Health Organization (WHO), gliomas are categorized as grade I–IV.6 Glioblastoma multiforme (GBM), also known as WHO grade IV glioma, is the most common and aggressive sub-type of primary brain tumors, with a median survival of merely 12–15 months.7,8 Despite significant progress in the diagnosis and treatment over the past several decades, the long-term prognosis of glioma patients remains very unfavorable. To date, effective therapeutic options for glioma are still limited. Hence, it is imperative to illuminate the molecular mechanisms of gliomagenesis and develop novel therapeutic strategies.
MicroRNAs (miRNAs) are a large group of endogenously expressed, evolutionarily conserved, and small non-coding RNA molecules with sizes of 17~25 nucleotides.9,10 Evidence is accumulating that miRNAs could exercise their post-transcriptional regulatory roles by directly binding to the 3ʹ-untranslated region (3ʹUTR) of corresponding target messenger RNAs (mRNAs) in a sequence-specific manner.11–13 Mounting evidence has demonstrated that the aberrant expression of miRNAs triggers tumorigenesis and tumor progression of a wide range of human malignancies.14–16 Recent studies have identified miR-410-3p, a novel cancer-related miRNA, as a tumor suppressor in human breast cancer and pancreatic ductal adenocarcinoma.17,18 Nonetheless, the biological role of miR-410-3p in glioma is still poorly understood.
Numerous studies reveal that up-regulation of oncogenic genes triggers leads to human carcinogenesis and cancer progression.19–21 It is well documented that Ras-related protein 1 (RAP1) is a member of the Ras oncogene family and acts like a crucial molecular switch in signal transduction.22,23 Previous studies showed that RAP1A, an isoform of RAP1, was frequently up-regulated and contributed to carcinogenesis and progression in multiple types of human malignant tumors, such as non-small cell lung cancer, esophageal squamous cell carcinoma, and ovarian cancer.24–26
In the current study, we found that miR-410-3p was significantly down-regulated in glioma tissues compared with matched non-tumorous tissues, and that its decreased expression was associated with poor prognosis. Functional investigations demonstrated that miR-410-3p suppressed glioma cell proliferation, migration, and invasion, and facilitated apoptosis. Mechanistic investigations revealed that miR-410-3p executed its tumor-suppressing functions in glioma via directly targeting RAP1A. Collectively, these findings may provide some evidence for miR-410-3p as a candidate therapeutic target in human glioma.
Materials and methods
Patients and tissue specimens
A total of 56 patients who were pathologically diagnosed with glioma and underwent surgical resection were recruited from Taihe Hospital Affiliated to Hubei University of Medicine (Shiyan, China) between January 2008 and November 2014. None of patients underwent chemotherapy or radiotherapy before surgery. Cancerous tissues were collected from 56 recruited glioma patients. Fresh normal non-neoplastic brain tissues were obtained from 56 donors who were injured in road traffic accidents. Glioma tissues and normal brain tissues were reviewed and identified by expert pathologists. All the clinical specimens were frozen in liquid nitrogen after surgical resection for further experiments. The clinicopathological characteristics of patients are listed in Table 1. Overall survival was defined as the time interval between the date of diagnosis and the date of death or last follow-up. This study was approved by the Ethics Committee of Taihe Hospital Affiliated to Hubei University of Medicine. This study was conducted in accordance with the Declaration of Helsinki. All the patients gave their written informed consent.
Table 1.
Relationship between miR-410-3p expression and clinicopathological characteristics of glioma patients
| Parameters | No. of cases | miR-410-3p expression | P-value | |
|---|---|---|---|---|
| High (n=21) | Low (n=35) | |||
| Age | 0.408 | |||
| <50 | 28 | 9 | 19 | |
| ≥50 | 28 | 12 | 16 | |
| Gender | 0.446 | |||
| Male | 25 | 8 | 17 | |
| Female | 31 | 13 | 18 | |
| Tumor size (cm) | 0.019 | |||
| <5 | 26 | 14 | 12 | |
| ≥5 | 30 | 7 | 23 | |
| KPS | 0.026 | |||
| <80 | 32 | 8 | 24 | |
| ≥80 | 24 | 13 | 11 | |
| WHO grade | 0.002 | |||
| I | 10 | 5 | 5 | |
| II | 11 | 9 | 2 | |
| III | 16 | 3 | 13 | |
| IV | 19 | 4 | 15 | |
Abbreviations: KPS, Karnofsky performance score; WHO, World Health Organization.
Cell culture
Normal human gliocyte HEB cells and four glioma cell lines (U87MG, SF126, LN229, and U251MG) were purchased from Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RIPM1640 medium supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA). Cells were maintained in a 5% CO2 humidified atmosphere at 37°C.
Cell transfection
Oligonucleotides for miR-410-3p mimics and miR-410-3p inhibitor were designed and purchased from GenePharma (Shanghai, China). The sequences of miR-410-3p mimics and miR-410-3p inhibitor were listed as followed: miR-410-3p mimics, 5ʹ-AAUAUAACACAGAUGGCCUGU-3ʹ; miR-410-3p inhibitor, 5ʹ-ACAGGCCAUCUGUGUUAUAUU-3ʹ. Cells were transfected with miR-410-3p mimics or miR-410-3p inhibitor using Lipofectamine (Invitrogen; Carlsbad, CA, USA) at a final concentration of 100 nM according to the manufacturer’s instructions. Cells were harvested at 48 hours post-transfection, and transfection efficiency was determined using PCR analysis.
Quantitative real time PCR (qRT-PCR)
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and reversely transcribed according to the manufacturer’s protocol. A Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) was used for the reverse transcription of miR-410-3p. A Taqman High-Capability cDNA Reverse Transcription Kit (Applied Biosystems) was used for the reverse transcription of RAP1A. The cDNA was used to analyze miR-410-3p and RAP1A expression. Real-time PCR analysis was conducted using a SYBR Premix Ex Taq Kit (Roche, Indianapolis, IN, USA) on the Applied Biosystems 7500 Real-time PCR system. The sequences of the specific primers were synthesized by GenePharma and listed as followed: RAP1A, forward 5ʹ-TGCCGAGAGGGAAATAAATG-3ʹ and reverse 5ʹ-GTCAAAGCCTCCAGCTGTTC-3ʹ; GAPDH, forward 5ʹ-AGAAGGCTGGGGCTCATTTG-3ʹ and reverse 5ʹ-AGGGGCCATCCACAGTCTTC-3ʹ; miR-410-3p, forward 5ʹ-GTCAGCGCAATATAACACAG-3ʹ and reverse 5ʹ-GAGAACAGCTCTGTGTTATAT-3ʹ; and U6, forward 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and reverse 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. The relative expression levels of miR-410-3p and RAP1A were calculated using the 2−△△Ct method. U6 and GAPDH were used as an endogenous control to normalize the expression of miR-410-3p and RAP1A, respectively. All the experiments were carried out in triplicate.
Cell proliferation assays
A cell proliferation kit (MTT; Roche) was used to examine glioma cell proliferation in accordance with the manufacturer’s instructions. Human glioma cells were seeded into each well of 96-well culture plates at a density of 2×103 and allowed to grow for 24 hours. Glioma cells were incubated for different periods of time (0, 24, 48, or 72 hours) at 37°C, the medium was then removed and incubated with MTT solution for another 4 hours at 37°C. Then the cells were treated with dimethyl sulfoxide (DMSO, Sigma-Aldrich). Absorbance at 570 nm was examined using a microplate reader (Thermo Fisher Scientific, Grand Island, NY, USA).
Cell apoptosis assays
An Annexin-V-FITC/PI Apoptosis Detection Kit (BD Bioscience, CA, USA) was used to evaluate cell apoptosis. Annexin-V and PI staining were carried out in accordance with the manufacturer’s instructions. In brief, 1×104 cells in 100 μl binding buffer were incubated with FITC Annexin V and 5 μLPI for 20 minutes at room temperature. Then stained cells were determined using flow cytometry (Becton Dickinson, San Jose, CA, USA).
Wound healing assays
The migration capabilities of glioma cells were evaluated using wound healing assays as previously described. Briefly, glioma cells from different groups were seeded into each well of 24-well plates at a density of 1×104 and allowed to grow until reaching confluency. Then the monolayer of glioma cells were manually scratched using a micropipette tip to generate a wound. Subsequently, the cells were allowed to grow for another 12 hours. Closure of scratch was monitored and photographed under an inverted microscope (Olympus IX50, Tokyo, Japan). All the experiments were conducted in triplicate.
Transwell invasion assays
The invasion capabilities of glioma cells were detected using Transwell invasion assays as previously described. In brief, glioma cells from different groups at a density of 1×105 cells were seeded into the upper chamber of the inserted pre-coated with Matrigel (Sigma-Aldrich) and allowed to invade for 48 hours. The medium in the upper chamber did not contain fetal bovine serum, whereas the lower chamber was filled with complete medium supplemented with 10% fetal bovine serum. Then the invaded cells were fixed using 4% polyformaldehyde and stained using crystal violet (Sigma-Aldrich) for 10 minutes. Finally, stained glioma cells in five randomly chosen fields were counted under an inverted microscope (Olympus IX50). All the experiments were conducted in triplicate.
Western blotting analysis
Total proteins were extracted using RIPA buffer containing protease inhibitor cocktail (Roche, Basel, Switzerland). The concentrations of extracted proteins were determined using BCA Protein Quantification Kit (Beyotime, Shanghai, China). A total of 20 μg protein from each treatment was separated using 10% SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred onto the polyvinylidene fluoride (PVDF) membranes. Then the membranes were incubated with primary antibodies against RAP1A, MMP9, vimentin, and GAPDH overnight at 4°C. Anti-RAP1A (ab115776), anti-MMP9 (ab58803), anti-vimentin (ab137321), and anti-GAPDH (ab181603) were purchased from Abcam (Cambridge, MA, USA) and used at the following dilutions: anti-RAP1A (1:500), anti-MMP9 (1:500), anti-vimentin (1:500), and anti-GAPDH (1:1,000). After being incubated with horseradish peroxidase-labeled secondary antibody at 37°C for 1 hour, the blots were developed using the enhanced chemiluminescence detection system (Amersham, Buckinghamshire, UK),and the blots were quantified using Image J software (NIH, Bethesda, MD, USA).
Luciferase reporter assays
The 3ʹUTR fragments of wild-type RAP1A containing the predicted binding site of miR-410-3p were amplified from a human cDNA library. The 3ʹUTR fragments of the wild-type or mutant RAP1A were inserted into the pmirGLO reporter vectors (Promega, Madison, WI, USA). The relative luciferase activity was determined using a luminescence reporter system (Promega) based on the manufacturer’s protocols at 48 hours post-transfection. Here, firefly luciferase activity functioned as an internal control to normalize the Renilla luciferase activity.
Transplantation tumor models in nude mice
BALB/c nude mice aged 5–6 weeks were purchased from Beijing HFK Bioscience Co. Ltd (Beijing, China). For tumor xenograft experiment in vivo, lentivirus-mediated stably transfected U251MG cells were established and used. In brief, murine xenograft models were established by subcutaneous injection of miR-410-3p mimics-treated-U251MG cells in the flank regions. The length (mm) and width (mm) of tumors were examined using a slide caliper every 5 days. At day 35 post-inoculation, all the animals were sacrificed, and the tumor nodules were surgically removed and weighed. Tumor size was calculated based on the following formula: tumor size (mm3)=length (mm)×width (mm)2/2. Animal experiments were approved by the Animal Care and Use Committee of Taihe Hospital Affiliated to Hubei University of Medicine, and performed in accordance with the guidelines of the Animal Welfare Committee of Hubei University of Medicine.
Immunohistochemistry (IHC)
IHC staining was conducted as previously described. In brief, paraffin-embedded sections were deparaffinized in xylene solution and rehydrated through gradient ethanol concentrations. Then, slides were subjected to antigen retrieval with citrate buffer. After being blocked with 3% hydrogen peroxide for 30 minutes, slides were then incubated with corresponding primary antibody at 4°C overnight. Subsequently, sections were incubated with streptavidin-horseradish peroxidase for 20 minutes. Finally, 3,3ʹ-diaminobenzidine (DAB) was used to visualize the stained cells. The positively stained cells were observed under a light microscope (Olympus).
Terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL)
Apoptotic cells were detected by TUNEL staining using the In Situ Cell Death Detection Kit (Roche) according to the manufacturer’s protocol. In brief, paraffin-embedded sections were deparaffinized and rehydrated for TUNEL staining. Then tissue sections were incubated with 50 μL TUNEL reaction mixture at 37°C in the dark for 1 hour, followed by being labeled with dTd labeling reaction mixture. Finally, a fluorescence microscope (Thermo Fisher, Waltham, MA, USA) was used to observe TUNEL-positive cells.
Statistical analysis
Data were expressed as mean±standard deviation (SD) from at least three independent experiments. SPSS 22.0 software (IBM, Chicago, IL, USA) was used to conduct statistical analysis. Differences between two groups were analyzed by Student’s t-test. Differences among three or more groups were compared using One-way analysis of variance (ANOVA). Survival analysis was performed using the log-rank test and Kaplan-Meier method. The correlation between miR-410-3p expression and clinicopathological characteristics was analyzed by Fisher’s exact test. The relationship between miR-410-3p expression and RAP1A mRNA expression in glioma tissues was evaluated by Pearson’s correlation analysis. A two-tailed P-value<0.05 was considered as statistically significant.
Results
Decreased expression of miR-410-3p predicts poor prognosis in glioma patients
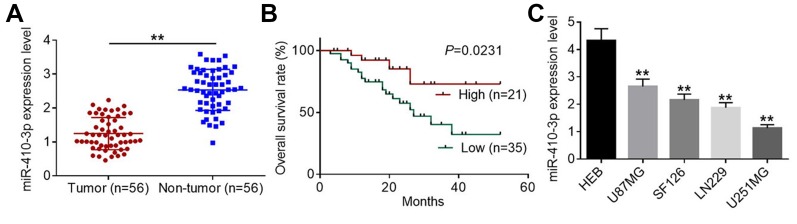
Even though previous studies have identified miR-410-3p as a tumor suppressor in human breast cancer and pancreatic ductal adenocarcinoma, its biological role in glioma is still unclear. To investigate the role of miR-410-3p, we firstly carried out qRT-PCR analysis to determine its expression patterns in glioma tissues and normal non-neoplastic brain tissues. As shown in Figure 1A, miR-410-3p was significantly down-regulated in glioma tissues compared with normal brain tissues. To analyze the correlation between miR-410-3p expression and clinicopathological characteristics of glioma patients, tumor tissues were divided into low miR-410-3p expression group and high miR-410-3p expression group according to the average value of its expression levels. Fisher’s exact test unveiled that decreased expression of miR-410-3p was associated with larger tumor size, lower KPS score, and higher WHO grade (Table 1). Kaplan-Meier analysis and log-rank test demonstrated that glioma patients with low miR-410-3p expression had a shorter survival than those with high miR-410-3p expression (Figure 1B). In addition, we found that miR-410-3p was markedly down-regulated in glioma cell lines (U87MG, SF126, LN229, and U251MG) compared with normal human gliocyte HEB cells (Figure 1C). These results indicate that decreased expression of miR-410-3p correlates with poor prognosis of glioma patients.
Figure 1.
Decreased expression of miR-410-3p predicts poor prognosis in glioma patients. (A) Relative expression levels of miR-410-3p in glioma tissues and normal non-neoplastic brain tissues were examined by qRT-PCR analysis. (B) Tumorous tissues were divided into high miR-410-3p expression group and low miR-410-3p expression group based on the average value of its expression levels. The log-rank test and Kaplan-Meier survival analysis were performed to evaluate the relationshiop between miR-410-3p expression and overall survival of glioma patients. (C) Relative expression levels of miR-410-3p in normal human gliocyte HEB cells and four glioma cell lines (U87MG, SF126, LN229, and U251MG) were examined by qRT-PCR analysis. **P<0.01.
Abbreviations: qRT-PCR, quantitative real-time polymerase chain reaction.
MiR-410-3p inhibits glioma cell proliferation and accelerates apoptosis
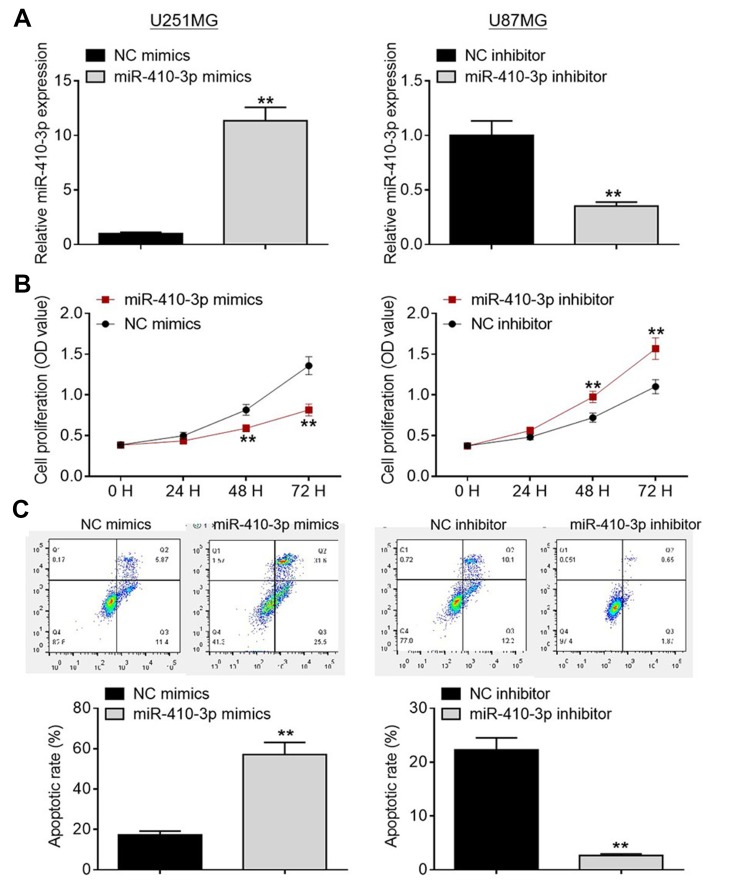
To explore the biological functions of miR-410-3p during glioma development and progression, we transfected glioma cells with miR-410-3p mimics or miR-410-3p inhibitor to manipulate its expression. Over-expression and knockdown studies were performed in U251MG cells (lowest endogenous miR-410-3p expression) and U87MG cells (highest endogenous miR-410-3p expression), respectively. We evaluated the transfection efficiency via qRT-PCR analysis (Figure 2A). As evident from MTT assays, miR-410-3p over-expression significantly inhibited U251MG cell proliferation compared with the negative control group, whereas miR-410-3p depletion notably promoted U87MG cell proliferation (Figure 2B). As demonstrated in Figure 2C, a significant increase in apoptosis was observed in miR-410-3p mimics-treated U251MG cells compared with the negative control group, whereas a notable decrease in apoptosis was found in miR-410-3p inhibitor-treated U87MG cells. These data indicate that miR-410-3p inhibits glioma cell proliferation and accelerates apoptosis.
Figure 2.
MiR-410-3p inhibits glioma cell proliferation and accelerates apoptosis. (A) Transfection efficiency was evaluated by qRT-PCR analysis. (B) Cell proliferation was determined by MTT assays after transfection of miR-410-3p mimics or miR-410-3p inhibitor. (C) Cell apoptosis was assessed by FCM analysis after transfection of miR-410-3p mimics or miR-410-3p inhibitor. **P<0.01.
Abbreviations: FCM, flow cytometry; NC, negative control; qRT-PCR, quantitative real-time polymerase chain reaction.
MiR-410-3p suppresses glioma cell migration and invasion
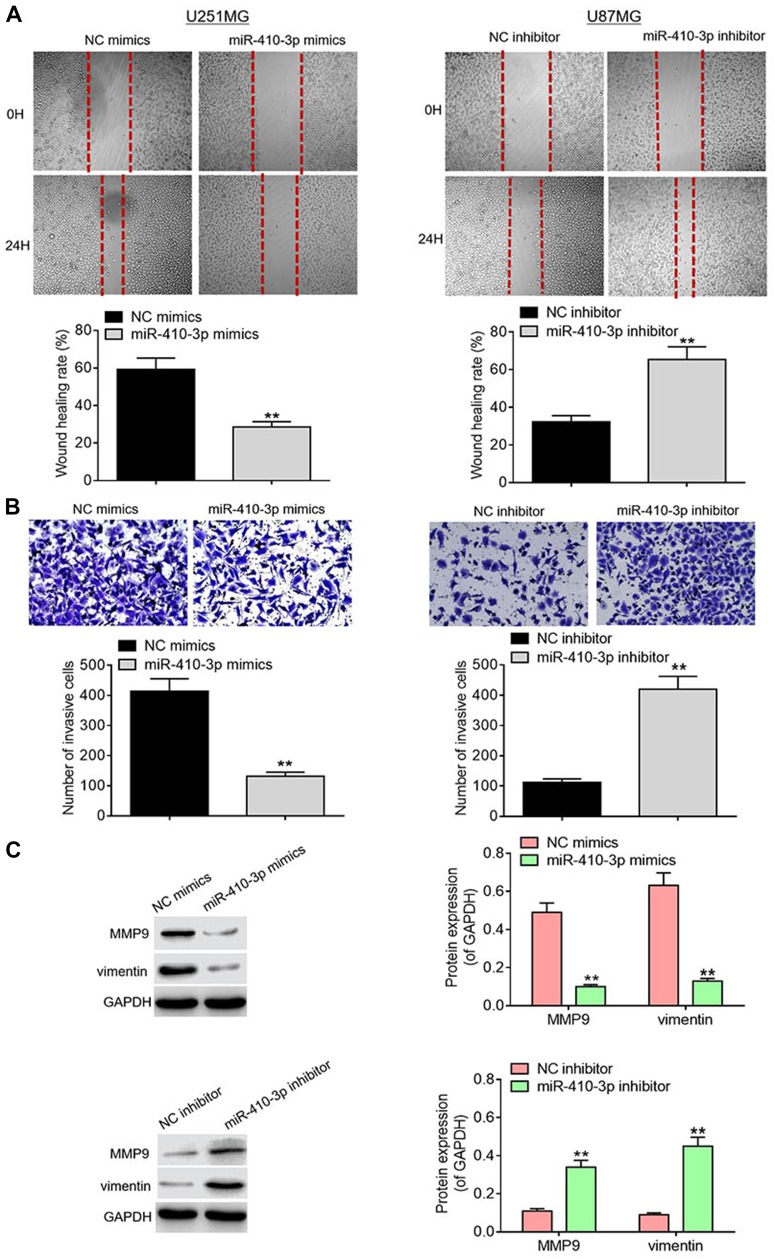
It is well documented that cell migration and invasion are critical events during tumor progression. To determine whether miR-410-3p affects the motility of glioma cells, we subsequently evaluated their migration and invasion capabilities. Wound healing assays showed that miR-410-3p over-expression significantly suppressed U251MG cell migration compared with the negative control group, whereas miR-410-3p ablation dramatically facilitated U87MG cell migration (Figure 3A). As evident from Transwell invasion assays, miR-410-3p over-expression significantly weakened the invasion capability of U251MG cells compared with negative control treatment, whereas miR-410-3p ablation markedly enhanced the invasion capability of U87MG cells (Figure 3B). In addition, lower protein expression levels of MMP9 and vimentin were observed in the miR-410-3p over-expression group compared with the negative control group, whereas miR-410-3p ablation significantly promoted the protein expression of MMP9 and vimentin (Figure 3C). These data indicate that miR-410-3p suppresses glioma cell migration and invasion.
Figure 3.
MiR-410-3p suppresses glioma cell migration and invasion. (A) Migration capability was analyzed by wound healing assays after transfection of miR-410-3p mimics in U251MG cells or miR-410-3p inhibitor in U87MG cells. (B) Invasion abiltiy was detected via Transwell invasion assays after transfection of miR-410-3p mimics in U251MG cells or miR-410-3p inhibitor in U87MG cells. (C) The expression levels of motility-associated proteins (MMP9 and vimentin) were examined by Western blotting analysis after transfection of miR-410-3p mimics in U251MG cells or miR-410-3p inhibitor in U87MG cells. **P<0.01.
Abbreviations: MMP9, matrix metalloproteinase 9; NC, negative control.
RAP1A is a direct target of miR-410-3p
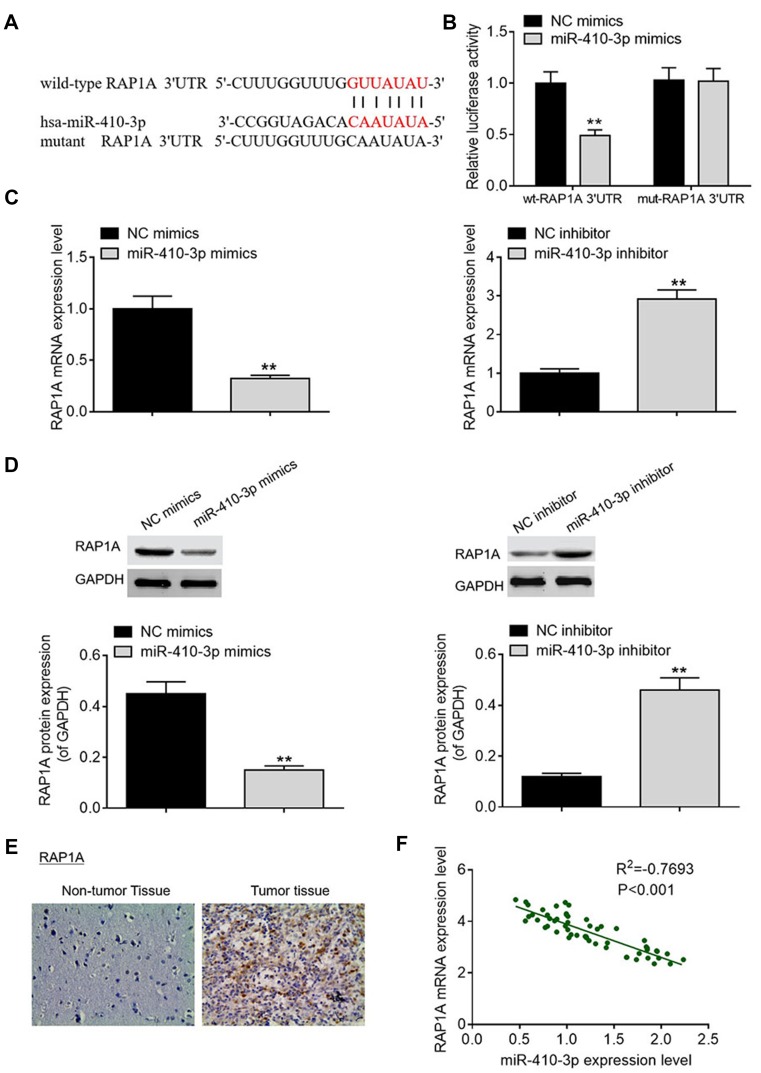
To elucidate the potential molecular mechanism underlying glioma tumorigenesis, we conducted bio-informatics analysis using TargetScan online software. Among all the putative targets, RAP1A drew our attention for its crucial roles in various cancers, and was selected as a candidate target of miR-410-3p (Figure 4A). To validate whether miR-410-3p could bind to 3ʹUTR of RAP1A mRNA, luciferase reporter assays were performed. As exhibited in Figure 4B, transfection of miR-410-3p mimics significantly reduced the luciferase activity of the reporter vectors carrying wild-type RAP1A 3ʹUTR fragments, whereas transfection of miR-410-3p did not elicit significant alterations in the luciferase activity of the reporter vectors carrying mutant RAP1A mRNA 3ʹUTR fragments. Furthermore, miR-410-3p over-expression was observed to decrease the mRNA and protein expression levels of RAP1A compared with negative control group, whereas miR-410-3p depletion increased the mRNA and protein expression levels of RAP1A (Figures 4C and D). As evident from IHC staining, glioma tissues displayed higher RAP1A expression than adjacent non-cancerous tissues (Figure 4E). Interestingly, Pearson’s correlation analysis revealed that miR-410-3p expression was negatively correlated with RAP1A mRNA expression in the tumorous tissues (Figure 4F). Taken together, our results suggest that miR-410-3p negatively regulates RAP1A expression by directly targeting its mRNA 3ʹUTR.
Figure 4.
RAP1 is a direct target of miR-410-3p. (A) A putative binding site of miR-410-3p in the 3ʹUTR of RAP1A mRNA was predicted by TargetScan online software. (B) Luciferase activity of the reporter vectors carrying wild-type or mutant RAP1A 3ʹUTR fragment was determined after transfection of NC mimics or miR-410-3p mimics. (C) RAP1A mRNA expression was examined by qRT-PCR analysis after transfection of miR-410-3p mimics in U251MG cells or miR-410-3p inhibitor in U87MG cells. (D) RAP1A protein expression was determined by Western blotting analysis after transfection of miR-410-3p mimics in U251MG cells or miR-410-3p inhibitor in U87MG cells. (E) IHC staining was performed to visualize RAP1A protein expression in glioma tissues and matched pericarcinomatous tissues. (F) Pearson’s correlation analysis was carried out to determine the correlation between miR-410-3p expression and RAP1A mRNA expression in the tumorous tissues. **P<0.01.
Abbreviations: 3ʹUTR, 3ʹ untranslated region; IHC, immunohistochemistry; mut, mutant; NC, negative control; qRT-PCR, quantitative real-time polymerase chain reaction; RAP1A, Ras-related protein 1A; wt, wild-type.
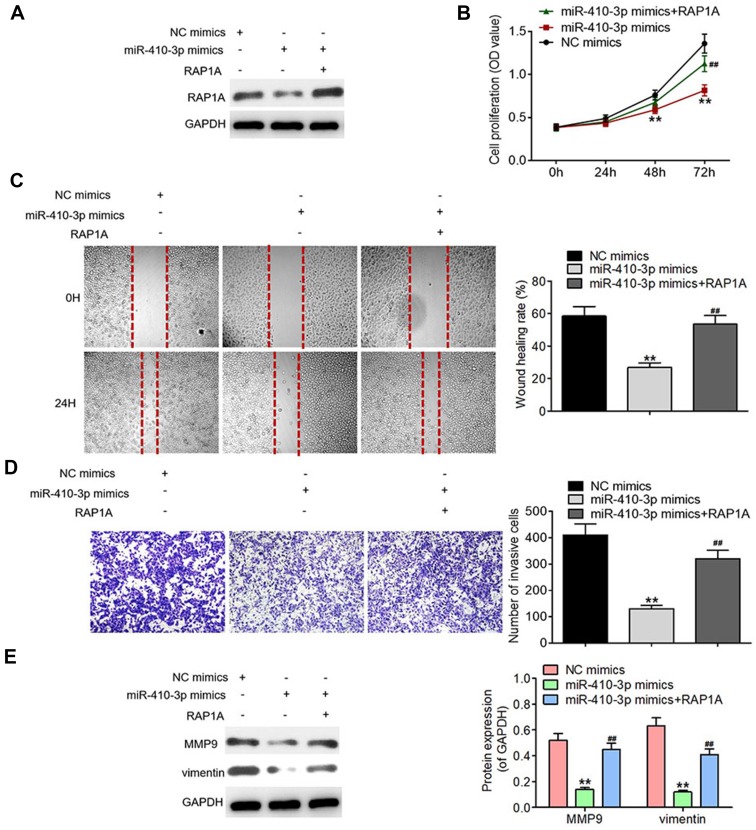
Rescue of RAP1A expression reverses the tumor-suppressing effects of miR-410-3p over-expression in glioma cells
To verify the functional link between miR-410-3p and RAP1A, we rescued the expression of RAP1A in the U251MG cells over-expressing miR-410-3p (Figure 5A). As displayed in Figures 5B–D, rescue of RAP1A expression accelerated proliferation, migration, and invasion of miR-410-3p mimics-treated U251MG cells, suggesting that miR-410-3p-dependent inhibition of proliferation, migration, and invasion was mediated by RAP1A. Additionally, Western blotting analysis demonstrated that rescue of RAP1A alleviated the suppressing effects of miR-410-3p over-expression on MMP9 and vimentin protein expression (Figure 5E). These findings indicate that miR-410-3p executes its inhibitory effects on glioma proliferation, migration, and invasion through targeting RAP1A.
Figure 5.
Rescue of RAP1A expression reverses the tumor-suppressing effects of miR-410-3p over-expression in glioma cells. (A) RAP1A protein expression was determined by Western blotting analysis after rescue of its expression in miR-410-3p mimics-treated U251MG cells. (B) Cell proliferation was evaluated by MTT assays after rescue of RAP1A expression in miR-410-3p mimics-treated U251MG cells. (C) Migration was assessed by wound healing assays after rescue of RAP1A expression in miR-410-3p mimics-treated U251MG cells. (D) Invasion was determined by transwell invasion assays after rescue of RAP1A expression in miR-410-3p mimics-treated U251MG cells. (E) The expression levels of motility-associated proteins (MMP9 and vimentin) are examined by Western blotting analysis after rescue of RAP1A expression in miR-410-3p mimics-treated U251MG cells. **P<0.01 vs NC mimics group, ##P<0.01 vs miR-410-3p mimics group.
Abbreviations: MMP9, matrix metalloproteinase 9; NC, negative control.
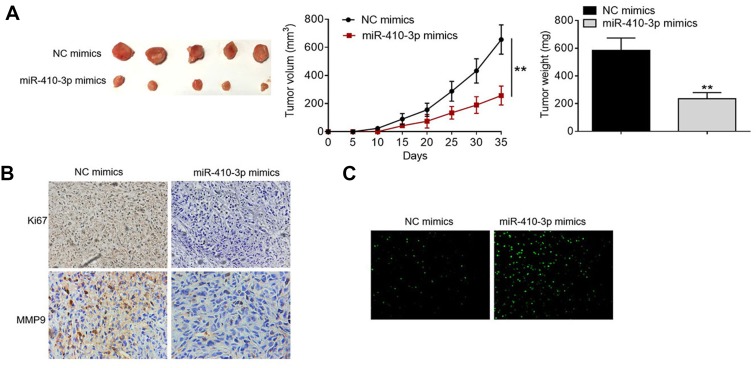
MiR-410-3p over-expression represses tumor growth in mouse xenograft models
To validate the findings obtained from in vitro studies, we established mouse xenograft tumor models via subcutaneous injection of NC mimics- or miR-410-3p mimics-treated U251MG cells. As shown in Figure 6A, tumors formed by miR-410-3p mimics-treated U251MG cells grow much slower than those formed by NC mimics-treated glioma cells (Figure 6A). Furthermore, tumors collected from the over-expression group weighed significantly lighter than those harvested from the negative control group. IHC staining demonstrated that miR-410-3p over-expression inhibited Ki67 and MMP9 protein expression in the tumors compared with the negative control group (Figure 6B). In addition, TUNEL analysis revealed that more apoptotic cells were observed in the tumors collected from the over-expression group compared with those from the negative control group (Figure 6C). Taken together, these results suggest that miR-410-3p over-expression represses tumor growth in vivo.
Figure 6.
MiR-410-3p over-expression tumor growth in mouse xenograft models. (A) Lentivirus-mediated stably transfected U251MG cells were established and used for tumor transplantation models. miR-410-3p mimics-treated U251MG cells were subcutaneously injected into the flanks of the nude mice (n=5). Tumor volumes were measured every 5 days; at day 35 post-implantation, all the mice were sacrificed under anesthesia and tumors were weighed. (B) Ki67 and MMP9 protein expression in the collected tumors was visualized by IHC staining. (C) Cell apoptosis in the collected tumors was detected by TUNEL analysis. **P<0.01.
Abbreviations: IHC, immunohistochemistry; MMP, matrix metalloproteinase 9; NC, negative control; TUNEL, terminal deoxynucleotidyl transferase-mediated nick end labeling.
Discussion
Accumulating evidence has demonstrated the crucial roles of miRNAs in the carcinogenesis and tumor progression of various types of human malignancies.13–16 Numerous studies unveiled that the ectopic expression of miRNAs was implicated in gliomagenesis and tumor progression.27–29 Glioma, one of the most frequently diagnosed human malignant tumors, has already posed serious threats to public health worldwide.30 Although considerable and encouraging progress has been made in the diagnosis and treatment of glioma, the therapeutic effects remain unfavorable. Hence, there is a pressing need to identify novel prognostic bio-markers and develop effective therapeutic strategies for glioma treatment.
Previous studies revealed that miR-410-3p was dysregulated in human breast cancer and pancreatic ductal adenocarcinoma. Zhang et al17 demonstrated that miR-410-3p was significantly down-regulated in breast cancer tissue samples and suppressed tumor progression via targeting snail. Xiong et al18 reported that miR-410-3p was markedly down-regulated in pancreatic ductal adenocarcinoma and attenuated chemotherapy resistance of cancer cells.Nonetheless, the biological role of miR-410-3p in glioma has not yet been characterized. In the present study, we found that miR-410-3p expression was significantly decreased in glioma tissues in comparison with normal brain tissues, which was consistent with the previous findings in breast cancer and pancreatic ductal adenocarcinoma. In addition, we noticed that decreased miR-410-3p expression was associated with the poor prognosis of glioma patients. Taken together, these data indicate a potential of miR-410-3p as a tumor suppressor in glioma progression.
To gain a better understanding of the biological role of miR-410-3p, we subsequently conducted functional investigations. Furthermore, functional studies demonstrated that miR-410-3p over-expression suppressed cell proliferation, migration, and invasion, and accelerated apoptosis; whereas miR-410-3p ablation promoted cell proliferation, migration, and invasion, and inhibited apoptosis. Interestingly, miR-410-3p over-expression was observed to repress tumor growth in mouse xenograft models. Collectively, these findings suggest that miR-410-3p possesses anti-tumor activity in vitro and in vivo.
To illuminate the potential molecular mechanism by which miR-410-3p exerts its tumor-suppressing functions in glioma, we then carried out bio-informatics analysis using TargetScan online software, and selected RAP1A as a candidate target. Additionally, RAP1A was validated as a direct target of miR-410-3p by the luciferase reporter assays. RAP1A belongs to the Ras family of small GTPases and serves as a pivotal molecular switch for signal transduction.31,32 It is widely documented that RAP1A play crucial roles in various biological processes, including cell proliferation, differentiation, adhesion, and motility.26,33 Furthermore, previous studies reported that RAP1A acted as an oncogene in diverse types of human tumors, including non-small cell lung cancer,24 esophageal squamous cell carcinoma,25 ovarian cancer,26 prostate cancer,34 and breast cancer.35
To verify the functional relationship between miR-410-3p and RAP1A, we rescued the expression of RAP1A in glioma LM3 cells over-expressing miR-410-3p. Interestingly, rescue of RAP1A expression was observed to reverse the inhibitory effects of miR-410-3p over-expression on proliferation and motility. Taken together, these findings indicate that miR-410-3p functions as a tumor suppressor in glioma through directly targeting RAP1A.
In conclusion, the present study unveiled for the first time that miR-410-3p was significantly down-regulated in glioma tissues, and that decreased miR-410-3p expression was associated with poor prognosis of glioma patients. Furthermore, our data indicate that miR-410-3p exerts tumor-suppressing functions in glioma by directly targeting RAP1A. This study may provide some new insights into the molecular mechanisms underlying gliomagenesis and implicate miR-410-3p as a potential prognostic bio-marker and a novel therapeutic target for glioma.
Acknowledgments
This study was supported by a grant from Shiyan Science and Technology Bureau (No.17Y14).
Disclosure
The authors have declared that they have no competing interests in this work.
References
- 1.Simon M, Hosking FJ, Marie Y, et al. Genetic risk profiles identify different molecular etiologies for glioma. Clin Cancer Res. 2010;16(21):5252–5259. doi: 10.1158/1078-0432.CCR-10-1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng T, Cui D, Gao L. Glioma: an overview of current classifications, characteristics, molecular biology and target therapies. Front Biosci. 2015;20(7):1104–1115. doi: 10.2741/4362 [DOI] [PubMed] [Google Scholar]
- 3.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci. 2009;100(12):2235–2241. doi: 10.1111/j.1349-7006.2009.01308.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanson M, Hosking FJ, Shete S, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum Mol Genet. 2011;20(14):2897–2904. doi: 10.1093/hmg/ddr192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shete S, Lau CC, Houlston RS, et al. Genome-wide high-density SNP linkage search for glioma susceptibility loci: results from the gliogene consortiu. Cancer Res. 2011;71(24):7568–7575. doi: 10.1158/0008-5472.CAN-11-0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai WC, Hueng DY, Nieh S, Gao HW. ARID4B is a good biomarker to predict tumour behaviour and decide WHO grades in gliomas and meningiomas. J Clin Pathol. 2017;70(2):162–167. doi: 10.1136/jclinpath-2016-203804 [DOI] [PubMed] [Google Scholar]
- 7.Liu M, Lin Y, Zhang XC, et al. Phosphorylated mTOR and YAP serve as prognostic markers and therapeutic targets in gliomas. Lab Invest. 2017;97(11):1354–1363. doi: 10.1038/labinvest.2017.70 [DOI] [PubMed] [Google Scholar]
- 8.Yoon DJ, Kwan BH, Chao FC, et al. Intratumoral therapy of glioblastoma multiforme using genetically engineered transferrin for drug delivery. Cancer Res. 2010;70(11):4520–4527. doi: 10.1158/0008-5472.CAN-09-4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Clark AG. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012;22(7):1243–1254. doi: 10.1101/gr.132514.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X. MicroRNA: function, detection, and bioanalysis. Chem Rev. 2013;113(8):6207–6233. doi: 10.1021/cr300362f [DOI] [PubMed] [Google Scholar]
- 11.Fu L, Shi Z, Luo G, et al. Multiple microRNAs regulate human FOXP2 gene expression by targeting sequences in its 3ʹ untranslated region. Mol Brain. 2014;7(1):71. doi: 10.1186/s13041-014-0056-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Carroll D, Schaefer A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology. 2013;38(1):39–54. doi: 10.1038/npp.2012.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang WX, Rajeev BW, Stromberg AJ, et al. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28(5):1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei R, Tang J, Zhuang X, et al. Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis. Oncogene. 2014;33(10):1287–1296. doi: 10.1038/onc.2013.65 [DOI] [PubMed] [Google Scholar]
- 15.Li BS, Zuo QF, Zhao YL, et al. MicroRNA-25 promotes gastric cancer migration, invasion and proliferation by directly targeting transducer of ERBB2, 1 and correlates with poor survival. Oncogene. 2015;34(20):2556–2565. doi: 10.1038/onc.2014.214 [DOI] [PubMed] [Google Scholar]
- 16.Zhao B, Han H, Chen J, et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342(1):43–51. doi: 10.1016/j.canlet.2013.08.030 [DOI] [PubMed] [Google Scholar]
- 17.Zhang YF, Yu Y, Song WZ, et al. miR-410-3p suppresses breast cancer progression by targeting snail. Oncol Rep. 2016;36(1):480–486. doi: 10.3892/or.2016.4828 [DOI] [PubMed] [Google Scholar]
- 18.Xiong J, Wang D, Wei A, et al. MicroRNA-410-3p attenuates gemcitabine resistance in pancreatic ductal adenocarcinoma by inhibiting HMGB1-mediated autophagy. Oncotarget. 2017;8(64):107500–107512. doi: 10.18632/oncotarget.22494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen N, Wang Y, Wen L, et al. Overexpression of FOXM1 predicts poor prognosis and promotes cancer cell proliferation, migration and invasion in epithelial ovarian cancer. J Transl Med. 2014;12(1):134. doi: 10.1186/1479-5876-12-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldie SJ, Mulder KW, Tan DW, Lyons SK, Sims AH, Watt FM. FRMD4A upregulation in human squamous cell carcinoma promotes tumor growth and metastasis and is associated with poor prognosis. Cancer Res. 2012;72(13):3424–3436. doi: 10.1158/0008-5472.CAN-12-0423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok WK, Ling MT, Lee TW, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65(12):5153–5162. doi: 10.1158/0008-5472.CAN-04-3785 [DOI] [PubMed] [Google Scholar]
- 22.Gao L, Feng Y, Bowers R, et al. Ras-associated protein-1 regulates extracellular signal-regulated kinase activation and migration in melanoma cells: two processes important to melanoma tumorigenesis and metastasis. Cancer Res. 2006;66(16):7880–7888. doi: 10.1158/0008-5472.CAN-06-0254 [DOI] [PubMed] [Google Scholar]
- 23.Daumke O, Weyand M, Chakrabarti PP, Vetter IR, Wittinghofer A. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature. 2004;429(6988):197–201. doi: 10.1038/nature02505 [DOI] [PubMed] [Google Scholar]
- 24.Du L, Subauste MC, Desevo C, et al. miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS One. 2012;7(6):e39167. doi: 10.1371/journal.pone.0039167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Li J, Guo H, et al. MiR-196a binding-site SNP regulates RAP1A expression contributing to esophageal squamous cell carcinoma risk and metastasis. Carcinogenesis. 2012;33(11):2147–2154. doi: 10.1093/carcin/bgs259 [DOI] [PubMed] [Google Scholar]
- 26.Lu L, Wang J, Wu Y, Wan P, Yang G. Rap1A promotes ovarian cancer metastasis via activation of ERK/p38 and notch signaling. Cancer Med. 2016;5(12):3544–3554. doi: 10.1002/cam4.946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28(17):5369–5380. doi: 10.1128/MCB.00479-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang J, Sun X, Wang W, et al. Tumor microRNA-335 expression is associated with poor prognosis in human glioma. Med Oncol. 2012;299(5):3472–3477. doi: 10.1007/s12032-012-0259-z [DOI] [PubMed] [Google Scholar]
- 29.Que T, Song Y, Liu Z, et al. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34(38):4952–4963. doi: 10.1038/onc.2014.419 [DOI] [PubMed] [Google Scholar]
- 30.Li M, Li J, Liu L, Li W, Yang Y, Yuan J. MicroRNA in human glioma. Cancers. 2013;5(4):1306–1331. doi: 10.3390/cancers5041306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Retta SF, Balzac F, Avolio M. Rap1: a turnabout for the crosstalk between cadherins and integrins. Eur J Cell Biol. 2006;85(3–4):283–293. doi: 10.1016/j.ejcb.2005.09.007 [DOI] [PubMed] [Google Scholar]
- 32.Zhang YL, Wang RC, Cheng K, Ring BZ, Su L. Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol Med. 2017;14(1):90–99. doi: 10.20892/j.issn.2095-3941.2016.0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma SK, Ganesan TS, Kishore U, Parker PJ. The tumor suppressor RASSF1A is a novel effector of small G protein Rap1A. Protein Cell. 2011;2(3):237–249. doi: 10.1007/s13238-011-1028-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang J, Bian C, Wang H, Huang S, Wu D. MiR-203 down-regulates Rap1A and suppresses cell proliferation, adhesion and invasion in prostate cancer. J Exp Clin Cancer Res. 2015;34(1):8. doi: 10.1186/s13046-015-0125-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang T, Jiang K, Zhu X, et al. miR-433 inhibits breast cancer cell growth via the MAPK signaling pathway by targeting Rap1a. Int J Biol Sci. 2018;14(6):622–632. doi: 10.7150/ijbs.24223 [DOI] [PMC free article] [PubMed] [Google Scholar]