ABSTRACT
The relation between microRNAs (miRNAs) and malignant melanoma has been demonstrated in previous studies, while there was little research about miR-139-5p and malignant melanoma. The aim of this study is to investigate the ability of miR-139-5p in malignant melanoma cells via the modulation of the PI3K/AKT signaling pathway by targeting IGF1R. MiR-139-5p expression in malignant melanoma tissues and 5 malignant melanoma cell lines was detected. The melanoma cells were transfected with miR-139-5p mimic negative control (NC) sequence, miR-139-5p mimic, IGF1R overexpressed recombinant plasmid NC or IGF1R overexpressed sequence. The expression of Akt signaling pathway-related protein was evaluated. The biological functions in malignant melanoma cells were evaluated by a string of experiments. MiR-139-5p expressed a poor level in tissues and cell lines of malignant melanoma. Overexpressed miR-139-5p suppressed the cell proliferation, migration, and invasion, and contributed to the promoted apoptosis of malignant melanoma cells by decreasing IGF1R. MiR-139-5p down-regulated the IGF1R expression, and IGF1R accelerated the activation of the PI3K/AKT signaling pathway. miR-139-5p reversed the promotive impacts of IGF1R on the PI3K/AKT signaling pathway. The study validates that miR-139-5p could suppress malignant melanoma progression through the repression of the PI3K/AKT signaling pathway by down-regulating IGF1R. Therefore, miR-139-5p could pave a new way for the treatment of malignant melanoma.
KEYWORDS: MicroRNA-139-5p, IGF1R, PI3K/AKT signaling pathway
Background
Malignant melanoma is a kind of cancer that typically occurs in skin, and the incidence has been rising throughout the world in recent years [1], while the mortality has been rising not so fast [2]. Some risk factors including sun exposure [3], both natural and artificial UV radiation, type of skin, ethnicity, number of melanocytic nevi, geographic location [4] and season of birth [5] were correlated with the progression of malignant melanoma. However, in the advanced stages of malignant melanoma, the curative effects of conventional treatments were constrained [6]. In recent years, many studies have researched on the pathogenesis of malignant melanoma, while there was a lack of studies about its molecular mechanism [7]. Recent discoveries led to the rapid development of new therapeutic approaches for malignant melanoma treatment, while the functions have been limited [8]. Thus, new indicators for prognosis and therapeutic targets of malignant melanoma are demanded.
It has been proved that microRNAs (miRs), such as miR-18b [7], miR-21 [9] and miR-124 [10] have been verified to be related with the progression and development of malignant melanoma. As a significant tumor inhibitor, miR-139-5phas been proved to be down-regulated in several kinds of cancers and was supposed to be the candidate for the cancer diagnosis as a promising biomarker [11]. It has been identified that miR-139-5p expression could be regulated by its target gene insulin-like growth factor receptor type 1 (IGF1R) [12], which has been considered to be a treatment for malignancies [13]. According to a current study, IGF1R has been proved to be not regulated by the activity of phosphatidylinositol 3-kinase (PI3K), and its expression is not dependent on phosphorylated protein kinase B (AKT) of melanoma cells [14]. PI3K pathway is a commonly activated signal transduction pathway in cancers, and AKT is one of its key downstream effector molecules [15], which can advance the progression of tumor [16]. Furthermore, Simon Caporali et al. have proved that melanoma cells could be excited by activating the PI3K/AKT/mTOR signaling pathway [17]. Nevertheless, there was little known about the mechanisms of miR-139-5p targets IGF1R to mediate the PI3K/AKT signaling pathway, hence, this study was conducted to focus on the capacity of miR-139-5p and IGF1R in malignant melanoma, and we speculated that miR-139-5p could cast effects on the development of malignant melanoma by down-regulating IGF1R, which could be a possible candidate for remedy of malignant melanoma.
Materials and methods
Ethics statement
The study was ratified by the Ethics Committee of The Sixth Affiliated Hospital of Sun Yat-sen University and based on the ethical principles for medical research involving human subjects of the Helsinki Declaration. Informed written consent was obtained from all patients.
Study subjects
A total number of 82 malignant melanoma tissues from patients (49 males and 33 females, with a mean age of 42.65 ± 5.67 years) who were diagnosed with malignant melanoma in The Sixth Affiliated Hospital of Sun Yat-sen University from January 2016 to January 2018 were collected. A number of 30 benign skin disease tissues that obtained from healthy controls were served as control (17 males and 13 females, with a mean age of 42.65 ± 3.26 years).
Cell culture, grouping, and transfection
Primary normal skin melanocytes PIG1 and five malignant melanoma cell lines (A375, SK-MEL-1, SK-MEL-2, SK-MEL-5, and SK-MEL-28) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). These cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (SP1355, Shifeng Biotechnology Co., Ltd., Shanghai, China) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin, and the cells were incubated at 37°C and 5% CO2in an incubator (DHP-9162, Chengjie Experimental Instrument Co., Ltd., Shanghai, China). Fresh medium was changed every 1–2 days, and the cell subculture was conducted when the cell confluence degree reached 80–90%. Cells in the third passage were conducted with miR-139-5p quantitative detection to screen cell lines.
Cells in the logarithmic growth phase were allocated into five groups: miR-139-5p mimic-negative control (NC) group (melanoma cells transfected with miR-139-5p mimic-NC), miR-139-5p mimic group (melanoma cells transfected with miR-139-5p mimic), overexpression (oe)-NC group (melanoma cells transfected with IGF1R overexpressed recombinant plasmid NC), oe-IGF1R group (melanoma cells transfected with IGF1R overexpressed recombinant plasmid), miR-139-5p mimic + oe-IGF1R group (melanoma cells transfected with overexpressed sequence of miR-139-5p and overexpressed sequence of IGF1R). MiR-139-5p mimic-NC, miR-139-5p mimic, oe-NC, and oe-IGF1R were all obtained from ShanghaiGenePharma Co., Ltd. (Shanghai, China), the information of these sequences was not provided here because of business factors. Cells in the logarithmic growth phase were seeded in six-well plates with a cell density of 30%-50%, then transfected on the basis of the protocols of lipofectamine 2000 (Invitrogen, Carlsbad, California, USA). There was no RNase solution after the centrifugation, the cells were diluted with 250 µL serum-free minima essential medium (MEM) and 5 µL lipofectamine 2000 was diluted with 250 µL serum-free MEM (Gibco, Grand Island, N.Y., USA) for 5 min; both two above were mixed up and put in cell culture wells for 20 min. Cells were cultured at 37°C with 5% CO2 for 6–8 h, then cultured in complete medium for 24–48 h for subsequent experiments.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The total RNA was extracted using the Trizol reagent (Takara Biotechnology Co., Ltd., Dalian, China) after the collection of the treated cells, and the concentration and purity were measured. Based on the protocols of the reverse transcription kit (K1621, Fermentas, Maryland, NY, USA), the total RNA was reversely transcripted into complementary DNA (cDNA). The primer sequences of miR-139-5p and IGF1R (Table 1) were synthetized by Shanghai Genechem Co., Ltd. (Shanghai, China). Fluorescent quantitative PCR kit (Takara Biotechnology Co., Ltd., Dalian, China) was employed to detect the mRNA expression of genes. U6 was taken as the internal reference for miR-139-5p and glyceraldehyde phosphate dehydrogenase (GAPDH) was used as the internal reference for IGF1R. The target gene expression was calculated by 2−ΔΔCt method.
Table 1.
Primer sequence.
| Gene | Primer sequence (5’ – 3’) |
|---|---|
| miR-139-5p | Forward: 5’-AGCGGCATGATCAACATTTGCTAAAG-3’ |
| Reverse: 5’-TCTGTATGGTTTTCCTTCCGGCTT-3’ | |
| IGF1R | Forward: 5’–CGAGCAAGTTCTTCGTTTCGT-3’ |
| Reverse: 5’-TGTACTGCCAGCACATGCG-3’ | |
| U6 | Forward: 5’-CTCGCTTCGGCAGCACA-3’ |
| Reverse: 5’-AACGCTTCACGAATTTGCGT-3’ | |
| GAPDH | Forward: 5’-TCCCATCACCATCTTCCA-3’ |
| Reverse: 5’-CATCACGCCACAGTTTTCC-3’ |
Note: miR-139-5p, microRNA-139-5p; IGF1R, insulin-like growth factor receptor type 1; GAPDH, glyceraldehyde phosphate dehydrogenase.
Western blot analysis
The total proteins were extracted using radio immunoprecipitation assay (RIPA) kit. The protein concentrations were determined using a bicinchoninic acid (BCA) kit. Subsequently, the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then were transferred onto the polyvinylidene fluoride membrane. The membrane was blocked with 5% no-fat milk powder for 1 h and then incubated at 4°C with the following diluted primary rabbit anti-human antibodies: IGF1R (ab39675, 1:1000), PI3K (1:600, ab86714), AKT (1:600, ab8805), p-PI3K (1:1000, ab182651), p-AKT (1:1000, ab38449) and GAPDH (ab9485, 1:2500). The above antibodies were purchased from Abcam Inc. (Cambridge, MA, USA). After washed with tris-buffered saline solution containing tween 20 (TBST) for three times, 10 min per time, the membrane was incubated with horseradish peroxidase (HRP)-labeled goat anti-rabbit immunoglobulin G (IgG) H&L (ab97051, 1:2000, Abcam, Cambridge, UK) for 1 h. The proteins were photographed by Bio-Rad (Bio-Rad Laboratories, Hercules, CA, USA) and analyzed by Quantity One v4.6.2 software (Bio-Rad Laboratories, Shanghai, China). The relative protein content was expressed as the gray value of relative protein band/the pray value of GAPDH protein band.
Immunohistochemical staining
The samples were embedded in paraffin and sectioned into 4 μm, then toasted at 70°C for 15 min, and the sections were dehydrated by ethanol and the endogenous enzyme was inactivated by H2O2, subsequently, the sections were rinsed by phosphate buffered solution (PBS) for 3 times, 5 min for each time, then incubated by normal goat serum (ZSGB-Bio, Beijing, China) for 15 min, appended with 20–30 μL IGF1R that diluted by PBS at 1:150 (Cell Signaling Technology, Beverly, MA, USA), and incubated at 4°C overnight. Next, the sections were washed by PBS for 3 times, incubated by biotin labeling secondary antibody for 20 min and rinsed by PBS, cultured by streptavidin that labeled by horseradish peroxidase at 37°C for 20 min. Rinsed by PBS, the sections were developed by alcohol ether carboxylate (AEC, ZSGB-Bio, Beijing, China) for 3–5 min and washed by tap water, then stained by hematoxylin for 5 min. Then, the sections were conducted with dehydration and xylene permeabilization, finally sealed. PBS was used to replace primary antibody and taken as blank control, the positive control that provided by ZSGB-Bio (Beijing, China) was taken as a positive control. The cells were counted by double blind method. The positive results of IGF1R performed as red cytoplasm, five random fields of view of each sections were selected to observed the positive expression of IGF1R.
Dual luciferase reporter gene assay
Human embryo kidney cell HEK293T was cultured in DMEM medium with 10% FBS at 37°C with 5%CO2. The IGF1R 3’-untranslated region (3’-UTR) fragment containing miR-139-5p binding sites was inserted in pmirGLO vector; IGF1R 3’-UTR fragment with mutation of binding sites was generated by method ofrite-directed mutagenesis, and then inserted into pmirGLO vector. PmirGLO-wild type (Wt) IGF1R and PmirGLO-mutant type (Mut) IGF1R recombinant vector and miR-139-5p mimic or miR-139-5p mimic-NC was cotransfected into HEK293T cells by the method of lipofection. The multifunctional microplate reader SpectraMax M5 was used to test the activity of renilla luciferase and firefly luciferase, respectively, the interval time was set as 2 s, and the test time was set as 10 s.
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-h-tetrazolium bromide (MTT) assay
Cells in the logarithmic growth phase were made to cell suspension of 1 × 105 cells/mL which was made by DMEM culture medium containing 10% FBS, then was seeded to 96-well plates, eight wells were set for every group with 100 μL in each well. Cells were cultured at 37°C with 5% CO2, the incubator was taken out at 4 h before the same time point on the 1st, 3rd, 5th and 7th d, and each well was added with 10 μL MTT solution (5 mg/mL, Sigma-Aldrich, St. Louis, MI, USA) to continue incubating for 4 h, then the incubation was ended and the supernatant was discarded, the optical density (OD) value of each well was tested by an automatic microplate reader (BIO-RAD, Hercules, CA, USA) at 492 nm. The experiment was repeated three times.
Bromodeoxyuridine (BrdU) staining
Cells in the logarithmic growth phase were selected to dissociated and centrifuged to cell suspension with 2 × 104cells/well, and was then seeded into wells with slides. At 12 h after cell adherence, the cells were changed to 1 mL serum-free DMEM medium for 72 h, and 10 μL BrdU (1 mg/mL) was added to the medium for incubation at 37°C for 1 h. Next, the 24-well plates were taken out and cells were fixed with polyformaldehyde for 30 min, treated with 2 mol/L HCL for 10 min, permeabilized with 1% Triton X-100 and sealed with 10% goat serum for 1 h. Subsequently, the cells were added with BrdU monoclonal antibody (1:300) and incubated at 37°C for 1.5 h, and the cells were then added with BrdU secondary antibody (1:300) in dark for 1 h, and washed three times with l × phosphate-buffered saline with Tween (PBST). The nucleus were stained by 4’,6-diamidino-2-phenylindole 2hci (DAPI) for 15 min, then a drop of anti-fluorescence quenching solution was added, and the cells were mounted in neutral balsam, then observed and captured under a fluorescent microscope.
Colony formation assay
As to the colony formation assay, the transfected cells were seeded to 6-well plates with the density of 400 cells each well and cultured for 7–14 d, then the culture was stopped when the colonies could be observed by eyes. The medium was washed twice by PBS, fixed with methanol for 30 min and stained with 0.1% crystal violet, then the imaging of bacterial colony was counted.
Hoechst 33258 staining
Cells in the logarithmic growth phase were detached and centrifuged into cell suspension, and was seeded in wells with slides at 2 × 104 cells/well. Twelve hours after adherence, the medium was changed to 1 mL serum-free DMEM medium and washed with PBS after stained for 12 h and 24 h separately. Afterward, the cells were fixed with 0.5 mL fixative solution for 10 min, then stained with 0.5 mL Hoechst 33258 staining solution for 5 min. Subsequently, the cells were added with a drop of anti-fluorescence quenching solution, sealed with neutral balsam, observed and photographed under a fluorescent microscope.
Flow cytometry
Propidium iodide (PI) single staining: cells were collected after 48-h transfection, which were then trypsinized with 0.25% trypsin at 4°C and centrifuged at 1000 r/min for 5 min with the supernatant discarded. The cells were resuspended with PBS, and the concentration of cells was regulated at 1 × 105 cells/mL. Next, the cells were fixed with 2 mL 75% ethanol at 4°C for 30 min, the cold ethanol was discarded after centrifugation, the supernatant was discarded after the cells were added with 100 µL RNase A without light exposure, water-bathed at 37°C for 30 min and added with 400 µL PI staining (P4170, Sigma-Aldrich, St. Louis, MI, USA). After avoidance of light at 4°C for 30 min, cell cycle which was detected by red fluorescence at excitation wavelength of 488 mm was recorded by flow cytometry (Beckman Coulter Fullerton, CA, USA).
Annexin V/PI double staining: after 48-h transfection, the cells were detached with 0.25% trypsin (no ethylene diaminetetraacetic acid) (PYG0107, Boster Biological Technology Co., Ltd., Wuhan, China), collected at a flow tube and centrifuged with the supernatant wan discarded. Annexin-V-fluoresceineisothiocyanate(FITC)/PI dye was made of Annexin-V-FITC, PI, and hydroxyethylpiperazineethanesulfonic acid (HEPES) buffer solution at 1:2:50 according to the instruction of Annexin-V-FITC apoptosis detection kit (K201-100, Biovision, Milpitas, CA, USA). Every 100 µL dye was resuspended with 1 × 106 cells, mixed up by shaking, incubated for 15 min, then added with 1 mL HEPES buffer solution and mixed up again. FITC and PI fluorescence was separately detected by the band-pass filter of 515 nm and 620 nm excited by 488 nm wavelength, and the apoptosis was observed.
Transwell assay
The chambers of Transwellthat were covered with matrigelwas preheated to 37°C, the transfected cells were diluted as the groups above and washed with serum-free culture solution twice, resuspended with serum-free culture solution and counted, with the density of cells adjusted to 1 × 105 cells/mL. The RPMI 1640 culture medium containing 2% FBS (600 µL) was added in basolateral chamber of Transwell. The cell suspension (200 mL) was added in apical chamber at 37°C for 48 h. The chamber was taken out and the cells at apical chamber membrane were erased, cells were fixed with 4% paraformaldehyde solution for 10 min, and finally, stained with crystal violet. The cells in randomly selected five visual fields were photographed, and counted under an optical microscope. Three replicate wells were set for each group.
Statistical analysis
All data analyses were conducted using SPSS 21.0 software (IBM Corp. Armonk, NY, USA). All data were subjected to normal distribution and homogeneity of variance test. The measurement data conforming to the normal distribution were expressed as mean ± standard deviation. The t-test was performed for comparisons between two groups and one-way analysis of variance (ANOVA) was used for comparisons among multiple groups, followed by Tukey's post hoc test. Statistical significance was set at p< 0.05.
Results
Mir-139-5p is poorly-expressed in malignant melanoma
The expression of melanoma tissues and adjacent normal skin tissues were detected by RT-qPCR to study the relation between miR-139-5p and malignant melanoma, the results of the experiment indicated that miR-139-5p was poorly expressed in malignant melanoma tissues in contrast to adjacent normal skin tissues (P < 0.05; Figure1(a)), indicating that there was differential expression of miR-139-5p in malignant melanoma. Additionally, the miR-139-5p expression of the primary normal skin melanocytes PIGI and five malignant melanoma cell lines (A375, SK-MEL-1, SK-MEL-2, SK-MEL-5, and SK-MEL-28) were evaluated by RT-qPCR, and the results implied that in comparison to PIGI cells, miR-139-5p was poorly expressed in the five malignant melanoma cell lines (all P< 0.05), and its expression was evidently declined in the A375 cell line, so the A375 cell line was adopted for the subsequent experiments (Figure 1(b)).
Figure 1.
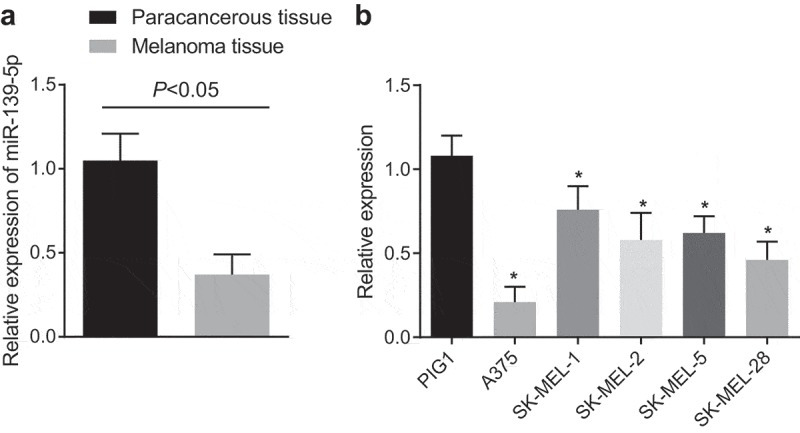
MiR-139-5p was down-regulated in malignant melanoma. (a), RT-qPCR was used to measure the miR-139-5p expression in malignant melanoma tissues and adjacent normal skin tissues; (b), RT-qPCR was used to screen the malignant melanoma cells; the data were all measurement data and expressed as mean ± standard deviation; the independent t-test was adopted for statistical analysis in figure (a); one-way ANOVA and Tukey’s post hoc test was adopted in figure (b); * P< 0.05vs PIGI cell line.
Overexpressed mir-139-5p suppresses the proliferation, migration and invasion, and enhances the apoptosis of malignant melanoma cells
MiR-139-5p was overexpressed (Figure 2(a)) to further understand whether miR-139-5p could take effects on malignant melanoma. MTT assay (Figure 2(b)), BrdU staining (Figure 2(c)) and colony formation assay (Figure 2(d)) were adopted to measure the development of malignant melanoma cells, and the results revealed that in A375 cells, the cell proliferation reduced in miR-139-5p mimic group (P < 0.05), which was relative to the miR-139-5p mimic-NC group. Flow cytometry (Figure 2(e,f)) and Hoechst 33258 staining (Figure 2(g)) suggested that the proportion of cells in G0/G1 phase was enhanced, the proportion of cells in S phase was significantly declined and the apoptosis was obviously increased (all P < 0.05) after miR-139-5p has been overexpressed. Besides, Transwell assay (Figure 2(h,i)) was adopted to detect the invasion and migration of cells, the results of which showed that invasion and migration of cells were suppressed in the miR-139-5p mimic group versus those in the miR-139-5p mimic-NC group (P < 0.05). Results above expressed for that the overexpressed miR-139-5p led to the suppressed proliferation, migration and invasion and induced apoptosis of malignant melanoma cells.
Figure 2.
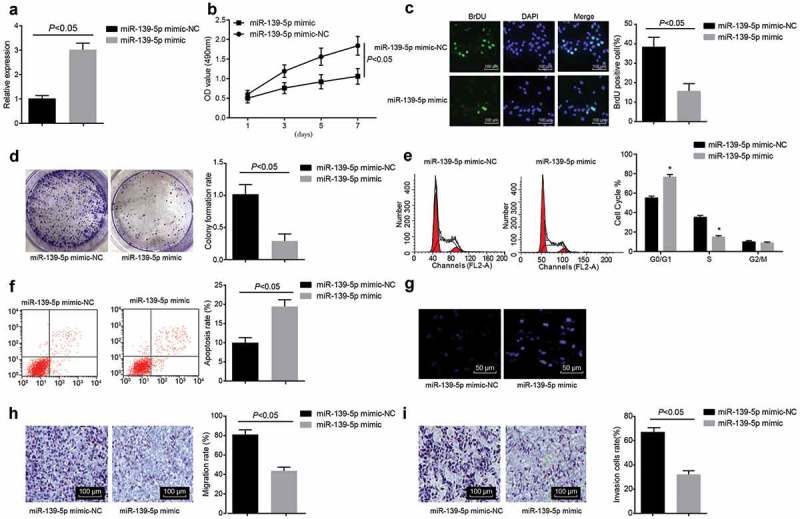
MiR-139-5p affected the biological functions of malignant melanoma cells. (a), RT-qPCR was used to evaluate the efficiency of miR-139-5p transfection (b-d), the cell proliferation was measured by MTT assay, BrdU staining and colony formation assay, respectively; (e), cell cycle distribution was detected by PI single staining; (f), cell apoptosis was evaluated by Annexin V/PI double staining; (g), morphological changes of cell apoptosis were observed by Hoechst 33,258 staining; (h-i), the changes of migration and invasion of cells were, respectively, detected by Transwell assay; data were all measurement data and expressed as mean ± standard deviation deviation, the independent t-test was adopted for statistical analysis.
Mir-139-5p targets and down-regulates the expression of IGF1R
We have found from the bioinformatics site (http://www.targetscan.org/vert_71/) that there were binding sites between miR-139-5p and IGF1R (Figure 3(a)). Meanwhile, dual-luciferase reporter gene assay was carried out to test whether IGF1R was the target gene of miR-139-5p (Figure 3(b)). The results unraveled that the luciferase activity of IGF1R-Wt was apparently inhibited in the miR-139-5p mimic group relative to the miR-139-5p mimic-NC group (P < 0.05), while there was no significant effect on luciferase activity of IGF1R-Mut (P > 0.05). Besides, RT-qPCR and Western blot analysis were used to measure the IGF1R expression in tissues, the results reflected that the expression level of IGF1R in malignant melanoma tissues was markedly enhanced (Figure 3(c,d)), which was relative to adjacent normal skin tissues. Results above implied that miR-139-5p could target and down-regulate the expression of IGF1R. The outcomes of immunohistochemical staining unraveled that IGF1R was mainly expressed in the cytoplasm of the malignant melanoma cells, and positive cells were in red cytoplasm. We could find that the cytoplasm was stained into red by AEC, while the nuclei and cytomembrane were not in red (Figure 3(e)), which was in broad contrast to the surrounding negative cells. In comparison to adjacent normal skin tissues, the rate of positive expression of IGF1R in malignant melanoma tissues was considerably elevated (Figure 3(f), P < 0.05).
Figure 3.
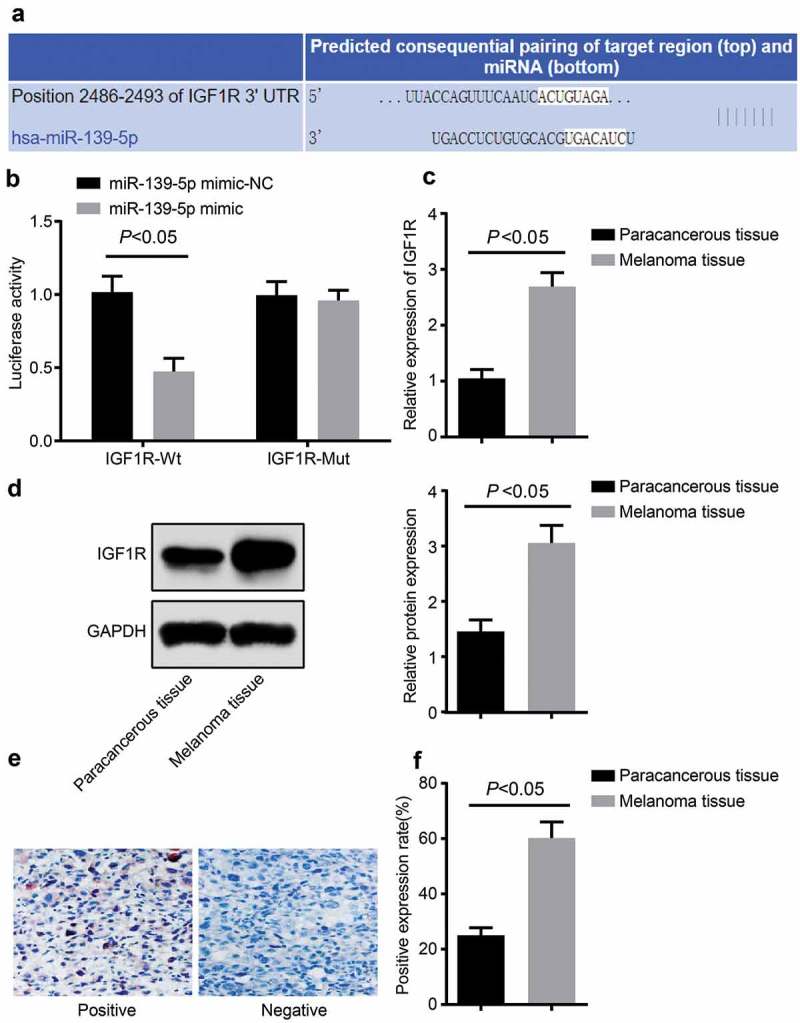
MiR-139-5p targeted and down-regulated IGF1R. (a), the bioinformatics site was used to forecast the binding sites of miR-139-5p and IGF1R; (b), the dual-luciferase reporter gene assay was used to detect the targeted relation between IGF1R and miR-139-5p; C, RT-qPCR was used to measure the mRNA expression level of IGF1R in tissues; (d), western blot analysis was used to detect the expression level of IGF1R protein in tissue; (e), immunohistochemical staining of IGF1R protein in tissues; (f), the rate of positive expression of IGF1R protein in tissues. Data were all measurement data and expressed as mean ± standard deviation; the independent t-test was adopted for statistical analysis; * P< 0.05vs the miR-139-5p mimic-NC group; # P< 0.05vsadjacent normal skin tissues.
Overexpression of mir-139-5p represses the proliferation, migration, and invasion and enhances the apoptosis of malignant melanoma cells through down-regulating IGF1R
To further investigate whether miR-139-5p laid effects on malignant melanoma by down-regulating IGF1R, and to intuitively explore the effects that the IGF1R expression laid on malignant melanoma, we overexpressed miR-139-5p and IGF1R simultaneously, the results implied that in A375 cells, the expression of IGF1R was increased (Figure 4(a)), and the cell proliferative capacity was markedly advanced (Figure 4(b,c)), the proportion of cells in G0/G1 phase inhibited, the proportion of cells in S phase was augmented (Figure 4(d)), the cell apoptosis was noticeably decreased (Figure 4(e)), while the cell invasion and migration capacity were observably amplified (Figure 4(f,g)) in the oe-IGF1R group in contrast to the oe-NC group (all P < 0.05). Compared with the oe-IGF1R group, the expression of IGF1R was decreased, the cell proliferation capacity was evidently alleviated, the proportion of cells in G0/G1 phase was apparently advanced and the proportion of cells in S phase was considerably repressed, the apoptosis was substantially accelerated, and the cell invasion and migration capacity were definitely suppressedin the miR-139-5p mimic + oe-IGF1R group (all P < 0.05). These results illuminated that miR-139-5p could down-regulate IGF1R and inhibit the development of malignant melanoma cells.
Figure 4.
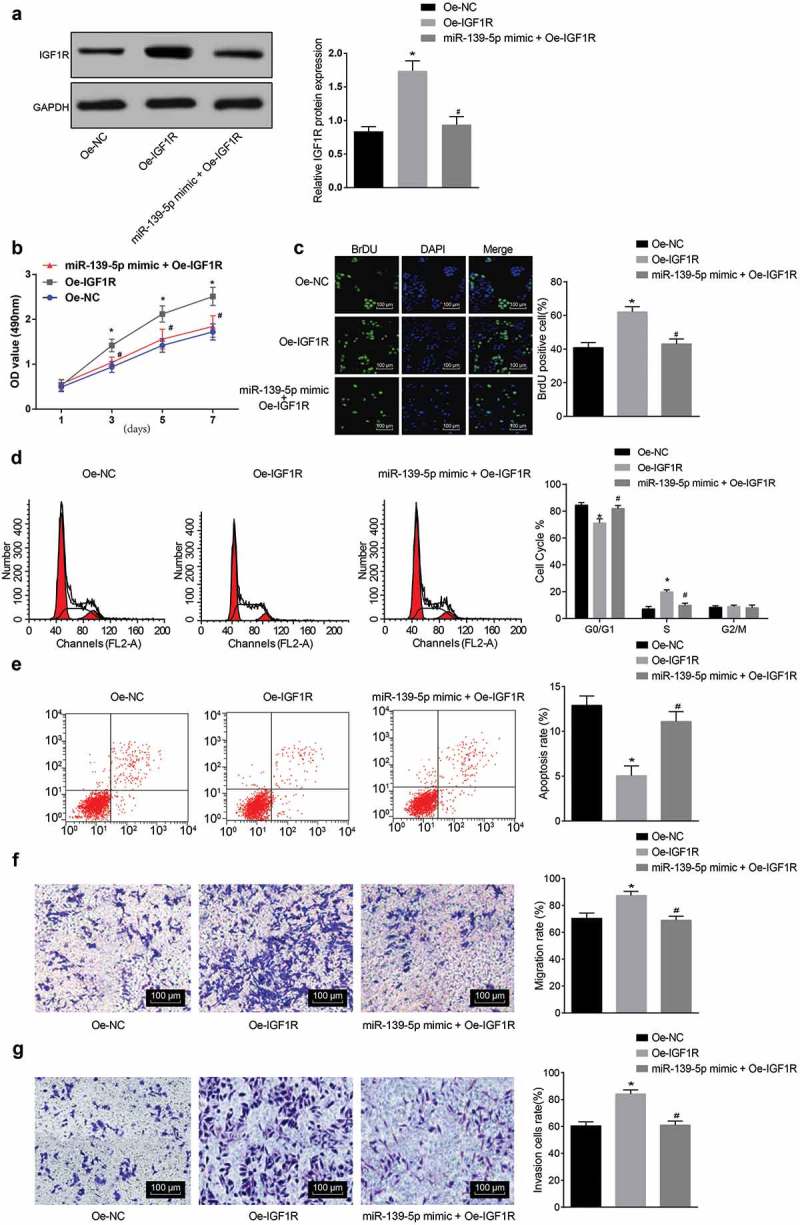
MiR-139-5p affected the biological functions of malignant melanoma cells by regulating IGF1R. (a), protein expression of IGF1R in cells of each group; (b-c), cell proliferation was evaluated by MTT assay and BrdU staining; (d), cell cycle distribution was detected with PI single staining; (e), cell apoptosis condition of groups was detected with Annexin V/PI double staining; (f-g), changes of cell migration and invasion capacity were, respectively, detected with Transwell assay; data were all measurement data and expressed as mean ± standard deviation; ANOVA was used for comparisons among multiple groups, followed by Tukey’s post hoc test;* P< 0.05vs the oe-NC group; # P< 0.05vs the oe-IGF1R group.
The activation of PI3K/AKT signaling pathway is advanced by IGF1R
Researches have proved that the PI3K/AKT signaling pathway was tightly associated with melanoma [18,19], so we discussed whether IGF1R could make effects on the PI3K/AKT signaling pathway and eventually affect the disease. We adopted Western blot analysis to determine the expression of PI3K, AKT and its phosphorylation products p-PI3K, p-AKT, which were relative factors of the PI3K/AKT signaling pathway. The obtained results revealed that in A375 and SK-MEL-1 cells, compared with the oe-NC group, expression of p-PI3K/t-PI3K and p-AKT/t-Aktwas broadly heightened in the oe-IGF1R group (P < 0.05). Results above demonstrated that IGF1R could activate the PI3K/AKT signaling pathway. To further elucidate whether miR-139-5p could reverse the promotive impacts of IGF1R on the PI3K/AKT signaling pathway, Western blot analysis was selected to determine the expression of PI3K, Akt, p-PI3K and p-AKT after overexpressed miR-139-5p was combined with overexpressed IGF1R. The outcomes indicated that in A375 and SK-MEL-1 cells, relative to the oe-IGF1R group, the expression of p-PI3K/t-PI3K and p-AKT/t-AKT was evidently lowered in the miR-139-5p mimic + oe-IGF1R group (Figure 5(a,b), P < 0.05).
Figure 5.
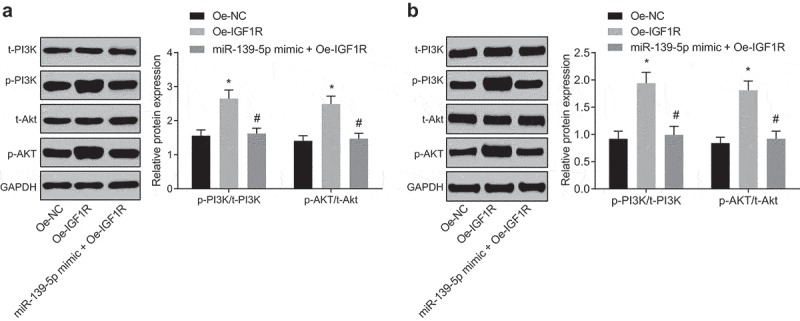
The expression of relative factors of the PI3K/AKT signaling pathway was measured by western blot analysis. (a), the expression of related factors of the PI3K/AKT signaling pathway in A375 cells of each group; (b), the expression of related factors of the PI3K/AKT signaling pathway in SK-MEL-1 cells of each group. Data were all measurement data and expressed as mean ± standard deviation; ANOVA was used for comparisons among multiple groups, followed by Tukey’s post hoc test; * P< 0.05vs the oe-NC group, # P< 0.05 vs the oe-IGF1R group.
Discussion
Malignant melanoma has an increasing tendency of poor survival rates, which is an aggressive skin cancer [20]. What has been verified is that the miRNAs, that were known as small non-coding RNAs, played a part of leading molecules in the RNA silencing [21]. Furthermore, there were many recent studies have revealed that the aberrant expression of particular miRNAs maybe related with human disease, such as breast cancer [22], lung cancer [23] and prostate cancer [24]. Particularly, miR-139-5p has been revealed to be instrumental in preventing malignant melanoma, which could be the candidates with enough sensitivity and specificity for cancer diagnosis in a clinical setting [11]. Nevertheless, there is still no literature about the connection between miR-139-5p and malignant melanoma, took which for the reason, this study was focused on the role of miR-139-5p, one of the miRNAs, and IGF1R in malignant melanoma. This study has provided the evidence to prove that the development of malignant melanoma could be suppressed by miR-139-5p through the PI3K/AKT signaling pathway by means of down-regulating IGF1R.
One of the most important findings in our study suggested that miR-139-5p was poorly expressed in malignant melanoma, indicating that there were an aberrant expression of miR-139-5p in malignant melanoma. Similar to our study, miR-139-5p was proved to be down-regulated in breast cancer [25] and bladder cancer [26]. Moreover, it was obvious in our study that the overexpressed miR-139-5p could apparently restrain proliferation, migration, and invasion of malignant melanoma cells, and promote its cell apoptosis. It also could be found in other literatures that the overexpressed miR-139-5p has the ability to reduce the cell migration and invasion in hepatocellular carcinoma [27] and colorectal cancer [28]. Meanwhile, Huang #60;italic#62;et al.#60;/italic#62; have stated that elevated miR-139-5p is capable of repressing cell migration and invasion in vitroalong with metastasis in vivo, and it has the ability to act as a molecular therapeutic target [29]. These evidence opened up a new way for pointing out the molecular mechanisms of miR-139-5pin malignant melanoma.
Additionally, our study has made it clear that miR-139-5p could target and down-regulate the expression of IGF1R, and the overexpressed miR-139-5pwas able to considerably restrain the cell proliferation, migration, and invasion of malignant melanoma, and advance the apoptosis of malignant melanoma cells by way of down-regulating IGF1R expression. As previously reported, IGF1R is upregulated by malignant melanomas in contrast to benign nevi, and could able to control proliferation, motility and apoptosis of malignant melanoma cells [30]. Wei Xu et al. have also proved that the cell proliferation, migration, and invasion in vitro of lung cancer could be inhibited by the miR-139-5p expression, and it has been revealed in the study that non-small cell lung cancer could be suppressed by miR-139-5p through down-regulating the IGF1R expression [31]. Similarly, another study has verified that function as a metastasis suppressor, miR-139 was corresponded with IGF1R, and cell invasion and metastasis might be suppressed by miR-139 through binding to IGF1R [31]. It has also been revealed that miR-139 is overexpressed in osteoarthritis and represses proliferation and migration of chondrocytes possibly via restricting IGF1R [32]. Another vital finding in our study was that IGF1R could enhance the activation of the PI3K/AKT signaling pathway, and miR-139-5p could inhibit the activation of the PI3K/AKT signaling pathway by means of down-regulating IGF1R. Consistent with our result, Zhang et al. have demonstrated that the PI3K/AKT signaling pathway could be activated by IGF1R/IGF1 to advance the survival of glioblastoma multi forme cells [33]. Furthermore, it has been verified that IGF1R binding its cognate ligand could promote a multistep activation process in the PI3K signaling pathway, by which the signaling could be transduced to P13K [34]. As to the relation between miR-139-5p and PI3K/AKT signaling, it has been identified that miR-139-5p could inhibit 3T3-L1 (mouse preadipocytes) differentiation the by IRS1/PI3K/AKT signaling pathway [35]. These data implied that miR-139-5p could regulate the development and metastasis of malignant melanoma cells through the PI3K/AKT signaling pathway by targeting IGF1R. Furthermore, we also found that miR-139-5p could reverse the promotive impacts of IGF1R on the PI3K/AKT signaling pathway, which was reflected by the outcomes that in A375 and SK-MEL-1 cells, relative to the oe-IGF1R group, the expression of p-PI3K/t-PI3K and p-AKT/t-AKT was evidently lowered in the miR-139-5p mimic + oe-IGF1R group.
Conclusion
To sum up, our study has proved that miR-139-5p could regulate the development and metastasis of malignant melanoma cells through the PI3K/AKT signaling pathway by targeting IGF1R. Yet more efforts, such as enlarging the sample size, supplementing the animal experiment or verify our results, are required to better elucidate the function mechanisms of miRNAs in the progression of malignant melanoma, which could undoubtedly enhance our understanding in the progression of malignant melanoma and further facilitate the development of microRNA-directed diagnosis and therapy for malignant melanoma.
Funding Statement
We would like to acknowledge the reviewers for their helpful comments on this paper, and the project was supported by the Department of Science and Technology of Guangdong Province (No: 2014A020212434).
Authors’ contributions
Guarantor of integrity of the entire study:Chaoying Yang, ZhaoxiaXia, Liangcai Wu
Study design:Chaoying Yang, ZhaoxiaXia, Liangcai Wu
Definition of intellectual content:Chaoying Yang, Zhaoxia Xia
Experimental studies:Lifei Zhu, Yanchang Li, ZhixinZheng
Data analysis:Lifei Zhu, Yanchang Li, ZhixinZheng
Manuscript editing:Chaoying Yang, Zhaoxia Xia, Jianying Liang
Disclosure statement
No potential conflict of interest was reported by the authors.
Ethical statement
The experiment was approved by The Sixth Affiliated Hospital of Sun Yat-senUniversity.
References
- [1].Kappelmann M, Bosserhoff A, Kuphal S.. AP-1/c-Jun transcription factors: regulation and function in malignant melanoma. Eur J Cell Biol. 2014;93(1–2):76–81. [DOI] [PubMed] [Google Scholar]
- [2].Dvorankova B, Szabo P, Kodet O, et al. Intercellular crosstalk in human malignant melanoma. Protoplasma. 2017;254(3):1143–1150. [DOI] [PubMed] [Google Scholar]
- [3].Higgins HW 2nd, Lee KC, Galan A, et al. Melanoma in situ: part I. Epidemiology, screening, and clinical features. J Am Acad Dermatol. 2015;73(2):181–90, quiz 191–2. [DOI] [PubMed] [Google Scholar]
- [4].Kulichova D, Dáňová J, Kunte C, et al. Risk factors for malignant melanoma and preventive methods. Cutis. 2014;94(5):241–248. [PubMed] [Google Scholar]
- [5].Crump C, Sundquist K, Sieh W, et al. Season of birth and other perinatal risk factors for melanoma. Int J Epidemiol. 2014;43(3):793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ding Z, Jian S, Peng X, et al. Loss of MiR-664 Expression Enhances Cutaneous Malignant Melanoma Proliferation by Upregulating PLP2. Medicine (Baltimore). 2015;94(33):e1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen Y, Zhang Z, Luo C, et al. MicroRNA-18b inhibits the growth of malignant melanoma via inhibition of HIF-1alpha-mediated glycolysis. Oncol Rep. 2016;36(1):471–479. [DOI] [PubMed] [Google Scholar]
- [8].Mimeault M, Batra SK. Novel biomarkers and therapeutic targets for optimizing the therapeutic management of melanomas. World J Clin Oncol. 2012;3(3):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jiao J, Fan Y, Zhang Y. Expression and clinicopathological significance of microRNA-21 and programmed cell death 4 in malignant melanoma. J Int Med Res. 2015;43(5):672–678. [DOI] [PubMed] [Google Scholar]
- [10].Zhang D, Han Y, Xu L. Upregulation of miR-124 by physcion 8-O-beta-glucopyranoside inhibits proliferation and invasion of malignant melanoma cells via repressing RLIP76. Biomed Pharmacother. 2016;84:166–176. [DOI] [PubMed] [Google Scholar]
- [11].Zhang HD, Sun DW, Li J, et al. MiR-139-5p: promising biomarker for cancer. Tumour Biol. 2015;36(3):1355–1365. [DOI] [PubMed] [Google Scholar]
- [12].Shen K, Mao R, Ma L, et al. Post-transcriptional regulation of the tumor suppressor miR-139-5p and a network of miR-139-5p-mediated mRNA interactions in colorectal cancer. Febs J. 2014;281(16):3609–3624. [DOI] [PubMed] [Google Scholar]
- [13].Sun J, Li W, Sun Y, et al. A novel antisense long noncoding RNA within the IGF1R gene locus is imprinted in hematopoietic malignancies. Nucleic Acids Res. 2014;42(15):9588–9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang J, Sinnberg T, Niessner H, et al. PTEN regulates IGF-1R-mediated therapy resistance in melanoma. Pigment Cell Melanoma Res. 2015;28(5):572–589. [DOI] [PubMed] [Google Scholar]
- [15].Martini M, De Santis MC, Braccini L, et al. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46(6):372–383. [DOI] [PubMed] [Google Scholar]
- [16].Bai M, Zhang M, Long F, et al. Circulating microRNA-194 regulates human melanoma cells via PI3K/AKT/FoxO3a and p53/p21 signaling pathway. Oncol Rep. 2017;37(5):2702–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Caporali S, Alvino E, Lacal PM, et al. Targeting the PI3K/AKT/mTOR pathway overcomes the stimulating effect of dabrafenib on the invasive behavior of melanoma cells with acquired resistance to the BRAF inhibitor. Int J Oncol. 2016;49(3):1164–1174. [DOI] [PubMed] [Google Scholar]
- [18].Brighton HE, Angus SP, Bo T, et al. New mechanisms of resistance to MEK inhibitors in melanoma revealed by intravital imaging. Cancer Res. 2018;78(2):542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Redmer T. Deciphering mechanisms of brain metastasis in melanoma - the gist of the matter. Mol Cancer. 2018;17(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sahni S, Valecha G, Sahni A. Role of Anti-PD-1 antibodies in advanced melanoma: the era of immunotherapy. Cureus. 2018;10(12):e3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. [DOI] [PubMed] [Google Scholar]
- [22].Egeland NG, Lunde S, Jonsdottir K, et al. The Role of MicroRNAs as Predictors of Response to Tamoxifen Treatment in Breast Cancer Patients. Int J Mol Sci. 2015;16(10):24243–24275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2015;34(27):3547–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fabris L, Ceder Y, Chinnaiyan AM, et al. The potential of MicroRNAs as prostate cancer biomarkers. Eur Urol. 2016;70(2):312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang H-D, Sun D-W, Mao L, et al. MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem Biophys Res Commun. 2015;465(4):702–713. [DOI] [PubMed] [Google Scholar]
- [26].Yonemori M, Seki N, Yoshino H, et al. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016;107(9):1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wong CC, Wong C, Tung EK, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140(1):322–331. [DOI] [PubMed] [Google Scholar]
- [28].Song M, Yin Y, Zhang J, et al. MiR-139-5p inhibits migration and invasion of colorectal cancer by downregulating AMFR and NOTCH1. Protein Cell. 2014;5(11):851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Huang -L-L, Huang LW, Wang L, et al. Potential role of miR-139-5p in cancer diagnosis, prognosis and therapy. Oncol Lett. 2014;14(2):1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yeh AH, Bohula EA, Macaulay VM. Human melanoma cells expressing V600E B-RAF are susceptible to IGF1R targeting by small interfering RNAs. Oncogene. 2006;25(50):6574–6581. [DOI] [PubMed] [Google Scholar]
- [31].Xu W, Hang M, Yuan C-Y, et al. MicroRNA-139-5p inhibits cell proliferation and invasion by targeting insulin-like growth factor 1 receptor in human non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(4):3864–3870. [PMC free article] [PubMed] [Google Scholar]
- [32].Hu W, Zhang W, Li F, et al. miR-139 is up-regulated in osteoarthritis and inhibits chondrocyte proliferation and migration possibly via suppressing EIF4G2 and IGF1R. Biochem Biophys Res Commun. 2016;474(2):296–302. [DOI] [PubMed] [Google Scholar]
- [33].Zhang M, Liu J, Li M, et al. Insulin-like growth factor 1/insulin-like growth factor 1 receptor signaling protects against cell apoptosis through the PI3K/AKT pathway in glioblastoma cells. Exp Ther Med. 2018;16(2):1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhou Y, Geng P, Liu Y, et al. Rotavirus-encoded virus-like small RNA triggers autophagy by targeting IGF1R via the PI3K/Akt/mTOR pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864(1):60–68. [DOI] [PubMed] [Google Scholar]
- [35].Jiang C, Tong Z, Fang W-L, et al. Microrna-139-5p inhibits epithelial-mesenchymal transition and fibrosis in post-menopausal women with interstitial cystitis by targeting LPAR4 via the PI3K/Akt signaling pathway. J Cell Biochem. 2018;119(8):6429–6441. [DOI] [PubMed] [Google Scholar]


