ABSTRACT
Preeclampsia (PE) is a pregnancy-specific syndrome that substantially leads to maternal and fetal mortality. Multiple factors contribute to the disease, but the exact pathogenesis still remains elusive. Here we explored the roles of lncRNA MALAT1 and miR-206 in PE. qRT-PCR was applied to measure mRNA levels of MALAT1 and miR-206 in the placenta of PE patients. Scratch wound healing assay and transwell invasion assay were conducted to test the effects of MALAT1 and miR-206 on migration and invasion of trophoblast cells. In addition, we validated MALAT1/miR-206 and miR-206/IGF-1 interactions with dual luciferase reporter assay. Western bot was used to detect protein expressions of IGF-1, p-PI3K, PI3K, p-Akt and Akt. We found that MALAT1 was decreased but miR-206 was increased in the placenta of patients with PE. Inhibition of MALAT1, knockdown IGF-1, or miR-206 mimics suppressed the trophoblast cells migration and invasion, while overexpression of MALAT1, IGF-1 or miR-206 inhibitors exhibited opposite effects. Further, miR-206 was confirmed as a direct target of MALAT1. Besides, miR-206 inhibited IGF-1 expression by directly binding to the 3’UTR. Mechanistically, our study demonstrated that MALAT1 regulates IGF-1/PI3K/Akt signaling via miR-206. Together, these results suggest that MALAT1 and miR-206 play important roles in PE. MALAT1 regulates miR-206/IGF-1 axis, thereby modulating trophoblast cells migration and invasion through PI3K/Akt signal pathway. These results show light on the underlying mechanisms of PE and provide potential targets for PE therapy.
Abbreviations: PE: Preeclampsia; lncRNA: Long-non-coding RNA; MALAT1: Metastasis-associated lung adenocarcinoma transcript 1; IGF-1: Insulin-like growth factor 1; PI3k: Phosphatidylinositol-4, 5-bisphosphate 3-kinase; Akt: Protein kinase B; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase; qRT-PCR: Quantitative Reverse Transcription polymerase chain reaction; shRNA: Short hairpin RNA; siRNA: Small interfering RNA; EMT: Epithelial-mesenchymal transition
KEYWORDS: PE, MALAT1, miR-206
Introduction
Preeclampsia (PE) is a multisystem pregnancy disorder usually developed after 20 wks of gestation with symptoms of hypertension and proteinuria or pulmonary edema [1,2]. It affects 3–5% of pregnancies worldwide and is one of the leading causes of maternal, fetal and neonatal morbidity and mortality. Currently, the most effective treatment for PE is delivery, but it depends on the maturity of fetus and may not be suitable in certain conditions [3]. The pathogenesis of PE is not fully elucidated although many factors are believed to contribute to PE, including trophoblastic dysfunction, such as inadequate migration and invasion, defective spiral artery remodeling, systematic inflammation [2]. Therefore, exploring the underlying molecular mechanisms is very necessary, which helps provide more effective treatments for PE.
Long-non-coding RNAs (lncRNAs) are a class of noncoding RNAs that are more than 200 nucleotides in length [4]. Many of them have been reported to play key roles in regulating gene expressions at both transcriptional and post-transcriptional levels, and thus were implicated in various cellular processes including cell proliferation and apoptosis, cell-cycle progression and chromatin regulation [5,6]. Moreover, they have been shown involved in many diseases such as cancers and neurological disorders as well as PE [7,8]. For example, lncRNA DLX6-AS1 was observed increased in PE patients [9] while lncRNA 00473 was shown decreased [10]. The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a large and ubiquitously expressed lncRNA that played crucial roles in various cancers via multiple mechanisms [11]. Recently, overexpression of MALAT1 was observed in placenta previa increta/percreta [12]. However, MALAT1 has been shown down-regulated in PE [13]. It implies that MALAT1 might has some crucial functions in PE, though the exact function of MALAT1 in trophoblast cells is largely unknown.
MicroRNAs (miRNAs) are a class of endogenous, small and non-coding RNAs (19–25 nucleotides) that regulate gene expressions preferentially by binding to the 3’-untranslated regions (3’UTR) of target genes [14]. It is well known that miRNAs play essential roles in diverse physiological processes as well as pathological processes. Recently, emerging studies have observed dysregulated miRNAs in PE patients, indicating that miRNAs are new players in the pathobiology of PE [15–17]. Importantly, a study from Delles group shows that miR-206 is significantly increased in PE patients [18], suggesting a potential role of miR-206 in PE, despite that how miR-206 contributes to PE remains elusive.
In this study, we fully investigated the functions of MALAT1 and miR-206 in PE. We confirmed that MALAT1 was decreased while miR-206 was increased in PE patients. Moreover, we showed that MALAT1 directly bound to miR-206 in trophoblast cells. Knockdown MALAT1 depressed trophoblast cells migration and invasion, while miR-206 inhibitor had opposite effects and reversed the effect of MALAT1 inhibition. Mechanistically, we found that IGF-1 was the downstream target of miR-206 and knockdown IGF-1 also suppressed invasion of trophoblast cells. MALAT1 modulated IGF-1/PI3K/Akt signaling via miR-206. Together, these findings reveal that lncRNA MALAT1 acts as a sponge of miR-206 to activate IGF-1/PI3K/Akt pathway, thereby regulates migration and invasion of trophoblast cells.
Materials and methods
Patients and placental tissues
A total of 60 pregnant women (30 patients diagnosed as preeclampsia and 30 age-matched normal pregnant women) hospitalized in Department of Obstetrics, Henan Provincial People’s Hospital between August 2017 and November 2018 were enrolled in this study. There was no significant difference in clinical manifestations between two groups. Patients were excluded from the study if they have the following symptoms or history: 1) smoking; 2) alcohol drinking; 3) substance abuse; 4) multiple pregnancy; 5) gestational stage < 38 wks; 6) body mass index (BMI) > 35; 7) congenital heart disease; 8) chronic hypertension; 9) chronic nephritis; 10) diabetes; 11) thyroidism dysfunction; 12) metabolic syndrome.
Human placental tissues were immediately collected when the placenta was delivered. About 1 cm3 placental tissues were cut from the middle of maternal side and calcification and bleeding sites were avoided. All samples were immediately snap-frozen in liquid nitrogen after resection. A written informed consent was signed and obtained from each patient. This study was reviewed and approved by the Ethics Committee of Henan Provincial People’s Hospital.
Cell culture
Trophoblast cell lines HTR-8/SVneoc, JEG-3 were purchased from American Type Culture Collection (ATCC; Manassas, USA). HTR-8/SVneo and JEG-3 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, MA, USA) and 1% pen-strep. All cells were maintained in a humidified atmosphere containing 5% CO2.
RNA extraction and qRT-PCR
Total RNA was extracted from the cells or placenta tissue (about 90 mg) using RNA extract kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. DNaseI was added to avoid potential DNA contamination. 1 μg total RNA from each sample was used to reverse transcription using the SuperScript kit (Invitrogen, Massachusetts, USA) followed by PCR amplification with Power SYBR Green PCR Master Mix (Thermo Fisher, MA, USA). qPCR analysis was then carried out. Briefly, SYBR Green qPCR assay (Takara, China) and gene-specific primers were used for qPCR with GAPDH or U6 used for normalization. The relative expression levels of RNAs were calculated through ΔΔCt. The following primers were used for analysis:
MALAT1 forward: 5′-AAAGCAAGGTCTCCCCACAAG-3′;
MALAT1 reverse: 5′ -GGTCTGTGCTAGATCAAAAGGCA-3′.
miR-206 forward: 5′-TGCGCTGGAATGTAAGGAAGT-3′;
miR-206 reverse: 5′-CGGCCCAGTGTTCAGACTAC-3′.
IGF-1 forward: 5′-GCTGAAGCCGTTCATTTAGC-3′;
IGF-1 reverse: 5′-GAGGAGGCCAAATTCAACAA-3′.
GAPDH forward: 5′-AGGTCGGTGTGAACGGATTTG-3′;
GAPDH reverse: 5′-GGGGTCGTTGATGGCAACA-3′.
U6 forward: 5′-TGCGTTCCCTTTGTCATCCT-3′;
U6 reverse: 5′-AACGCTTCACGAATTTGCGT-3′.
Western blot
Cultured cells were lysed with RIPA buffer with protease inhibitor cocktail to extract proteins. Protein concentration was measured by BCA protein Assay (Thermo Fisher Scientific, Massachusetts, USA). Equal amount of protein from each sample was loaded into the polyacrylamide gel and separated by SDS-PAGE and then transferred to PVDF membranes (Millipore, Massachusetts, USA). The membranes were blocked with 3% BSA for half an hour and then incubated with primary antibodies overnight. The following antibodies were used: IGF-1 (1: 1000; Abcam, USA), PI3K (1: 1000; Invitrogen, USA), p-PI3K (Tyr458) (1:1000; Thermofisher, USA), Akt (1:2000, Cell signal, USA), p-Akt (Ser473) (1:2000, Cell Signal, USA), GAPDH (1:5000, Abcam, USA). The membranes were incubated with secondary antibodies for 1 h at room temperature after 3 times wash with Tris-buffered saline with 0.1% (vol/vol) Tween-20 (TBST). Blots were washed again and developed with film.
Plasmid construction and cell transfection
MALAT1 shRNA, miR-206 mimics, negative control (NC) mimic and IGF-1 si-RNA were synthesized by Genepharm (Shanghai, China). Their corresponding sequences are listed as follows. For overexpression of MALAT1, IGF-1, full-length of the MALAT1 or IGF-1 was subcloned into the Sall and Haml sites of pcDNA3.1 (Invitrogen, USA) vector (pcDNA3.1-MALAT1 [p-MALAT1]) and the empty vector pcDNA3.1-NC was used as control.
MALAT1 shRNA sense: 5-GAGGUGUAAAGGGAUUUAUTT-3′;
MALAT1 shRNA anti-sense: 5-AUAAAUCCCUUUACACCUCTT-3;
IGF-1 siRNA sense: 5′-TGAGACCTGAAAGGAAGCGGAGATTC-3′;
IGF-1 siRNA anti-sense: 5’-GAATCTCCGCTTCCTTTCAGGTCTCA-3′;
si-NC sense: 5-GGCCUAAAGUAGUAGCUAUTT-3;
si-NC antisense: 5-AUAGCUACUACUUUAGGCCTT-3.
Transfection was conducted using Lipofectamine 2000 (Invitrogen, Missouri, USA) according to manufacturer’s instruction. Briefly, cells were grown until 60~80% confluence. 1 μg of plasmid was used for transfection together with 1μl Lipofectamine 2000 and subsequent experiments were conducted after 24 h of transfection.
Transwell invasion assay
Transfected or control non-transfected cells were plated on the upper part of a transwell chamber (pre-coated with Matrigel) in serum-free DMEM, and DMEM plus 10% FBS was added in the lower part of the chamber as chemoattractant. After 24 h of incubation at 37°C, cells inside the upper part of the chamber were removed away. Migrated cells on the lower part of the chamber were analyzed by crystal violet staining with the microscope.
Scratch wound healing assay
Transfected or control non-transfected cells were seeded in a 6-well plate and grown to about 70%-80% confluence. The monolayer cells were scratched by a plastic tip in a straight line and washed with PBS subsequently to remove cell debris. The scratched cells were then cultured in a serum-free medium and put back in the incubator. Images were taken with a phase-contrast microscope at 0 h and 24 h after scratch.
Luciferase report assay
To explore the potential binding partners of MALAT, we used StarBase (http://starbase.sysu.edu.cn/index.php) and confirmed the complementary binding sites between MALAT1 and miR-206. To find the potential downstream targets of miR-206, we employed Targetscan (http://www.targetscan.org/vert_72/) and found that IGF-1 was one of the targets. The amplified fragments of cDNAs containing the wild type (WT) or mutant (MUT) binding sites of miR-206 in MALAT1 or IGF-1 3’ UTR were amplified by PCR and were cloned into the Sall and bamHl restriction sites (Promega, USA) of the luciferase report gene of pmirGLO. Mutants were created using KOD Plus Mutagenesis kit (TOYOBO, Osaka, Japan) according to the manufacturers’ instructions. Trophoblast cells were cultured in 24-well culture plates for 12 h and then recombinant plasmids together with miR-206 mimic or NC were co-transfected into trophoblast cells. The co-transfected cells were lysed using the Reporter Lysis Buffer, and the luciferase activity was measured by the Luciferase Reporter Gene Assay Kit (Dual-GloTM Luciferase Assay System, Promega, Madison, WI, USA).
Statistical analysis
Three replicate wells were used per sample in each experiment and experiments were replicated three times. All statistical analyzes were performed with GraphPad Prism 7 (Graphpad Software Inc., USA). Statistical significances were determined by unpaired two-tailed Student t test for two groups or one-way ANOVA followed by Tukey’s post hoc test for multiple groups. Spearman’s correlation analysis was used for the determination of correlation between two parameters. *P < 0.05, **P < 0.01, ***P < 0.001. Data were presented as Mean ± SD (standard deviation).
Results
MALAT1 is down-regulated while miR-206 is up-regulated in placenta of PE patients
To evaluate the roles of MALAT1 and miR-206 in PE, we first examined expressions of MALAT1 and miR-206 in PE patients and normal pregnant women. Using qRT-PCR, we found that MALAT1 level was significantly decreased in the placenta of PE patients compared to normal pregnant women (Figure 1(a)). However, there was a significantly higher level of miR-206 in patients with PE (Figure 1(b)). These results indicate that MALAT1 is down-regulated but miR-206 is up-regulated in PE. Moreover, we also examined the relationship between MALAT1 level and miR-206 level in PE patients, and a strong negative correlation was detected, suggesting that MALAT1 and miR-206 might have a negative regulatory relationship.
Figure 1.
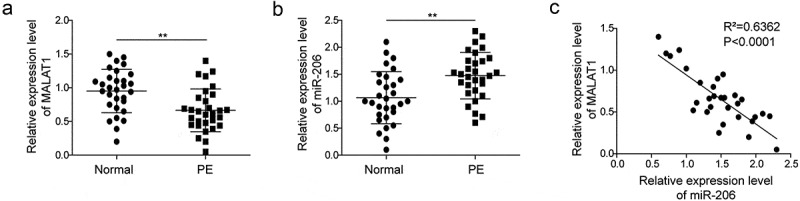
MALAT1 is down-regulated while miR-206 is up-regulated in the placenta of PE patients.
(a) qRT-PCR analysis of MALAT1 level in the placenta of normal pregnant women (n = 30) and patients with PE (n = 30). (b) qRT-PCR analysis of miR-206 level in the placenta of normal pregnant women and patients with PE. (c) Correlation between MALAT1 level and miR-206 level in PE patients. Three replicate wells were used per sample in each experiment and experiments were replicated three times. **P < 0.01
Inhibition of MALAT1 suppresses migration and invasion of trophoblasts while overexpression enhances
To further investigate the functions of MALAT1 in PE, human trophoblast cell lines, HTR-8/Svneo and JEG-3 were employed in this study [19]. We manipulated MALAT1 level through shRNA and overexpression and examined their effects on trophoblast cell migration and invasion. First, we confirmed that cells transfected with MALAT-1 shRNA had much lower expressions of MALAT1 compared with non-transfected cells and cells transfected with sh-NC, while cells transfected with p-MALAT1 had higher levels of MALAT1. (Figure 2(a,b)). Using scratch would healing assay, we observed that cells transfected with MALAT1 shRNA had significantly smaller migration areas than control cells, but cells with overexpression of MALAT1 showed larger migration areas (Figure 2(c,d)). Moreover, results from the transwell assay showed that knockdown MALAT1 significantly decreased the percentage of invasion cells compared to control cells, while overexpression of MALAT1 greatly increased (Figure 2(e,f)). All together, these data demonstrate that inhibition of MALAT1 restrains migration and invasion of trophoblast cells, whereas ectopic expression of MALAT1 enhances.
Figure 2.
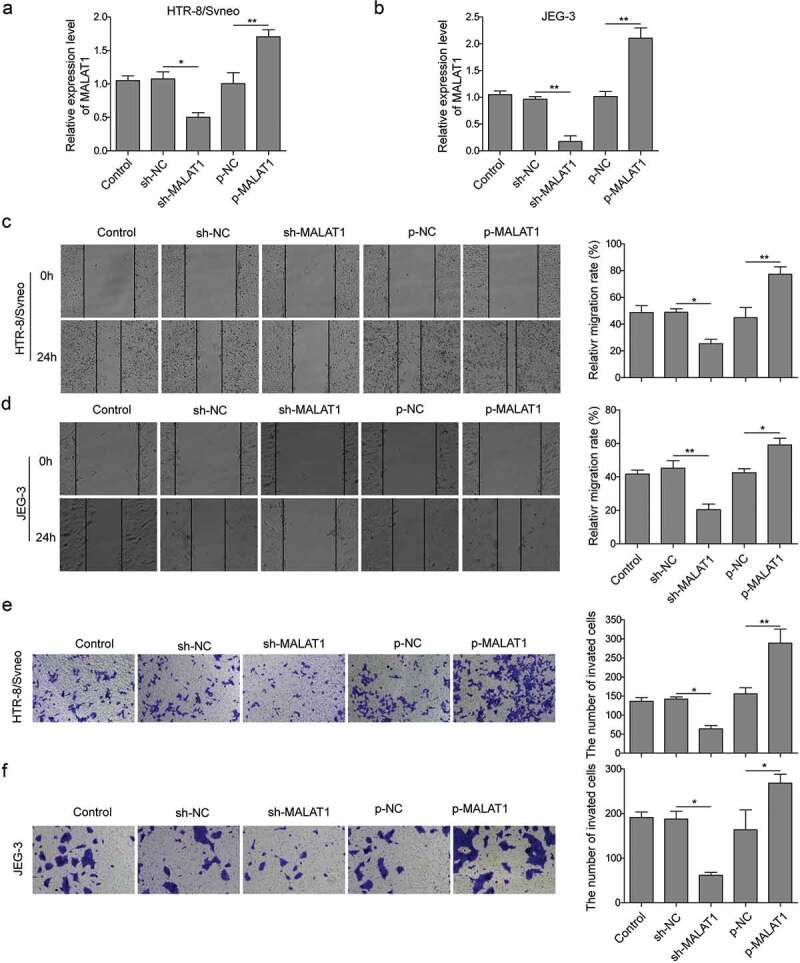
Inhibition of MALAT1 suppresses migration and invasion of trophoblast cells while overexpression enhances.
HTR-8/Svneo and JEG-3 were transfected with sh-NC or sh-MALAT1, p-NC, p-MALAT1, and subsequent experiments were conducted after 24 h of transfection. (a-b) MALAT1 expression was detected by qRT-PCR analysis. (c-d) Scratch wound assay was conducted to measure cells migration. (e-f) Transwell migration assay was used to determine cell invasion. Three replicate wells were used per sample in each experiment and experiments were replicated three times. *P < 0.05; **P < 0.01
MALAT1 binds and negatively regulates miR-206 expression
LncRNAs can regulate gene expression through different mechanisms and one of them is to act as sponges of miRNAs, leading to dis-inhibition of target gene expression. Previous studies have indicated that MALAT1 regulates miR-206 expression in various cancers [20,21]. To verify whether MALAT1 and miR-206 have the similar binding in trophoblast cells, we first confirmed that there were complementary binding sites between MALAT1 and miR-206 using bioinformatics tools (StarBase: http://starbase.sysu.edu.cn/index.php) (Figure 3(a)). Next, dual-luciferase reporter assay was performed to further investigate the interaction. Both MALAT1 WT and mutant fragments (the binding site was replaced with its complementary sequence) were cloned into the pmirGLO vector. We found that the luciferase activity in trophoblasts transfected with miR-206 mimics was significantly lower and miR-206 inhibitors was significantly higher than that in cells transfected with NC. Nevertheless, miR-206 had little effects on MALAT1 mutant (Figure 3(b,c)). Further, expression of miR-206 in cells transfected with MALAT1 shRNA was much higher than in control cells or cells transfected with sh-NC (Figure 3(d)). Together, these results suggest that MALAT1 directly binds and inhibits miR-206 expression in trophoblast cells.
Figure 3.
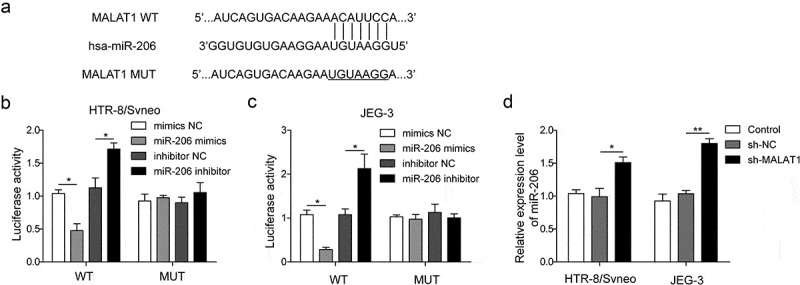
MALAT1 binds and negatively regulates miR-206 expression.
(a) The binding site between MALAT1 and miR-206. (b-c) Relative luciferase activity was measured in cells co-transfected with miR-206 mimic and WT MALAT1 or MUT MALAT1 (*P < 0.05). (d) miR-206 expression was detected in cells transfected with sh-MALAT1 or sh-NC by qRT-PCR analysis. Three replicate wells were used per sample in each experiment and experiments were replicated three times. *P < 0.05; **P < 0.01
miR-206 inhibition promotes migration and invasion of trophoblasts
To determine the role of miR-206 in PE, we transfected trophoblasts with miR-206 inhibitor and accessed its effects on migration and invasion of trophoblasts. Compared to cells transfected with NC or non-transfected control cells, cells transfected with miR-206 inhibitor had greatly lower levels of miR-206, while miR-206 mimic had greatly higher levels of miR-206 (Figure 4(a,b)). With wound scratch assay, we found that miR-206 inhibitor significantly increased the migration ability of trophoblasts (Figure 4(c,d)). However, knockdown MALAT1 reversed the effects of miR-206 inhibitor (Figure 4(c,d)). Additionally, miR-206 inhibitor increased the number of invasion cells while MALAT1 shRNA prevent that increase induced by miR-206 inhibitor (Figure 4(e,f)). We also confirmed that the level of miR-206 recovered to the control level after MALAT1 knockdown (Figure 4(a,b)). As an alternate strategy, we next increased the function of miR-206 with miR-206 mimics and examined its effects on migration and invasion abilities of trophoblast. As expected, we found that cells transfected with miR-206 mimics had lower migration and invasion capabilities while overexpression of MALAT rescued the effects of miR-206 mimics (Figure 4(c,f)). These data, together with aforementioned results, demonstrate that miR-206 inhibition has opposite functions of MALAT1 inhibition and promotes migration and invasion of trophoblast cells.
Figure 4.
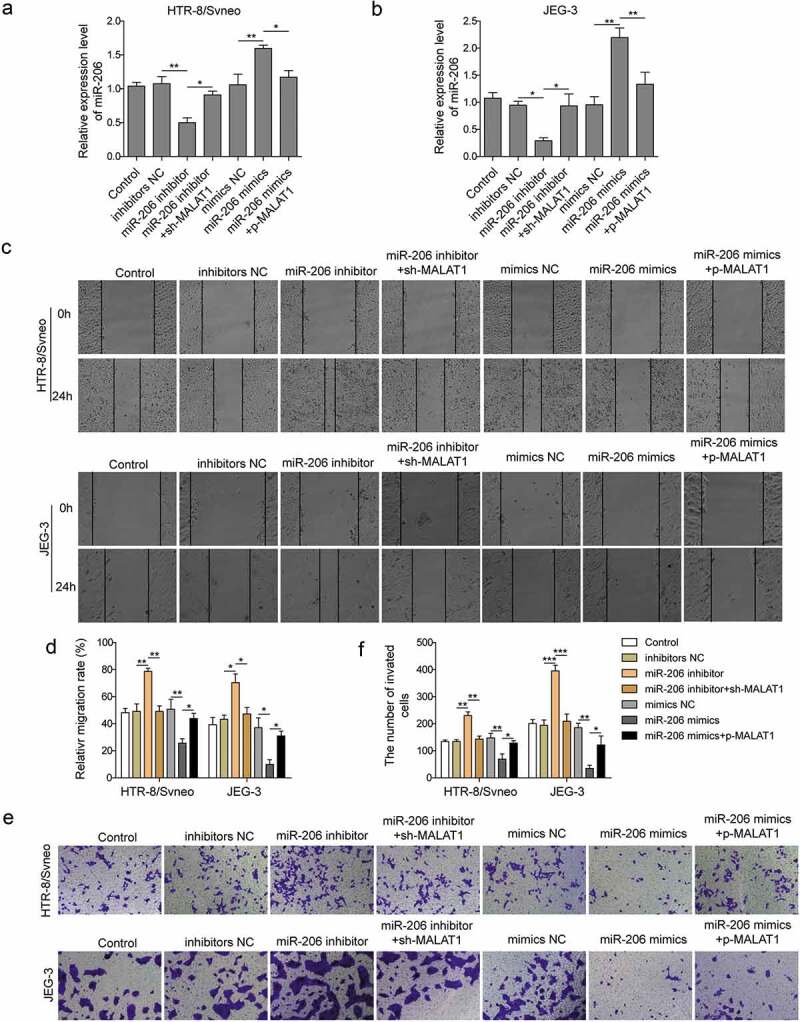
miR-206 promotes migration and invasion of trophoblast cells.
HTR-8/Svneo and JEG-3 were transfected with miR-206 inhibitor NC, miR-206 inhibitor and/or sh-MALAT1, mimics NC, miR-206 mimics and/or p-MALAT1, and subsequent experiments were conducted after 24 h of transfection. (a-b) miR-206 expression was detected by qRT-PCR. (c-d) Scratch wound assay was used to measure cells migration. (e-f) Transwell migration assay was conducted to measure cell invasion of transfected cells. Three replicate wells were used per sample in each experiment and experiments were replicated three times. *P < 0.05; **P < 0.01; ***P < 0.001
miR-206 directly targets IGF-1
miRNAs exert their functions via binding to mRNA of specific target genes to suppress their expressions. We next tried to identify the downstream target of miR-206 in PE. Previous studies have shown that miR-206 contributes to Parkinson’s diseases by targeting IGF-1 [22]. Further, circulating IGF-1 level has been reported low in PE patients [23]. Therefore, we hypothesized that IGF-1 might be an important downstream target of miR-206 during PE. We first examined the expression of IGF-1 in the placental of PE patients and found that IGF-1 level was significantly reduced compared to normal pregnancy (Figure 5(a)). Moreover, IGF-1 level was positively correlated with MALAT1 but negatively correlated with miR-206 (Figure 5(b,c)). These data highly suggest that IGF-1 might be regulated by miR-206 during PE.
Figure 5.
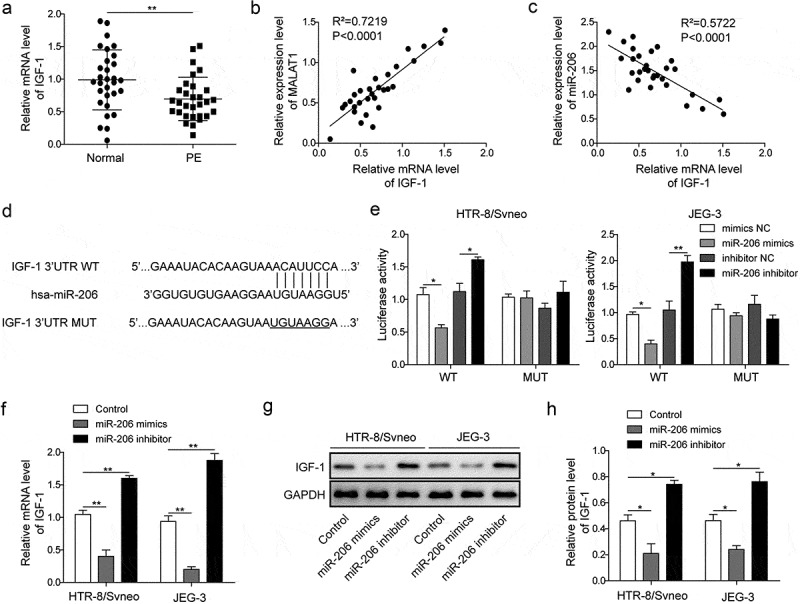
miR-206 directly targets IGF-1.
(a) qRT-PCR analysis of IGF-1 level in the placenta of normal pregnant women (n = 30) and patients with PE (n = 30). (b) Correlation between IGF-1 level and MALAT1 level in PE patients. (c) Correlation between IGF-1 level and miR-206 level in PE patients. (d) The binding sites between miR-206 and IGF-1. (e) Relative luciferase activity was measured in cells co-transfected with miR-206 mimic or miR-206 inhibitor and IGF-1-WT-3’UTR or IGF-1-MUT-3’UTR. (f) IGF-1 mRNA levels were measured in cells transfected with miR-206 mimics or miR-206 inhibitor by qRT-PCR analysis. (g-h) IGF-1 protein levels were measured in cells transfected with miR-206 mimics or miR-206 inhibitor by Western blot analysis. Three replicate wells were used per sample in each experiment and experiments were replicated three times. *P < 0.05; **P < 0.01
To directly examine the binding between miR-206 and IGF-1, we used Targetscan (http://www.targetscan.org/vert_72/) bioinformatics tool and found that IGF-1 was one of the potential targets of miR-206 (Figure 5(d)). We then performed a dual-luciferase assay to confirm the interaction. miR-206 significantly decreased the luciferase activity of WT-IGF-1 but not Mut-IGF-1 in which the binding sites were mutated, whereas miR-206 inhibitor greatly enhanced the luciferase activity of WT-IGF-1 but not Mut-IGF-1 (Figure 5(e)). Further, miR-206 mimics greatly reduced IGF-1 mRNA level as well as protein level in trophoblast cells while miR-206 inhibitor increased IGF-1 mRNA and protein expressions (Figure 5(f–h)). These results strongly support that miR-206 directly binds to IGF-1 mRNA and modulates its expression in trophoblast cells.
miR-206 regulates migration and invasion of trophoblasts via IGF-1
To further investigate the role of miR-206/IGF-1 signaling in PE, we transfected trophoblasts with IGF-1 si-RNA together with or without miR-206 inhibitor, or p-NC, p-IGF-1 with or without miR-206 mimics, and evaluated their effects on migration and invasion of cells. qRT-PCR results showed that expression of IGF-1 was significantly suppressed by si-IGF-1 while it significantly by upregulated p-IGF-1 (Figure 6(a,b)). The results of scratch assay manifested that knockdown IGF-1 decreased the migration of trophoblast cells while miR-206 inhibitor rescued the effect of si-IGF-1 (Figure 6(c,d)). Similarly, si-IGF-1 significantly reduced the invasion of trophoblast cells but miR-206 inhibitor prevented that reduction (Figure 6(e,f)). In addition, we also examined the effects of IGF-1 overexpression on migration and invasion abilities of trophoblasts. We observed higher migration and invasion capabilities of trophoblast cells transfected with p-IGF-1 compared to cells transfected with p-NC, whereas miR-206 mimics reversed the effects of IGF-1 overexpression (Figure 6(c–f)). Therefore, we conclude that miR-206 regulates migration and invasion of trophoblasts via targeting IGF-1.
Figure 6.
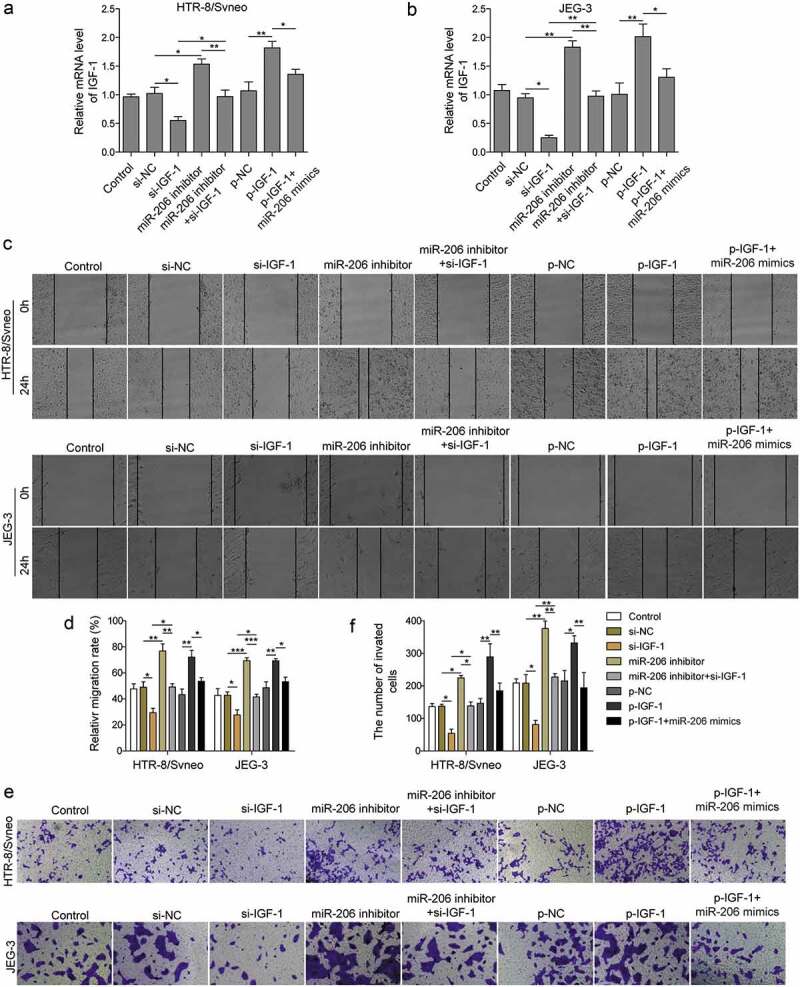
miR-206 regulates migration and invasion of trophoblast cells via IGF-1.
HTR-8/Svneo and JEG-3 were transfected with si-NC, si-IGF-1 with or without miR-206 inhibitor, or p-NC, p-IGF-1 with or without miR-206 mimics, and subsequent experiments were conducted after 24 h of transfection. (a-b) IGF-1 mRNA levels was detected by qRT-PCR analysis (*P < 0.05; **P < 0.01). (c-d) Scratch wound assay was used to measure cells migration. (e-f) Transwell migration assay was performed to measure cell invasion of transfected cells. Three replicate wells were used per sample in each experiment and experiments were replicated three times. *P < 0.05; **P < 0.01; ***P < 0.001
MALAT1 modulates IGF-1 mediated PI3K/Akt signaling via miR-206
In the end, we attempted to further examine the signaling pathway involved in PE. IGF-1 mediated PI3K-Akt signaling pathway is a highly conserved pathway and plays essential roles in cell migration and invasion [24]. We thus examined how MALAT1 and miR-206 regulate IGF-1/PI3K/Akt pathway in PE. As expected, knockdown IGF-1 in trophoblast cells suppressed the expressions of p-PI3K and p-Akt (T308 and S473) (Figure 7(a,b)). However, miR-206 inhibitor partially reversed the effect of si-IGF-1 (Figure 7(a,b)). Moreover, knockdown MALAT1 through shRNA also significantly decreased the levels of p-PI3K and p-Akt while miR-206 inhibitors reversed the effects of MALAT1 shRNA on IGF-1/PI3K/Akt pathway (Figure 7(a)). To further evaluate the regulatory relationship between miR-206 and PI3K/Akt signaling, we employed a specific PI3 kinase inhibitor LY294002. We found that LY294002 inhibited the increase of p-PI3K and p-Akt induced by miR-206 inhibitor (Figure 7(a)). Moreover, LY294002 reversed the positive effects of miR-206 inhibitor on migration and invasion capabilities of trophoblast cells (Figure 7(b–e)). Together, these data indicate that MALAT1 regulates IGF-1/PI3K/Akt signaling via miR-206.
Figure 7.
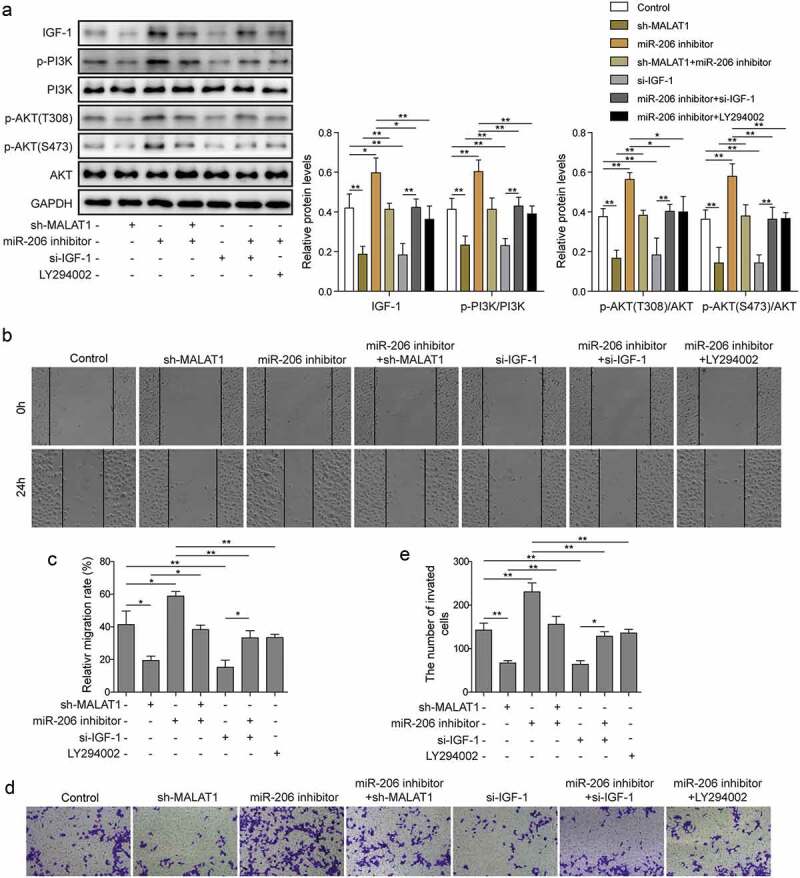
MALAT1 modulates IGF-1 mediated PI3K/Akt signaling via miR-206.
(a) Western blot analysis was performed to detect protein levels of IGF-1, p-PI3K, PI3K, p-Akt (S473), p-AKT (S473), Akt levels in control or transfected cells, and subsequent experiments were conducted after 24 h of transfection. (b-c) Scratch wound assay was used to measure cells migration. (d-e) Transwell migration assay was performed to measure cell invasion of transfected cells. Three replicate wells were used per sample in each experiment and experiments were replicated three times. *P < 0.05; **P < 0.01s
Discussion
Despite advances in research and therapy, the exact pathogenesis of PE remains largely unknown [25]. Hence, it is of great important to understand the underlying mechanisms on the development and progression of PE. Emerging evidence has implicated dysregulated miRNAs in PE and the functions of these differentially expressed miRNAs in PE have been gradually revealed [15,16]. Here, we show that lncRNA MALAT1 is down-regulated in placentas from PE patients and contributes to PE by regulating miR-206/IGF-1 axis.
MALAT1 was first identified as a prognostic marker for stage I lung adenocarcinoma [26] and later has been shown involved in many physiological and pathological processes [27]. Numerous studies have linked MALAT1 to cancer progression and metastasis [28,29], but it was until recent that MALAT1 was observed decreased in PE patients [13]. Consistent with the previous study, we confirmed that MALAT1 was significantly reduced in placentas of PE patients. Moreover, we showed that MALAT1 inhibition suppressed the migration and invasion of trophoblasts. The role of MALAT1 in PE is similar to its function in cancers. Indeed, many molecular pathways have been shared in common between trophoblasts and cancer cells and this probably is related to the invasive nature inherited in both cell types [30,31].
Another shared molecule between trophoblasts and cancer cells is miR-206, which plays key roles in various cancers [32,33] by regulating various downstream targets, such as PI3k/Akt, TGF-β signaling, miR-206 has been shown to regulate the epithelial-mesenchymal transition (EMT) process, thereby modulating invasion and migration of cancer cells. However, the function of miR-206 in PE is elusive although higher level of miR-206 has been observed in placenta of patients with PE [18]. In our study, we demonstrate that silencing miR-206 expression promotes the motility and invasiveness of trophoblast cells. Moreover, MALAT1 has been shown to promote cancer progression by targeting miR-206. In PE, we observed similar roles of MALAT1/miR-206 and their levels were negatively correlated. LncRNA MALAT1 regulates the migration of trophoblast by acting as a molecular sponge of miR-206.
It is well known that miRNAs function through binding to mRNAs of downstream targets, resulting in mRNA degradation or inhibition of translation [14]. Here, we found that IGF-1 was the downstream target of miR-206. In PE patients, IGF-1 level was reduced and its expression was negatively correlated with miR-206. Also, we showed that miR-206 directly bound to IGF-1 mRNA and regulated its expression in PE. Further, knockdown IGF-1 could rescue the effects of miR-206 inhibitor on migration and invasion of trophoblasts, indicating that miR-206 functions via targeting IGF-1. IGF-1 is a hormone that has a similar structure to insulin. It is the primary mediator of the effects of growth hormone and functions to promote growth of almost every cell. Aberrant IGF-1 signaling has been shown to promote the proliferation and invasion of cancer cells. Similarly, previous studies indicate that IGF-1 can also regulate the invasion and apoptosis of trophoblast cells although its upstream signaling is largely unknown [34–36]. We showed that MALAT1/miR-206 acted as upstream modulators of IGF-1/PI3K/Akt signaling pathway in PE. There might be other mediators and it remains to be further explored.
In conclusion, we demonstrate that lncRNA MALAT1 is reduced but miR-206 is increased in the placenta of PE patients. MALAT1 acts as a sponge for miR-206 to depress the expression of downstream target gene IGF-1, thereby promoting trophoblast cells migration and invasion. Collectively, our research reveals the roles of MALAT1 and miR-206 in PE and suggests that MALAT1/miR-206/IGF-1 serve as a potential therapeutic target for PE.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
All data generated or analyzed during this study are included in this published article.
References
- [1].Mol BWJ, Roberts CT, Thangaratiman S, et al. Pre-eclampsia. Lancet. 2016;387(10022):999–1011. [DOI] [PubMed] [Google Scholar]
- [2].Sircar M, Thadhani R, Karumanchi SA.. Pathogenesis of preeclampsia. Curr Opin Nephrol Hypertens. 2015;24(2):131–138. [DOI] [PubMed] [Google Scholar]
- [3].Esteve-Valverde E, Ferrer-Oliveras R, Gil-Aliberas N, et al. Pravastatin for preventing and treating preeclampsia: a systematic review. Obstet Gynecol Surv. 2018;73(1):40–55. [DOI] [PubMed] [Google Scholar]
- [4].Sun Q, Hao Q, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34(2):142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31(5):239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18(1):206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77(15):3965–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang X, Chen Y, Du L, et al. Evaluation of circulating placenta-related long noncoding RNAs as potential biomarkers for preeclampsia. Exp Ther Med. 2018;15(5):4309–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tan Y, Xiao D, Xu Y, et al. Long non-coding RNA DLX6-AS1 is upregulated in preeclampsia and modulates migration and invasion of trophoblasts through the miR-376c/GADD45A axis. Exp Cell Res. 2018;370(2):718–724. [DOI] [PubMed] [Google Scholar]
- [10].Wu D, Xu Y, Zou Y, et al. Long noncoding RNA 00473 is involved in preeclampsia by lsd1 binding-regulated tfpi2 transcription in trophoblast cells. Mol Ther Nucleic Acids. 2018;12:381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wu Y, Conrad ND, Ntini E, et al. Long noncoding RNA MALAT1: insights into its biogenesis and implications in human disease. Curr Pharm Des. 2015;21(34):5017–5028. [DOI] [PubMed] [Google Scholar]
- [12].Tseng JJ, Hsieh YT, Hsu SL, et al. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod. 2009;15(11):725–731. [DOI] [PubMed] [Google Scholar]
- [13].Chen H, Meng T, Liu X, et al. Long non-coding RNA MALAT-1 is downregulated in preeclampsia and regulates proliferation, apoptosis, migration and invasion of JEG-3 trophoblast cells. Int J Clin Exp Pathol. 2015;8(10):12718–12727. [PMC free article] [PubMed] [Google Scholar]
- [14].Moran Y, Agron M, Praher D, et al. The evolutionary origin of plant and animal microRNAs. Nat Ecol Evol. 2017;1(3):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lv Y, Lu C, Ji X, et al. Roles of microRNAs in preeclampsia. J Cell Physiol. 2019;234(2):1052–1061. [DOI] [PubMed] [Google Scholar]
- [16].Bounds KR, Chiasson VL, Pan LJ, et al. MicroRNAs: new players in the pathobiology of preeclampsia. Front Cardiovasc Med. 2017;4:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lykoudi A, Kolialexi A, Lambrou GI, et al. Dysregulated placental microRNAs in early and late onset preeclampsia. Placenta. 2018;61:24–32. [DOI] [PubMed] [Google Scholar]
- [18].Akehurst C, Small HY, Sharafetdinova L, et al. Differential expression of microRNA-206 and its target genes in preeclampsia. J Hypertens. 2015;33(10):2068–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Martinez-Fierro ML, Hernández-Delgadillo GP, Flores-Morales V, et al. Current model systems for the study of preeclampsia. Exp Biol Med (Maywood). 2018;243(6):576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tang Y, Xiao G, Chen Y, et al. LncRNA MALAT1 promotes migration and invasion of non-small-cell lung cancer by targeting miR-206 and activating Akt/mTOR signaling. Anticancer Drugs. 2018;29(8):725–735. [DOI] [PubMed] [Google Scholar]
- [21].Wang SH, Zhang WJ, Wu XC, et al. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget. 2016;7(25):37857–37867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim W, Lee Y, McKenna ND, et al. miR-126 contributes to Parkinson’s disease by dysregulating the insulin-like growth factor/phosphoinositide 3-kinase signaling. Neurobiol Aging. 2014;35(7):1712–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Halhali A, Díaz L, Barrera D, et al. Placental calcitriol synthesis and IGF-I levels in normal and preeclamptic pregnancies. J Steroid Biochem Mol Biol. 2014;144 Pt A:44–49. [DOI] [PubMed] [Google Scholar]
- [24].Kasprzak A, Kwasniewski W, Adamek A, et al. Insulin-like growth factor (IGF) axis in cancerogenesis. Mutat Res Rev Mutat Res. 2017;772:78–104. [DOI] [PubMed] [Google Scholar]
- [25].Armaly Z, Jadaon JE, Jabbour A, et al. Preeclampsia: novel mechanisms and potential therapeutic approaches. Front Physiol. 2018;9:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. [DOI] [PubMed] [Google Scholar]
- [27].Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu J, Peng WX, Mo YY, et al. MALAT1-mediated tumorigenesis. Front Biosci (Landmark Ed). 2017;22:66–80. [DOI] [PubMed] [Google Scholar]
- [29].Amodio N, Raimondi L, Juli G, et al. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Louwen F, Muschol-Steinmetz C, Reinhard J, et al. A lesson for cancer research: placental microarray gene analysis in preeclampsia. Oncotarget. 2012;3(8):759–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang Z, Wang X, Zhang L, et al. Wnt/beta-catenin signaling pathway in trophoblasts and abnormal activation in preeclampsia (Review). Mol Med Rep. 2017;16(2):1007–1013. [DOI] [PubMed] [Google Scholar]
- [32].Watt K, Newsted D, Voorand E, et al. MicroRNA-206 suppresses TGF-beta signalling to limit tumor growth and metastasis in lung adenocarcinoma. Cell Signal. 2018;50:25–36. [DOI] [PubMed] [Google Scholar]
- [33].Hesari Z, Nourbakhsh M, Hosseinkhani S, et al. Down-regulation of NAMPT expression by mir-206 reduces cell survival of breast cancer cells. Gene. 2018;673:149–158. [DOI] [PubMed] [Google Scholar]
- [34].Ma M, Zhou QJ, Xiong Y, et al. Preeclampsia is associated with hypermethylation of IGF-1 promoter mediated by DNMT1. Am J Transl Res. 2018;10(1):16–39. [PMC free article] [PubMed] [Google Scholar]
- [35].Niu ZR, Han T, Sun XL, et al. MicroRNA-30a-3p is overexpressed in the placentas of patients with preeclampsia and affects trophoblast invasion and apoptosis by its effects on IGF-1. Am J Obstet Gynecol. 2018;218(2):249 e1–249 e12. [DOI] [PubMed] [Google Scholar]
- [36].Dubova EA, Pavlov KA, Lyapin VM, et al. Expression of insulin-like growth factors in the placenta in preeclampsia. Bull Exp Biol Med. 2014;157(1):103–107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


