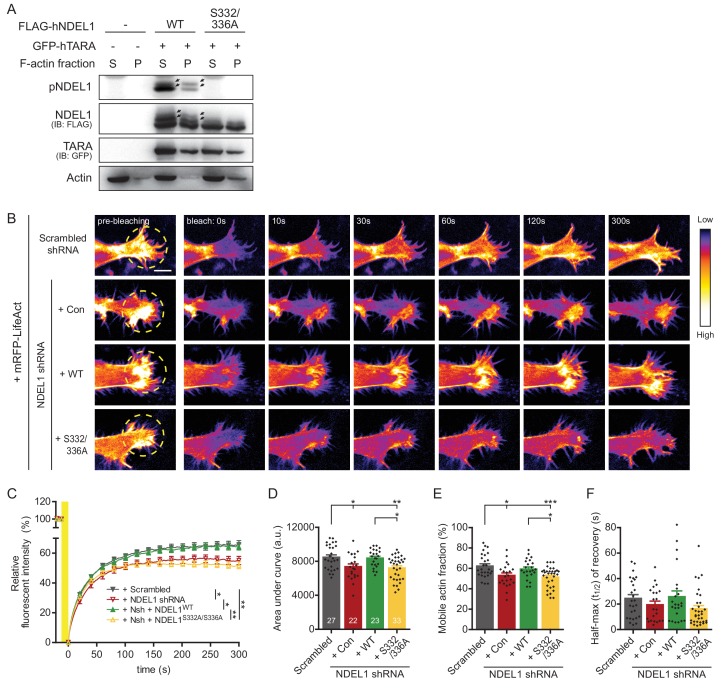
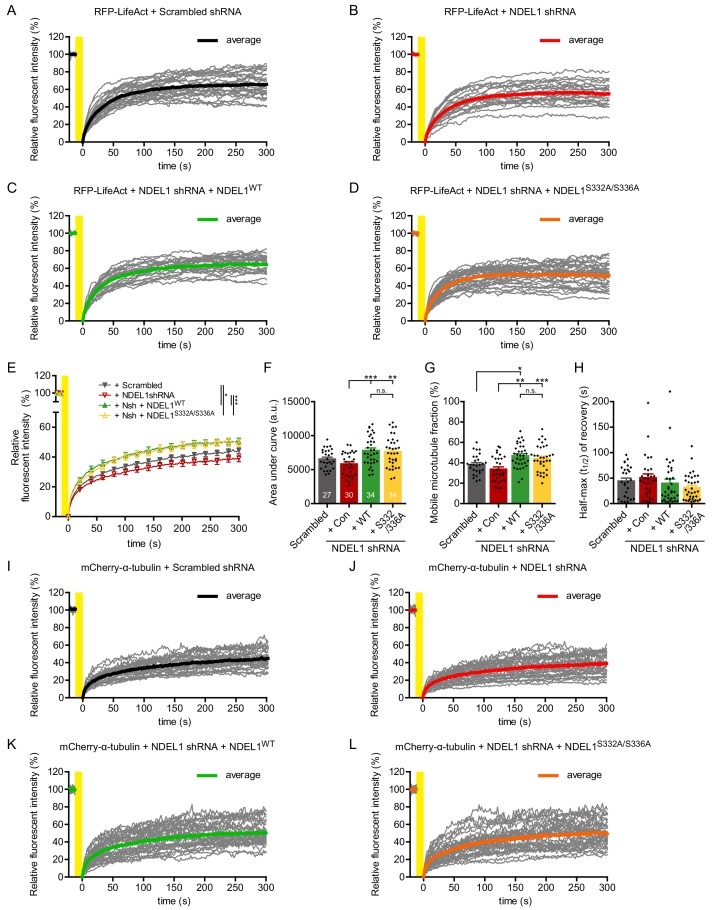
Figure 6. Phosphorylation of NDEL1 S336/S332 modulates F-actin dynamics.
(A) F-actin fractionation assay of NDEL1 and TARA. NDEL1 S336/S332 phosphorylation induced by TARA was observed at both G-actin in the supernatant fraction (S) and F-actin in pellet fraction (P). (B–F) Decreased F-actin dynamics by suppression of NDEL1 S336/S332 phosphorylation. (B) Representative time-lapse images of FRAP assay to measure F-actin dynamics at differentiating SH-SY5Y cells expressing RFP-LifeAct. All of NDEL1 over-expressing constructs here contain an shRNA-resistant mutation. A yellow-dashed circle indicates region-of-interest used for bleaching. Bleaching was given by stimulating with 10% 568 nm laser for 10 s. (C) Time-dependent fluorescence recovery graph. Comparisons of the area under FRAP curves (D), the percentage of mobile F-actin fraction calculated by the amount of eventual fluorescence recovery (E), and the average half-max (t1/2) of RFP-LifeAct fluorescence recovery (F). NDEL1 knockdown cells had decreased fluorescence recovery meaning more immobile fraction of F-actin and could not be rescued by NDEL1S332/336A implying its phosphorylation dependency. Each n number is shown at the bottom of the bar of the graph. The scale bar at (B) represents 10 μm. All results are presented as means ± SEM. *p<0.05, **p<0.01, and ***p<0.001 from two-way ANOVA for (C) and one-way ANOVA for (D–F). See also Figure 6—figure supplements 1, 2 and 3, Figure 6—video 1, and Figure 6—source data 1.