Abstract
Background
Practices in somatic variant interpretation and classification vary between Canadian clinical molecular diagnostic laboratories, and understanding of current practices and perspectives is limited. To define gaps and future directions, including consensus guideline development, the Somatic Curation and Interpretation Across Laboratories (social) project examined the present state of somatic variant interpretation in Canadian molecular laboratories, including testing volumes and methods, data sources and evidence criteria, and application of published classification guidelines.
Methods
Individuals who perform somatic variant interpretation in Canadian centres were invited to participate in an online survey. Invitees included laboratory directors (certified as Fellows of the Canadian College of Medical Geneticists or the American College of Medical Geneticists), md or md and phd molecular pathologists, and other phd experts, including phd specialists in variant annotation or bioinformatics. Current testing methods, volumes, and platforms in next-generation sequencing, use of variant annotation resources and evidence criteria, and preference for variant classification schemes were evaluated.
Results
Responses were received from 37 participants in 8 provinces. A somatic variant classification scheme jointly supported by the Association for Molecular Pathology (amp), the American Society of Clinical Oncology (asco), and the College of American Pathologists (cap) was used by 47% of respondents; an alternative guideline or a combination of published guidelines was used by 35% of respondents. The remaining 18% did not use a published scheme. Only 41% of respondents used a published scheme without alteration. Although all respondents indicated that there is a need for Canadian laboratories to adopt a somatic variant classification guideline, only 38% of respondents felt that it should be mandatory to adopt the amp/asco/cap–endorsed guideline.
Conclusions
Data from the social project identified high variability in current practice, yet strong support for standardization of solid-tumour somatic variant interpretation across Canadian institutions. Aligning classification methods will reduce variation in cross-institutional classification and reporting practices, aiding in consistent practice nationwide.
Keywords: Classification guidelines, classification schemes, national surveys, solid tumours, somatic variant interpretation, variant annotation
INTRODUCTION
Molecular profiling to inform drug treatment choice or patient management by identifying somatic variants in tumour tissue is an important tool in current clinical care for Canadian patients with solid tumours. Clinical molecular diagnostic laboratories performing molecular testing have a significant role in that process, particularly in the interpretation of the numerous variants identified in next-generation sequencing (ngs) tests of tumours. Consensus guidelines for somatic tumour variant interpretation are important tools in clearly and consistently translating the clinical importance of genomic variants identified during profiling. Agreement by Canadian laboratories on a classification scheme will enable harmonization between laboratories, allow for longitudinal comparisons of particular variants, and offer a structure to define clinically significant findings.
In recent years, a number of publications have defined standardized protocols for the interpretation of somatic variants1–7. In contrast, guidelines for interpreting germline inherited variants are established8 and have been endorsed by the Canadian College of Medical Geneticists9. Of the available somatic interpretation guidelines, the most broadly recognized scheme is the guideline endorsed by the Association for Molecular Pathology (amp), the American Society of Clinical Oncology (asco), and the College of American Pathologists (cap), which was developed based on a survey, a literature review, and committee member expertise1. However, the guideline is heavily therapy-based, with limitations for interpreting variants of diagnostic or prognostic relevance. In addition, the extent of its clinical applicability within the context of the Canadian health care system is unclear, because of differences in test and drug funding, legislation, and laboratory accreditation requirements. Health care in Canada is publicly funded under the Canada Health Act, with provincial funding agencies assessing the application of molecular profiling as a balance between patient benefit and associated test and treatment costs. Furthermore, legislation, regulations, and laboratory accreditation requirements are governed at the provincial level, which can influence site-specific laboratory practices.
Given the foregoing issues, we initiated the Somatic Curation and Interpretation Across Laboratories (social) project to examine the current state of somatic variant interpretation in Canadian molecular laboratories as a first-step gap analysis in understanding the environment for standardization on a Canadian guideline for tumour somatic variant interpretation.
METHODS
Participant Cohort
A survey was distributed to individuals at Canadian molecular clinical laboratories. A comprehensive catalog of clinical laboratories that perform somatic molecular testing within Canada does not exist, and so individuals were asked to participate and to further distribute the survey to relevant colleagues and members of their clinical laboratories. The target individuals included those who currently perform variant annotations as a component of their role, and the survey was primarily directed to individuals with experience in somatic variant interpretations for solid tumours. The survey was circulated to laboratory directors certified as Fellows of the Canadian College of Medical Geneticists or of the American College of Medical Geneticists, to md or md and phd molecular pathologists, and to individuals with a phd who have expertise in any one or a combination of variant annotation, molecular oncology, or bioinformatics.
Survey, Confidentiality, and Consent
Responses to an online survey (SurveyMonkey, San Mateo, CA, U.S.A.) were collected between June 2018 and February 2019. The survey included a confidentiality disclaimer, and participants were asked to voluntarily consent to participate with the understanding that de-identified data collected for the purpose of the survey would be published and presented broadly. Participants were not required to submit a response for every question. The survey questionnaire is available in supplementary Appendix A
Data Analysis
In a comparative analysis, non-ngs-based and ngs-based sequencing techniques and solid-tumour testing volumes at the participating institutions were evaluated. Current approaches to somatic variant interpretation were reviewed, including the use of published guidelines with and without modification, use of variant classification criteria and evidence sources, and opinions related to adoption of available published guidelines.
RESULTS
Participation and Demographics
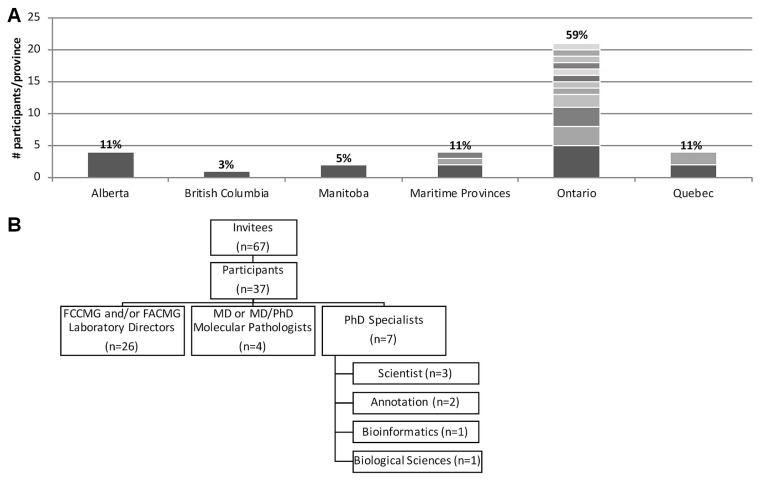
Survey responses were received from 37 participants, representing 8 provinces in Canada. Most survey respondents, 59% (n = 22, distributed across 12 sites), resided in Ontario; 11% (n = 4) came from each of Alberta, Quebec, and the Maritime provinces; 5% (n = 2) came from Manitoba; and 3% (n = 1) came from British Columbia [Figure 1(A)]. Individuals from 20 Canadian institutions participated, with no more than 5 individuals from any single institution participating. Participants identified primarily as Canadian laboratory directors certified by the Canadian College of Medical Geneticists or the American College of Medical Geneticists (70%, n = 26); an additional 11% (n = 4) identified as md or md and phd molecular pathologists. The remaining 19% (n = 7) identified as non-certified phd specialists, including scientists and annotation, bioinformatics, or biologic sciences specialists [Figure 1(B)].
FIGURE 1.
Participation and demographics. (A) Survey responses were received from 37 participants at 20 Canadian clinical laboratories, with most respondents residing within Ontario. Bar divisions indicate the number of participants per site. Respondents from Alberta are grouped under a single provincial organization. (B) Participants self-identified as laboratory directors certified as Fellows of the Canadian College of Medical Geneticists (FCCMG) or the American College of Medical Geneticists (FACMG), MD or MD and PhD molecular pathologists, and other PhD experts, including PhD scientists and specialists in variant annotation or bioinformatics, or both.
Non-NGS-Based and NGS-Based Sequencing Platforms and Volumes
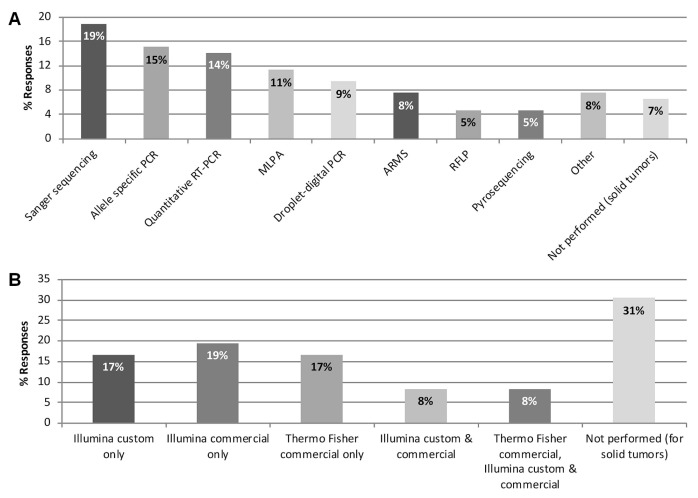
Participants were surveyed about the current use of nonngs-based and ngs-based sequencing methods at their site for the detection of somatic variants in solid tumours. A range of non-ngs-based sequencing techniques were identified as being in use to varying degrees, including Sanger sequencing, allele-specific polymerase chain reaction, quantitative reverse-transcription polymerase chain reaction, multiplex ligation-dependent probe amplification, and droplet-digital polymerase chain reaction, among others [Figure 2(A)].
FIGURE 2.
Sequencing platforms used for the detection of somatic variants in solid tumours. (A) Non–next-generation sequencing techniques (33 responses from 20 sites). (B) Next-generation sequencing platforms (35 responses from 19 sites). PCR = polymerase chain reaction; RT = reverse-transcription; MLPA = multiplex ligation-dependent probe amplification; ARMS = amplification-refractory mutation system; RFLP = restriction fragment length polymorphism; Illumina = Illumina, San Diego, CA, U.S.A.; Thermo Fisher = Thermo Fisher, Waltham, MA, U.S.A.
Next-generation sequencing was used for the detection of solid-tumour somatic variants by 69% of the 36 respondents to this question (n = 25); the remaining 31% (n = 11) were not performing ngs for solid tumours at the time of the survey [Figure 2(B)]. Of the 25 respondents performing ngs, 76% (n = 19) were using custom or commercial panels for the Illumina platform (Illumina, San Diego, CA, U.S.A.), and 36% (n = 9) were using commercial panels for the Thermo Fisher platform (Thermo Fisher, Waltham, MA, U.S.A.). Of the 25 respondents performing ngs, 3 (12%) had access to both platforms. Of the 11 respondents who did not perform ngs for solid tumours at the time of the survey, 4 indicated that ngs was in the process of test work-up or validation, and 4 performed ngs for hematologic malignancies.
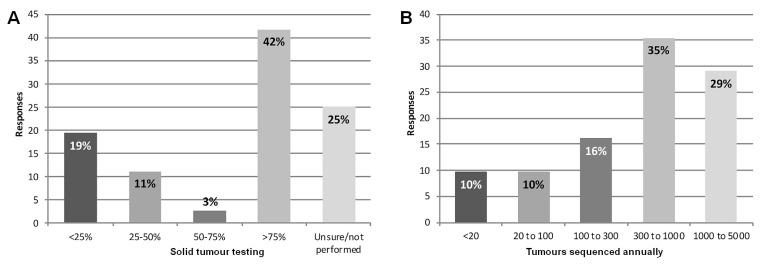
To evaluate experience with the detection of non-hotspot somatic variants (that is, the variants most likely to require classification), survey respondents were polled about testing volumes at their institution. Overall, 42% of the 36 respondents to the question about the proportion of tumour somatic testing being performed at their site using ngs (n = 15) indicated that it was more than three quarters, and 30% (n = 11) indicated that it was less than half [Figure 3(A)]. Furthermore, more than 61% of the 31 respondents to the question about the number of tumours being tested using ngs (n = 19) indicated that the number was more than 300 annually, with 19% (n = 6) indicating that the number was fewer than 100 annually [Figure 3(B)].
FIGURE 3.
Somatic testing volumes using next-generation sequencing (NGS). (A) Percentage of solid-tumour testing performed using NGS (36 responses from 20 sites). (B) Number of solid tumours sequenced annually using NGS (31 responses from 18 sites).
Current Somatic Variant Interpretation Practices
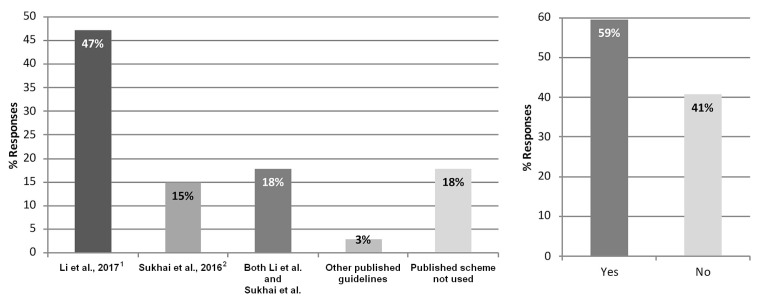
Methods for somatic variant interpretation were investigated, with survey participants being asked to indicate whether a published somatic variant interpretation scheme or guideline was in use [Figure 4(A)]. Of 34 respondents to that question, 18% (n = 6) indicated that a published guideline was not used at their site, although 6% (n = 2) noted that a laboratory-developed scheme was used instead. Of 34 respondents who used a published interpretation scheme, 47% (n = 16) exclusively used the amp/asco/cap–endorsed guideline, and 15% (n = 5) exclusively used the Sukhai et al.2 guideline. Furthermore, 18% (n = 6) indicated the use of both the amp/asco/cap–endorsed guideline and the Sukhai et al. guideline as a framework for routine variant interpretations, with comments specifying that certain schemes were in use for particular scenarios, such as for hematologic malignancies compared with solid tumours. Several reasons were given for the site-specific selection of a preferred scheme: it was endorsed by multiple societies; it was used by colleagues; it provided a framework viewed by the site to be suitable based on variant actionability; or the scheme was the most recently published of those available. Interestingly, of 32 respondents to a question about how they classified variants, 59% (n = 19) indicated using a custom or laboratory-developed guideline, a modified version of a currently available published guideline, or a third-party commercial vendor. Only 41% (n = 13) indicated using a published scheme without alteration [Figure 4(B)].
FIGURE 4.
Current somatic variant interpretation practices. (A) Frequency of application of a published somatic variant interpretation scheme as a guideline for solid-tumour variant interpretations (34 responses from 20 sites). (B) Whether a custom or laboratory-developed guideline, a modified currently available published guideline, or a third-party commercial vendor is used to classify variants (32 responses from 19 sites).
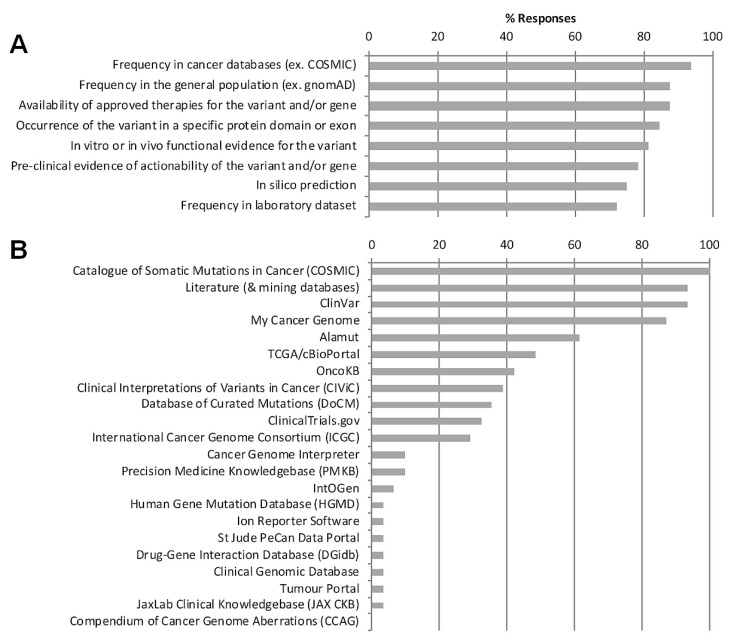
Participants were polled on the criteria and data sources used to collect evidence for the interpretation of somatic variants [Figure 5(A)]. Most respondents selected frequency of the variant in cancer as the key criterion; also mentioned were the frequency of the variant in the general population, the availability of approved therapies, preclinical evidence, and predicted or demonstrated functional effect using any or all of in silico, in vitro, and in vivo evidence. Sources and databases used to collect evidence for somatic interpretation included the Catalogue of Somatic Mutations in Cancer database, literature and literature-mining databases, The Cancer Genome Atlas, and My Cancer Genome, among others [Figure 5(B)].
FIGURE 5.
Evidence criteria and resources used for somatic variant classification. Most frequently applied somatic variant classification (A) evidence criteria (33 responses from 18 sites) and (B) resources or databases (31 responses from 18 sites). gnomAD = Genome Aggregation Database.
Preferences for Endorsement of Current Somatic Interpretation Guidelines
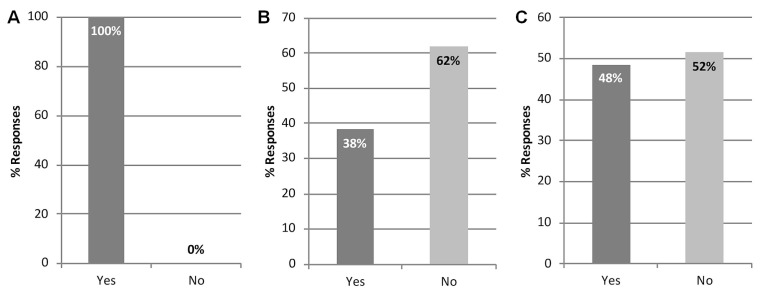
Participants were polled about their institutional or individual preferences for endorsement of a particular published scheme, with 100% of the 36 respondents to that question indicating a need for Canadian laboratories to endorse a somatic variant interpretation guideline [Figure 6(A)]. When asked whether it should be mandatory for Canadian laboratories to adopt the amp/asco/cap–endorsed guideline, 62% of the 34 respondents to that question (n = 21) said that it should not be mandatory [Figure 6(B)]. Comments related to that question suggested that a standard approach should be adopted, but not necessarily the amp/asco/cap–endorsed guideline, and that Canadians should have their own endorsed guideline because of differences in health care systems. Others suggested that it would be beneficial to align with non-Canadian laboratories using the amp/asco/cap–endorsed guideline.
FIGURE 6.
Preference for endorsement of a published somatic variant interpretation scheme. Participant perspectives about (A) whether there is a need for Canadian laboratories to endorse a somatic variant classification guideline (36 responses from 20 sites); (B) whether it should be mandatory for Canadian laboratories to adopt the guideline endorsed by the Association for Molecular Pathology, the American Society of Clinical Oncology, and the College of American Pathologists (AMP/ASCO/CAP) (34 responses from 19 sites); and (C) whether there is a need to implement consensus guidelines for somatic variant interpretation in Canada other than the AMP/ASCO/CAP–endorsed guideline (33 responses from 19 sites).
Lastly, participants were asked whether there is a need to implement a consensus guideline for somatic variant interpretation in Canada other than the amp/asco/cap–endorsed guideline. Interestingly, no agreement emerged with respect to the implementation of an alternative classification guideline, with 48% of the 33 respondents to that question (n = 16) indicating that they saw a need to implement a consensus guideline other than the amp/asco/cap–endorsed guideline, and 52% (n = 17) indicating that they did not see such a need [Figure 6(C)]. Respondents commented that various schemes should be evaluated and a consensus developed across Canada, that now would be an appropriate time to consider what the right scheme for Canadian laboratories would be, and that modification of an existing guideline (for example, the amp/asco/cap–endorsed guideline) would be preferred to adoption without revision.
DISCUSSION
The survey described here investigated the current state of practice in solid-tumour somatic variant interpretation at Canadian clinical laboratories, including testing volumes and methods, criteria and data sources used to gather evidence for variant annotation, and preference for available published classification schemes. Survey results spanned 8 Canadian provinces and 20 institutions, with the largest group of respondents being laboratory directors certified by the Canadian College of Medical Geneticists or the American College of Medical Geneticists, or both. Most respondents hailed from Ontario, in part because of the larger number of accredited laboratories in Ontario compared with other provinces.
The survey demonstrated that implementation of ngs for somatic tumour molecular profiling varies between laboratories in Canada, with more than 30% of respondents indicating that ngs is not currently performed for solid tumours at their institutions. For laboratories that do currently use ngs for solid-tumour profiling, sites vary in their choice of ngs technology and platform. Notably, limited use of ngs by many participating laboratories might have contributed to the narrow adoption of available somatic variant interpretation schemes as shown in the survey results. To manage small samples or low dna yield, to ensure rapid turnaround time, or to save costs, solid-tumour test algorithms in use at many Canadian institutions include recurrent (“hotspot”) variant testing before ngs. More than one third of respondents indicated that ngs testing is used for less than 50% of somatic tests performed at their institution, and one third indicated that fewer than 300 solid-tumour samples are sequenced annually. In the context of testing known hotspot variants, the need for classification of variants is irrelevant, because those variants are already known to be actionable for patient care. Thus, although laboratories are performing tumour molecular testing, they might still have limited experience with somatic interpretation if not performing ngs-based molecular profiling that would identify non-hotspot somatic variants. This issue of limited experience might also in part be attributable to differences in laboratory focus (on somatic or other testing priorities) and the availability of funding for staffing at each institution. The expanding adoption of larger panels for somatic testing might affect laboratory staffing requirements, increasing demand for more complex bioinformatic and analytic pipelines and appropriately trained staff to be incorporated into the somatic testing workflow. As the use of ngs panels for molecular profiling of solid tumours increases exponentially across Canada10, the need for a classification guideline increases, given complexity in the interpretation of the range of non-hotspot variants and heterogeneous mutational profiles identified using non-targeted sequencing approaches.
The survey identified unanimous support for standardized processes in somatic variant interpretation, and all respondents indicated that there is a need for Canadian laboratories to endorse a somatic variant classification guideline. As anticipated, no consensus about the approach to variant annotation was observed. With respect to the adoption of variant classification schemes, nearly 20% of respondents indicated that a published classification scheme was not used for solid-tumour variant interpretations. That result might reflect a focus on testing only for hotspot variants, as already discussed. In addition, only 41% of respondents used a published scheme without alteration, highlighting clear limitations and reservations at Canadian institutions for the adoption of available guidelines. Survey results suggest that the somatic interpretation guideline endorsed by amp/asco/cap does not sufficiently fit the needs of Canadian laboratories, because only 47% of respondents exclusively applied that guideline, 38% indicated that the guideline should not be mandatory to adopt, and approximately 50% felt that there is a need to adopt a guideline other than that endorsed by amp/asco/cap. The limited use of the guideline might be attributable to the fact that it relies heavily on classifying variants for therapeutic targetability, is disease-site agnostic, and applies strict criteria for the clinical relevance of variants that offer diagnostic utility. Furthermore, clinical significance at the gene, domain, or exon level is not considered, and the ambiguity in minor allele frequency cut-offs and definitions of clinical evidence might lead to variance in application from user to user. Those features of the amp/asco/cap–endorsed guideline restrict its applicability for diagnosis and prognostication in certain solid-tumour profiling scenarios. In addition, application of the scheme to hematologic malignancies, in which mutational profiles are highly relevant for diagnosis and disease stratification, was noted as a challenge.
CONCLUSIONS
The social project survey highlighted robust national support for adoption of a standardized guideline for solid-tumour somatic variant interpretation across institutions, although opinions about the appropriate guideline varied. Narrow adoption of available guidelines might be attributable to the somewhat limited use of ngs for molecular profiling of solid tumours or might reflect the limited applicability of existing guidelines for use in the context of the Canadian health care system. Therefore, there might be a need to adopt a guideline distinct from the ones applied in other jurisdictions because of fundamental differences in health care systems. While recognizing that a publicly funded health care system in which clinical tests and drugs funded at the provincial level could present barriers to achieving a national consensus, nationwide consistency would offer a structured approach to somatic annotation, reduce discrepancy between laboratories, and support the standardization of classification and reporting practices across institutions. With the growing use of ngs panels for tumour profiling, now is the time to critically evaluate the advantages and limitations of available classification schemes and to develop consensus somatic interpretation recommendations for adoption across Canada.
Supplementary Information
Footnotes
Supplemental material available at http://www.current-oncology.com
CONFLICT OF INTEREST DISCLOSURES
We have read and understood Current Oncology’s policy on disclosing conflicts of interest, and we declare that we have none.
REFERENCES
- 1.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sukhai MA, Craddock KJ, Thomas M, et al. A classification system for clinical relevance of somatic variants identified in molecular profiling of cancer. Genet Med. 2016;18:128–36. doi: 10.1038/gim.2015.47. [DOI] [PubMed] [Google Scholar]
- 3.Chakravarty D, Gao J, Phillips S, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00011. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meric-Bernstam F, Johnson A, Holla V, et al. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv098. pii:djv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Allen EM, Wagle N, Stojanov P, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–8. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andre F, Mardis E, Salm M, Soria JC, Siu LL, Swanton C. Prioritizing targets for precision cancer medicine. Ann Oncol. 2014;25:2295–303. doi: 10.1093/annonc/mdu478. [DOI] [PubMed] [Google Scholar]
- 7.Carr TH, McEwen R, Dougherty B, et al. Defining actionable mutations for oncology therapeutic development. Nat Rev Cancer. 2016;16:309–29. doi: 10.1038/nrc.2016.35. [DOI] [PubMed] [Google Scholar]
- 8.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson W, Stavropoulos J, Sinasac D, McCready E, Mahmutoglu, Nelson TN. CCMG Statement on Germline Variant Classification. Kingston, ON: CCMG; 2017. [Google Scholar]
- 10.Yip S, Christofides A, Banerji S, et al. A Canadian guideline on the use of next-generation sequencing in oncology. Curr Oncol. 2019;26:e241–54. doi: 10.3747/co.26.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.