Abstract
Crystal structures of recombinant Lactococcus lactis 6-phosphogluconate dehydrogenase (LlPDH) in complex with substrate, cofactor, product and inhibitors have been determined. LlPDH shares significant sequence identity with the enzymes from sheep liver and the protozoan parasite Trypanosoma brucei for which structures have been reported. Comparisons indicate that the key residues in the active site are highly conserved, as are the interactions with the cofactor and the product ribulose 5-phosphate. However, there are differences in the conformation of the substrate 6-phosphogluconate which may reflect distinct states relevant to catalysis. Analysis of the complex formed with the potent inhibitor 4-phospho-d-erythronohydroxamic acid, suggests that this molecule does indeed mimic the high-energy intermediate state that it was designed to. The analysis also identified, as a contaminant by-product of the inhibitor synthesis, 4-phospho-d-erythronamide, which binds in similar fashion. LlPDH can now serve as a model system for structure-based inhibitor design targeting the enzyme from Trypanosoma species.
Keywords: African trypanosomiasis, enzyme inhibition, Lactococcus lactis, pentose phosphate pathway, 6-phosphogluconate dehydrogenase
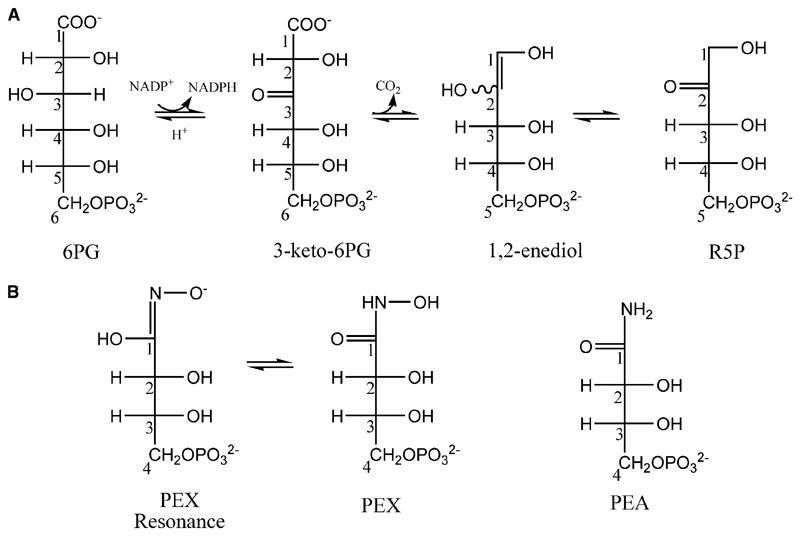
The pentose phosphate pathway is an anabolic pathway, the major functions of which are production of ribose 5-phosphate, utilized in the biosynthesis of nucleotides, and to maintain a pool of NADPH [1]. The NADPH serves to alleviate the oxidative stress of aerobic metabolism and participates in varied biosynthetic processes [2]. The third enzyme in the pathway, 6-phosphogluconate dehydrogenase (PDH; EC 1.1.1.44), converts 6-phosphogluconate (6PG) to ribulose 5-phosphate (RU5P). Loss of the enzyme activity is lethal, as a high concentration of 6PG is toxic to eukaryotic cells including Drosophila melanogaster [3,4], Saccharomyces cerevisae [5] and Trypanosoma brucei [2]. 6PG inhibits phosphoglucose isomerase [6] and it has been proposed that this disrupts the main glycolytic pathway, and establishes a positive feedback loop, although definitive results that identify the precise mechanism leading to cell death have yet to be obtained. Extensive kinetic studies have been carried out on PDH [7–11] and crystal structures from two species have been elucidated [12,13]. The catalytic conversion of 6PG to RU5P is considered as a three-step mechanism, with two possible reaction intermediates (Fig. 1A). Studies using 13C isotope, deuterium substitution [14] and different oxidants [15] have established that oxidative decarboxylation of 6PG occurs in a stepwise fashion with oxidation preceding decarboxylation.
Fig. 1.
(A) Catalytic reaction of PDH and two intermediate states. (B) Structures and numbering of the inhibitors PEX (two resonance forms) and PEA.
RNAi technology has established that PDH is essential in the causal agent of African trypanosomiasis, the protozoan parasite T. brucei [2]. This observation suggests that the enzyme is a potential target for the development of improved drugs. The characterization of potential substrate mimics has revealed potent inhibitors which, for reasons that are not understood, display good selectivity for the T. brucei enzyme (TbPDH) over a mammalian PDH [16–18]. In particular 4-phospho-d-erythronohydroxamic acid (PEX; Fig. 1B) has a Ki of 0.01 μm against TbPDH, which is 250 times more potent against the parasite enzyme than against sheep liver PDH (Ovis aries; OaPDH). We set out to study the mode of binding of PEX to determine if it was acting as a high-energy-state mimic. In our hands, TbPDH is unstable and has proven troublesome for crystallographic studies to characterize ligand binding and inhibition; therefore, an improved system was sought.
We identified that PDH from Lactococcus lactis (LlPDH) is highly suited for structural studies. This enzyme has been overexpressed and preliminary kinetic data reported [19]. LlPDH shares 38 and 58% sequence identity with TbPDH and the mammalian counterpart OaPDH, respectively [13]. Our experience is that, unlike other forms of PDH and in particular TbPDH, recombinant LlPDH is stable, amenable to crystallographic studies and therefore provides a good model for ligand-binding studies.
Here we report crystallographic studies of LlPDH complexes with physiological ligands, the substrate, the product of the enzyme reaction, and also with the cofactor. These represent the first structures of a bacterial PDH. In addition, the first PDH complex with an inhibitor is also detailed. These structures provide insight into the key features of PDH specificity and mode of inhibition of the enzyme.
Results and Discussion
Structural analysis and model quality
Crystal structures of three different complexes of LlPDH have been determined. Diffraction from the crystals was anisotropic and one unit cell length (> 240 Å) was significantly longer than the others (Table 1). Our data collection and processing strategy was designed to provide as much of the highest resolution data as possible, minimizing reflection overlap in certain crystal orientations and, although the outer shells of data are not complete, we were content to include these diffraction terms and trust to the benefits of maximum-likelihood weighting (see below). The approach appears to have been successful given that the refinement statistics and stereochemical parameters indicate that the coordinates represent acceptable medium resolution models (Table 1).
Table 1. Data and refinement statistics. Values in parentheses pertain to the highest resolution shell (width = 0.1 Å).
| Structure | Complex I | Complex II | Complex IIIa (In-house) | Complex IIIb (Synchrotron) |
| Protein Data Bank code | 2IYO | 2IYP | 2IZ0 | 2IZ1 |
| Space group | P3212 | C2 | C2 | C2 |
| Unit cell | ||||
| a, b, c (Å) | 60.58, 60.58, 243.13 | 71.06, 105.06, 240.48 | 71.06, 104.81, 240.52 | 71.07, 104.82, 240.52 |
| β (°) | 98.3 | 98.5 | 98.3 | |
| Resolution range (Å) | 20–2.4 | 30–2.8 | 45–2.6 | 35–2.3 |
| Unique reflections/Redundancy | 19588/5.5 | 39483/1.8 | 52468/2.9 | 73076/6.1 |
| Completeness (%) | 95.7 (74.8) | 90.7 (81.9) | 97.5 (83.4) | 94.5 (67.3) |
| < I/ σ(I) > /Mosaicity (°) | 13.7 (6.7)/0.4 | 6.5 (3.0)/0.6 | 41.7 (11.4)/0.3 | 32.9 (11.2)/0.3 |
| R-sym (%)a | 6.5 (14.6) | 7.9 (25.0) | 7.0 (18.0) | 5.7 (12.7) |
| Wilson B (Å2) | 40.5 | 50.9 | 36.9 | 31.8 |
| No. of protein residues/solvent molecules | 470/313 | 1407/429 | 1406/1708 | 1406/1963 |
| R-work b/R-free c (%) | 15.9/22.1 | 18.3/26.3 | 12.3/19.3 | 13.7/19.7 |
| Average B (Å2) | ||||
| Overall/protein/solvent | 27.6/27.2/32.1 | 36.8/36.9/31.7 | 16.7/15.3/24.9 | 18.9/16.4/32.8 |
| rmsd | ||||
| Bond lengths (Å)/bond angles (°) | 0.009/1.165 | 0.010/1.315 | 0.008/1.124 | 0.010/1.234 |
| Cruickshank’s DPId (Å) | 0.24 | 0.41 | 0.25 | 0.19 |
| Ramachandran plot (%) | ||||
| Most favoured region | 94.2 | 91.7 | 94.0 | 93.7 |
| Additional allowed regions | 5.3 | 7.8 | 5.4 | 5.6 |
| General allowed regions | 0.0 | 0.2 | 0.1 | 0.2 |
| Disallowed region | 0.5 | 0.3 | 0.5 | 0.5 |
R-sym = ΣhΣi||(h,i) - < I(h) > I/ΣhΣi I(h,i), where |(h,i) is the intensity of the ith measurement of reflection h and < |(h) > is the mean value of |(h,i) for all i measurements.
R-work = Σhkl||Fo| − |Fc||/Σ|Fo|, where Fo is the observed structure-factor amplitude and Fc the structure-factor amplitude calculated from the model.
R-free is the same as R-work except only calculated using a subset, 5%, of the data that are not included in any least-squares refinement calculations.
DPI = diffraction-component precision index [35].
The three structures are (complex I) LlPDH in complex with substrate 6PG, (complex II) in ternary complex with the cofactor and the product RU5P and (complex III) inhibited by PEX and by a contaminant 4-phospho-d-erythronamide (PEA). Complex I is a trigonal crystal form, space group P3212 with a monomer in the asymmetric unit. A two-fold crystallographic axis of symmetry forms the functional dimer. Complexes II and III are in the monoclinic space group C2 and are isomorphous. Three subunits constitute the asymmetric unit, one noncrystallographic symmetry related dimer is formed by subunits A and B and the remaining monomer (subunit C), in similar fashion to complex I, forms a dimer via the crystallographic twofold axis. Two structures corresponding to complex III have been determined, first using in-house diffraction data (IIIa) and then synchrotron data to extend the resolution (IIIb).
In complexes II, IIIa and IIIb only the active site of subunit A is occupied by ligands. In the cofactor-binding site, a complete NADP+ molecule is present in only one subunit of complex II and complex IIIa, whereas for the other subunit although the electron density is well defined for the adenine, ribose and two phosphate groups of NADP+, the nicotinamide and a-phosphate are missing. Hydrolysis may have occurred or there is disorder. In complex IIIb, all subunits present the same fragment of cofactor although in subunit A there is diffuse electron density, suggestive of low-occupancy nicotinamide. Different crystals were used to obtain the inhibitor complex structures although they were grown at the same time. The period between the data collections was several weeks and the time lapse may have allowed hydrolysis to occur. For completeness details of both structures are reported. In the active site, the electron and difference density maps clearly indicated ordered binding of PEX. However, a strong feature of positive density was observed, too close to PEX to be an associated water molecule. Our interpretation was that the smaller compound PEA (Fig. 1B) and a water molecule are present and PEX and PEA refined satisfactorily with occupancies of 0.7 and 0.3, respectively. Subsequent chemical analysis of the PEX sample by MS (data not shown) identified the presence of PEA, a contaminant carried through from the synthesis of PEX, due to some cleavage of the N–O bond, during the final hydrogenolysis step. The different ligands present in the complexes are summarized in Table 2, together with their respective average isotropic thermal parameters (B-factors) and, where appropriate, the ligand occupancies.
Table 2. Thermal parameters and occupancies of ligands in LlPDH complexes.
| Structure | Complex I | Complex II | Complex III In-house/Synchrotron | ||||
|---|---|---|---|---|---|---|---|
| Subunit | A | A | B | C | A | B | C |
| Substrate 6PG | 26.3 | ||||||
| Product RU5P | 38.7 | ||||||
| NADP+ | 49.3 | 21.7/– | |||||
| A2P | 53.9 | 44.7 | –/37.8 | 22.7/29.8 | 20.7/26.4 | ||
| Inhibitor | |||||||
| PEX(0.7)a | 13.7/13.3 | ||||||
| PEX(0.3)a | 15.2/15.2 | ||||||
Occupancy of the ligand
Superposition of subunit A on B and C (468 Cα atoms) in complex II and III of LlPDH gives an rmsd of 1.6 Å in each case, however, superposition of B on C gives an rmsd of only 0.3 Å. We analysed this difference using the program dyndom [20] and identified a movement of the cofactor-binding domain of subunit A relative to subunits B and C (not shown). This domain alteration involves a rotation of 5° and translation of 0.7 Å. Five segments of polypeptide within the cofactor-binding domain (Fig. 2) constitute the bending or hinge regions. These involve residues 76–77, 82–89, 98–101, 111–127 and 148–153. A similar difference is observed when comparing the cofactor domains of the OaPDH and TbPDH structures [13]. The superposition of subunit A of complex I onto subunits A, B, C of complex II and III yield rmsd-values of 0.8 Å for subunit A and 1.2 Å for subunit B and C, respectively (468 Cα atoms). The superposed coordinates of NADP+ in complex I have allowed us to model a functional ternary complex when considered with the substrate.
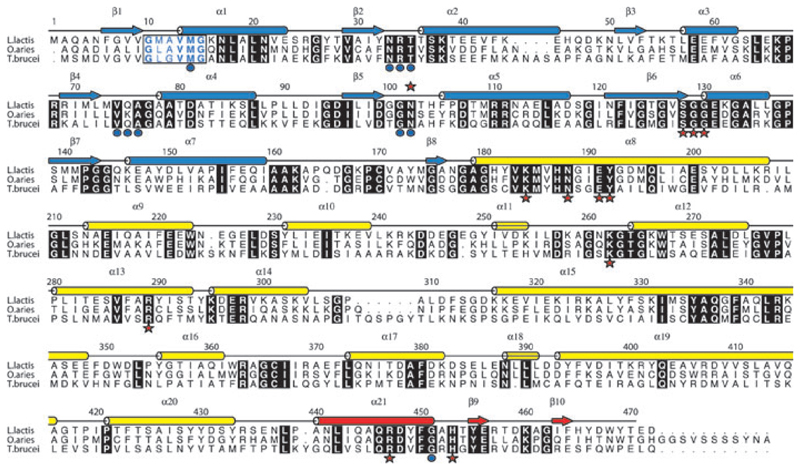
Fig. 2.
Amino acid sequence and secondary structure of LlPDH. Arrows depict β strands, cylinders depict α helices and these are labelled β1–β10 and α1–α21. The elements of secondary structure are coloured according to the domain in which they occur; blue for domain I, yellow for domain II and red for domain III. Aligned sequences of OaPDH and TbPDH are also shown. Most of the amino acids conserved in all three sequences are shown as white letters in black boxes. The exception is the NADP+ fingerprint region (residues 10–15 in LlPDH) where the letters are coloured blue and bold indicates conservation. Red stars identify active site residues that form direct hydrogen bonding interaction with ligands, blue dots identify those residues that interact with cofactor.
Topology of LlPDH and comparison with TbPDH and OaPDH
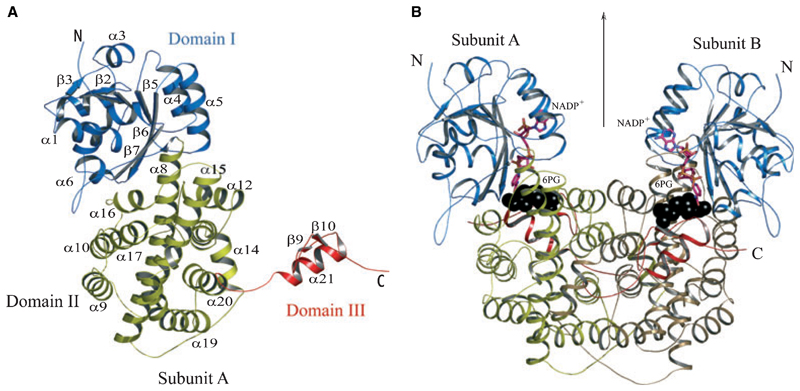
The LlPDH subunit is constructed from three domains (Figs 2,3A). Residues 1–177 form domain I, the cofactor-binding domain, which shows the typical dinucleotide binding Rossmann fold with an additional α–β–α unit. Six parallel β strands in the order β3, β2, β1, β4, β5, β6 and one β strand β7 from the α–β–α unit run antiparallel with respect to others forming a buried β sheet. Six helices, including two small 310 helices, surround the buried sheet. Residues 178–433 form domain II, the helical domain. Two large helices α8 and α14, antiparallel to each other, form the core of this domain and they are enclosed on either side by a set of four helices (α9– α10–α16–α17). Helices α12–α13–α14–α20 are placed at the dimer interface. Domain III, residues 434–469, is assigned as the tail domain. A single helix α21, and two short β strands, β9–β10, extend like an arm through the helical domain of the partner subunit and terminate near the active site of that subunit.
Fig. 3.
(A) Ribbon diagram of an LlPDH subunit. Elements of secondary structure are coloured according to domain as described in Fig. 2 and labelled. The N- and C-termini are marked. (B) The LlPDH dimer viewed perpendicular to the molecular twofold axis of symmetry, which is marked by an arrow. Black spheres depict the position of the substrate (6PG) at the catalytic centre, a stick model is shown for NADP+ and the cofactor is colored according to atom type; C is pink, N is blue, O is orange and P is yellow.
Sequence alignments of LlPDH with TbPDH and OaPDH, based on the automated procedures in clustal w [22], are shown in Fig. 2 together with the secondary structure assignment from LlPDH. TbPDH shares 38% and OaPDH 58% sequence identity with LlPDH, respectively. Differences arise from a deletion of 11 residues and an insertion of four residues in comparison with TbPDH. With respect to OaPDH, there are three insertions and two deletions. LlPDH is truncated by 11 residues compared with OaPDH and is three residues shorter than TbPDH. A least-squares fit of 468 Cα atoms of LlPDH with TbPDH and OaPDH results in an rmsd of 1.2 Å in both cases (not shown). These values indicate a level of structural conservation similar to that of LlPDH in the different crystal forms and we can conclude that the enzymes adopt highly similar folds and dimers.
The dimerization of PDH involves the tail domain of one subunit threading through the helical domain of the neighbouring subunit (Fig. 3B). There are 134 residues that form the dimer interface in LlPDH, 45 of which form stabilizing hydrogen bonding and salt-bridge interactions. There is a larger number of hydrogen-bonding interactions formed between monomers in TbPDH (63) compared with either LlPDH (45) or OaPDH (42). Approximately 25% of the accessible surface area of a monomer contributes to LlPDH dimer formation. Whereas LlPDH and OaPDH have a buried surface area of ≈ 5500 Å2, TbPDH has a larger interface surface area of 6200 Å2.
The cofactor-binding site
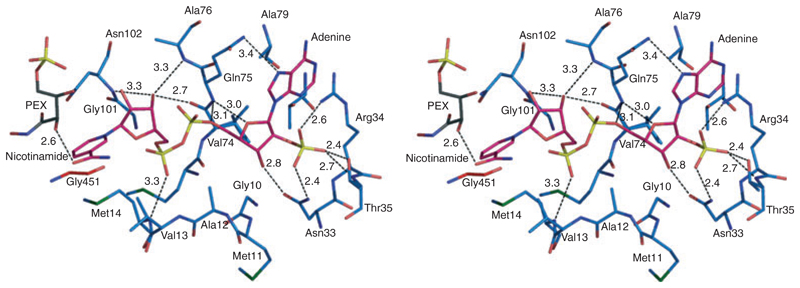
The cofactor binds on the periphery of domain I (Figs 3B,4) with the adenine ribose approaching the β1–α1 turn that carries the fingerprint motif GxAxxG [12]. The fingerprint Ala12 protrudes into the NADP+-binding pocket and restricts the binding depth of cofactor in a similar fashion to that observed for OaPDH [12]. In TbPDH, the alanine in this motif is replaced by glycine suggesting less steric influence on cofactor binding. The adenine stacks against the Arg34 guanidinium group. This arginine, essential for NADP+ binding [19], becomes well ordered in the ternary complex with NADP+. On the other side, the adenine forms hydrophobic interactions with Ala78 and Ala79. The hydrogen bonding of the enzyme with the adenine part of the cofactor is predominantly with the 2′-phosphate and ribose. The 2′-phosphate forms hydrogen bonds with Asn33, Arg34 and Thr35. As a consequence of this, the main chain of α2 adjusts position by ≈ 1 Å, in comparison with complex I. The ribose hydroxyl and phosphate groups participate in hydrogen-bonding interactions with the main chain amide of Gln75 and side chain of Asn33. Met14 is an important residue for positioning the nicotinamide. The cofactor pyrophosphate is hydrogen bonded to the main chain amide of Met14, whereas the side chain is placed against and serves to orient the nicotinamide ring so it is placed to participate in hydride transfer. The importance of the conserved methionine has been noted previously in OaPDH [10]. The nicotinamide ribose is positioned by hydrogen bonds to the carbonyl group of Val74 and main chain amides of Asn102 and Ala76. The nicotinamide carbonyl interacts with substrate/product, forming a hydrogen bond with the C3-hydroxyl of RU5P, C2-hydroxyl of PEX or the C4-hydroxyl of 6PG (in the modelled ternary complex).
Fig. 4.
Stereoview of the cofactor-binding site in complex III. NADP+ is shown as in Fig. 3. LlPDH C atoms are blue, PEX C atoms are black and NADP+ C atoms are pink. All N positions are dark blue, O are red, P are yellow and S are green. Potential hydrogen bonding interactions are depicted as black dashed lines with interatomic distances given in Å.
Some 84 residues of the cofactor-binding domain of LlPDH are strictly conserved in TbPDH, 89 in OaPDH. There are 17 residues within 4 Å of the cofactor, of which 15 are identical in the bacterial, trypanosomal and mammalian PDH. The binding of NADP+ is similar to that in OaPDH, however, differences do exist. First, at the adenine-binding site, Phe83 of OaPDH is replaced by Thr83 in LlPDH. Second, Lys75 of OaPDH is replaced by Gln75 in LlPDH. In OaPDH, the Lys75 side chain adopts different conformations when binding the oxidized and the reduced cofactor [12]. On binding NADP+, Lys75 is directed towards the active site forming a hydrogen bond with the nicotinamide ribose. In LlPDH the side chain of Gln75, adopts a similar conformation, where NE2 donates a hydrogen bond to the adenine N7 and the nicotinamide ribose is hydrogen bonded to the carbonyl group of Val74 and amide of Ala76 and Asn102. The nicotinamide adopts a different conformation in OaPDH compared with LlPDH. In OaPDH, the nicotinamide carbonyl group is hydrogen bonded to the main chain amide of Val12 (Val13 in LlPDH). Furthermore, to accommodate the carboxamide, the main chain of Val12 moves 2.5 Å with respect to LlPDH (data not shown). Also the C4 position, the site of hydride provision and acceptance, is directed away from the active site, towards the carbonyl oxygen of Gly450 of the partner subunit. In LlPDH the nicotinamide is oriented with C4 positioned near C2 of RU5P in the ternary complex, C3 of 6PG in the modelled ternary complex and C1 of PEX/PEA in the inhibitor complex (see below for further discussion).
The active site and catalytic mechanism
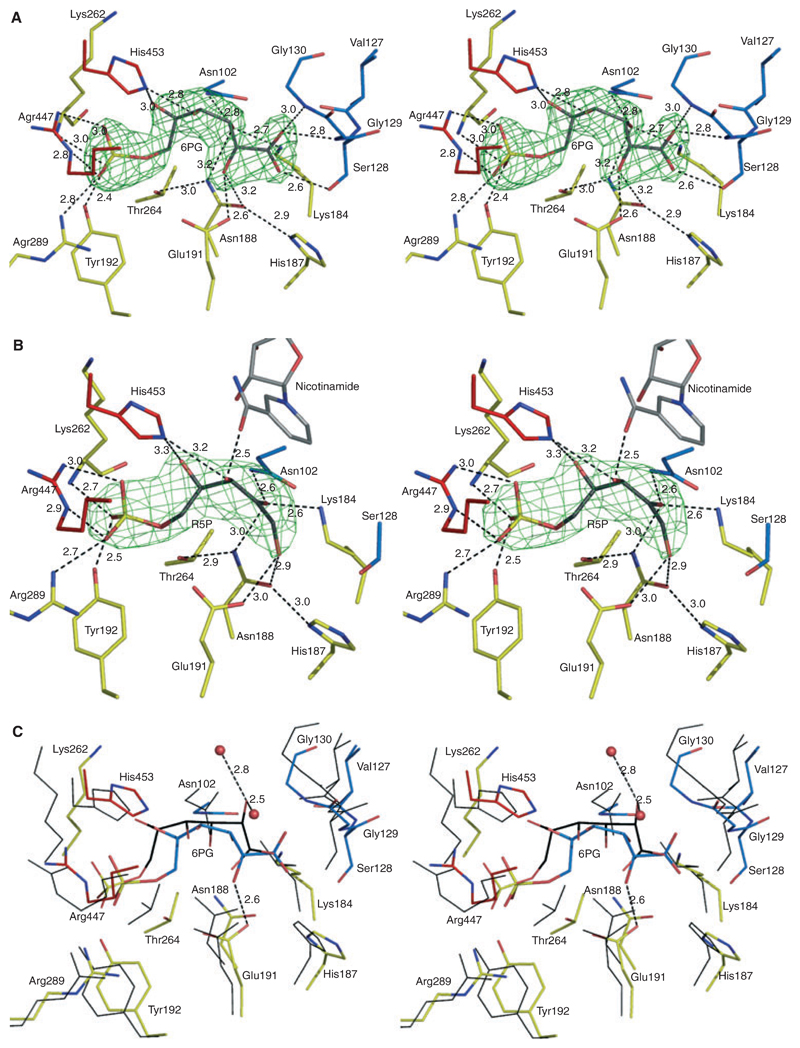
PDH is a homodimer with one active site per monomer. Figure 5A shows the conformation of 6PG in the active site of LlPDH based on the interpretation of well-defined electron density. The average B-factor of the substrate (26.3 Å2) is less than the overall temperature factor of the protein (27.5 Å2).
Fig. 5.
Stereoviews showing interactions at the catalytic centre of LlPDH. (A) The omit difference density map (green mesh) for the substrate 6PG is shown. The map was calculated with coefficients |Fo − Fc|, αcalc and contoured at 4σ. Fo and Fc represent observed and calculated structure-factor amplitudes, respectively, αcalc phases calculated on the basis of atomic coordinates of the model but excluding the substrate. 6PG atomic positions are coloured are follows: C, grey; O, red; P, yellow. The amino acid C atoms are coloured by domain assignment as in Fig. 2. Domain I is blue, domain II is yellow and domain III is red. O positions are red, N are blue. Black dashed lines represent potential hydrogen bonds with distances given in Å. (B) Binding of product, RU5P with the associated omit map. (B) is similar to (A), though note the presence of NADP+ with C atoms grey. (C) The superposition of LlPDH : 6PG and OaPDH : 6PG complexes based on Cα positions of residues shown. LlPDH : 6PG is shown as in (A) except that the C positions of 6PG are coloured cyan. Thin black lines represent OaPDH residues and the associated 6PG is shown as a stick model with C atoms in black.
The active site is a deep cleft surrounded by residues from all three domains, here 6PG lies across α8, which forms the floor of the active site. The C1 of 6PG is placed near to the cofactor-binding domain with the phosphate directed to the loop between α10–α11 and the tail domain of the partner subunit. There are 19 residues within 4 Å of 6PG, of which 14 are absolutely conserved in all known PDH sequences. Eleven of the substrate neighbours are contributed from the helical domain, of which five residues are on α8. These are His187, Asn188, Tyr192, and the catalytically important Lys184 [23] and Glu191 [24]. The cofactor-binding domain contributes five residues (Asn102, Val127, Ser128, Gly129, Gly130), and the tail domain of the partner subunit provides three (Arg447, Arg450, His453) for substrate binding. In addition, there are four water molecules mediating interactions between 6PG and the enzyme (not shown).
The phosphate group of 6PG forms hydrogen bonds with Tyr191, Arg289, Arg447 and the main chain amide of the highly conserved Lys262. Once the phosphate has bound, Lys262 covers the active site. This basic residue is placed between two glycines, in a conserved GxKGT motif, where x is serine in TbPDH, glutamine in OaPDH and asparagine in LlPDH (Fig. 2). In addition, a conserved water molecule in all complexes, mediates interaction between the phosphate and Gly263 and Thr264 of the motif. The 4-OH of 6PG interacts with His453 and a water molecule. The C4 position cannot have an R-conformation due to steric clash with Asn102, which in turn is hydrogen bonded to C3-OH. This is consistent with binding studies on Candida utilis PDH using substrates with different conformation at C4, 6-phosphogalactose in particular, which was unable to bind the enzyme [7,8]. The water molecule with which C4-OH interacts is replaced by the carbonyl group of the nicotinamide in the modelled ternary complex. Whereas C4-OH dictates the binding specificity of the substrate 6PG, C5-OH has only a weak interaction with His453 and a water molecule. The C3-OH forms hydrogen-bonding interactions with the catalytic Lys184 and two asparagines (Asn188 and Asn102). Lys184 also interacts with the carbonyl oxygen of Val129 and the carboxylate oxygen O1. Furthermore three residues Ser128, Gly129 and Gly130 interact with the carboxylate. The carboxylate oxygen O1A accepts hydrogen bond from main chain amides of Gly129 and Gly130. Ser128 is hydrogen bonded in a chain with carboxylate O1 and NE2 of His187. The side chain OE1 of the catalytic Glu191 interacts with 2-OH with OE2 hydrogen bonded to a water molecule. RU5P binds in a similar fashion as 6PG in complex II (Fig. 5B). The difference compared with the 6PG complex is the loss of hydrogen bonds with Ser128, Gly129 and Gly130, because the carboxylate group is absent in the product.
That we observe a difference in the cofactor-binding domains of each subunit in the homodimeric PDH, as outlined earlier is, in the context of a previous hypothesis, worth further comment. Hanau et al. [21] showed that 6PG activates decarboxylation of a substrate mimic, 6-phospho-3-keto-2-deoxygluconate, suggesting that occupancy of one active site has an influence on the other. The reduced cofactor is necessary for the enzyme to carry out this decarboxylation yet is not required to participate as a redox partner. To explain these observations a model was proposed in which the two active sites of PDH are engaged in different reactions during catalysis. One active site will be primed to carry out decarboxylation, whereas the other is oxidizing the substrate. The subunits then reverse their roles during turnover. Our structural models indicate that PDH is not a fixed entity but that the cofactor-binding domain has a capacity to adjust position and such movements may contribute to cooperativity in this enzyme.
The catalytic residues of PDH are absolutely conserved in the bacterial, trypanosomatid and mammalian enzymes. The overlay of the active site residues of the sheep liver enzyme onto LlPDH gave an rmsd of Å positional deviation (19 residues at 4 Å distance). The second neighbours of 6PG (at 6 Å distance) are also fully conserved with respect to the sheep homologue. Thus, previously known active site differences between the sheep and TbPDH enzyme also applies here for LlPDH [13]. However, despite the fact that the catalytic residues of LlPDH are the same as those of the OaPDH, contrary to expectation, there are significant differences in the conformation of 6PG (Fig. 5C).
In OaPDH and LlPDH, the phosphate group binds in a similar fashion, approaching the triplet Arg447, Arg289, Tyr191. The C5-OH and C3-OH positions are conserved, although we note that C3-OH forms an additional hydrogen bond with Asn102, an interaction that is absent in OaPDH. However C4-OH, C2-OH and the carboxylate show significant differences between the two structures. In OaPDH, the C4-OH and C2-OH do not form any hydrogen bonds with the enzyme. In LlPDH, C4-OH accepts a hydrogen bond from His453 and also interacts with a water molecule. The C2-OH forms a hydrogen bond with the proposed catalytic residue Glu191 and with Asn188. This interaction of C2-OH with Glu191 is essential, because the tautomerization step of the catalytic reaction requires a general acid to donate a proton to the C1 carbon of the 1,2-enediol, whereas C2 accepts a proton from Lys184 in the conversion to RU5P. The structure of the LlPDH structure in complex with 6PG, is entirely consistent with previous mechanistic studies [8–11,14]. In contrast, in OaPDH, the carboxylate oxygen occupies the C2-OH position and is hydrogen bonded with the glutamate. Furthermore the conserved Gly129 and Gly130 have no hydrogen bonds with the substrate as seen in LlPDH. It is unclear why two conformations of 6PG are observed in a highly conserved PDH active site though it may be significant because large differences in affinity for substrate analogues were noted in comparing the sheep and trypanosomal enzymes. Different crystallization conditions were employed for the two structure determinations and these may have contributed in some way to isolating the different structures. Although the structure of the trypanosomal enzyme bound to substrate has not been resolved, analogous differences in binding potential, in spite of conservation of key residues, may offer an explanation for those results.
Inhibition by PEX/PEA
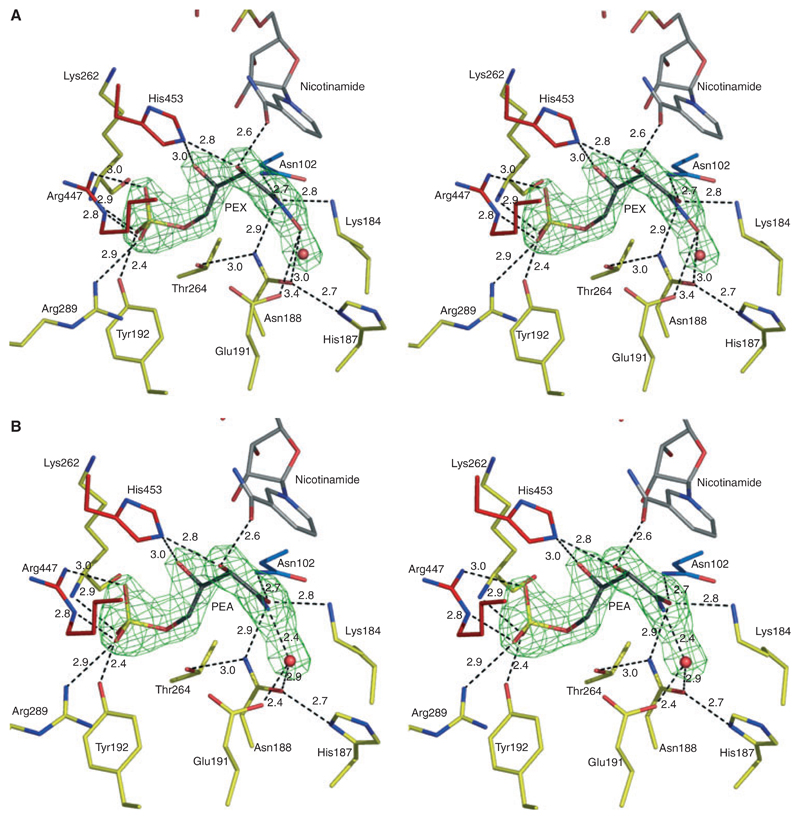
Well-defined electron density in the active site of complex III was modelled as a mixture of PEX (occupancy 0.7; Fig. 6A) and PEA (occupancy 0.3; Fig. 6B). The mean atomic B-factor of the PEX/PEA combination (13.3 Å2/15.2 Å2) is less than the overall B-factor (19.0 Å2) of the protein. The mode of binding of PEX/PEA in the active site of LlPDH is similar to that of 6PG/RU5P, where the phosphate is recognized by interactions with Tyr192, Arg289 and Arg447. All of the functional groups of PEX/PEA participate in hydrogen bonding with the enzyme either directly or via a solvent mediated network. PEX/PEA also interacts with NADP+. The C2-OH of PEX/PEA donates a hydrogen bond to the nicotinamide carbonyl and accepts one from the side chain of His453 of the partner subunit. The C3-OH interacts with the nicotinamide ribose via a water-mediated interaction. The planar sp2 hybridized C1 is near to the nicotinamide ring and the C1 carbonyl oxygen potentially accepts hydrogen bonds donated from the catalytic Lys184, Asn102 and Asn188. The PEX/PEA amide is linked to the main chain amide of Gly130 via a water molecule. The PEX N1 hydroxyl is hydrogen bonded to the catalytic Glu191. However PEA, lacking this hydroxyl, interacts with a water molecule that bridges over to Glu191. As observed in the complexes with substrate and product, the phosphate group of PEX/PEA has solvent mediated interactions with the catalytic glutamate Glu191, Arg289, Gly263 and Thr264 (not shown).
Fig. 6.
Stereoviews depicting inhibition of LlPDH. (A) Omit difference density map (green mesh) in the active site calculated as described in Fig. 5 by ignoring the scattering contributions from the ligands and the water (red sphere) in estimating αcalc, the map is contoured at 4σ. The PEX model is shown with C atoms in black. (B) PEA model.
An overlay of PEX/PEA with 6PG and RU5P (not shown) indicates that the inhibitors adopt similar conformations in the active site. The PEX hydroxamate mimics the enol-keto resonance structure of the 2-cis-enediol high-energy intermediate proposed for the 6PGDH reaction and so effectively inhibits the enzyme. When PEX is bound, Glu191 accepts a hydrogen bond from the hydroxamate but when PEA is present then a water molecule is sequestered to satisfy the hydrogen-bonding capacity of the functional groups. The presence of the terminal hydroxyl group of PEX is important because the additional hydrogen bond interactions compared with PEA results in improved binding and inhibition. PEX has a Ki value of 10 nm, PEA a Ki value of 1520 nm against TbPDH [18]. It has not yet been possible to extend inhibition analysis using pure PEX and PEA against LlPDH.
Experimental procedures
Purification, crystallization, data collection and processing
LlPDH was obtained following an established protocol [25], then concentrated to 20 mg·mL−1 in a buffer containing 50 mm Tris/HCl pH 7.2 and 200 mm NaCl. The high purity of the sample was confirmed with SDS/PAGE and MALDI-TOF MS. Protein concentration was determined spectrophotometrically using a theoretical extinction coefficient of 61895 m−1·cm−1 (280 nm). 6PG, NADP+ and RU5P were purchased from Sigma Chemicals (St Louis, MO); the inhibitor PEX was synthesized [18]. Complexes I and II, were prepared by incubating protein with ligands in a 1 : 5 ratio. Complex III was prepared by mixing the protein, NADP+ and inhibitor in a ratio of 1 : 5 : 10. Crystals were grown in hanging drops constructed from 2 μL of protein solution and 2 μL of reservoir containing 0.1 m sodium cacodylate pH 6.5, 300 mm ammonium acetate, 25% w/v polyethylene glycol 3350.
Prior to X-ray exposure, crystals were cryoprotected in 20% glycerol and cooled in a stream of gaseous nitrogen at 100 K. Diffraction data were measured in-house using a Micromax 007 rotating anode generator (Cu-Kα λ = 1.5418 Å, 40 kV, 20 mA) and R-AXIS IV++ image plate detector (Rigaku-Europe, Sevenoaks, UK). A second dataset for complex III was measured at the Daresbury synchrotron on beam-line ID14.1 (λ = 1.488 Å), using a QUANTUM detector (Area Detector Systems Corp., Powey, CA). The denzo/scalepack programs [24] were used to index and process the data (Table 1).
Structure determination and refinement
LlPDH is a homodimer and each subunit comprises 472 amino acids of molecular mass 52 kDa. A monomer constitutes the asymmetric unit of the binary substrate complex, whereas three monomers form the asymmetric unit for the ternary complexes. Molecular replacement (amore) [27,28], using a polyalanine model of OaPDH, solved the binary complex structure. Density modification (dm) [27] then produced a map of excellent quality. Graphics inspection of electron density maps, together with model fitting was carried out using the program o [29]. Refinement was carried out in refmac5 [30], with a random selection of reflections (5%) flagged for the calculation of R-free. A particular strength of this refinement program is the use of maximum-likelihood weighting which allows weak or incomplete higher resolution data to be incorporated into the calculations. The benefit is that both the number and resolution of observations are increased. The refined model provided the subunit template in MR calculations (phaser) [31] to determine the structures of the ternary complexes by placement of the three subunits. The program coot [32] was used for map inspection and model building and structure refinements were completed by inclusion of solvent positions, and the relevant ligands. In the 6PG complex, cacodylate is observed interacting with the side chain of His453. In the PEX complex, a strong electron density feature at the interface of subunit A and C has been identified as polyethylene glycol, derived from the crystallization conditions. Four chloride ions are also modelled in this structure. The electron density is continuous for main chain atoms from residue 1–469 in all subunits with only three C-terminal residues missing. In addition, in complex III, remnants of the affinity tag used to facilitate purification were also defined. In all subunits, two residues Asn177 and Thr454 are well defined in the density but present ϕ/ψ combinations in the disallowed region of a Ramachandran plot. Stereochemistry was assessed using procheck [33], molecular images were prepared with pymol [34] and Fig. 2 with the program aline (provided by CS Bond, unpublished). Coordinates and diffraction data have been deposited in the Protein Data Bank and codes are given in Table 1.
Acknowledgements
JI thanks CAPES for fellowship number BEX 2000/04-0. WNH, IHG and MPB thank the Wellcome Trust and WNH thanks the Biotechnology and Biological Sciences Research Council, Swindon, UK (Structural Proteomics of Rational Targets) for support. We gratefully acknowledge provision of synchrotron beam time at the Synchrotron Radiation Source, Daresbury Laboratory, for data collection, and for preliminary crystal characterization at the European Synchrotron Radiation Facility, Grenoble.
Abbreviations
- PDH
6-phosphogluconate dehydrogenase
- PEA
4-phospho-d-erythronohydroxamide
- PEX
4-phospho-d-erythronohydroxamic acid
- 6PG
6-phosphogluconate
- RU5P
ribulose 5-phosphate
References
- 1.Barrett MP. The pentose phosphate pathway and parasitic protozoa. Parasitol Today. 1997;13:11–16. doi: 10.1016/s0169-4758(96)10075-2. [DOI] [PubMed] [Google Scholar]
- 2.Hanau S, Rinaldi E, Dallocchio F, Gilbert IH, Dardonville C, Adams MJ, Gover S, Barrett MP. 6-Phosphogluconate dehydrogenase: a target for drugs in African trypanosomes. Curr Med Chem. 2004;11:2639–2650. doi: 10.2174/0929867043364441. [DOI] [PubMed] [Google Scholar]
- 3.Gvozdev VA, Gerasimova TI, Kogan GL, Braslavskaya O. Role of the pentose phosphate pathway in metabolism of Drosophila melanogaster elucidated by mutations affecting glucose 6-phosphate and 6-phosphogluconate dehydrogenases. FEBS Lett. 1976;64:85–88. doi: 10.1016/0014-5793(76)80255-4. [DOI] [PubMed] [Google Scholar]
- 4.Hughes MB, Lucchesi JC. Genetic rescue of a lethal ‘null’ activity allele of 6-phosphogluconate dehydrogenase in Drosophila melanogaster. Science. 1977;196:1114–1115. doi: 10.1126/science.404711. [DOI] [PubMed] [Google Scholar]
- 5.Lobo Z, Maitra PK. Pentose phosphate pathway mutants of yeast. Mol Gen Genet. 1982;185:367–368. doi: 10.1007/BF00330815. [DOI] [PubMed] [Google Scholar]
- 6.Marchand M, Kooystra U, Wierenga RK, Lambeir AM, Van Beeumen J, Opperdoes FR, Michels PA. Glucosephosphate isomerase from Trypanosoma brucei. Cloning and characterization of the gene and analysis of the enzyme. Eur J Biochem. 1989;184:455–464. doi: 10.1111/j.1432-1033.1989.tb15038.x. [DOI] [PubMed] [Google Scholar]
- 7.Berdis AJ, Cook PF. Chemical mechanism of 6-phosphogluconate dehydrogenase from Candida utilis from pH studies. Biochemistry. 1993;32:2041–2046. doi: 10.1021/bi00059a022. [DOI] [PubMed] [Google Scholar]
- 8.Berdis AJ, Cook PF. Overall kinetic mechanism of 6-phosphogluconate dehydrogenase from Candida utilis. Biochemistry. 1993;32:2036–2040. doi: 10.1021/bi00059a021. [DOI] [PubMed] [Google Scholar]
- 9.Rippa M, Giovannini PP, Barrett MP, Dallocchio F, Hanau S. 6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from different species. Biochim Biophys Acta. 1998;1429:83–92. doi: 10.1016/s0167-4838(98)00222-2. [DOI] [PubMed] [Google Scholar]
- 10.Cervellati C, Dallocchio F, Bergamini CM, Cook PF. Role of methionine-13 in the catalytic mechanism of 6-phosphogluconate dehydrogenase from sheep liver. Biochemistry. 2005;44:2432–2440. doi: 10.1021/bi0476679. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Dworkowski FS, Cook PF. Importance in catalysis of the 6-phosphate-binding site of 6-phosphogluconate in sheep liver 6-phosphogluconate dehydrogenase. J Biol Chem. 2006;281:25568–25576. doi: 10.1074/jbc.M601154200. [DOI] [PubMed] [Google Scholar]
- 12.Adams MJ, Ellis GH, Gover S, Naylor CE, Phillips C. Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism. Structure. 1994;2:651–668. doi: 10.1016/s0969-2126(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 13.Phillips C, Dohnalek J, Gover S, Barrett MP, Adams MJ. A 2.8 Å resolution structure of 6-phosphogluconate dehydrogenase from the protozoan parasite Trypanosoma brucei: comparison with the sheep enzyme accounts for differences in activity with coenzyme and substrate analogues. J Mol Biol. 1998;282:667–681. doi: 10.1006/jmbi.1998.2059. [DOI] [PubMed] [Google Scholar]
- 14.Hwang CC, Cook PF. Multiple isotope effects as a probe of proton and hydride transfer in the 6-phosphogluconate dehydrogenase reaction. Biochemistry. 1998;37:15698–15702. doi: 10.1021/bi981288w. [DOI] [PubMed] [Google Scholar]
- 15.Hwang CC, Berdis AJ, Karsten WE, Cleland WW, Cook PF. Oxidative decarboxylation of 6-phosphogluconate by 6-phosphogluconate dehydrogenase proceeds by a stepwise mechanism with NADP and APADP as oxidants. Biochemistry. 1998;37:12596–12602. doi: 10.1021/bi980611s. [DOI] [PubMed] [Google Scholar]
- 16.Pasti C, Rinaldi E, Cervellati C, Dallocchio F, Hardre R, Salmon L, Hanau S. Sugar derivatives as new 6-phosphogluconate dehydrogenase inhibitors selective for the parasite Trypanosoma brucei. Bioorg Med Chem. 2003;11:1207–1214. doi: 10.1016/s0968-0896(02)00650-8. [DOI] [PubMed] [Google Scholar]
- 17.Dardonville C, Rinaldi E, Hanau S, Barrett MP, Brun R, Gilbert IH. Synthesis and biological evaluation of substrate-based inhibitors of 6-phosphogluconate dehydrogenase as potential drugs against African trypanosomiasis. Bioorg Med Chem. 2003;11:3205–3214. doi: 10.1016/s0968-0896(03)00191-3. [DOI] [PubMed] [Google Scholar]
- 18.Dardonville C, Rinaldi E, Barrett MP, Brun R, Gilbert IH, Hanau S. Selective inhibition of Trypanosoma brucei 6-phosphogluconate dehydrogenase by high-energy intermediate and transition-state analogues. J Med Chem. 2004;47:3427–3437. doi: 10.1021/jm031066i. [DOI] [PubMed] [Google Scholar]
- 19.Tetaud E, Hanau S, Wells JM, Le Page RW, Adams MJ, Arkison S, Barrett MP. 6-Phosphogluconate dehydrogenase from Lactococcus lactis: a role for arginine residues in binding substrate and coenzyme. Biochem J. 1999;338:55–60. [PMC free article] [PubMed] [Google Scholar]
- 20.Hayward S, Berendsen HJC. Systematic analysis of conformational domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 21.Hanau S, Dallocchio F, Rippa M. Is there an alternating site cooperativity between the two subunits of lamb liver 6-phosphogluconate dehydrogenase? Biochem J. 1993;291:325–326. doi: 10.1042/bj2910325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Chooback L, Cook PF. Lysine 183 is the general base in the 6-phosphogluconate dehydrogenase-catalyzed reaction. Biochemistry. 1999;38:11231–11238. doi: 10.1021/bi990433i. [DOI] [PubMed] [Google Scholar]
- 24.Karsten WE, Chooback L, Cook PF. Glutamate 190 is a general acid catalyst in the 6-phosphogluconate-dehydrogenase-catalyzed reaction. Biochemistry. 1998;37:15691–15697. doi: 10.1021/bi9812827. [DOI] [PubMed] [Google Scholar]
- 25.Tetaud E, Hall DR, Gourley DG, Leonard GA, Arkison S, Barrett MP, Hunter WN. Crystallization and preliminary X-ray diffraction studies of 6-phosphogluconate dehydrogenase from Lactococcus lactis. Acta Crystallogr D Biol Crystallogr. 1998;54:1422–1424. doi: 10.1107/s0907444998004909. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr, Part A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Navaza J. Amore – an automated package for molecular replacement. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 29.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck – a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 34.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. http://pymol.sourceforge.net. [Google Scholar]
- 35.Cruickshank DW. Remarks about protein structure precision. Acta Crystallogr D Biol Crystallogr. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]