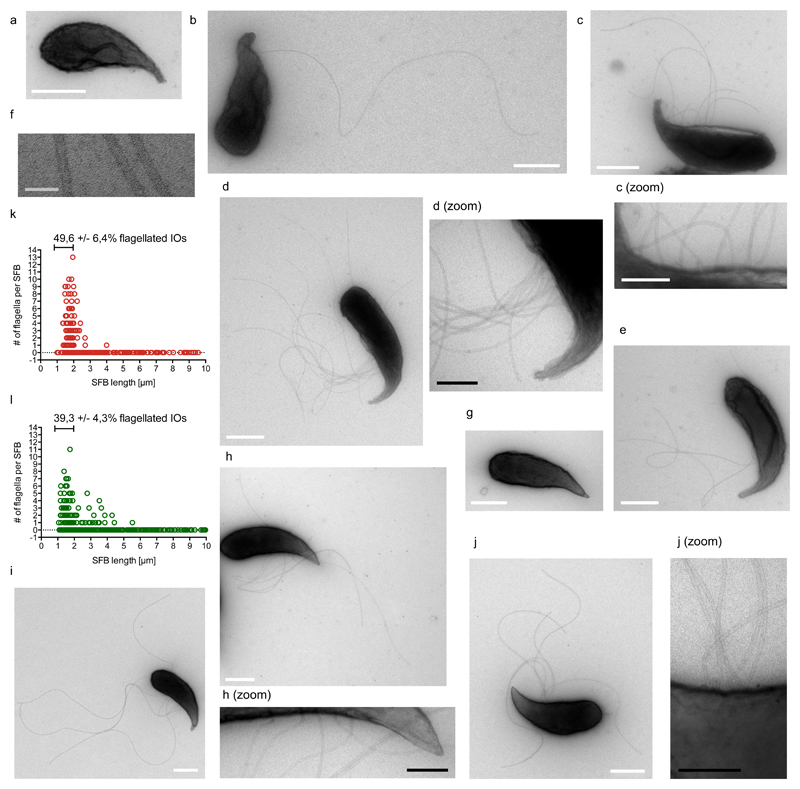
The gut commensal Segmented Filamentous Bacterium (SFB) attaches to the ileal epithelium and potently stimulates the host immune system1. Using transmission electron microscopy (TEM), we show that mouse and rat SFB are flagellated above the concave tip at the unicellular intracellular offspring (IO) stage and that flagellation occurs prior to full IO differentiation and release of IOs from SFB filaments. This finding adds a missing link to the SFB life-cycle.
The Clostridia-related anaerobe SFB is found in many vertebrate species and has emerged as a key member of the gut microbiota2. Through its attachment, SFB stimulates innate and adaptive immune responses, including strong IgA and Th17 responses1,3. SFB promotes colonization resistance both in and outside of the gut and exerts a complex adjuvant effect on systemic immune responses that can aggravate pathologies in models of colitis, encephalitis, arthritis and neurodevelopmental abnormalities1,4,5. Much about the complex life-cycle of SFB remains unexplored as SFB is difficult to culture and instead is routinely propagated in germfree mice6.
The description of the SFB life-cycle is largely based on TEM images of mouse and rat ileal sections from the 1970s7,8. Single-cell teardrop-shaped IOs were shown to intimately attach to host cells using their pointed tip and to grow out into long filaments. Filament segments then undergo a symmetric followed by an asymmetric division leading to the engulfment of daughter cells by their mother cells. Division of the daughter cell produces two IOs that are released upon breakdown of the filament end. Alternatively, daughter cells may become spores for host-to-host transmission2,4.
Surprisingly, genome analysis of mouse SFB (mSFB) and rat SFB (rSFB) identified a full set of flagella biosynthesis genes, including up to four genes for FliC, the main structural component of flagella9–12. SFB flagellation was hypothesized to be part of the IO stage11,12, and to possibly mediate attachment11. Yet, multiple studies in various host species failed to detect a flagellated SFB stage2,9,13,14, and as SFB flagellin transcript abundance was very low10 and flagellin could not be detected in feces of SFB monocolonized mice15, it suggested that flagellation may not be prevalent or possibly absent9. More recently, mass spectrometry analysis revealed SFB-specific FliC peptides in mouse and human fecal samples16,17 and, based on immunofluorescent analysis, flagella were proposed to be located along long SFB filaments in an irregular fashion17.
Here, we explored the existence of a flagellated phase in the SFB life-cycle using TEM of mSFB and rSFB purified from monoassociated mice. As previously shown3, mSFB were adherent in germfree mice while rSFB were non-adherent and hence not detected in PBS-flushed ileal preparations (Supplementary Figures 1a/b and 2a/b). SFB were isolated from the caecal and intestinal contents (Supplementary Figures 1c and 2c), purified (Supplementary Figures 1d and 2d), fixed, and processed for negative contrast staining. Flagella-like structures were identified on mSFB and rSFB of up to 4,0 μm and 5,5 μm in length, respectively (Figure 1a-j, Supplementary Figures 3a-g and 4a-i), while 1-3 μm-long SFB accounted for 99% of all flagellated mSFB and 89% of all flagellated rSFB (Figure 1k/l). None of the flagellated SFB appeared to have a septum and, consistent with their small size and pointed tip, therefore fit the definition of unicellular IOs. Flagella emanated from above the concave side of the pointed tip and were absent from the posterior region. Flagella filaments were found clustered (Figure 1d/j, Supplementary Figure 4d) and spread out along the IO body (Figure 1c/h, Supplementary Figure 4a/e). On average, flagella were located between approximately the 38th and 52nd percentile of the total SFB length, starting from the pointed tip, for 1-3 μm-long mSFB and rSFB (Supplementary Figures 3f and 5a-d, Supplementary Table 1). The maximum number of flagella identified was 13 flagella for mSFB and 11 flagella for rSFB (Figure 1k/l). When considering only small IOs (1-2 μm), 49,6 +/- 6,4 % of mSFB and 39,3 +/- 4,3 % of rSFB were flagellated (Figure 1k/l). Flagella filaments exhibited a continuous lumen, averaged 17,7 +/- 1,0 nm in diameter (Figure 1f), and were up to 5,5 μm and 5,2 μm long for mSFB and rSFB, respectively (Supplementary Figures 3g and 4i), although debris occasionally prevented full flagella length measurements (Supplementary Figure 3b/c). Together, these results show that SFB flagellation is restricted to the unicellular IO stage and that flagellation is conserved for SFB from different host species.
Figure 1. SFB from mouse and rat are flagellated at the intracellular offspring stage.
a-f. TEM images of in vivo-grown mSFB without (a) and with (b-e) flagella; (f) shows high magnification of three flagella filaments. g-j. TEM images of in vivo-grown rSFB without (g) and with (h-j) flagella. k/l. mSFB (k) and rSFB (l) summary of flagella number for in vivo-grown SFB including mean percent, +/- standard error (SE), of 1 to 2 μm-long SFB that are flagellated. Scale bars: white: 500 nm; black: 200 nm; grey: 50 nm. TEM images and summaries are from four mSFB (n=320) and three rSFB (n=468) independent experiments with similar results.
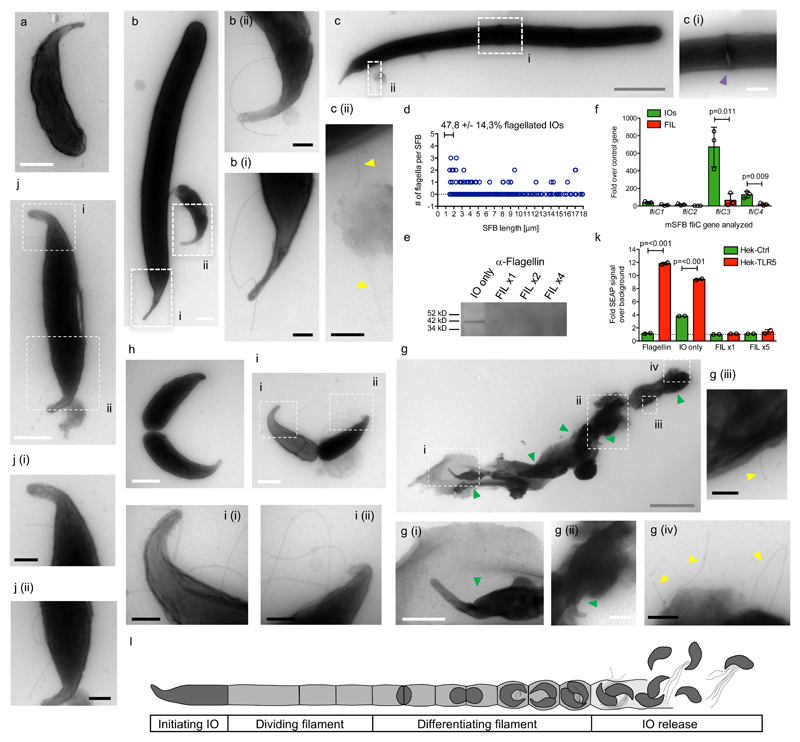
SFB flagellation was then assessed during in vitro growth. SFB from monocolonized mice were purified, filtered through a 5-μm filter (Supplementary Figure 6a), and IOs were grown on colonic epithelial cells under low oxygen conditions in rich medium for three days (Supplementary Figure 6b)6. SFB were purified by selective centrifugation (Supplementary Figure 6c), fixed, and processed for TEM. For in vitro-grown mSFB, 47,8 +/- 14,3 % of 1-2 μm-long SFB were flagellated (Figure 2a-d; Supplementary Figure 7a-e); yet, flagellation could be observed on mSFB up to 17 μm in length (Figure 2b-d, Supplementary Figure 7c-f). Furthermore, flagellated SFB above 14 μm in length exhibited one or multiple septa (Figure 2c; Supplementary Figure 7d/f), indicating that flagellation is not restricted to the unicellular SFB stage during in vitro growth. Flagella were found on average between the 47th and 51st percentile of the total SFB length in 1-3 μm-long mSFB, while this percentage decreased sharply with increasing SFB length (Supplementary Figure 5e/f, Supplementary Table 1), reflecting the outgrowth of the bacterium from its distal end. Together, these results demonstrate that IO flagellation occurs during both in vitro and in vivo growth while flagellation dynamics differ in the two conditions.
Figure 2. SFB flagellation occurs during in vitro growth and IO development.
a-c. TEM images of in vitro-grown mSFB without (a) and with (b-c) flagella and (c) a septum. d. Summary for in vitro-grown mSFB with mean percent, +/- SE, of 1 to 2 μm-long flagellated SFB that are flagellated. e. Anti-flagellin western blot of IO-only and filament-enriched (FIL) mSFB fractions. The full gel blot from which this panel was generated is available as Supplementary Figure 8c. f. qPCR analysis of fliC genes of in vivo-grown mSFB fractions. TEM images of (g) in vitro-grown mSFB with flagella in the vicinity of IOs released from a ruptured filament (n=2) and (h-j) in vivo-grown mSFB without (h) and with (i-j) flagella during IO development (n=5 and n=8, respectively). k. TLR5 stimulation by mSFB fractions using HEK reporter cell lines. l. Schematic overview of the SFB replicative life-cycle. Arrows: yellow: flagella; green: IOs. In (e/k), “x” denotes fold increase in SFB numbers assayed. Scale bars: white: 500 nm; black: 200 nm; grey: 2 μm. (e, f, k) Data from one of two (e), from three (f), and one of three (k) independent experiments with similar results. (f/k) Mean +/- standard deviation and two-sided t-test statistical analysis. TEM images are from four in vitro (n=278) and four in vivo (n=320) independent mSFB experiments with similar results.
To confirm SFB flagellation through additional methods, purified SFB were separated by filtration through a 5-μm filter into an IO-only fraction and, through reverse filtration, a filament-enriched fraction. Using a commercially available anti-FliC antibody that shows high specificity to recombinant SFB FliCs (Supplementary Figure 8a/b), Western blot analysis of SFB lysate revealed only one band, present exclusively in the IO-only fraction, at the expected molecular weight for full-length SFB flagellin (42 kD) (Figure 2e). In addition, surface proteins of SFB were labelled with biotin, purified using streptavidin-coated beads, and analysed by mass spectrometry. FliC peptides were exclusively identified in the IO-only fraction; FliC3 was the major flagellin, followed by FliC4 (Supplementary Figure 8d). In agreement, transcriptional analysis showed that fliC transcripts are enriched in the IO-only fraction and identified fliC3 as the most abundant fliC transcript, followed by fliC4 (Figure 2f). These findings validate the IO stage as the major flagellated stage in the SFB life-cycle and identifies FliC3 as the dominant flagellin.
IO differentiation is morphologically associated with a broadening and bulbous appearance of the filament end (Supplementary Figure 9a) and TEM images previously revealed that the breakdown of the septa dividing newly-formed IOs precedes IO release from the filament end4,7,8. In agreement, the fixable membrane dye FM4-64FX stained septa along the mSFB filament until the filament end where discrete septa disappeared and IOs were discernible (Supplementary Figure 9b/c). Breakdown of the filament leads to empty filament remnants visible at the filament end (Supplementary Figure 9d)6. Using TEM, flagella could be found emanating from filament remnants surrounding clusters of IOs (Figure 2g), suggesting that IOs are flagellated when released from the filament. As two IOs are produced from one daughter cell during IO development, TEM grids were scanned for SFB resembling stages of IO development. For both mSFB and rSFB, IOs at the late stage of septation could be found non-flagellated and flagellated (Figure 2h/i, Supplementary Figures 10a-f and 11a-c). Furthermore, flagellation could be seen on IOs at an earlier stage of IO development, prior to septum formation (Figure 2j, Supplementary Figures 10g and 11d). These findings reveal that IO flagellation can occur during IO development before IO septation.
Finally, the immunostimulatory potential of flagellin expression by SFB was assessed. Control and TLR5-expressing HEK reporter cell lines were challenged with SFB lysate and exclusively the IO-only fraction, and not the filament-enriched fraction, significantly stimulated the TLR5 reporter cell line (Figure 2k). Together with data showing that recombinantly-expressed SFB FliCs stimulate TLR5 (Supplementary Figure 8e)9,17, these data demonstrate that IOs can stimulate TLR5 signaling.
Using quantitative TEM, we hereby show that flagella are unique to the unicellular IO stage of SFB from different host species and that flagella are located above the concave part of the pointed tip that mediates attachment. In addition, through the separation of IOs from SFB filaments, we show that IOs, but not filaments, express high fliC transcript numbers, express FliCs on their surface, and stimulate a TLR5 reporter cell line. We furthermore provide evidence that flagellation starts during IO development and that IOs are released in a flagellated state from the filament as part of their regular replicative life-cycle (Figure 2l). We thereby place SFB flagellation in the proper context of the SFB life-cycle.
In the SFB genome, flagella synthesis genes are clustered together with methyl-accepting chemotaxis proteins9,10 and IOs may thus use chemoattractant sensing and flagella-mediated motility to cross the mucus layer and reach the epithelial surface. Notably, flagella dynamics differed during in vitro growth condition. As gravity alone promotes SFB interaction with host cells, we speculate that this, in addition to the rich growth medium, may perturb flagella regulation. Interaction of flagellated SFB IOs with intestinal epithelial cells, and potential uptake of IOs by antigen presenting cells, may therefore induce TLR5 signalling or be sensed in the host cell cytosol to stimulate the NLRC4 inflammasome18. Future work will need to address the impact of SFB flagellation on the host, including potential roles in Th17 activation and differentiation, IgA responses and microbiota composition15,19,20.
Material and Methods
SFB monocolonization
Germ-free C57BL/6 were bred at the Institut Pasteur animal facility accredited by the French Ministry of Agriculture. Animals were maintained in germ-free isocages and fed ad libitum on commercial diet (R03-40, SAFE) sterilized by gamma irradiation (40 kGy). Monocolonization of 7 to 11-week old germfree mice with either the mouse SFB-NL or rat SFB-YIT strain was achieved by gavage with previously frozen fecal pellets of monocolonized mice resuspended in water. Male and female mice were used randomly, based on availability. Blinding was not performed and number of mice used depended on quantities of purified SFB required for individual experiments. Monocolonization was verified by Gram strains, wet mounts and culturing techniques. Mice were killed between 5 and 21 days post colonization.
Purification of SFB from monoassociated mice
All liquids were pre-equilibrated overnight in an anaerobic chamber. SFB-monoassociated mice were killed aseptically and dissected in an anaerobic cabinet. The intestinal contents were resuspended in 40 ml PBS, homogenized by gentle pipetting and 10 ml of suspension was layered onto 5 ml of 50% Nycodenz (Progen, 1002424) in PBS in 15-ml tubes and centrifuged at 4,000xg in a swinging bucket for ten minutes at room temperature (RT). The interphase was recovered, pooled and layered onto 2x4 mL 30% Nycodenz, and centrifuged. All but the top 3 ml and pellet were collected, pooled and washed in 50ml PBS. Samples were centrifuged for 15 min, pellets were resuspended in PBS and cleaned through additional 30% Nycodenz/PBS cushions as needed. Washed bacteria were fixed in 4% paraformaldehyde (PFA) for 15 min and stored in 1% PFA at 4°C. Gram stains were performed using the Gram-Hücker kit (RAL Diagnostics, 361520).
TEM
Washed, PFA-fixed IOs were spotted onto glow discharged grids (FCF-200-Cu50, Delta Microscopy) for 15 min in a humid chamber, washed 3x in PBS for 1 min and post-fixed with 2.5% glutaraldehyde for 5 min. Samples were contrasted with a mixture of 4% uranyl acetate/2% methyl cellulose (ratio 1:9) (Sigma:M6385) and imaged on a Tecnai Spirit 120 Kv microscope equipped with EAGLE 4Kx4K camera. SFB length, flagella number and length were quantified using ImageJ software.
TC7 and SFB growth medium
The Caco-2 sub-clone TC7 was originally obtained from A. Servin and grown as indicated by the ATCC. SFB growth medium was prepared as previously described6, with minor modification. Iron and nucleotides were made the day before every experiment and hemin and additional sugars were not included.
In vitro culture of SFB, SFB purification and lysate preparation
The in vitro culturing of SFB was performed at 2% oxygen as described previously6. On day 3, the supernatant was collected and centrifuged at 100xg for 3 min. Bacteria in the supernatant were recovered, pelleted at 4,000xg for 10 min and again cleaned through a 100xg spin for 3 min. For TEM, bacteria were fixed in 4% PFA for 15 min and stored in 1% PFA at 4°C.
For lysate used in Western and TLR5 reporter cell stimulation, bacteria were separated into IO-only and filament-enriched fractions using a 5-μm filter. Bacterial suspensions were centrifuged for 15 min at 4000xg and pellets were resuspended in PBS, quantified by qPCR, diluted in PBS for equal SFB numbers and frozen at -80°C until use.
SFB quantitation
SFB DNA was prepared using the QIAamp PowerFecal kit (Qiagen) and quantitated using SFB-specific 16S primers (F:5’-tgtgggttgtgaataacaat-3’; R:5’-gcgagcttccctcattacaagg-3’).
SFB fliC transcript analysis
The caecal and intestinal content of mSFB monocolonized mice was resuspended in RNAprotect Bacterial (Qiagen) under anaerobic conditions, incubated for 15 min and washed in PBS. SFB purification was performed using a 35% Nycodenz/PBS cushion. SFB were separated into IO-only and filament-enriched fractions using 5 μm-filtration, digested for 15 min with 10 mg/ml mutanolysin and 2 mg/ml proteinase K, and lysed with 3x20 sec pulses at minimal speed with a FastPrep dissociator (MP Bio) and 0.1 mm beads (MP Bio). RNA was isolated with the RNAeasy Mini Kit (Qiagen) and cDNA synthetized with M-MLV (Invitrogen).
Mass spectrometry sample preparation
Purified SFB fractions were labeled at RT with 2 mM sulfosuccinimidyl biotin NHS-LC-LC-Biotin (ThermoFisher, 21343) in PBS for 45 min, quenched with 500 mM glycine, washed twice in PBS and frozen at -80°C. Thawed samples were lysed in PBS with protease inhibitors (Roche, Complete Mini) by sonication (5x30 sec rounds, 30 Khz sonicator at 80%, Hielscher UP50H), and ultracentrifuged at 100,000xg for one hour at 4°C. Pellets were resuspended in CHAPS(4%)/ASB-14(4%) (Sigma), dissociated by 3x20 sec rounds with the FastPrep dissociator (MPBio, 0.1 mm beads), and incubated for 20 min on ice. The streptavidin bead purification was performed as per vendor’s manual (Dynabeads MyOne Streptavidin C1, ThermoFisher). Proteins were eluted at 95°C for 5 min in 1x Laemmli and digested in S-Trap™ micro spin columns (Protifi) as per vendor’s protocol. After elution, peptides were injected in a nanoRSLC-Q Exactive PLUS (RSLC Ultimate 3000) (ThermoScientific) as previously described (PMID:31042281). MS files were processed with Proteome Discoverer software (v1.4) and searched with Mascot search engine against SFB proteins.
TLR5 stimulation assay
Equal numbers of IO-only and filament enriched SFB fractions, based on 16S rDNA qPCR analysis, from 3-day old in vitro-grown SFB cultures were sonicated for 10 min (Bioblock, 88154) and 1/5 dilutions of lysate were added to control and mTLR5-expressing HEK cells in HEK-Blue detection medium (Invivogen, hkb-null1v, hkb-mtlr5, hb-det2) seeded in 96-well plates for 16 hours before alkaline phosphatase activity was measured at 620 nm using an ELISA plate reader (Labsystem Multiscan RC). Purified Salmonella Typhimurium flagellin (Invivogen, tlrl-stfla) served as the positive control.
Western blots
Equal numbers of bacteria, based on OD600 for E. coli and SFB-specific 16S rDNA qPCR for SFB, were immunoblotted with the rabbit antibody raised against Fla1 of Roseburia hominis (Antibodies Online, ABIN1107247) and anti-GST antibody (Upstate, 05-311).
Scanning electron microscopy
Terminal ileum of mSFB and rSFB-monocolonized mice were washed with PBS, opened and pinned onto 3% agarose cushion in 12 well-plates before being fixed overnight at 4°C in 0.1 M sodium cacodylate buffer (pH 7.2) containing 2.5% glutaraldehyde. Washed samples were glued onto 13 mm glass coverslips and kept in 0.1 M sodium cacodylate buffer at 4°C until processing. Samples were post-fixed with 1% osmium tetroxide at RT for 1 hour in the dark, dehydrated in a series of graded ethanol baths, dried using the critical-point drying (CPD300, Leica), coated with gold/palladium and observed with a SEM JEOL 6700F microscope.
Primers used for SFB flic quantitation
The following primers were used to assess relative fliC abundance: mSFB fliC1: F-ATGAAAAGGATATTATTCAGAATGAGG, R- GTTAATACACTTATTAATTCTGGATTACTG; mSFB fliC2: F- TAATGGTAGTAAACTTATCAATGGGAG, R- TTTCATAACCTCATTAATTGAATTTGG; mSFB fliC3: F- AGAAGGAGCATTGTCCGAG, R- CTCTATCTTCATCTGTATAAGTTCCG; mSFB fliC4: F- ATTAATCAAGAGTTTGGACAATTAAGAAG, R- CAGTTCCAGTACTTGCAACA; mSFB control primers (from Prakash et al. 2011): F- GACGCTGAGGCATGAGAGCAT, R- GACGGCACGGATTGTTATTCA.
In vitro SFB growth quantification
SFB was collected from the culture supernatant, centrifuged at 8,000xg for 5 min and resuspended in 70 μl PBS. 20 μl were used for Gram stain and 50 μl for DNA extraction using a QIAamp DNA Stool Mini Kit (Qiagen) following the manual. DNA was diluted 1/25 and 6 μl mixed with 2 μl H2O, 2 μl 4 mM SFB-specific 16S primer pair and 10 μl SybrGreen Mix (ThermoFisher) and run on a 96-well plate on an Applied Biosystems QuantStudio Flex 7 qPCR machine. SFB growth was calculated based on qPCR results for input samples.
Expression of recombinant SFB flagellins in E.coli
The genes for mSFB Flic2 and Flic3 were amplified from DNA isolated from feces of mSFB-monocolonized mice with primers PS597-ATTGGATCCATGATAATAAATCACAATATTAATGCGATGAACGC / PS598-AATGTCGACTTATTTTAAAATAGAAATAACTTGTTGCGGTGATTGATTTG and PS599-ATTGGATCCATGATAATTAACCACAATATGAATGCGATGAAC / PS600-AATGTCGACTTATCTCAATATAGAAAGTACTTGTTGTGGTGC, respectively, and cloned into the pGEX-4T2 expression vector using the BamHI and SalI restriction enzyme sites. Constructs were verified by Sanger sequencing and transformed into E. coli Rosetta (DE3) (Novagen 70954). E. coli Rosetta (DE3) expressing either GST alone or GST-FliC2 or GST-FliC3 fusions were grown shaking overnight at 30°C in 8 ml Luria broth (LB) with 25 μg/ml chloramphenicol and 100 μg/ml Ampicillin and then subcultured at 1/80 in 20 ml LB+antibiotics at 37°C for 1.5 hours while shaking. At an OD600 of 0.6, uninduced samples were taken and the remaining cultures were induced with 1 mM final IPTG for 2.5 hours shaking at 37°C.
Fluorescent in situ hyridization of SFB
For fluorescent in situ hyridization, the terminal ileum of mice monocolonized with rat or mouse SFB for 21 days was washed with PBS, dissected, fixed in 4% PFA overnight, quenched for 1 hour in 100 mM glycine in PBS, incubated in 15% sucrose for 3 hours at 4°C and 30% sucrose overnight at 4°C, embedded in OCT blocks and frozen in methylbutanol on a dry ice/100% ethanol bath before being stored at -80°C. 8 μm sections were rinsed with PBS and fixed in 4% PFA for 10 minutes, washed with PBS and incubated with 10 mg/ml lysozyme (Sigma L6876) and 40 U/ml mutanolysin (Sigma M9901) for 30 min at 37ºC. After two PBS washes, pre-hybridization was performed at 50ºC for 30 min in hybridization buffer (20 mM Tris-HCl [pH8], 0.9 M NaCl, 0.01% SDS) while hybridization with 50 nM of a SFB-specific 16S Cy3-labeled probe (5'-GGGTACTTATTGCGTTTGCGACGGCAC-3') and a A488-labeled all bacteria EUB338 probe (5’GCTGCCTCCCGTAGGAGT-3) was performed overnight at 50ºC in a humid chamber. After two washes with the hybridization buffer, slides were stained for 1 min with 0.125 μg/ml 4',6-diamidino-2-phenylindole (DAPI) and mounted in ProLong Gold (Thermo Fisher P36930). Images were taken on an Olympus IX81 fluorescence microscope equipped with a CCD camera and processed using ImageJ.
SFB membrane staining and structured illumination microscopy
For fluorescence staining of SFB, purified mSFB from monocolonized mice were stained for 5 min with DAPI (0.2 mg/ml) and the membrane dye FM4-64FX (5 mg/ml) (Thermo Fisher F34653). Samples were washed twice in PBS and fixed in 4% PFA for 15 min at RT. Samples were again washed twice with PBS, spotted onto slides, mixed with Floromount-G mounting medium (Abcam ab104135), covered with a No. 1.5H cover slip and sealed with nail polish. For structured illumination microscopy acquisition, images (13 images/plane/channel) were taken on a Zeiss LSM780 Elyra PS1 microscope (Carl Zeiss, Germany) using a 63x/1.4 oil Plan Apo objective and an EMCCD Andor Ixon 887 1 K camera. Images were analyzed with ZEN and ImageJ software while the SIMcheck plugin in ImageJ was used to evaluate the acquisition and processing parameters. Brightness/contrast was applied to reduce the background signal.
Statistical analysis
Statistical analysis was performed using Prism® 5 (GraphPad) software.
Supplementary Material
Acknowledgements
We are grateful to the members of the Centre for Gnotobiology of the Insitut Pasteur (Thierry Angélique, Eddie Maranghi, Martine Jacob and Marise Gabriela Lopez Dieguez) for technical support with the gnotobiotic mice and also thank Christine Schmitt and Regis Tournebize for technical assistance. We thank the UTechS PBI and UBI (center for resources and research in technology, Institut Pasteur, Paris), and the France-BioImaging infrastructure network supported by the ANR (ANR-10-INSB-04; Investments for the Future), and the Région Ile-de-France (program DIM-Malinf) for the use of the Zeiss LSM 780 Elyra PS1 microscope.
This work was supported by the ERC Advanced Grant DECRYPT (339579) to PJS (a Howard Hughes Medical Institute Foreign Scholar), a Pasteur LabEx IBEID Doctoral fellowship to IN, the ERC Advanced Grant IMMUNOBIOTA (339407) to NCB and a Bill and the Melinda Gates Foundation Grand Challenge grant (OPP1141322) to PS. OG, AS, MB, MNM, GN, JLK and PJS were supported by Pasteur Institute; NCB and PS were supported by Paris Descartes University and NCB, PJS and PS were also supported by INSERM.
Footnotes
Author Contributions
Conception of project by PS and PJS. Monocolonizations overseen by MB and VGR. In vitro experiments by IN. SFB sample preparations for TEM, SEM, SIM, FISH, Western and TLR5 assays by IN and PS. TEM imaging by OG and PS. SEM imaging by MMN. Ultrastructural work overseen by JKL. SIM imaging by AC and AS. TEM image analysis, RNA analysis and mass spec preparations by IN. Mass spectrometry performed by VJ and overseen by ICG. Western blots and FISH by CM. TLR5 reporter cell assay by IN. Technical help by GN. Rat SFB from TS and YU. Technical input for TEM grid preparation and first visualization of flagellated IOs using TEM by GPD with assistance from OG. Funding for IN and AC by NCB, for GPD by PJS, and for CM by PS. Manuscript preparation by PS. The position of GPD in the author list is disputed by GPD. The published order of authors has been approved by the Institut Pasteur Comité d’Intégrité Scientifique et de Conciliation, by the INSERM Délégation à l’Intégrité Scientifique and by all authors except GPD.
Competing interests
The authors declare no competing financial interests.
Ethics statement
Animal experiments were approved by the Ethics Committee #89 (CETEA Institut Pasteur, reference 2013-0030) and performed in compliance with French and European regulations on the care and protection of laboratory animals (EC Directive 2010/63 and French Law 2013-118, February 6, 2013).
Data availability
The data relative to in vivo growth of mSFB shown in this manuscript are representative of 4 independent experiments with n=320 bacteria imaged, including 91 flagellated SFB. Figure 1 shows 5 representative images, with additional images (n=3, n=5 and n=6) shown in Figure 2 and Supplementary Figures 3 and 10, respectively. Data relative to in vivo growth of rSFB shown in this manuscript are representative of 3 independent experiments with n=468 bacteria imaged, including 108 flagellated SFB. Figure 1 shows 3 representative images, with additional images (n=8 and n=4) shown in Supplementary Figures 4 and 11, respectively. The data relative to in vitro growth of mSFB shown in this manuscript are representative of 4 independent experiments with n=278 bacteria imaged, including 38 flagellated SFB. Figure 2 shows 4 representative images, with additional images (n=5 and n=1) shown in Supplementary Figures 7 and 9, respectively. Raw data and summary of the TEM analysis is made available in the Supplementary Data, as are the raw data for Figures 1k, 1l, 2d, 2f and 2k as well as Supplementary Figures 3g, 4i, 5a-f, 6b, 7f and 8e, including replicas where appropriate. Any additional images not included in the paper or its Supplementary Information can be made available by the corresponding author upon reasonable request. No custom computer code has been used in this study.
References
- 1.Schnupf P, Gaboriau-Routhiau V, Sansonetti PJ, Cerf-Bensussan N. Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Curr Opin Microbiol. 2017;35:100–109. doi: 10.1016/j.mib.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Klaasen HL, Koopman JP, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria. FEMS Microbiol Rev. 1992;8:165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- 3.Atarashi K, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Host interactions with Segmented Filamentous Bacteria: an unusual trade-off that drives the post-natal maturation of the gut immune system. Seminars in Immunology. 2013;25:342–351. doi: 10.1016/j.smim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017;549:528–532. doi: 10.1038/nature23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnupf P, et al. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015;520:99–103. doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chase DG, Erlandsen SL. Evidence for a complex life cycle and endospore formation in the attached, filamentous, segmented bacterium from murine ileum. J Bacteriol. 1976;127:572–583. doi: 10.1128/jb.127.1.572-583.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson DJ, Birch-Andersen A. Electron microscopy of a filamentous, segmented bacterium attached to the small intestine of mice from a laboratory animal colony in Denmark. Acta Pathol Microbiol Scand B. 1979;87:247–252. doi: 10.1111/j.1699-0463.1979.tb02434.x. [DOI] [PubMed] [Google Scholar]
- 9.Kuwahara T, et al. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 2011;18:291–303. doi: 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prakash T, et al. Complete Genome Sequences of Rat and Mouse Segmented Filamentous Bacteria,a Potent Inducer of Th17 Cell Differentiation. Cell Host Microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Sczesnak A, et al. The Genome of Th17 Cell-Inducing Segmented Filamentous Bacteria Reveals Extensive Auxotrophy and Adaptations to the Intestinal Environment. Cell Host Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Research. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamauchi KE, Snel J. Transmission electron microscopic demonstration of phagocytosis and intracellular processing of segmented filamentous bacteria by intestinal epithelial cells of the chick ileum. Infect Immun. 2000;68:6496–6504. doi: 10.1128/iai.68.11.6496-6504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyerholz DK, Stabel TJ, Cheville NF. Segmented filamentous bacteria interact with intraepithelial mononuclear cells. Infect Immun. 2002;70:3277–3280. doi: 10.1128/IAI.70.6.3277-3280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullender TC, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B, et al. Presence of Segmented Filamentous Bacteria in Human Children and Its Potential Role in the Modulation of Human Gut Immunity. Front Microbio. 2018;9:1403. doi: 10.3389/fmicb.2018.01403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Yin Y, Wang Y, Wang X, Xiang C. Host Specificity of Flagellins from Segmented Filamentous Bacteria Affects Their Patterns of Interaction with Mouse Ileal Mucosal Proteins. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.01061-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijay-Kumar M, Gewirtz AT. Flagellin: key target of mucosal innate immunity. Mucosal Immunology. 2009;2:197–205. doi: 10.1038/mi.2009.9. [DOI] [PubMed] [Google Scholar]
- 19.Uematsu S, Akira S. Immune responses of TLR5(+) lamina propria dendritic cells in enterobacterial infection. J Gastroenterol. 2009;44:803–811. doi: 10.1007/s00535-009-0094-y. [DOI] [PubMed] [Google Scholar]
- 20.Vijay-Kumar M, Carvalho FA, Aitken JD, Fifadara NH, Gewirtz AT. TLR5 or NLRC4 is necessary and sufficient for promotion of humoral immunity by flagellin. Eur J Immunol. 2010;40:3528–3534. doi: 10.1002/eji.201040421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data relative to in vivo growth of mSFB shown in this manuscript are representative of 4 independent experiments with n=320 bacteria imaged, including 91 flagellated SFB. Figure 1 shows 5 representative images, with additional images (n=3, n=5 and n=6) shown in Figure 2 and Supplementary Figures 3 and 10, respectively. Data relative to in vivo growth of rSFB shown in this manuscript are representative of 3 independent experiments with n=468 bacteria imaged, including 108 flagellated SFB. Figure 1 shows 3 representative images, with additional images (n=8 and n=4) shown in Supplementary Figures 4 and 11, respectively. The data relative to in vitro growth of mSFB shown in this manuscript are representative of 4 independent experiments with n=278 bacteria imaged, including 38 flagellated SFB. Figure 2 shows 4 representative images, with additional images (n=5 and n=1) shown in Supplementary Figures 7 and 9, respectively. Raw data and summary of the TEM analysis is made available in the Supplementary Data, as are the raw data for Figures 1k, 1l, 2d, 2f and 2k as well as Supplementary Figures 3g, 4i, 5a-f, 6b, 7f and 8e, including replicas where appropriate. Any additional images not included in the paper or its Supplementary Information can be made available by the corresponding author upon reasonable request. No custom computer code has been used in this study.