Abstract
ParABS systems facilitate chromosome segregation and plasmid partitioning in bacteria and archaea. ParB protein binds centromeric parS DNA sequences and spreads to flanking DNA. We show that ParB is an enzyme that hydrolyzes cytidine triphosphate (CTP) to diphosphate (CDP). parS DNA stimulates cooperative CTP binding by ParB and CTP hydrolysis. A nucleotide co-crystal structure elucidates the catalytic center of the dimerization-dependent ParB CTPase. Single-molecule imaging and biochemical assays recapitulate features of ParB spreading from parS in the presence but not absence of CTP. The findings suggest that centromeres assemble by self-loading of ParB DNA sliding clamps at parS. ParB CTPase is not related to known nucleotide hydrolases and might be a promising target for developing new classes of antibiotics.
Sister chromosomes separate from one another and distribute within bacterial cells by means of the ParABS system and the SMC complex. Bacterial centromeres – assembled from ParB proteins and 16 bp palindromic parS sites – initiate chromosome segregation soon after replication initiation by recruiting SMC condensin and by moving along ParA ATPase gradients (1–5). Multiple ParB proteins localize to a given parS site thereby forming distinctive clusters in the cell (6, 7). ParB recognizes parS and enriches in ~15 kb wide DNA regions flanking parS (6, 8–10). ParB spreading is hindered by DNA-binding protein ‘roadblocks’ engineered next to parS (10–12). Several models for ParB spreading have been proposed (6, 13). However, despite being critical for chromosome segregation, spreading has not yet been faithfully reconstituted in vitro, indicating the lack of an essential component.
ParB proteins harbor three globular domains (Fig. 1). A helix-turn-helix (HTH) motif in the middle domain (M) recognizes parS DNA. The C-terminal domain (C) homodimerizes and in chromosomally encoded ParB also supports sequence-non-specific DNA binding (14). The N-terminal domain (N) harbors conserved residues that produce strong phenotypes when mutated (5–7, 9, 15) but its function is unclear. Domain N shares marginal sequence similarity with a functionally unrelated eukaryotic enzyme, called sulfiredoxin (Srx) (fig. S1A) (16). Srx catalyzes the repair of peroxiredoxin by transferring ATP γ-phosphate groups onto hyper-oxidized cysteine moieties. A conserved GxxRxxA motif forms the ATP binding pocket in Srx (16, 17). Mutations in corresponding ParB residues (G77S and R80A) lead to loss of function and subcellular localization defects in Bacillus subtilis (Bsu) (5–7). Those subtle similarities invoke the possibility that ParB proteins are not merely DNA-binding proteins but enzymes.
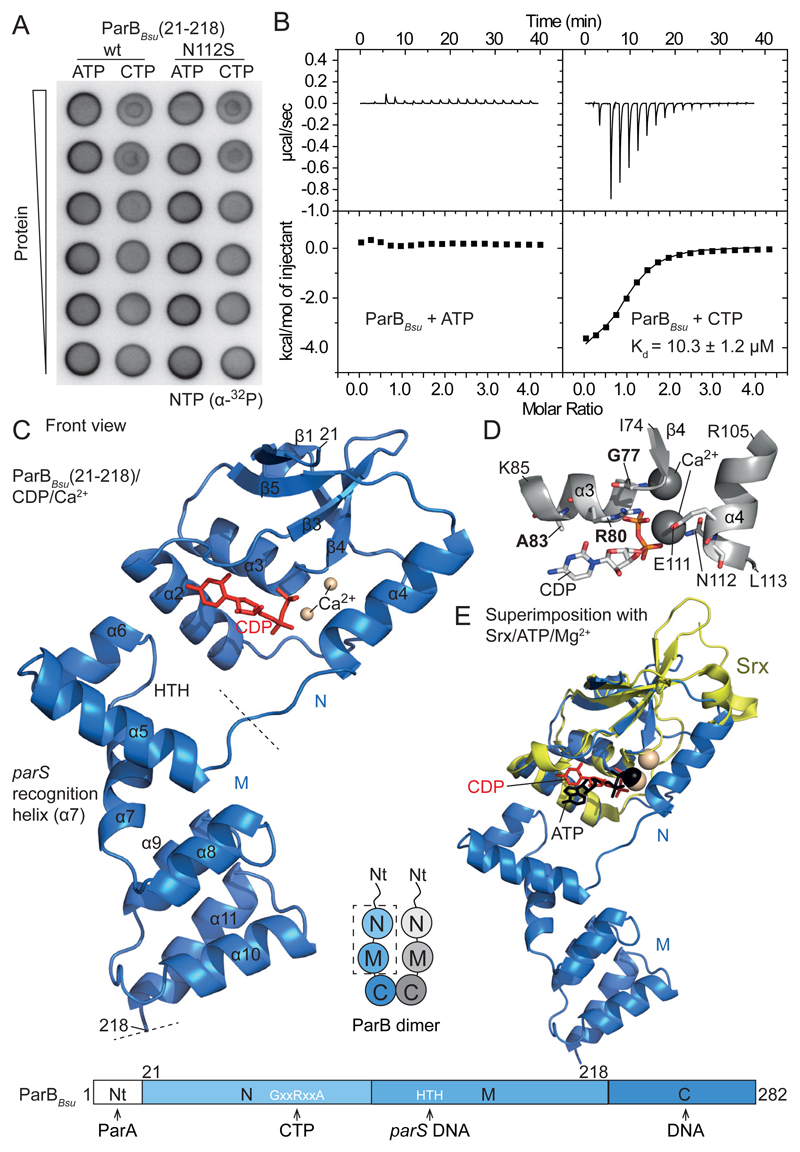
Fig. 1. ParB CTP binding.
(A) Membrane-spotting assay using radiolabeled nucleotides. (B) ITC measurements with full-length ParBBsu in the presence of Ca2+. The Kd obtained in a typical experiment is given. The interval indicates deviations of data points from the fit. (C) Crystal structure of a single chain of ParBBsu(21-218)/with CDP/Ca2+. (D) Nucleotide binding pocket in ParBBsu/CDP/Ca2+. GxxRxxA residues marked in bold. (E) Superimposition of ParBBsu(21-218)/CDP/Ca2+ with human Srx/ATP/Mg2+ (PDB: 3CYI).
Membrane-spotting assays with radiolabeled NTPs (18) surprisingly indicated that full-length ParBBsu and the fragment ParBBsu(21-218) bind CTP rather than ATP (Fig. 1A and fig. S1B). CTP binding was abolished in the ParBBsu(R80A) mutant (fig. S1C). Isothermal titration calorimetry (ITC) confirmed ParBBsu binding to CTP yielding dissociation constants (Kd) in the range of 10-50 μM. In contrast, other nucleotides produced little response in ITC (Fig. 1B and fig. S2A-B). CTP was also bound by plasmid-encoded ParB proteins (F plasmid ParBF and P1 prophage ParBP1) implying broad evolutionary conservation (fig. S1B-D) (10, 19).
We then solved the atomic structure of ParBBsu(21-218) (comprising domains N and M) using crystals grown with CTP and Ca2+ ions. The unit cell contained two ParBBsu(21-218) dimers. Each chain associated with a CDP molecule and two coordinated Ca2+ ions (Fig. 1C). CTP must thus have been hydrolyzed during crystallization. In the structure, the CDP β phosphate group contacts G77 and R80, explaining the strict conservation of the GxxRxxA motif (Fig. 1D) and implying that phenotypes associated with GxxRxxA mutations are caused by defective nucleotide binding (or hydrolysis). Individual domains N and M superimposed well with published ParB structures of bacterial and archaeal origin (with few exceptions) (fig. S2C) (20–22). Domain N also superimposed fairly well with human Srx/ATP/Mg2+ (17) yielding a close overlap of α and β phosphate groups (Fig. 1E). This striking resemblance implies that Srx ATP phosphotransferases and ParB CTP phosphohydrolases evolved from a common ancestor.
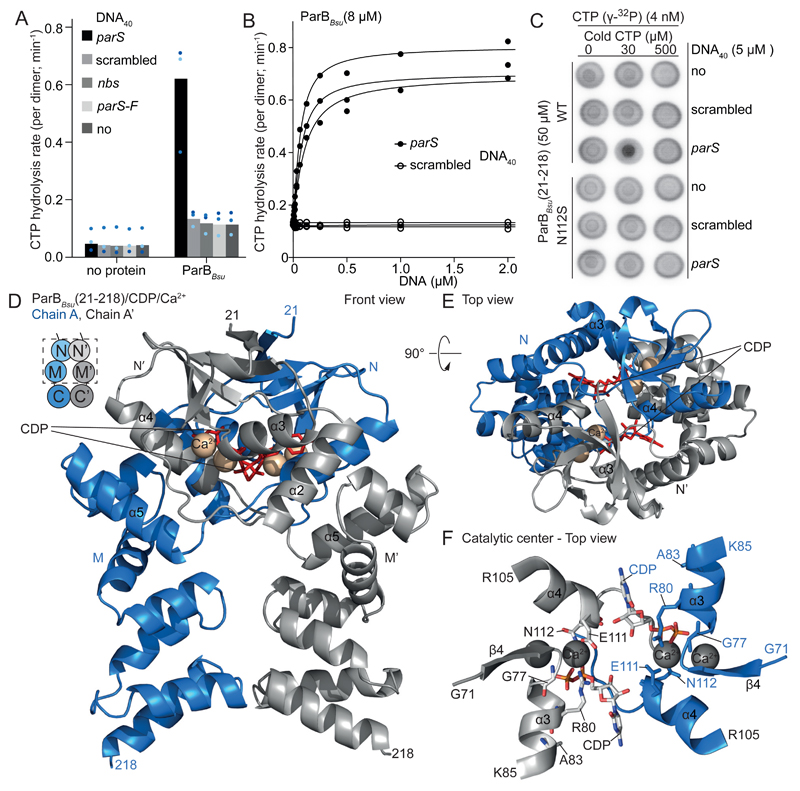
We next investigated ParB CTPase activity by measuring the accumulation of inorganic phosphate (Fig. 2A and fig. S3A). ParBBsu dimers hydrolyzed about five CTP molecules per hour. CTP hydrolysis was abolished in the CTP-binding mutant R80A and also in the CTP-binding proficient mutant N112S (fig. S2B, S3B, S4A) (5, 23). Importantly, addition of 40-mer parS duplex (parSDNA40) strongly increased CTP turn-over to about 36 molecules per hour (Fig. 2A). Sub-stoichiometric amounts of parSDNA40 (ratio ~40:1) were sufficient to saturate enzymatic activity (Fig. 2B), implying that parS only transiently associates with ParB. Noncognate DNA sequences did not alter the activity, even at elevated concentrations (Fig. 2A). ParBF and the paralogue NocBsu (24) also displayed CTPase activity which was stimulated by cognate DNA (fig. S3A).
Fig. 2. ParB CTP hydrolysis.
(A) CTP hydrolysis measured by colorimetric detection of inorganic phosphate using malachite green. Mean values from three repeat measurements. Data points are shown as dots. (B) CTP hydrolysis by ParBBsu dimers with increasing DNA concentrations. Data points and fits of three repeat measurements. (C) Membrane-spotting showing parS-stimulated cooperative CTP binding. (D, E) Crystal structure of a ParBBsu(21-218)/CDP/Ca2+ dimer. (F) Catalytic center in ParBBsu(21-218)/CDP/Ca2+.
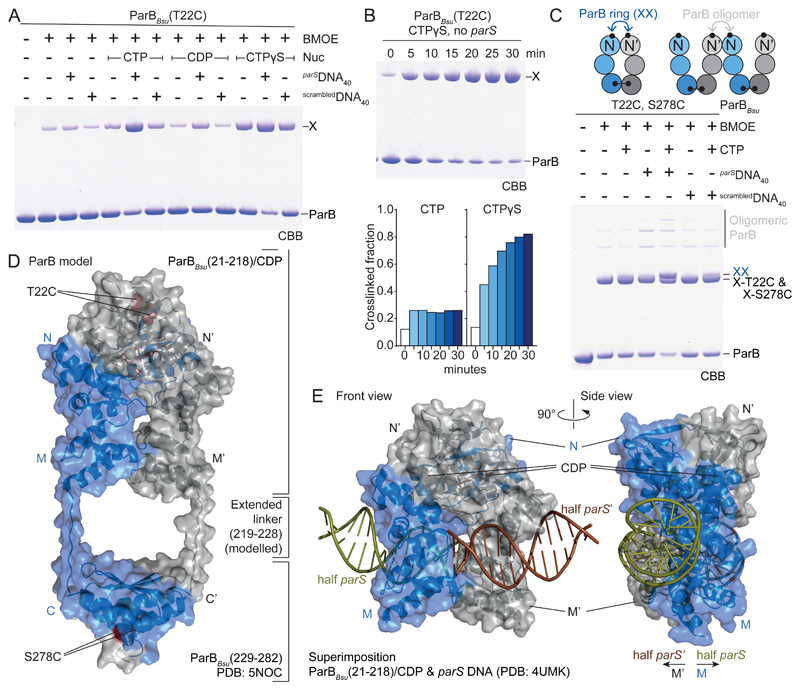
Addition of parSDNA40 promoted cooperative CTP binding by ParBBsu in membrane-spotting assays, suggesting that the functional unit harbors at least two CTP molecules (Fig. 2C and fig S4C). Indeed, two nucleotides are sandwiched by a ParB dimer in the ParB/nucleotide structure (Fig. 2D-E) being analogous to dimerization-activated NTPases of the large family of P-loop proteins (e.g. SIMIBI and ABC proteins including ParA and SMC, respectively) (25, 26). Residues E111 to E116 are located next to the nucleotide at the N dimer interface and appear critical for N engagement (Fig. 2F). They are conserved in ParB proteins but absent from monomeric Srx (fig. S1A). Consistent with a role in N engagement, N112S impairs parS-stimulated cooperative but not basal CTP binding (Fig. 2C). To detect N engagement in solution, we employed chemical cross-linking of purified ParB(T22C). T22 is ideally positioned at the symmetry axis of the ParB/nucleotide structure to support dimer cross-linking (Fig. 3D). ParBBsu(T22C) cross-linking was robust only in the presence of both CTP and parSDNA40 (Fig. 3A). As expected for a N engagement-defective mutant, N112S abolished parS-stimulated T22C cross-linking (fig. S5C). Stimulation by parSDNA40 occurred at sub-stoichiometric concentrations again implying that parS does not stably bind ParB and indicating that parS catalyzes the N engagement step (fig. S5D). The non-hydrolysable analog CTPγS also promoted efficient T22C cross-linking with parSDNA40 but CDP failed to do so (Fig. 3A). In contrast to CTP, CTPγS supported robust cross-linking even without parS after extensive incubation periods (Fig. 3B and fig. S6B). This demonstrates that N engagement is energetically favorable, but the transition to the engaged state is very slow without parS. We conclude that parS catalyzes CTP-dependent engagement of domain N in ParB.
Fig. 3. ParB gate closure.
(A) Gel analysis of cross-linking products of purified ParBBsu(T22C). X denotes cross-linked species. CBB, Coomassie Brilliant Blue. (B) Time-course of ParBBsu(T22C) cross-linking with CTPγS without parS. Aliquots were taken at the indicated time points and mixed with BMOE. Quantification of cross-linked fraction also shown for CTP (see fig. S6B). (C) As in A using ParBBsu(T22C, S278C). X-T22C and X-S278C denote single cross-linked dimer species. XX marks double cross-linked ParB rings. Other species are oligomeric forms. (D) Model of ParB built from ParBBsu(21-218)/CDP/Ca2+ and ParBBsu(229-282) (PDB: 5NOC) dimers. Linker at the M-C junction is manually modelled as fully extended peptide. (E) Superimposition of domain M in ParBBsu(21-218)/CDP/Ca2+ and a ParB/parS structure (PDB: 4UMK). Only DNA is shown for the latter.
To determine whether N engagement occurs within or between ParB dimers, we purified ParBBsu(T22C, S278C) (Fig. 3C). In the presence of CTP and parSDNA40, double cross-linked ParB dimers emerged as predominant species, demonstrating that N engagement produces mostly ring-shaped ParB dimers (and few ParB oligomers). This invokes the possibility that domain N serves as a nucleotide-operated DNA gate in ParB rings which closes at parS.
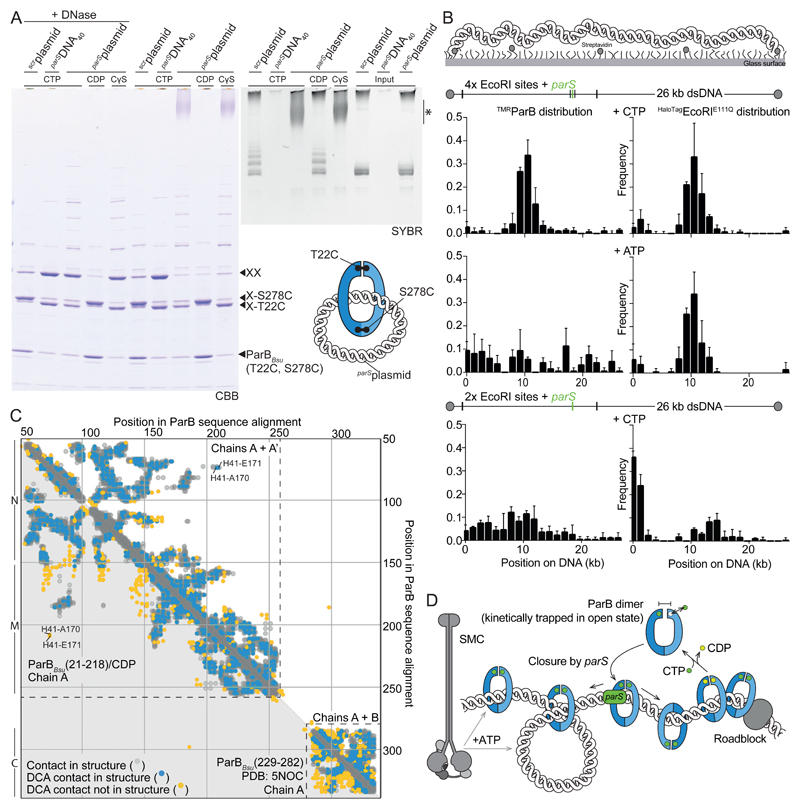
Comparing the ParB/nucleotide structure with a ParB/parS DNA co-crystal structure, it became apparent that parS DNA associating with one HTH motif clashes with the opposing HTH motif (Fig. 3E and fig. S6C). This indicates that parS DNA duplexes may detach from the HTH motif upon N engagement (or CTP hydrolysis) possibly explaining the transient nature of ParB/parS association. ParB dimers may, however, remain associated with chromosomal DNA even after release from parS if they were to entrap the DNA backbone. To test this, we next used circular DNA for in vitro DNA loading experiments (Fig. 4A). ParBBsu(T22C, S278C) was incubated with plasmid DNA (1873 bp) or linear DNA40 in the presence of CTP and cross-linked with BMOE to preserve topological interactions under protein-denaturing conditions (5). Control samples were nuclease-treated to release proteins from DNA. As expected, ParBBsu(T22C, S278C) generated large fractions of covalently closed ParB rings with parS DNA only (Fig. 4A). When nuclease digestion was omitted, the ParB ring species was retained in the parSDNA40 sample but was eliminated from the parSplasmid sample containing CTP or CTPγS. Instead a slowly migrating species was observed near the top of the gel, presumably corresponding to parSplasmid interlocked with multiple covalent ParB rings. DNA staining of polyacrylamide and agarose gels supported the notion of multiple ParB rings entrapping parSplasmid (Fig. 4A and fig. S7). The efficient stimulation of ParB-DNA entrapment by CTPγS implies that CTP hydrolysis is dispensable for all steps of DNA loading. CTP hydrolysis might instead play a role in unloading and recycling of ParB thereby determining the residence time of ParB on chromosomes (and the extent of ParB spreading).
Fig. 4. ParB DNA loading and sliding.
(A) Polyacrylamide gel analysis of ParBBsu(T22C, S278C) protein species (CBB) and DNA species (SYBR) from BMOE-cross-linked DNA loading reactions. A presumed ParB-XX/parSplasmid adduct is marked by asterisk. CDP and scrplasmid (scrambled) only support limited ParB loading. (B) Single-molecule imaging (SMI) of DNA-bound TMRParB in the presence of HaloTagEcoRIE111Q roadblocks. Mean and standard deviation were calculated from three repeat measurements of a sample. (C) Residue contact maps by coevolutionary analysis (DCA; blue and yellow dots) and residue proximities (grey dots) in monomeric (below diagonal) and dimeric chains (above diagonal) of ParBBsu(21-218)/nucleotide and ParBBsu(229-282) (PDB: 5NOC). (D) Model for ParB spreading by self-loading of DNA sliding clamps at parS.
We next used single-molecule imaging to elucidate how CTP binding may relate to ParB spreading (27). The ends of linear 26 kb DNA molecules containing a single parS sequence were immobilized in flow chambers. Increased DNA occupancy of labeled ParB (TMRParB) was detected in presence of CTP (fig. S8A). However, no local enrichment of TMRParB on DNA was observed, potentially due to ParB spreading from parS. We therefore used a labeled and catalytically inactive variant of the EcoRI restriction enzyme to test if this HaloTagEcoRIE111Q protein could function as a roadblock, which might constrain ParB spreading. Intriguingly, under these conditions TMRParB became locally enriched on DNA between two EcoRI sites flanking parS (Fig. 4B), indicating that HaloTagEcoRIE111Q had indeed restrained ParB spreading. TMRParB localized more broadly when these two parS flanking EcoRI sites were removed (Fig. 4B). Local enrichment of TMRParB was also lost when ATP was used instead of CTP or when HaloTagEcoRIE111Q was added at the end of the experiment, despite normal localization of HaloTagEcoRIE111Q (Fig. 4B, S8B). These results demonstrate that ParB enrichment at parS DNA is enhanced by CTP and indicate that ParB translocates onto parS flanking DNA, unless it is restrained by roadblocks. These findings support the notion that ParB forms self-loading DNA clamps whose sliding on chromosomal DNA is hampered by protein roadblocks. On the chromosome, ParB may frequently encounter roadblocks formed by nucleoid associated proteins (NAPs) and RNA polymerase. ParB diffusion might be locally restricted in a stochastic manner thus producing the characteristic gradient distribution patterns observed by ChIP-Seq (6, 13).
ParBBsu(T22C) cross-linking was rather inefficient in vivo (fig. S9), possibly indicating that levels of N engagement are low in the cell. Alternatively, cross-linking might be hampered for technical reasons or by steric blockage by other proteins such as ParA or SMC. To obtain information on ParB architecture by independent means, we extracted residue co-evolution signatures from sequence data by direct coupling analysis (DCA) (28). We obtained excellent agreement between the top 900 DCA contacts predicted from an alignment of 157776 ParB sequences and residue proximities observed in the ParB/nucleotide dimer (Fig. 4C). In contrast, the ParB/nucleotide monomer and published ParB structures failed to explain numerous DCA contacts or produced proximities not observed by DCA (fig. S10). These results strongly suggest that the dimer in the ParB/nucleotide structure closely resembles a state of ParB that is relevant in many bacteria.
In summary, our work shows that the widespread family of ParB proteins are enzymes that bind the atypical nucleotide CTP to promote spreading from cognate DNA sequences. CTP utilization enables robust enrichment of ParB at the rare recognition sites (~1 cognate per 106 noncognate sites) even with little or no sequence-specificity in DNA binding (14). The CTPase domain is found in a large variety of protein sequences with diverse domain organizations (> 100; PFAM: PF02195), implying that it also plays prominent roles in other cellular processes.
Supplementary Materials
Acknowledgments
We thank Daniela Barillà, Marie-Laure Diebold-Durand and Barbara Funnell for providing purified proteins, Luke Lavis for JF646 ligand, Duccio Malinverni for sharing DCA code, and Martin Thanbichler for exchanging unpublished results. We are grateful to Roberto Vazquez Nunez and Diogo Tavares for technical help and to Sophie Martin, Aleksandar Vjestica and members of the Gruber lab for comments and discussions.
Funding: The authors acknowledge financial support from the European Research Council (724482 to S.G. and 693949 to J.-M.P.), the Swiss National Science Foundation (170242 to S.G.), the Austrian Research Promotion Agency (FFG-834223, FFG-852936 to J.-M.P.), Boehringer Ingelheim and the Human Frontier Science Program grant RGP0057/2018 (to J.-M.P.).
Footnotes
Author contributions: Initial finding Y.-M.S., S.G., J.-W.V., and M.T.; Crystallography, Y.-M.S. and J.B.; In vivo cross-linking Y.-M.S. and F.P.B.; SMI, I.F.D. and J.-M.P.; DCA, S.Z. and P.D.L.R.; Other experiments, Y.-M.S.; Writing and visualization, Y.-M.S., S.G.; Supervision, S.G.
Competing interests: None declared.
Data and materials availability: Raw data is available at Mendeley Data (doi: 10.17632/n5jjdrr55r.1). Structure coordinates are deposited at the Protein Data Bank (PDB: 6SDK). DCA code at GitHub (http://gitlab.com/LBS-EPFL/code/lbsDCA/tree/v1.0).
References
- 1.Surovtsev IV, Jacobs-Wagner C. Subcellular Organization: A Critical Feature of Bacterial Cell Replication. Cell. 2018;172:1271–1293. doi: 10.1016/j.cell.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 3.Vecchiarelli AG, Hwang LC, Mizuuchi K. Cell-free study of F plasmid partition provides evidence for cargo transport by a diffusion-ratchet mechanism. Proc Natl Acad Sci U S A. 2013;110:E1390–1397. doi: 10.1073/pnas.1302745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim HC, et al. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. Elife. 2014;3:e02758. doi: 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilhelm L, et al. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. Elife. 2015:4. doi: 10.7554/eLife.06659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham TG, et al. ParB spreading requires DNA bridging. Genes Dev. 2014;28:1228–1238. doi: 10.1101/gad.242206.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Autret S, Nair R, Errington J. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol Microbiol. 2001;41:743–755. doi: 10.1046/j.1365-2958.2001.02551.x. [DOI] [PubMed] [Google Scholar]
- 8.Minnen A, et al. Control of Smc Coiled Coil Architecture by the ATPase Heads Facilitates Targeting to Chromosomal ParB/parS and Release onto Flanking DNA. Cell Rep. 2016;14:2003–2016. doi: 10.1016/j.celrep.2016.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breier AM, Grossman AD. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol. 2007;64:703–718. doi: 10.1111/j.1365-2958.2007.05690.x. [DOI] [PubMed] [Google Scholar]
- 10.Rodionov O, Lobocka M, Yarmolinsky M. Silencing of genes flanking the P1 plasmid centromere. Science. 1999;283:546–549. doi: 10.1126/science.283.5401.546. [DOI] [PubMed] [Google Scholar]
- 11.Murray H, Ferreira H, Errington J. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol Microbiol. 2006;61:1352–1361. doi: 10.1111/j.1365-2958.2006.05316.x. [DOI] [PubMed] [Google Scholar]
- 12.Rodionov O, Yarmolinsky M. Plasmid partitioning and the spreading of P1 partition protein ParB. Mol Microbiol. 2004;52:1215–1223. doi: 10.1111/j.1365-2958.2004.04055.x. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez A, et al. Stochastic Self-Assembly of ParB Proteins Builds the Bacterial DNA Segregation Apparatus. Cell Syst. 2015;1:163–173. doi: 10.1016/j.cels.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Fisher GL, et al. The structural basis for dynamic DNA binding and bridging interactions which condense the bacterial centromere. Elife. 2017;6 doi: 10.7554/eLife.28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song D, Rodrigues K, Graham TGW, Loparo JJ. A network of cis and trans interactions is required for ParB spreading. Nucleic Acids Res. 2017;45:7106–7117. doi: 10.1093/nar/gkx271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu MK, Koonin EV. Evolution of eukaryotic cysteine sulfinic acid reductase, sulfiredoxin (Srx), from bacterial chromosome partitioning protein ParB. Cell Cycle. 2005;4:947–952. doi: 10.4161/cc.4.7.1786. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson TJ, Murray MS, Johnson LC, Lowther WT. Reduction of cysteine sulfinic acid in peroxiredoxin by sulfiredoxin proceeds directly through a sulfinic phosphoryl ester intermediate. J Biol Chem. 2008;283:23846–23851. doi: 10.1074/jbc.M803244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci U S A. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouet JY, Ah-Seng Y, Benmeradi N, Lane D. Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol Microbiol. 2007;63:468–481. doi: 10.1111/j.1365-2958.2006.05537.x. [DOI] [PubMed] [Google Scholar]
- 20.Leonard TA, Butler PJ, Lowe J. Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol Microbiol. 2004;53:419–432. doi: 10.1111/j.1365-2958.2004.04133.x. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher MA, et al. Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages. Science. 2015;349:1120–1124. doi: 10.1126/science.aaa9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen BW, Lin MH, Chu CH, Hsu CE, Sun YJ. Insights into ParB spreading from the complex structure of Spo0J and parS. Proc Natl Acad Sci U S A. 2015;112:6613–6618. doi: 10.1073/pnas.1421927112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 24.Wu LJ, et al. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–1952. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 26.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 27.Davidson IF, et al. Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 2016;35:2671–2685. doi: 10.15252/embj.201695402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malinverni D, Barducci A. Coevolutionary Analysis of Protein Sequences for Molecular Modeling. Methods Mol Biol. 2019;2022:379–397. doi: 10.1007/978-1-4939-9608-7_16. [DOI] [PubMed] [Google Scholar]
- 29.Grimm JB, et al. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods. 2017;14:987–994. doi: 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.