Abstract
Excitatory neurons of the mammalian cerebral cortex are organized into six functional layers characterized by unique patterns of connectivity, as well as distinctive physiological and morphological properties. Cortical layers appear after a highly regulated migration process in which cells move from the deeper, proliferative zone toward the superficial layers. Importantly, defects in this radial migration process have been implicated in neurodevelopmental and psychiatric diseases. Here we report that during the final stages of migration, transcription factor Neurogenic Differentiation 2 (Neurod2) contributes to terminal cellular localization within the cortical plate. In mice, in utero knockdown of Neurod2 resulted in reduced numbers of neurons localized to the uppermost region of the developing cortex, also termed the primitive cortical zone. Our ChIP-Seq and RNA-Seq analyses of genes regulated by NEUROD2 in the developing cortex identified a number of key target genes with known roles in Reelin signaling, a critical regulator of neuronal migration. Our focused analysis of regulation of the Reln gene, encoding the extracellular ligand REELIN, uncovered NEUROD2 binding to conserved E-box elements in multiple introns. Furthermore, we demonstrate that knockdown of NEUROD2 in primary cortical neurons resulted in a strong increase in Reln gene expression at the mRNA level, as well as a slight upregulation at the protein level. These data reveal a new role for NEUROD2 during the late stages of neuronal migration, and our analysis of its genomic targets offers new genes with potential roles in cortical lamination.
Subject terms: Developmental neurogenesis, Lamination
Introduction
The cerebral cortex of higher animals is composed of numerous excitatory and inhibitory neuron types organized into six anatomical and functional layers. Developmentally, excitatory neurons originate from neural stem cells that divide in the ventricular zone of the anterior neural tube, then migrate radially to the overlying cortical plate. While neural stem cells initially divide symmetrically during an increase of the proliferative population, the division mode transitions to asymmetric division as embryogenesis progresses. One daughter cell from each cell division remains at the ventricular zone, while the other daughter exits cell cycle and initiates a complex mode of radial migration to the border between the cortical plate (CP) and the marginal zone (MZ). As development progresses, each wave of migrating neurons reaches the surface by passing through the nascent cortex in an inside-out manner, eventually giving rise to the layered structure of the cerebral cortex1,2. Consequently, early-born neurons populate the deep layers, and late-born neurons settle in the superficial layers. The precise control of the radial migration process is essential for the proper formation of the six cortical layers. Importantly, in mice and humans, mutations, infections or environmental agents that perturb cortical migration cause cortical structural malformations, intellectual disability, neuropsychiatric diseases, and seizures3–5.
A closer examination of the radial migration process reveals a series of different modes of cellular motility, as opposed to a uniform directed movement from the ventricular zone toward the superficial marginal zone. Initially neurons are multipolar in morphology as they emerge from the ventricular zone, which is followed by a transition to a bipolar morphology and locomotion on radial glia fibers through the intermediate zone and much of the cortical plate6,7. Upon approaching the border between the cortical plate and the marginal zone, they terminally translocate from the radial glia fibers. Terminal translocation is accompanied by positioning of neuronal cell bodies in the uppermost layer of the cortex, also termed as the primitive cortical zone8. During this developmental period, the marginal zone is populated by Cajal-Retzius cells which secrete the extracellular glycoprotein REELIN, considered a stop signal for migrating neurons9. Migrating neurons express the REELIN receptors VLDLR and LRP8 (APOER2), as well as the intracellular adaptor molecule DAB1, which conveys receptor activation to downstream signaling pathways. Ligand binding induces the endocytic internalization of REELIN and phosphorylation of DAB19. Activation of REELIN signaling in neurons induces anchorage of nascent primary dendrites to the extracellular matrix of the marginal zone, stabilization of the neuronal skeleton, and termination of migration10–15. Loss of REELIN, VLDLR, APOER2 or DAB1 function causes profound defects in cortical lamination; the cortical layers develop in an inverted pattern, with early- and late-born neurons being located in superficial and deep layers, respectively13,16–21.
Individual steps of radial migration are controlled by the combinatorial expression of specific transcription factors (TFs)2. Prominent among these TFs are the bHLH family members, including the Neurogenin and the NeuroD proteins. In addition to their roles in neuronal cell fate specification and neuronal differentiation, these factors have also been shown to control specific steps of cortical radial migration described above22–24. A key member of the NeuroD family of neurogenic bHLH TFs is NEUROD2. The Neurod2 gene is highly expressed in the developing cortex, and its expression persists, albeit at low levels, into adulthood in cortical excitatory neurons25,26. Interestingly, several recent studies by our group and others have implicated NEUROD2 in the radial migration process of cortical neurons and have provided a general overview of its downstream genetic targets27,28. However, how NEUROD2 regulates the expression of its downstream targets, and how this regulation impacts cortical migration, remain largely unknown.
We previously characterized the genetic targets of NEUROD2 in the cerebral cortex at two developmental timepoints: embryonic day 14.5 (E14.5), representing the peak of neurogenesis and migration in the mouse cortex; and postnatal day 0 (P0), representing the onset of neuronal differentiation, dendritic growth and synaptogenesis26,27. Here, we carry out a comparative analysis of these two datasets and overlay it with transcriptomics analysis of primary neurons in which Neurod2 expression is knocked down. We find that NEUROD2 exhibits quantitative and qualitative differences in target-selectivity at these two developmental timepoints. From our postnatal dataset, we identify several gene targets with known roles in neuronal projection development, such as the Ca2+/Calmodulin-dependent Kinase IV (Camk4) gene, and we demonstrate that suppressing Neurod2 expression in primary cortical neurons causes dendritic differentiation defects. Using our embryonic dataset, we identify numerous NEUROD2 targets with known functions in the Reelin signaling pathway, such as Reln, Dab1 and Lrp8. Furthermore, suppressing Neurod2 expression by in utero shRNA electroporation caused a defect in cellular positioning of neurons to the primitive cortical zone. Our results point to NEUROD2 as a regulator of the terminal stage of radial migration that acts, at least in part, by regulating genes functioning in the Reelin pathway. Our dataset also provides novel candidate genes that might have functions in different aspects of cortical radial migration. Future experiments rescuing individual target genes with roles in neuronal migration in a Neurod2-deficient background will uncover the precise mechanism by which this TF orchestrates critical pathways for proper cortical lamination.
Results
NEUROD2 binds to overlapping and unique gene-sets in embryonic and postnatal cerebral cortex tissue
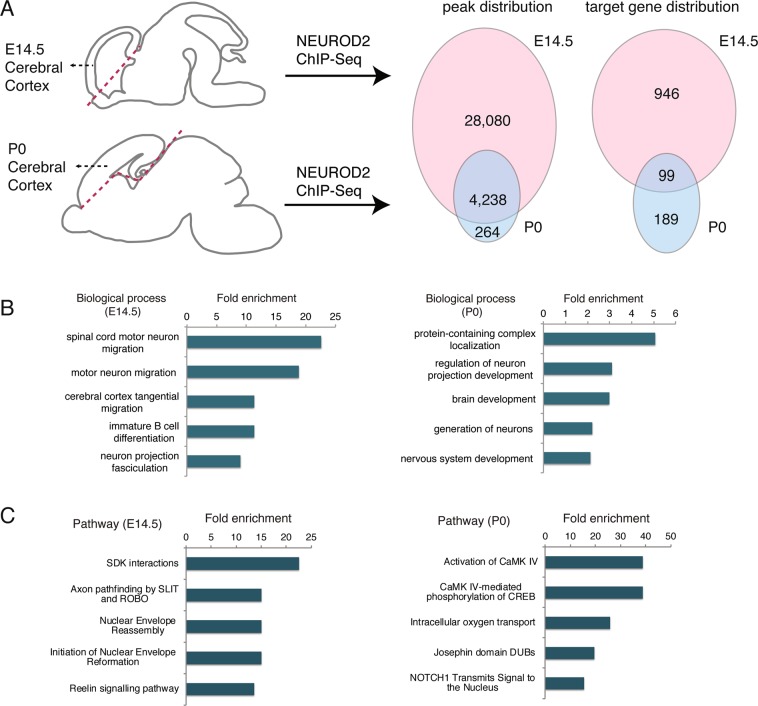
To evaluate whether NEUROD2 interactions with specific genomic sites are temporally regulated, we compared NEUROD2 binding profiles between embryonic day 14.5 (E14.5) and postnatal day 0 (P0). We retrieved binding scores from our previously published NEUROD2 ChIP-Seq datasets that represent the amount of relative TF binding to target sequences, then carried out a comparative analysis using the E14.5 and P0 results26,27. Based upon this comparison, we identified a large number of binding sites that were exclusively targeted at E14.5, and fewer sites that were targeted at both developmental stages or were specific to P0 (Supplemental Material 1) (Fig. 1A). For both datasets, we had previously reported that a majority of NEUROD2 binding sites were not located at promoters, but rather at distant regulatory elements. Therefore, we decided to employ a target gene prediction approach, called the Closest Gene Method, which generates a score based on the number of TF binding sites and their proximity to the transcription start site of each gene29. We calculated Closest Gene scores for the P0 dataset and compared it to the E14.5 dataset that we had previously published (Supplemental Material 2)27. Genes with a Closest Gene score above a threshold value were predicted as targets. Comparison of these predicted targets between two developmental stages revealed a large number of E14.5-specific genes, as well as a smaller number of target genes expressed only in the P0 developmental stage or at both stages (Fig. 1A).
Figure 1.
NEUROD2 binds to overlapping and unique target sites in embryonic and postnatal cerebral cortex. (A) NEUROD2 binding sites and target genes were compared between embryonic day 14.5 (E14.5) and postnatal day 0 (P0) cerebral cortical tissue. Numbers of genome-wide binding sites and target genes are based upon previously acquired ChIP-Seq data26,27. Target genes were identified based on the total number of binding sites proximal to the transcription start sites of individual genes29. (B) Gene ontology analysis of NEUROD2 target genes from P0 and E14.5 datasets (geneontology.org). Fold enrichment of categories with FDR < 0.01 are plotted. (C) Reactome pathway analysis of NEUROD2 targets from P0 and E14.5 datasets (reactome.org). Fold enrichment of categories with p < 0.05 are plotted.
In order to determine whether NEUROD2 gene targets at embryonic and postnatal timepoints represented functionally distinct gene groups, we carried out gene ontology analysis. Not surprisingly, at both tested developmental stages, genes related to nervous system development were significantly enriched (Fig. 1B). However, genes associated with migration of different types of neurons were specifically enriched at E14.5. To gain better insight into the signaling pathways regulated by NEUROD2, we performed pathway analysis at the two stages using the Reactome database30,31. In addition to pathways whose significance is not immediately clear to us, our results also demonstrated that embryonically expressed NEUROD2 might control pathways related to cellular adhesion, synaptic connectivity, migration, and postnatal neuron projection development. Furthermore, NEUROD2 appeared to impinge upon calcium-mediated transcription, including CaMKIV transactivation of CREB (cAMP-response element binding protein) (Fig. 1C, Supplementary Material 3). Collectively these data suggested that NEUROD2, a TF that is expressed throughout the lifetime of cortical excitatory neurons, may regulate distinct functions during different developmental events.
NEUROD2 is required for dendritic differentiation of cortical neurons
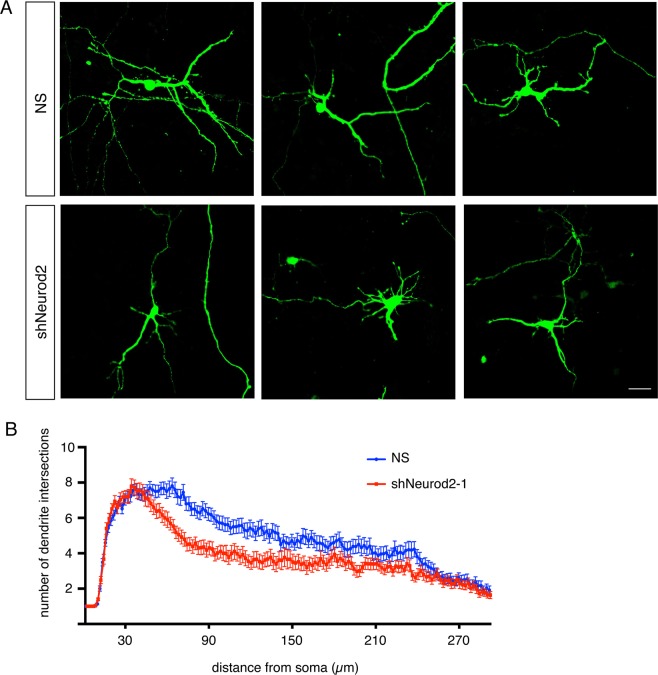
Since our analysis of our postnatal dataset suggested a role for NEUROD2 in neuronal projection development, as well as of CaMKIV-mediated transcription, a known regulator of dendritic differentiation32, we asked whether neurons silenced for Neurod2 also exhibited generalized dendritic arborization defects. Toward this aim, we cultured primary cortical neurons and transfected them with a short hairpin RNA targeting NeuroD2 transcripts (shNeurod2-1) at low efficiency to achieve knockdown in isolated neurons (Fig. 2A). We then quantified dendritic arborization of transfected cells, also marked by EGFP expression, by Sholl analysis, an approach that reports upon the number of dendrite branches as a function of distance from the neuronal soma33. Our results demonstrated that while the number of primary dendrites protruding from the soma were comparable between the two conditions, a significant reduction in higher order dendritic branches was observed in neurons where Neurod2 expression was silenced (Fig. 2A,B). While this experiment pointed to a requirement for NEUROD2 in dendritic arborization in cortical neurons, future experiments will uncover which specific target genes are functioning in dendrite arbor development.
Figure 2.
NEUROD2 is required for normal dendrite development in primary cortical neurons. (A) Primary cortical neurons from E14.5 embryos were transfected with NS (non-silencing) shRNA or shNeurod2-1 at 2 days DIV and fixed at 5 DIV. Images were captured by confocal microscopy, scale bar: 20 µm. (B) Dendrite development was quantified by Sholl analysis. n = 65 neurons per condition derived from two separate cultures. Bars indicate S.E.M. p = 0.0062 by two-way ANOVA.
RNA-Seq analysis reveals NEUROD2-regulated target genes in primary cortical neurons
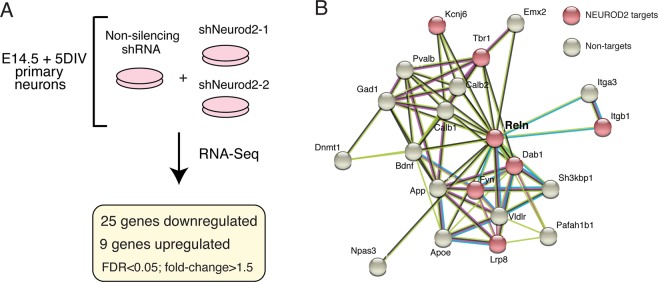
To further focus our attention upon genes for which expression is regulated by NEUROD2, we silenced Neurod2 expression in primary cortical cultures using two validated shRNAs26 and analyzed gene expression changes. When gene expression was compared to cells treated with a control shRNA, we found that 25 genes were down-regulated and 9 genes were up-regulated upon silencing of Neurod2 (Supplemental Material 4) (Fig. 3A). As expected, we detected Neurod2 as one of the significantly downregulated genes. Genes perturbed by Neurod2 depletion included those expressing DNA binding proteins (Arid5a, Ddit4, Ubn2, Trim66, Zglp1, and Sp8); regulators of extracellular matrix, adhesion and synaptogenesis (Thbs1, Reln, Col3a1, Col1a1, and Ank1); metabolic genes (Shmet2, Tkt, Pck2, and Pde12); signaling molecules (Chac1 and Sesn2), and a translational regulator (Rps6ka4). Next, we asked whether any of the regulated targets were also directly bound by NEUROD2 as revealed by our ChIP-Seq data. We found that 14/34 genes harbored binding sites for NEUROD2, with the remaining (20/34) potentially represented secondary or off-target effects (Supplemental Material 4). Among the direct and regulated targets of NEUROD2, we were intrigued to identify Reln, a gene with essential roles in neuronal migration, dendrite development and synaptogenesis. Our previous findings had also implicated NEUROD2 in regulating the signaling pathway activated by this extracellular matrix protein27.
Figure 3.
RNA-seq analysis of primary cortical neurons silenced for Neurod2 expression. (A) RNA-Seq analysis was carried out on primary cortical neurons electroporated with a NS (non-silencing) shRNA or one of the two independent shRNAs against Neurod2. 25 genes were down-regulated and 9 were up-regulated upon knockdown of Neurod2. fold-change > 1.5, FDR < 0.05. (B) A protein interactome network of REELIN revealed additional genetic targets of NEUROD2 that also interact with REELIN. Interactome analysis was based on the String Database34,35.
Next we predicted the protein interactome of REELIN using the String database34,35 and asked whether additional NEUROD2 targets were found in this network. As well as the previously reported targets, REELIN receptor Lrp8, intracellular adaptor molecule Dab1, and membrane-associated tyrosine kinase Fyn27, we found that we had also identified additional REELIN-interacting NEUROD2 targets, such as Itgb1, Tbr1 and Kcnj6 (Fig. 3B). Furthermore, in order to achieve a comprehensive view of NEUROD2 targets with roles in cortical lamination, we manually searched the literature with our list of target genes and compiled a list of 116 genes with previously demonstrated roles in neuronal migration (Supplemental Material 5). These results raise the possibility that NEUROD2 might regulate cortical neuron migration via fine-tuning the expression of several downstream targets. However, given that Reln was the only target that displayed gene expression changes in our RNA-Seq analysis following Neurod2 depletion, we decided to further investigate the relationship between NEUROD2 and REELIN expression.
NEUROD2 regulates the expression of Reln gene in cortical neurons
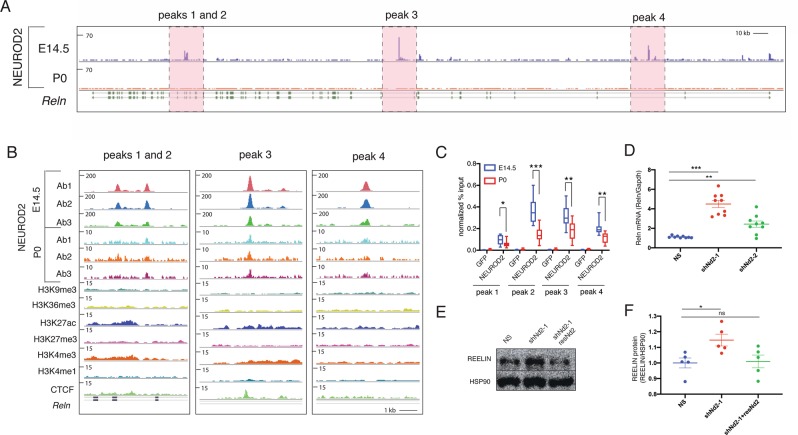
In mice, Reln gene consists of 65 exons, spanning approximately 460 kb of genomic sequence36. Upon inspection of NEUROD2 binding sites along this gene we identified four prominent peaks at introns 21, 22, 55 and 63 (Fig. 4A,B). Initially, we confirmed binding to all four peak sequences by ChIP followed by quantitative PCR (ChIP-qPCR) using cortical tissue from E14.5 and P0 pups (Fig. 4C). We normalized our qPCR signal individually from all four peaks to the amount of input DNA and compared to negative control samples where immunoprecipitation was carried out with an antibody against GFP. Our results suggested quantitatively more binding to all four intronic sequences from E14.5 cortex when compared to samples from P0 cortex (Fig. 4C).
Figure 4.
NEUROD2 binding to introns along the Reln gene is associated with suppression of Reln gene expression. (A,B) Four prominent intronic NEUROD2 binding sites are plotted on the Reln gene. Ab1, Ab2 and Ab3 represent ChIP experiment carried out with three separate NEUROD2 antibodies. Several different histone modifications corresponding to promoters (H3K4me3), enhancers (H3K4me1), actively transcribed (H3K27ac and H3K36me3) or repressed (H3K9me3 and H3K27me3) chromatin are not enriched in NEUROD2 binding sites. A slight enrichment of CTCF binding within intron 3 of Reln is detected. (C) NEUROD2 binding to all four intronic regions is confirmed by ChIP-qPCR. Immunoprecipitation with an unrelated GFP antibody is used as a negative control. n = 12 (three biological x four technical replicates). Bars represent S.E.M. *p < 0.05, **p < 0.005, ***p < 0.0001 by two-tailed unpaired t-test. (D) RT-qPCR analysis of Reln gene expression in primary cortical neurons reveals significant upregulation upon knockdown of Neurod2 with two separate shRNAs. Gapdh mRNA was used as a normalization control. n = 9 (three biological x three technical replicates). p < 0.0001 by one-way ANOVA. ***p < 0.0001, **p < 0.01 by post hoc Sidak multiple comparison tests. Bars represent S.E.M. (E) Immunoblotting of REELIN from 5 DIV primary cortical cultures electroporated with NS, shNeurod2-1 or with shNeurod2-1 together with a plasmid expressing an shRNA resistant Neurod2 cDNA (resNd2). The protein blot has been probed with REELIN antibody and HSP90 antibody as a loading control. Cropped parts of the gel are indicated with a line. (F) Quantification of REELIN protein amounts from five independent experiments. Data are presented after normalizing to HSP90 loading control. *p = 0.02 by two-tailed unpaired t-test. Abbreviations: NS (non-silencing), shNd2-1 (shNeurod2-1), shNd2-2 (shNeurod2-2), resNd2 (rescue Neurod2), ns (non-significant).
In order to gain insight into the transcriptional activity of these sites in the embryonic cortex, we overlaid our NEUROD2 binding profiles with those representing different histone marks associated with promoters (H3K4me3), enhancers (H3K4me1), actively transcribed (H3K27ac and H3K36me3) or silenced (H3K9me3 and H3K27me3) chromatin (Fig. 4B). We did not observe a significant enrichment of either of these histone marks at NEUROD2 binding sites. Our observation might be indicative of a true lack of enrichment of specific histone marks at these loci, or alternatively, might be due to the dilution of ChIP-Seq signals of histone marks acquired from bulk cortical tissue containing many cells that do not express Neurod2. Interestingly, we did observe a modest enrichment of CTCF (CCCTC-binding factor) at the third peak we analyzed (Fig. 4B). This TF has previously been shown to act as a chromatin insulator and as either a transcriptional repressor or activator in a context-dependent manner37–39. Hence, beyond functioning as a typical promoter-associated TF, NEUROD2 might have additional roles in formation of higher-order chromatin structures. Future experiments conducted with purified, Neurod2-expressing neurons will resolve how the binding of post-translationally modified histones and other factors regulating chromatin structure are localized to and regulated at NEUROD2 binding sites.
During radial migration, Reln is expressed and secreted by a transient group of neurons located in the marginal zone called Cajal-Retzius (CR) cells16,21, which do not express Neurod240. On the other hand, migrating neurons express Neurod2, genes encoding REELIN receptors Lrp8 (Apoer2) and Vldlr, as well as components of their downstream signaling pathway, such as the adaptor protein Dab1. Recent single cell RNA-Seq analysis also point to a mutually exclusive pattern of expression of Reln and Neurod2 within the CR cells and within excitatory neurons of the developing cortex41. Interestingly, our RNA-Seq results also suggested NEUROD2 as a negative regulator of Reln expression (Supplemental Material 4). To further explore the possibility that NEUROD2 represses Reln, we knocked down Neurod2 by two different shRNAs in primary cortical neurons. Indeed, we observed a significant upregulation in Reln mRNA expression (Fig. 4D) with the more efficient shNeurod2-1 RNA26 leading to a more robust upregulation in Reln transcript levels compared to those neurons transfected with shNeurod2-2 (Fig. 4D). Next, by immunoblotting we determined how REELIN protein levels were affected upon knockdown of Neurod2 in our primary cortical cultures. To our surprise, we observed a much weaker (approximately 15% of the control) but significant (p = 0.02) increase in REELIN at the protein level. To demonstrate the specificity of this effect, we rescued the increase in REELIN levels by co-expressing a previously characterized26 shRNA-resistant Neurod2 cDNA (Fig. 4E,F). Taken together, our results suggest that NEUROD2 strongly suppresses expression of Reln mRNA in cortical neurons. However, this regulation of gene expression at the mRNA level only modestly affects levels of REELIN protein.
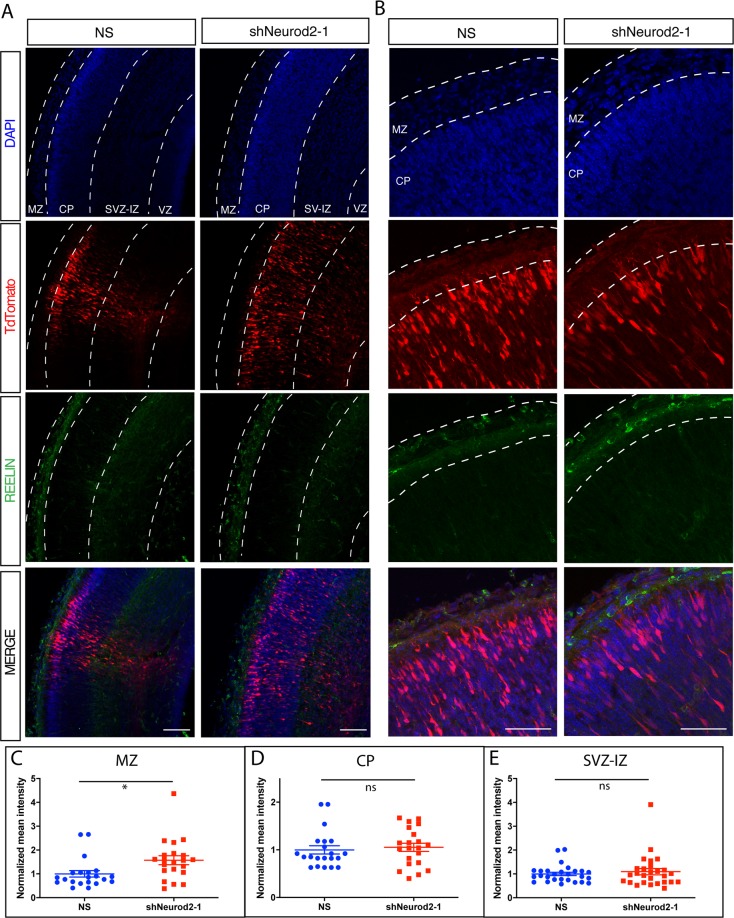
Next we determined how REELIN protein levels were affected in the embryonic cortex upon silencing Neurod2 expression via in utero electroporation of shNeurod2-1. We electroporated E14.5 embryos, fixed the cortices at E17.5 using paraformaldehyde, and carried out immunofluorescence staining against REELIN in cortical sections. In agreement with previous findings, we observed high levels of REELIN protein in the MZ, but not in the CP or the SVZ-IZ of control samples (Fig. 5A,B). Our quantification of immunofluorescence signal intensity revealed no difference within the CP and SVZ-IZ when comparing control and Neurod2 knockdown samples (Fig. 5D and E). Interestingly, in embryos electroporated with shNeurod2-1, we observed a small but significant upregulation of REELIN protein in the MZ, generally localized to the cell body (Fig. 5A–C). These results raised questions about the cellular source of this ectopic REELIN expression. Even though we observe a small number of neurons over-migrating into the MZ in neurons transfected with shNeurod2-1, the vast majority of neurons are localized to the CP. Whether this additional REELIN protein in the MZ is derived from the projecting dendrites of cortical neurons, from over-migrating neurons, or instead is a secondary response of the CR cells to reduction of Neurod2 expression in cortical neurons is currently unclear, but this topic is worthy of future investigation.
Figure 5.
In vivo quantification of REELIN protein levels upon Neurod2 silencing in the embryonic cortex. (A) Embryos were electroporated with non-silencing or shNeurod2-1 shRNA and TdTomato at E14.5, cortices were harvested at E17.5 and confocal images were acquired after REELIN immunofluorescent staining. REELIN levels were not significantly altered within the CP, SVZ-IZ and VZ. However we observed a slight but significant increase in REELIN protein within the MZ. Scale bar: 100 µm. (B) Higher magnification images of those displayed in (A). Scale bar: 50 µm. (C–E) Quantification of images displayed in (A,B). A total of 21 images acquired from n = 3 embryos per group were used for quantification. *p = 0.02 by unpaired two-tailed t-test. Abbreviations: MZ (marginal zone), SVZ-IZ (subventricular-intermediate zone), CP (cortical plate), NS (non-silencing), ns (non-significant).
NEUROD2 is required for the terminal stages of cortical radial migration
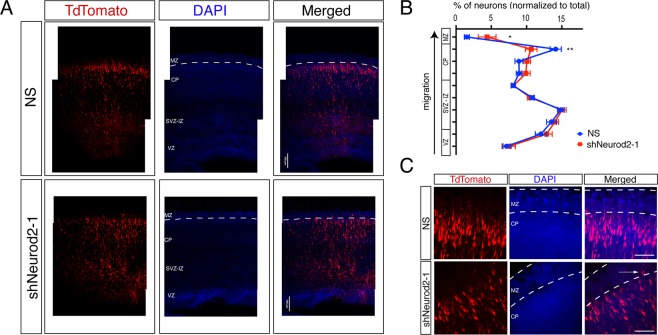
Our comprehensive analysis uncovering NEUROD2 target genes functioning in neuronal migration, along with focused experiments on regulation of Reln gene expression, prompted us to test whether cortical migration is affected upon suppressing Neurod2 expression. Toward this goal, we silenced Neurod2 in neural progenitor cells by in utero transfection of shNeurod2-1 and a tdTomato marker in E14.5 embryos, then quantified the number of tdTomato-positive neurons in different cortical layers at E17.5 by confocal microscopy (Fig. 6A,B). In control samples, we observed tdTomato-positive neurons at expected locations within the ventricular zone, along the subventricular and intermediate zones, and at the cortical plate. In samples in which we silenced Neurod2 expression, we did not observe gross defects in migration, and neurons generally appeared to migrate from the ventricular zone toward the cortical plate in a manner comparable to control samples. Specifically, multipolar migration and glial-guided migration appeared intact. However, we observed a reduction in the number of tdTomato-positive neurons in the primitive cortical zone (located immediately below the marginal zone), suggesting that NEUROD2 depletion leads to a defect specific to the terminal stage of migration (Fig. 6A,B). In Neurod2 knockdown neurons, concomitant with a significant reduction of neurons localized to the primitive cortical zone, we occasionally also observed neurons which had over-migrated into the MZ (Fig. 6B,C). Taken together, our results point to a mechanism where NEUROD2 regulates the positioning of somas at the primitive cortical zone. Future experiments will clarify how NEUROD2 target genes regulate this critical step during cortical lamination.
Figure 6.
NEUROD2’s role in neuronal soma positioning to the primitive cortical zone. (A) Embryos (at E14.5) were in utero electroporated with tdTomato fluorescent marker and with NS (non-silencing) shRNA or with shNeurod2-1. Embryos were retrieved at E17.5, coronally sectioned, and tdTomato signal was subsequently imaged by confocal microscopy. VZ (ventricular zone), SVZ-IZ (subventricular-intermediate zone), CP (cortical plate), and MZ (marginal zone) are labeled. Scale bar: 100 µm. (B) Transfected neurons (a total of n = 16,184 for NS and n = 19,339 for shNeurod2-1) derived from in utero electroporation of seven littermate embryo pairs from five independent pregnant mice were counted. Normalized percentages of tdTomato-positive neurons counted in individual zones are plotted. Significantly less neurons are localized to the upper cortical plate in neurons where Neurod2 is knocked down. (C) Higher magnification images of the MZ and the CP in NS and shNeurod2-1 electroporated cortices. White arrow points to occasional neurons that over-migrate into the MZ of the shNeurod2-1 transfected samples. Scale bar: 50 µm. Bars represent S.E.M. p < 0.0001 by two-way ANOVA and *p = 0.0165 and **p = 0.0015 by post hoc Sidak’s test.
Discussion
Here we report a comparative analysis of the genetic targets of NEUROD2 and reveal a new role for this TF in the terminal stage of cortical radial migration. We had previously reported NEUROD2 targets identified by ChIP-Seq experiments both in embryonic and postnatal cerebral cortices26,27, and here we have complemented these earlier studies by a comparative analyses of these two developmental timepoints. Furthermore, in order to focus upon targets whose expression is regulated by NEUROD2, we silenced its expression in primary cortical neurons and carried out an RNA-Seq analysis. Our comprehensive and comparative analyses of these two ChIP-Seq and RNA-Seq datasets converged upon the Reln gene. We show that in mice, NEUROD2 binds to several different introns of Reln at both embryonic and postnatal stages, and this binding correlates with reduced levels of Reln mRNA. Even though NEUROD2 has typically been considered as a transcriptional activator, our studies point to a potentially dual function for NEUROD2; in addition to its previously reported transcription activating function25,42,43, it may also act as a repressor in a context-dependent manner. In support of our findings, Reln expression was also previously shown to be upregulated in the embryonic cortex of Neurod2/6 double knockout mice44. However, whether this effect is a direct consequence of NEUROD2 binding to Reln introns, or instead is a secondary response, awaits confirmation by site-directed genome editing of these binding sites.
NEUROD2’s potential capacity to function as a transcriptional repressor agrees well with our previous results demonstrating that NEUROD2 inhibits expression of the Stim1 gene to control calcium signaling in cortical neurons26. Furthermore, several of the NEUROD2 binding peaks both in Reln and Stim1 genes have also been identified in ChIP-Seq experiments carried out for the chromatin insulating factor CTCF (encodeproject.org), suggesting that NEUROD2 might be involved in establishing or maintaining higher order architecture of chromatin loops. In support of this hypothesis, NEUROD2 was recently shown to function as a major organizer of cell-type specific chromatin folding in cortical neurons, suggesting that its transcription activating or repressing functions might in fact be directly related to its role in establishing or maintaining transcriptionally isolated domains45. The mechanistic details of how NEUROD2 regulates transcriptional activity in distinct genetic loci are of great interest for future investigations.
Past studies have suggested that NEUROD2 might have a role in regulating radial migration of excitatory cortical neurons27,28,46. In addition to our current finding demonstrating NEUROD2 regulation of Reln mRNA expression, our past work had shown that it also binds to promoters of several genes encoding downstream signaling proteins that transduce REELIN signals, such as Dab1 and Fyn, as well as the receptor gene Lrp827. To our surprise, upregulation of Reln mRNA levels upon Neurod2 silencing does not correlate well with changes at the protein level. We observe a small but significant increase of REELIN in primary neurons, but not in embryonic sections, after Neurod2 suppression. Immunofluorescence staining is a semi-quantitative method and might not report accurately upon small changes in protein levels of REELIN. Taken together, we conclude that the major effect of NEUROD2 is most prominent at the mRNA level of Reln.
The control of Reln gene expression within a precise spatiotemporal window in the developing cortex appears to be a conserved and tightly regulated mechanism that depends upon several TFs. For example, double-knockout mice for the TFs Pbx1/2 also exhibit ectopic Reln expression, leading to a dramatic inversion of cortical layering reminiscent of mouse models with defective REELIN signaling47. Similarly, knockdown of the histone methyltransferase Ezh2, as well as deletion of Foxg1, result in derepression of Reln gene expression and are associated with radial migration and cortical development defects48,49. Finally, ectopic expression of Reln cDNA by in utero electroporation into neural progenitors causes formation of neuronal aggregates in deep layers that are incapable of migrating towards the cortical plate50. Taken together, these findings highlight the importance of maintaining a precise level of Reln gene expression for the proper development of cortical layers. Our results suggest that NEUROD2 is an additional transcriptional regulator of the Reln gene. Furthermore, deficiencies in NEUROD2 function might remove one layer of control against ectopic REELIN expression and may potentially sensitize migrating neurons to synthetic genetic interactions with other genes functioning in REELIN signaling.
Beyond our analysis of Reln expression, we also generated a curated list of NEUROD2 binding targets with known roles in neuronal migration based on published literature. Interestingly, we identified a number of additional targets, including components involved in Ephrin signaling, members of the Cdk5 pathway, and additional TFs. Finally, in order to test whether NEUROD2-regulated transcription functions during radial migration of cortical neurons, we silenced Neurod2 expression in ventricular neural progenitors by in utero electroporation in mice and revealed that NEUROD2 is important for proper positioning of the neuronal soma upon arrival at the primitive cortical zone. These findings suggest that NEUROD2 might impact cortical lamination by regulating multiple components of the REELIN signaling pathway or other pathways. Finally, our dataset provides novel candidate genes that may function in different aspects of cortical lamination.
Methods
Bioinformatic analyses of ChIP-Seq data
Previously published NEUROD2 ChIP-Seq data were used to identify differential NEUROD2 binding to genomic regions at timepoints E14.5 and P026,27. These data were generated using three separate NEUROD2 antibodies (Abcam ab168932, ab104430 and ab109406). Histone ChIP-Seq datasets from E14.5 mouse embryonic cortex were produced by Bing Ren’s Laboratory, UCSD, USA51 (www.encodeproject.org). ChIP-Seq peaks were visualized using Trackster52 embedded within the Galaxy Project53. Details of Closest Gene score calculations were previously described27.
Primary cortical cultures and Sholl analysis
Primary cultures were prepared from E14.5 embryos as described26. For Sholl analysis, primary cortical neurons were transfected with non-silencing or shNeurod2-126 at 2 DIV with Lipofectamine 2000 transfection reagent (Invitrogen). Neurons were fixed with 4% PFA at 5 DIV, immunostained with an antibody recognizing EGFP (Aves Lab, GFP 10–20), and imaged with a Nikon 90i Eclipse confocal microscope affixed with a 60x oil objective. Dendritic development was quantified by Sholl analysis found within ImageJ33,54.
ChIP-qPCR
Chromatin immunoprecipitation was carried out using cortical tissue collected from E14.5 and P0 pups of both sexes and an antibody raised against NEUROD2 (Abcam, ab109406) and against GFP as a negative control (Santa Cruz, sc-8334). Formaldehyde-crosslinking, followed by chromatin DNA isolation from input and immunoprecipitated samples, was performed as previously described27. Cq values from immunoprecipitated DNA were normalized to those from input DNA as described26. Primer sequences used for qPCR experiments were as follows: Reln-peak1-F (AATGGAAACTGGCTCGCATG); Reln-peak1-R (ATCCTGAGCAATGAGTGGCT); Reln-peak2-F (TGTTTCTGTGGTCTGCTTGC); Reln-peak2-R (AAAACAATCACAGGCGAGCC); Reln-peak3-F (GCCGGACTACCCTGATGATT); Reln-peak3-R (TGGAGGAAATGGATGGCTCT); Reln-peak4-F (GGCCTCCTGTCTTACTAGCC); Reln-peak4-R (CCTGACAGATGGAGCGTTTG).
RNA-Seq and RT-qPCR
Primary cultures were prepared from E14.5 embryos as described26. Cortical neuron suspensions were electroporated with shNeurod2-1, shNeurod2-2, or a non-silencing plasmid immediately before plating (Amaxa™ P3 Primary Cell 4D-Nucleofector X Kit L; Lonza, CU133 program). The generation and knockdown efficiencies of shRNA plasmids were described previously26. Total RNA was isolated with Absolutely RNA Microprep Kit (Agilent Technologies) at 5 d in vitro (DIV). The RNA samples were further used for either RNA-seq-directed library construction or for RT-qPCR. Library construction and sequencing were carried out at Genewiz Inc. (New Jersey, USA). RNA-Seq libraries were sequenced on the Illumina HiSeq 2500 platform using a 50 bp single-end format. After quality filtering of reads, sequences were mapped onto the genome with TopHat55, assembled onto transcripts with Cufflinks56 and differential expression was determined with Cuffdiff56. Raw and processed RNA-Seq data are available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE131494. For RT-qPCR, cDNA was synthesized with Transcriptor high-fidelity cDNA synthesis kit (Roche) and RT-qPCR was performed using Luminaris HiGreen qPCR Master Mix (Thermo Scientific) in a CFX Connect Real-Time PCR Detection System (Bio-Rad). The 2−ΔΔCT method was used for calculating Reln mRNA levels as normalized to Gapdh mRNA. The primer sequences were as follows: Reln-F (TTTACACTGAGGCTGGGGAG); Reln-R (TGCCACCATCTGAACTGGAT); Gapdh-F (AATGTGTCCGTCGTGGATCTGACGTGC); Gapdh-R (TTGCTGTTGAAGTCGCAGGAGACAACC).
Immunoassays
An anti-REELIN antibody (Abcam, cat no. ab78540) was used at a 1:1000 dilution for immunoblotting and 1:500 dilution for immunofluorescent staining. For immunoblotting, protein lysates were prepared using primary cortical cultures from E14.5 embryos and electroporated with the appropriate shRNA plasmid. At 5 DIV cells were lysed in ice-cold RIPA Buffer (0.05 M Tris-HCl pH 7.5, 0.15 M NaCl, 1% Triton-X 100, 1% Na-DOC, 0.1% SDS) containing freshly added protease inhibitors, incubated on ice for 15 min. and centrifuged for 20 min at 14,000 rpm at 4 °C. Protein concentrations of lysates were measured by Bradford protein assay and 10 μg of total protein was loaded per well. Band intensities were quantified by ImageJ54. For immunofluorescent staining, embryonic brains were fixed at 4% PFA overnight, treated with 30% sucrose for one day, and sectioned to 50 µm with a Leica CM 1950 cryostat. Anti-REELIN antibody incubation was carried out in blocking buffer (3% BSA, 0.3% Triton X-100 in PBS, pH 7.4) overnight in 4 °C. Images were captured using a Leica DMI8 confocal microscope. Quantification of MZ, CP and SVZ-IZ regions were defined based on DAPI staining, and quantification was performed with ImageJ54.
In utero electroporation and image analysis
Our in utero electroporation method was based upon a previously published protocol57. Briefly, 14.5 day pregnant mice were anesthetized by inhalation of isoflurane. 1 µl of DNA solution (composed of 500 ng of shRNA plasmid, 500 ng of pCAGGS-IRES-tdTomato and 0.1% Fast Green FCF) was injected unilaterally into the lateral ventricular of each embryo. 0.5 pulses at 30 V with 400 ms intervals were applied with a 5 mm tweezer electrode using an electroporator (BTX, Harvard Apparatus, ECM830). Following electroporation, embryos were placed back into the uterine cavity, and the pregnant female was sutured and placed into an incubation chamber for recovery. Embryos were retrieved at developmental age E17.5, followed by brain removal and placement into 4% PFA. Embryos were coronally sectioned to 50 µm with a cryostat (Leica CM1950). Confocal images of tdTomato-positive neurons were captured with Nikon Eclipse 90i (20X objective) or Leica DMI8 (63X oil objective) and maximum intensity projection images were generated using Nikon NIS Elements AR or Leica LAS X Life Science software. A grid of ten equal bins were overlaid onto the cortical cross-section images, and the number of tdTomato-positive neurons in each bin was counted using the cell counting tool integrated into ImageJ54. These results were expressed as a percentage of total number of tdTomato-positive cells. In each experiment, littermate embryos from the same pregnant mouse were electroporated with non-silencing and shNeurod2-1, respectively.
Animals
All animal experiments were performed in accordance with the guidelines for the care and use of laboratory animals of Koç University. The study was approved by the Institutional Animal Care and Use Committee of Koç University with license number 2014–07. For in utero electroporations (CD-1 strain) and primary neuronal cultures (Balb/c strain), timed-pregnant mice were generated by The Koç University Experimental Animal Laboratory.
Supplementary information
Acknowledgements
We thank Ali C. Taşkın, Ahmet Kocabay, Muhammed Reşat Oguzkan Yaralı and Mehmet Yücel (Koç University Animal Research Facility, Turkey) for their help in providing timed pregnant mice; Efil Bayam (Institute of Genetics and Cellular and Molecular Biology, Strasbourg, France) for assistance with the in utero electroporation method; Tamer Önder (Koç University School of Medicine, Turkey) for generously sharing the Lonza nucleofector equipment, and the Molecular Imaging Core Facility of Koç University Research Center for Translational Medicine (KUTTAM) for assistance with confocal microscopy. This work was funded by following grants to G. Ince-Dunn: European Commission FP7 International Reintegration Grant (PIRG07-GA-2010-268433), Turkish Academy of Sciences Young Scientist Program (TUBA-GEBIP), and Koç University Seed Fund; to C. Akkaya: TUBITAK-BIDEB 2211/E Scholarship Program; and to C. Dunn: EMBO Installation Grant 2138.
Author contributions
G.G. carried out ChIP-qPCR and RNA-Seq experiments (Figs. 1,3 and 4), and primary neuronal cultures, and Sholl analysis (Fig. 2). C.A. carried out all in utero electroporation experiments, confocal microscopy and data analysis, and D.A. assisted these experiments (Figs. 5 and 6). A.K., C.D.D. and N.O. contributed to data analysis (Figs. 1,3 and 4). G.I.D. conceived the hypothesis, designed research, analyzed data and wrote the paper. All authors discussed the results and reviewed the manuscript.
Data availability
RNA-Seq data can be downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE131494.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gizem Guzelsoy and Cansu Akkaya.
Supplementary information
is available for this paper at 10.1038/s41598-019-56171-x.
References
- 1.Tan X, Shi SH. Neocortical neurogenesis and neuronal migration. Wiley Interdiscip Rev Dev Biol. 2013;2:443–459. doi: 10.1002/wdev.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu WF, Chahrour MH, Walsh CA. The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet. 2014;15:195–213. doi: 10.1146/annurev-genom-090413-025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchsbaum Isabel Yasmin, Cappello Silvia. Neuronal migration in the CNS during development and disease: insights from in vivo and in vitro models. Development. 2019;146(1):dev163766. doi: 10.1242/dev.163766. [DOI] [PubMed] [Google Scholar]
- 5.Romero DM, Bahi-Buisson N, Francis F. Genetics and mechanisms leading to human cortical malformations. Semin Cell Dev Biol. 2018;76:33–75. doi: 10.1016/j.semcdb.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 6.Ohtaka-Maruyama C, Okado H. Molecular Pathways Underlying Projection Neuron Production and Migration during Cerebral Cortical Development. Front Neurosci. 2015;9:447. doi: 10.3389/fnins.2015.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hippenmeyer S. Molecular pathways controlling the sequential steps of cortical projection neuron migration. Adv Exp Med Biol. 2014;800:1–24. doi: 10.1007/978-94-007-7687-6_1. [DOI] [PubMed] [Google Scholar]
- 8.Sekine K, Honda T, Kawauchi T, Kubo K, Nakajima K. The outermost region of the developing cortical plate is crucial for both the switch of the radial migration mode and the Dab1-dependent “inside-out” lamination in the neocortex. J Neurosci. 2011;31:9426–9439. doi: 10.1523/JNEUROSCI.0650-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirota Y, Nakajima K. Control of Neuronal Migration and Aggregation by Reelin Signaling in the Developing Cerebral Cortex. Front Cell Dev Biol. 2017;5:40. doi: 10.3389/fcell.2017.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekine K, et al. Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin alpha5beta1. Neuron. 2012;76:353–369. doi: 10.1016/j.neuron.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson EC, Kim S, Walsh CA. Impaired neuronal positioning and dendritogenesis in the neocortex after cell-autonomous Dab1 suppression. J Neurosci. 2006;26:1767–1775. doi: 10.1523/JNEUROSCI.3000-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Muller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanada K, Gupta A, Tsai LH. Disabled-1-regulated adhesion of migrating neurons to radial glial fiber contributes to neuronal positioning during early corticogenesis. Neuron. 2004;42:197–211. doi: 10.1016/S0896-6273(04)00222-3. [DOI] [PubMed] [Google Scholar]
- 14.Chai X, et al. Reelin Induces Branching of Neurons and Radial Glial Cells during Corticogenesis. Cereb Cortex. 2015;25:3640–3653. doi: 10.1093/cercor/bhu216. [DOI] [PubMed] [Google Scholar]
- 15.Matsuki T, et al. Reelin and stk25 have opposing roles in neuronal polarization and dendritic Golgi deployment. Cell. 2010;143:826–836. doi: 10.1016/j.cell.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Arcangelo G, et al. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 17.Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- 18.Trommsdorff M, et al. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/S0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 19.Caviness VS, Jr., Sidman RL. Time of origin or corresponding cell classes in the cerebral cortex of normal and reeler mutant mice: an autoradiographic analysis. The Journal of comparative neurology. 1973;148:141–151. doi: 10.1002/cne.901480202. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez JL, et al. Birthdate and cell marker analysis of scrambler: a novel mutation affecting cortical development with a reeler-like phenotype. J Neurosci. 1997;17:9204–9211. doi: 10.1523/JNEUROSCI.17-23-09204.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawa M, et al. The reeler gene-associated antigen on Cajal-Retzius neurons is a crucial molecule for laminar organization of cortical neurons. Neuron. 1995;14:899–912. doi: 10.1016/0896-6273(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 22.Pacary E, et al. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron. 2011;69:1069–1084. doi: 10.1016/j.neuron.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heng JI, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–118. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- 24.Hand R, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 25.Olson JM, et al. NeuroD2 is necessary for development and survival of central nervous system neurons. Dev Biol. 2001;234:174–187. doi: 10.1006/dbio.2001.0245S0012-1606(01)90245-3. [DOI] [PubMed] [Google Scholar]
- 26.Guner Gokhan, Guzelsoy Gizem, Isleyen Fatma Sadife, Sahin Gulcan Semra, Akkaya Cansu, Bayam Efil, Kotan Eser Ilgin, Kabakcioglu Alkan, Ince-Dunn Gulayse. NEUROD2 Regulates Stim1 Expression and Store-Operated Calcium Entry in Cortical Neurons. eneuro. 2017;4(1):ENEURO.0255-16.2017. doi: 10.1523/ENEURO.0255-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayam E, et al. Genome-wide target analysis of NEUROD2 provides new insights into regulation of cortical projection neuron migration and differentiation. BMC Genomics. 2015;16:681. doi: 10.1186/s12864-015-1882-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Telley L, et al. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science. 2016;351:1443–1446. doi: 10.1126/science.aad8361. [DOI] [PubMed] [Google Scholar]
- 29.Sikora-Wohlfeld W, Ackermann M, Christodoulou EG, Singaravelu K, Beyer A. Assessing computational methods for transcription factor target gene identification based on ChIP-seq data. PLoS computational biology. 2013;9:e1003342. doi: 10.1371/journal.pcbi.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joshi-Tope G, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein, L. D. Using the Reactome database. Curr Protoc Bioinformatics Chapter 8, Unit 8 7, 10.1002/0471250953.bi0807s7 (2004). [DOI] [PubMed]
- 32.Redmond, L., Kashani, A. H. & Ghosh, A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron34, 999–1010, doi:S0896627302007377 (2002). [DOI] [PubMed]
- 33.Ferreira TA, et al. Neuronal morphometry directly from bitmap images. Nat Methods. 2014;11:982–984. doi: 10.1038/nmeth.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen D, Lei EP. Function and regulation of chromatin insulators in dynamic genome organization. Curr Opin Cell Biol. 2019;58:61–68. doi: 10.1016/j.ceb.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arzate-Mejía Rodrigo G., Recillas-Targa Félix, Corces Victor G. Developing in 3D: the role of CTCF in cell differentiation. Development. 2018;145(6):dev137729. doi: 10.1242/dev.137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prickett AR, et al. Genome-wide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions. Genome Res. 2013;23:1624–1635. doi: 10.1101/gr.150136.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki H, Sekiguchi M, Takamatsu M, Tanabe Y, Nakanishi S. Distinct ontogenic and regional expressions of newly identified Cajal-Retzius cell-specific genes during neocorticogenesis. Proc Natl Acad Sci USA. 2004;101:14509–14514. doi: 10.1073/pnas.0406295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan X, et al. Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis. Cell Res. 2018;28:730–745. doi: 10.1038/s41422-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ince-Dunn, G. et al. Regulation of thalamocortical patterning and synaptic maturation by NeuroD2. Neuron49, 683–695, doi:S0896-6273(06)00088-2.10.1016/j.neuron.2006.01.031 (2006). [DOI] [PubMed]
- 43.Aizawa H, et al. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845.303/5655/197. [DOI] [PubMed] [Google Scholar]
- 44.Bormuth, I. Roles of bHLH transcription factors Neurod1, Neurod2 and Neurod6 in cerebral cortex development and commissure formation Ph.D. thesis, Georg-August Universität, (2015).
- 45.Bonev B, et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572 e524. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bormuth I, et al. Neuronal basic helix-loop-helix proteins Neurod2/6 regulate cortical commissure formation before midline interactions. J Neurosci. 2013;33:641–651. doi: 10.1523/JNEUROSCI.0899-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golonzhka O, et al. Pbx Regulates Patterning of the Cerebral Cortex in Progenitors and Postmitotic Neurons. Neuron. 2015;88:1192–1207. doi: 10.1016/j.neuron.2015.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao L, et al. Ezh2 is involved in radial neuronal migration through regulating Reelin expression in cerebral cortex. Sci Rep. 2015;5:15484. doi: 10.1038/srep15484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cargnin F, et al. FOXG1 Orchestrates Neocortical Organization and Cortico-Cortical Connections. Neuron. 2018;100:1083–1096 e1085. doi: 10.1016/j.neuron.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kubo K, et al. Ectopic Reelin induces neuronal aggregation with a normal birthdate-dependent “inside-out” alignment in the developing neocortex. J Neurosci. 2010;30:10953–10966. doi: 10.1523/JNEUROSCI.0486-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goecks J, Coraor N, Galaxy T, Nekrutenko A, Taylor J. NGS analyses by visualization with Trackster. Nat Biotechnol. 2012;30:1036–1039. doi: 10.1038/nbt.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data can be downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE131494.