Abstract
Mytilus mussels have been the object of much research given their sentinel role in coastal ecosystems and significant value as an aquaculture resource appreciated for both, its flavour and nutritional content. Some of the most-studied Mytilus species are M. edulis, M. galloprovincialis, M. chilensis and M. trossulus. As species identification based on morphological characteristics of Mytilus specimens is difficult, molecular markers are often used. Single-locus markers can give conflicting results when used independently; not all markers differentiate among all species, and the markers target genomic regions with different evolutionary histories. We evaluated the concordance between the PCR-RFLP markers most commonly-used for species identification in mussels within the Mytilus genus (Me15-16, ITS, mac-1, 16S rRNA and COI) when used alone (mono-locus approach) or together (multi-locus approach). In this study, multi-locus strategy outperformed the mono-locus methods, clearly identifying all four species and also showed similar specimen identification performance than a 49 SNPs panel. We hope that these findings will contribute to a better understanding of DNA marker-based analysis of Mytilus taxa. These results support the use of a multi-locus approach when studying this important marine resource, including research on food quality and safety, sustainable production and conservation.
Subject terms: Genetic markers, Haplotypes
Introduction
Marine mussels within the Mytilus genus are benthic organisms inhabiting the intertidal temperate and cold waters of both hemispheres1,2. As of April 2019, the Integrated Taxonomic Information System (ITIS) (http://www.itis.gov)3 listed four taxa within the genus: M. edulis Linnaeus,1758; M. galloprovincialis Lamarck, 1819; M. californianus Conrad, 1837 and M. trossulus Gould, 1850. The World Register of Marine Species (http://www.marinespecies.org)4 includes the above plus four additional taxa: M. planulatus Lamarck, 1819; M. platensis d’Orbigny, 1842; M. chilensis Hupé, 1854 and M. unguiculatus Valenciennes, 1858. As genetic similarity among these taxa is high, mussels interbreed when they coexist spatially, forming hybrid zones that have been studied by various authors, e.g. Crego-Prieto et al.5, Inoue et al.6, Kartavtsev et al.7, Mathiesen et al.8, Oyarzún et al.9, Rawson et al.10, Riginos & Cunningham11, Väinölä & Hvilsom12 and Wilhelm & Hilbish)13.
Mytilus bivalves are an object of research as cosmopolitan inhabitants of high-latitude coastal marine ecosystems in the Northern and Southern Hemispheres, serving as sensitive pollution bioindicators with great utility in ecotoxicology14–16. Mytilus bivalves are also of interest in invasion ecology, with M. galloprovincialis listed among the 100 most invasive species in the world17,18. Finally, mussels are highly valued as a flavorful and nutritious food. Mussels are extensively cultured and commercialized in many countries, representing an important economic activity for coastal communities. The FAO reported that the smooth-shelled blue mussel species M. chilensis, M. edulis and M. galloprovincialis represented 91.8% of Mytilus landings worldwide in 201619.
Species-level identification based on morphological traits is problematic within the Mytilus genus20, as shell shapes are fairly uniform and show environmentally-influenced phenotypic plasticity6,21,22. Furthermore, the shell is typically removed from processed foods, further hampering appearance-based identification23. Alternatively, mussels are characterized using genetic markers, with approaches relying on allozymes24–27, sequencing of mitochondrial genes2,28–30 and PCR-based DNA markers, such as amplified fragment length polymorphisms (AFLP)22, random amplification of polymorphic DNA (RAPD)31, forensically informative nucleotide sequencing (FINS)23 and fragment length polymorphisms (FLP). To enhance the specificity of FLP analysis, an enzymatic restriction step can be added to produce restriction fragment length polymorphism (RFLP) markers. Some RFLP banding patterns are easy to score and are almost fixed in allopatric populations. Therefore, these patterns are regarded as diagnostic loci and are widely used for Mytilus species identification32–34. Nowadays, is also possible to perform a trustworthy specimen identification with highly informative SNP panels35–40. However, no all laboratories have access to this genomic technology and PCR-RFLP markers are still used in recently valuable studies41–44.
PCR-RFLP analyses of mitochondrial loci have targeted genes such as the large 16S subunit of the rRNA gene45,46, the control region of maternally transmitted mtDNA and the cytochrome oxidase subunit I region (COI)20 of maternally transmitted mtDNA47. Nuclear DNA restriction FLP in anonymous and coding regions have been developed for species identification48, targeting at least seven nuclear loci: The internally transcribed spacer regions between the 18S and 28S rDNA nuclear coding regions (ITS)33, the protamine-like sperm packaging protein (PLIIa)33, an intron-length polymorphism at the actin gene locus (mac-1)49, a length polymorphism in the elongation factor 1 cDNA (EFbis)50, a coding locus designated as Mytilus anonymous locus-I (MAL-I)34,51, the acrosomal sperm protein M7 lysin52,53 and the polyphenolic adhesive protein gene, which encodes for a highly-conserved protein that allows mussels to adhere to bedrock.
The polyphenolic adhesive protein gene has been widely used for Mytilus species identification. Inoue et al.54 developed the Me15-16 primer set for genetic identification of three species (M. edulis, M. trossulus and M. galloprovincialis). Santaclara et al.23 added a restriction step to differentiate M. chilensis from the Northern-Hemisphere M. galloprovincialis. Rawson et al.32 described two markers: Glu-5′, to identify the three Northern Hemisphere blue mussel species, M. edulis, M. galloprovincialis and M. trossulus, and Glu-3′, which distinguishes M. edulis from M. galloprovincialis. Fernández-Tajes et al.20 used the primers Myti-F/R and further digestion with restriction enzymes (Aci I and Acl I) to differentiate commercial Mytilus species, while Jilberto et al.55 used the primers PAPM F/R followed by high-resolution melting analysis to differentiate M. chilensis M. edulis and M. galloprovincialis and their hybrids. Interestingly, Me15-16, Glu-5′, Myti and PAPM target the same region in the gene.
According to a search of the Science Direct and Web of Science databases for research using any PCR-RFLP DNA analysis to identify Mytilus species published from 1995 to date, ~80% of studies used a marker that targets the polyphenolic adhesive protein gene. Most works (~55%) used a single-marker diagnostic test, while others used two (~20%) or three (~20%), and a few (~5%) applied four or more markers. However, in studies with multiple markers, results sometimes varied by marker, as is showed in Mytilus populations from Europe56–59, Tasmania and the Kerguelen Islands60,61 and the Pacific North American coast62. These findings have led to divergent descriptions of the species distribution, especially in the Southern Hemisphere. Therefore, applying a combination of markers has been proposed as a more reliable identification method58.
Our aim was to compare the performance in Mytilus specimen identification obtained using single- and multi-locus approaches, and also to evaluate concordance among the PCR-RFLP markers most commonly used for Mytilus mussel identification (Mytilus spp.): Me15-16, ITS, mac-1, 16S rRNA and COI to contribute to a better understanding of the previous works (pre-genomic era) based mostly in the application of mono-locus PCR- RFLP markers to identify Mytilus taxa. Considering the irruption of genomic markers (SNPs), we also aim to contrast the performance in specimen identification of multi-locus panels composed by SNPs used by Larraín et al.38 with the five above mentioned PCR-RFLP markers.
Methods
Sampling and DNA extraction
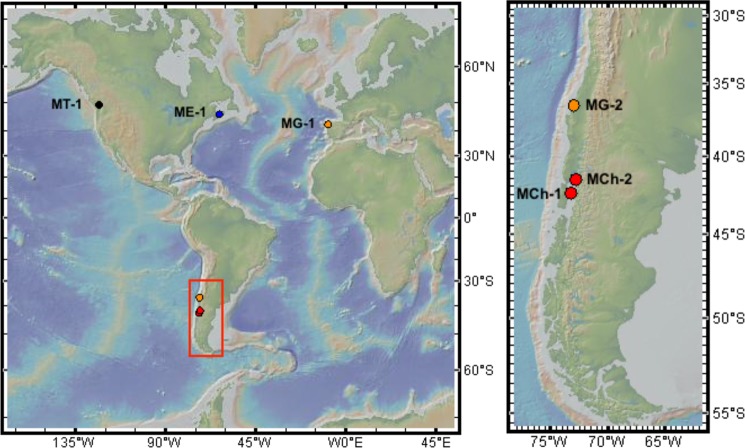
Mussel samples (n = 298) were obtained between 2008 and 2017 from six locations (Fig. 1). Four locations represented putatively pure populations outside of described hybrid zones: pacific mussels (M. trossulus) (MT-1) from West Vancouver, Canada (n = 50); blue mussels (M. edulis) (ME-1) from the Northwest Atlantic Ocean near Prince Edward Island, Canada (n = 50); Mediterranean mussels (M. galloprovincialis) (MG-1) from Galicia along the Atlantic coast of Spain (n = 49) and Chilean mussels (M. chilensis) (MCh-1) from Putemun on Chiloé Island (n = 50). The latter three are the most commercialized Mytilus species. Two other Chilean populations were sampled from locations where hybrid individuals have been found: Quillaipe (MCh-2) (n = 49) on the Gulf of Reloncaví63 and the northern coast of the Gulf of Arauco (MG-2) (n = 50), where the Mediterranean mussel (M. galloprovincialis) has also been found30,64. Detailed sample data is included in Supplementary Information (Table S1). Approximately 50–100 mg of ethanol-fixed mantle edge tissue was used for DNA extraction with a modified phenol-chloroform method63. Extracted DNA was quantified with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific).
Figure 1.
Locations and codes for the six sampling sites. Codes for locations can be found in Table S5. Color indicates species as determined using the PCR-RFLP Me15-16 AciI assay: red for Mytilus chilensis, orange for Mytilus galloprovincialis, blue for Mytilus edulis and black for Mytilus trossulus. Background topographic map from GeoMapApp (http://www.geomapapp.org).
Specimen identification
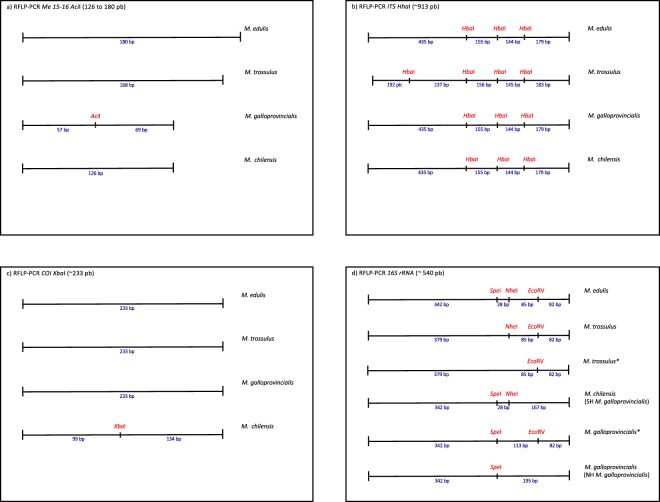
To avoid the unintended presence of other genera (i.e. Aulacomya or Choromytilus) in the Chilean samples, genus assignment was performed with PCR-RFLP using 18S rDNA and the enzyme BsaHI23. Species identification was performed for Mytilus specimens with PCR-RFLP using the nuclear DNA marker Me15-16 and the enzyme AciI23,54. This assay targets the polyphenolic adhesive protein gene, producing PCR products 180 bp in length for M. edulis, 168 bp for M. trossulus and 126 bp for both M. chilensis and M. galloprovincialis. To differentiate between these last two species, amplicons were digested with AciI, producing 77 and 49 bp fragments in M. galloprovincialis and leaving the M. chilensis amplicon uncut23 (Fig. 2a). These classifications were used as the reference for comparisons with the markers below.
Figure 2.
Restriction map of markers RFLP-PCR (a) Me15-16 AciI, (b) ITS HhaI, (c) COI XbaI and (d) 16S rRNA EcoRV, NheI and SpeI. *Is used to identify the new haplotypes found in this work. For clarity, we will conserve the name M. galloprovincialis to refer the former Northern Hemisphere haplotype and use M. chilensis for the former Southern Hemisphere haplotype.
The multi-allelic marker nuclear locus mac-1 has a size polymorphism49, allowing for discrimination among M. edulis, M. trossulus, and M. galloprovincialis according to the frequency of “synthetic” (pooled) alleles60,65,66.
The nuclear marker PCR-RFLP ITS HhaI produces a ~913 bp fragment. After fragment restriction with HhaI endonuclease, the RFLP patterns from M. edulis, M. galloprovincialis and M. chilensis consisted of four fragments with modal sizes of 435, 179, 155 and 144 pb, respectively, with only minor variations in size by species. Five fragments were observed for M. trossulus, with modal sizes of 237, 192, 183, 156 and 145 (Fig. 2b). As in Heath et al.33 and Toro67, this marker therefore only differentiated between M. trossulus and the other three species, although those authors obtained somewhat different fragment sizes using agarose gels.
Mitochondrial marker PCR-RFLP COI XbaI targets the cytochrome oxidase subunit I region with the primers COIXbaF and COIXbaIR, designed to distinguish M. chilensis from other mussels. The 233 bp amplicon was restricted with the XbaI enzyme, generating two fragments (134 and 99 bp) in M. chilensis20 only (Fig. 2c).
The mitochondrial marker PCR-RFLP 16S rRNA produces a ~540 bp fragment with universal primers 16sar-L/16sbr-H68. After digestion with the enzymes EcoRV, NheI and SpeI, M. edulis (342, 85, 82 and 28- bp fragments) and M. trossulus (379, 85 and 82 bp fragments) showed fixed haplotypes. Northern Hemisphere M. galloprovincialis individuals showed an exclusive haplotype (342 and 195 bp fragments) as well as the M. edulis haplotype (342, 85, 82 and 28 bp fragments). Westfall et al.46 and Zardi et al.69 described a Southern Hemisphere M. galloprovincialis haplotype with three fragments (342, 167 and 28 bp) (Fig. 2d). For clarity, we will conserve the name M. galloprovincialis to refer the former Northern Hemisphere haplotype and use M. chilensis for the former Southern Hemisphere haplotype (Fig. 2d).
DNA amplifications were performed in a Techne TC-412 (Bibby Scientific Ltd, UK) thermocycler and Palm-CyclerTM (Corbett Life Science, Australia) with recombinant Taq DNA polymerase (RBC Bioscience® and Thermo Fisher®) using 40 ng of template DNA in a final volume of 25 μL. The PCR primer pairs described were used as reported by the above authors with no modifications. Details regarding the PCR reaction conditions and amplification profiles for each marker are shown in Table 1. All experiments included a negative control (with no template DNA added) and a positive control consisting of template DNA that was previously extracted and successfully amplified by conventional PCR with Me15-1654. Digestion with AciI (New England Biolabs), HhaI and XbaI (Thermo Scientific) was performed separately in a final volume of 20 μL, using 15 μL of PCR product and 4, 10 and 10 units of each enzyme with 1x NEB4 and Tango buffers, respectively. Triple digestion with EcoRV, NheI and SpeI (New England Biolabs) was performed in the same reaction using 10, 5 and 5 units of each enzyme, respectively, with 1x NEB2 buffer. All incubations were carried out overnight at 37 °C. The size of amplified fragments resolved in PAGE was obtained by log-linear interpolation of the 10 bp DNA ladder (Invitrogen®) or HyperLadder V (Bioline®) on the gel.
Table 1.
PCR and enzymatic digestion conditions.
| Marker | ||||
|---|---|---|---|---|
| RFLP-PCR Me15-16 Aci I | mac-1 | RFLP-PCR ITS Hha I | RFLP-PCR COI Xba I | |
| Component [concentration] | ||||
| (40 ng DNA in 25 µL final reaction volume) MgCl2 [mM] | 2 | 1.5 | 2 | 1.5 |
| dNTP (each) [µM] | 50 | 60 | 100 | 200 |
| Primer F and R (each) [µM] | 0.4 | 0.14 | 0.2 | 0.6 |
| Taq [U] | 0.75 | 0.5 | 1 | 1.5 |
| Annealing conditions | ||||
| Temperature [°C] | 56 | 46 | 55 | 52 |
| Time [s] | 30 | 30 | 20 | 30 |
| Enzimatic digestion conditions (20 µL final reaction volume) | ||||
| Enzime - [U] | Aci I - 4 | — | Hha I - 10 | Xba I -10 |
Genotyping
Genotypes were scored using polyacrylamide gel electrophoresis (PAGE) (8%) and silver staining for the markers 18S rDNA, Me15-16, mac-1, COI and 16S rRNA, except for MT-1 specimens. In these individuals, mac-1 and 16S rRNA were genotyped with a Fragment AnalyzerTM instrument (Advanced Analytical Technologies, Ames, IA), using the dsDNA 905 Reagent Kit (35–500 bp) following manufacturer instructions. This kit resolves 2 bp differences in DNA fragments and alleles. The data were normalized to 35 bp and 500 bp lower and upper markers and calibrated to the 75 to 400 bp range using PRO Size 2.0 software (Advanced Analytical Technologies, Inc.). Correspondence between the allele sizes obtained using the two methods was established by constructing an allele ladder, sizing all alleles obtained from polyacrylamide-scored genotypes and genotyping this allele ladder with a Fragment AnalyzerTM instrument. Finally, the ITS marker was genotyped with the Fragment AnalyzerTM in all individuals. Quality was verified by including negative controls in each run and re-genotyping a randomly-selected 5% of individuals.
Data analysis
Mono-locus approach
The mitochondrial markers COI and16S rRNA might show two haplotypes, due to the double uniparental mitochondrial inheritance (DUI) observed in mussels, thus somatic cells carry the female and, in less frequency, the male mitochondrial genome possibly giving different haplotypes70. To avoid affecting the species identification, individuals with two haplotypes were excluded from further analysis. Mitochondrial markers with only one haplotype were analyzed as homozygous diploid genotypes following Narum et al.71 Numbers and frequencies of alleles and haplotypes were estimated with the R package strataG72.
Individuals were assigned to a species based on each marker separately (nuclear Me15-16, nuclear ITS and mitochondrial COI) according the allele sizes and patterns defined for each species, as described above (Fig. 2). These markers are considered diagnostic in allopatric populations7. In the case of multiallelic markers mac-1 and 16S rRNA, species was determined using a leave-one-out (LOO) algorithm with the Bayesian method described by Rannala & Mountain73 using GeneClass2 software74. Individuals were allocated to a species with an assignment threshold of 0.05. This procedure avoids the subjectivity of pooling species-specific compound alleles into synthetic alleles75,76.
Each marker was evaluated through re-allocation analysis with the software package GeneClass2 as described above. A re-assignment was considered correct if it matched the classification by PCR-RFLP Me15-16 AciI. Numbers and percentages of matching and mismatching assignments were determined for each marker.
The concordance between PCR-RFLP Me15-16 AciI and the other markers was evaluated graphically using a heatmap as well as classical diagnostic test performance statistics for each species. (i) Sensitivity (S), calculated as the number of individuals correctly assigned to the species divided by the total number of individuals sampled from that species, reflects how well the test correctly assigns individuals to a species77. (ii) Specificity (E), calculated as the number of individuals appropriately excluded from the species divided by the total number of individuals who do not belong to the species, reflects how well the test correctly excludes individuals from a species78,79. Sensitivity vs. specificity for each marker in each species was plotted. (iii) Positive likelihood ratio (LR+), calculated as S/(1-E), summarizes how many times more likely it is that individuals of a species will be assigned to the species as compared to specimens of other species80. When specificity is 1.0, LR+ will be undefined, therefore, we added 0.5 to all counts in the table to calculate an approximate LR+ value81,82. The 95% of confidence intervals (95% CI), to determine if the diagnostics statistics values (S, E and LR+) were significantly different from zero, were estimated with the R package epiR (https://cran.r-project.org/web/packages/epiR/).
Multi-locus approach
To visualize the separation of species using four markers (excluding PCR-RFLP Me15-16 AciI) and all five markers simultaneously. Two-dimensional factorial correspondence analysis (2D-FCA) and principal component analysis (PCA) were performed with the R package adegenet83. As with the mono-locus approach, the performance of the four and five markers together was evaluated using re-allocation analysis with the software package GeneClass2 (described above). We use the non-parametric Wilcoxon signed rank test to compare the assignment performance of the panel composed by the five RFLP-PCR markers obtained in this study (Table S4f) with that obtained with a 49 SNP panel in 338 mussels: M trossulus (17), M. edulis (27), M. galloprovincialis (105) and M chilensis (189), from previous work (Larraín et al.38) and summarized in Table S2. In both studies, PCR-RFLP Me15-16 Acil was used as reference marker to perform specimen identification.
Results
All 298 individuals were successfully genotyped with Me15-1623,54 as pure M. trossulus (50), M. edulis (50), M. galloprovincialis (99) or M. chilensis (99), producing the expected allele size (Fig. 2a). ITS and COI amplified in all individuals with PCR-RFLP. The loci mac-1 and 16S rRNA could not be amplified after two attempts each in five individuals. The global genotyping success rate was 98.32% across populations, with a 100% match rate in re-tested individuals (n = 15). On the other hand, only 16S rRNA marker showed two mitochondrial haplotypes in seven M. chilensis from MCh-1, 30 and three M. galloprovincialis from MG-2 and MG-1 populations, respectively. These 40 individuals were excluded from subsequent analysis, as was described in the data analysis section.
Mono-locus approach
First intron in the Mytilus actin protein gene: nuclear locus mac-1
Locus mac-1 was polymorphic in all locations, with 27 alleles ranging from 164 to 494 bp in length among individuals (Table S3). Two alleles (255 and 266 bp) were present in all four species and all six locations. Two alleles (303 and 328 bp) were exclusive to M. galloprovincialis. The frequency of the 328 bp allele was ten-fold higher in the Southern (0.220) than the Northern Hemisphere (0.021), while the reverse was true for the 303 bp allele. The M. chilensis populations (MCh-1 and MCh-2) and M. edulis sample from Canada (ME-1) were less diverse, with a maximum of three alleles (255, 266 and 298). M. trossulus had numerous private alleles with low frequencies except for two higher-frequency alleles (487 and 494 bp).
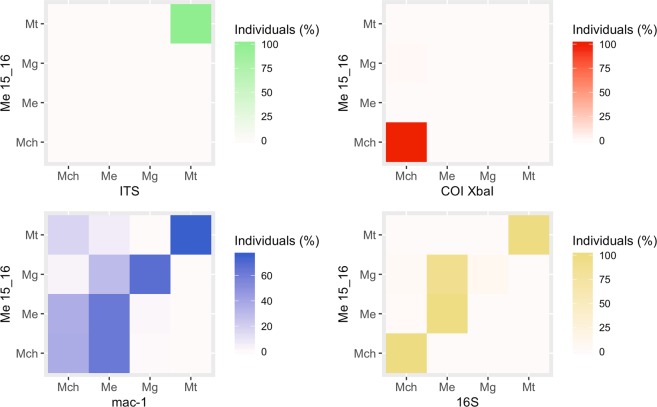
When the mac-1 locus genotypes were used to assign individuals to the species determined by PCR-RFLP Me15-16 AciI, 144 of 253 individuals (56.9%) had matching results for both markers. mac-1 correctly reassigned 67.7% of M. galloprovincialis (44 of 65) but only 37.0% of M. chilensis individuals (34 of 92) (Fig. 3, Table S4a), with 57 of 92 individuals (62.0%) from the latter species mis-reassigned to M. edulis. For M. edulis, 31 of 50 (62.0%) individuals were correctly reassigned, with 18 (36.0%) wrongly classified as M. chilensis.
Figure 3.
Heatmaps indicating concordance (% of individuals) in species identification between PCR-RFLP Me15-16 AciI and each of the other PCR-RFLP markers evaluated (ITS, COI and 16S rRNA) and mac-1.
Concordance between mac-1 and PCR-RFLP Me15-16 AciI is shown in Fig. 3. Sensitivity (number of individuals from each species correctly assigned by mac-1 divided by total number of individuals from that species) was high for M. trossulus and M. galloprovincialis (0.76 and 0.68) but only 0.37 for M. chilensis (Table S5). This marker accurately excluded individuals from M. trossulus and M. galloprovincialis, with specificities of 1.00 and 0.99, respectively. The positive likelihood ratio indicated that M. galloprovincialis individuals were 63.63 times more likely than other individuals to be assigned to the species.
Nuclear marker PCR-RFLP ITS HhaI
The RFLP assay clearly differentiated M. trossulus from M. chilensis, M. edulis and M. galloprovincialis but, as expected, did not distinguish among the latter three (Fig. 2b). All 50 M. trossulus individuals were correctly re-assigned and the other 208 individuals correctly excluded (Fig. 3, Table S4b). Sensitivity and specificity for M. trossulus were optimal, with full concordance between ITS and Me15-16 (Fig. 4, Table S5).
Figure 4.
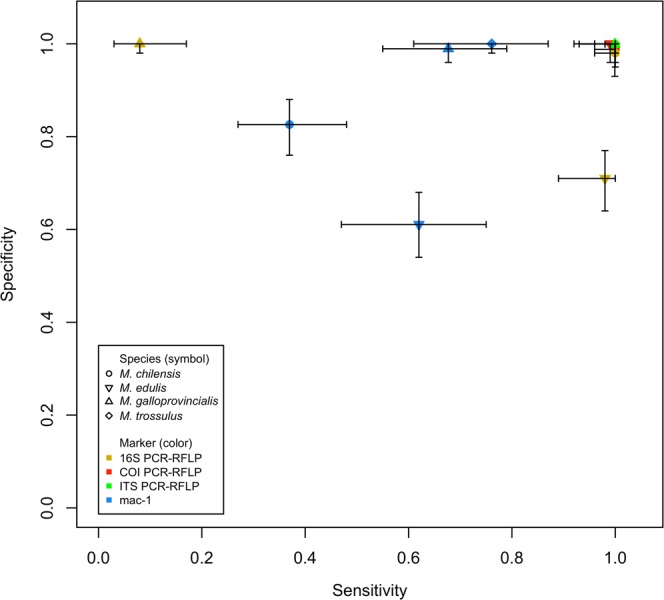
Sensitivity vs. specificity by species and marker, along with the respective 95% confidence intervals (raw data in Table S5). The species are represented by the symbols (°) Mytilus chilensis, (∇) Mytilus edulis, (Δ) Mytilus galloprovincialis and (⬨) Mytilus trossulus. The markers are represented by the color: gold for PCR-RFLP16S rRNA, red for PCR-RFLP COI, green for PCR-RFLP ITS, and blue for mac-1.
Cytochrome oxidase subunit I gene: mitochondrial marker PCR-RFLP COI XbaI
As expected, species identification results matched for COI and Me15-16 in 100% of M. chilensis individuals (MCh-1 and MCh-2), indicating optimal sensitivity (1.00) (Figs. 3 and 4). In two (B12 and B42) of the 50 M. galloprovincialis individuals from Dichato, Chile (MG-2), the amplicon was digested, producing the two-fragment pattern characteristic of M. chilensis (Fig. 2c. Table S4c), resulting in a specificity of 0.99 for M. chilensis (Table S5). This mitochondrial marker did not distinguish among M. trossulus, M. edulis and M. galloprovincialis but correctly excluded all M. chilensis specimens (specificity = 1.00).
16S rRNA gene: mitochondrial marker PCR-RFLP 16S rRNA
After triple enzymatic digestion, we found other two haplotypes not previously described for this locus (shown with asterisk in Fig. 2d). The first one was present in one MT-1 individual (0.022) that was missing the NheI site in contrast to the standard M. trossulus haplotype. The second was present in three MG-2 individuals (0.15), each with the EcoRV site present and NheI site missing in contrast to the standard M. galloprovincialis haplotype (Fig. 2d).
The M. chilensis haplotype (the species previously named Southern Hemisphere M. galloprovincialis) was fixed (1.0) in MCh-1 and MCh-2. The frequency of this haplotype was of 0.10 in the third Chilean population (MG-2), identified as M. galloprovincialis by PCR-RFLP Me15-16 AciI. In the Northern Hemisphere populations, this haplotype was present at very low frequencies (0.02) only in ME-1, and absent in MG-1 and MT-1 (Table S6).
As expected, the M. edulis haplotype was frequent in ME-1 (0.980) but also in MG-1 (0.957), identified as M. galloprovincialis by PCR-RFLP Me15-16 AciI. In this latter population from Galicia, the low frequency of the M. galloprovincialis haplotype (0.043) was unexpected (Table S6).
When species identification results for PCR-RFLP 16S rRNA and PCR-RFLP Me15-16 AciI were compared (Fig. 3, Table S4d), 191 of 253 individuals (75.5%) had matching results. 16S rRNA correctly reassigned 100, 98 and 100% of M. trossulus, M. edulis and M. chilensis individuals respectively, but only 5 of 66 (7.6%) M. galloprovincialis individuals. Moreover, 59 of 66 (89.4%) from this latter species were mis-reassigned to M. edulis and 2 of 66 (3.0%) to M chilensis.
Concordance between 16S rRNA and Me15-16 is shown in Fig. 3. The sensitivity of PCR-RFLP 16S rRNA was optimal for M. trossulus and M. chilensis (1.00) and high for M. edulis (0.98) but very low for M. galloprovincialis (0.08) (Fig. 3, Table S3). In general, the method correctly excluded individuals from M. trossulus and M. galloprovincialis, with specificities of 1.00 and also M. chilensis (0.98).
Multi-locus approach
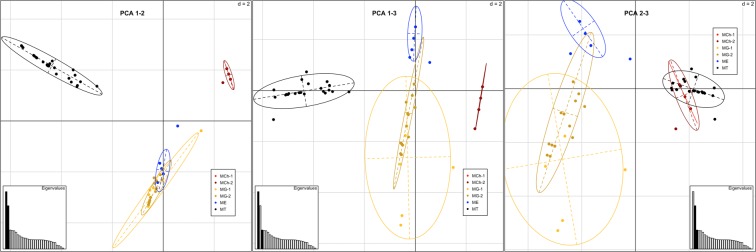
This analysis uses the genetic information provided by all five markers simultaneously. The 2D-FCA multi-locus approach separated all four species, as shown in Fig. 5. As expected, ME-1, MG-1 and MG-2 mapped very closely. The Northern Hemisphere populations (MG-1 and ME-1) overlapped in all of the PCA plots (Fig. 6). These plots also showed an overlap between the Chilean M. galloprovincialis population (MG-2) and the same species from Spain (MG-1) but no overlap with ME-1. On the other hand, the M. chilensis populations (MCh-1and MCh-2) were clearly separated from the other Mytilus species, including M. trossulus (MT-1).
Figure 5.
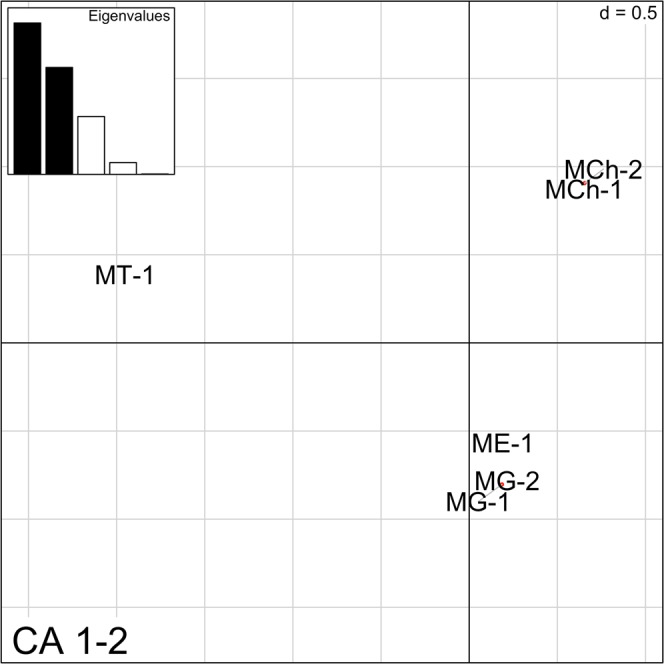
Two-dimensional factorial correspondence analysis (2D-FCA) constructed using the information from all five markers simultaneously. Eigenvalues corresponding to the represented components are filled in black.
Figure 6.
Principal component analysis (PCA) constructed using the information from all five markers simultaneously. Eigenvalues corresponding to the represented components are filled in black. Points represent genotypes and inertia ellipses are placed in 95%. Sampling locations are represented by the colors: red for MCh-1, dark red for MCh-2, blue for ME-1, yellow for MG-1, gold for MG-2 and black for MT-1.
Assignments performed using all five markers simultaneously showed 100% concordance with PCR-RFLP Me15-16 AciI (Table S4f). When Me15-16 was excluded, the percentage of correct species re-assignment remained at 100% for M. trossulus and M. chilensis but dropped to 96% for M. edulis and 65% for M. galloprovincialis (Table S4e). Sensitivity remained high when four markers were used (excluding Me15-16). Specificity was 1.00 for M. trossulus and M. chilensis, 0.96 for M. edulis but dropped to 0.65 for M. galloprovincialis (Table S5).
The comparison of assignment performance obtained with the RFLP-PCR multi-locus panel and the SNP panel did not show significantly differences (p-value = 1.00), indicating the same performance of both kind of markers sets.
Discussion
The use of genetic methods to assign taxonomic names to unknown Mytilus individuals, called specimen identification84, is useful for: increasing genetic knowledge of mussels, studying populations and hybrid zones, establishing taxonomy and systematics, identifying evolutionary relationships and phylogeny within the genus, performing ecological studies and verifying food authenticity.
As the Mytilus genus contains several taxa, researchers may need to authenticate the target species using molecular markers. Species assignment is typically performed using a single locus independently, called the mono-locus approach, likely because this method is relatively fast and cheap85. However, not all markers can differentiate all of the species in the genus. Furthermore, because the various markers target different regions in the genome, they often produce nonequivalent classification results38,86.
The 65 pb intron length polymorphism in the actin gene mac-1, used for genotyping in Mytilus population studies49, systematically fails to amplify in some individuals (~2%), possibly due to mutations in priming sites producing null alleles, as in microsatellites87. This phenomenon hinders allele scoring, limiting the accuracy of the mono-locus approach with this marker. mac-1 correctly excluded M. trossulus and M. galloprovincialis from other species but showed a weak ability to identify the four Mytilus species, with only 56.9% of assignments matching the results produced using PCR-RFLP Me15-16 (Table S4a). The poor performance of mac-1 is attributable to its limited ability to discriminate between M. edulis and M. chilensis: 18 of 50 M. edulis individuals were assigned to M. chilensis and 57 of 92 M. chilensis to M. edulis. Therefore, several studies that have used this marker in Chilean blue mussels (M. chilensis) may have been affected by this bias2,65. This nuclear marker has multiple alleles, some of which are shared across Mytilus species, and is therefore not fully diagnostic (Table S3). This multiallelic characteristic led others to propose the use of “synthetic alleles,” in which several similarly-sized alleles are pooled for specimen identification76. Pooling improved performance, but several of the compound synthetic alleles were still not exclusive to any one species76. Because the polymorphism in mac-1 is located in an intronic region, is quite variable even within a given species, as well as technically difficult to score, limiting performance.
ITS targets the internally transcribed spacer sectors between the 18S and the 28S genes from the nuclear rDNA coding region. The genomic organization of rDNA consists of a variable number of tandem repeats that is sufficient to provide a DNA template for PCR88. ITS successfully amplified all individuals and allowed for definitive discrimination of M. trossulus. Although ITS could not separate the other three species, concordance with Me15-16 AciI was optimal for M. trossulus. This result is consistent with other works comparing ITS to other markers58. For example, Toro67 discriminated M. trossulus from M. chilensis and M. edulis but could not distinguish between the latter two. In the same work, these three species were clearly differentiated using the Glu-5′ marker targeting the polyphenolic adhesive protein gene, confirming that ITS only can distinguish M. trossulus specimens. Due to its multicopy nature, authors have warned that ITS should not be considered a codominant single-copy Mendelian marker56,59. However, Heat et al.33 observed a Mendelian-like inheritance pattern when using ITS to genotype progeny from two test crosses (M. edulis x hybrids M. edulis/M. trossulus).
The mitochondrial PCR-RFLP COI marker showed full concordance with Me15-16 AciI marker, identifying the Chilean mussel in all locations sampled. However, two individuals (B12 and B42) from the Dichato population (MG-2) in the Arauco Gulf, that were classified as M. galloprovincialis by Me15-16 AciI, were identified by COI as M. chilensis. The Arauco Gulf is a sympatric zone where the presence of the non-indigenous M. galloprovincialis has been described29,63,64,89,90. Also, in this zone, our group found a frequency of ~4–7% for hybrids of the two species as part of a routine analysis using the PAPM marker55, equivalent to Me15-16 (unpublished data). The low-frequency presence of individuals carrying the nuclear M. galloprovincialis genotype (scored by Me15-16) and mitochondrial M. chilensis haplotype (scored by COI) shows introgression of the mitochondrial genome from the indigenous M. chilensis into the non-indigenous M. galloprovincialis, as described in Steinert et al.91 and Rawson & Hilbish92.
The mitochondrial PCR-RFLP 16S rRNA showed full concordance with the nuclear PCR-RFLP Me15-16 AciI marker for identifying M. trossulus and M chilensis, but somewhat lower sensitivity for M. edulis (0.98). This statistic decreased to 0.08 for the Mediterranean mussel (Table S5, Fig. 4). Most M. galloprovincialis specimens were classified as M. edulis (59 of 66). This inconsistency between the markers is likely due to the fact that all Mediterranean mussel populations that have been tested in Europe45 and the Southern Hemisphere64,69 also carry the M. edulis haplotype. Therefore, PCR-RFLP 16S rRNA is only semi-diagnostic for M. galloprovincialis and M. edulis.
PCR-RFLP 16S rRNA exhibits an evident DUI of mitochondrial DNA93 in 40 samples that present two mtDNA haplotypes, these individuals were not used in further species identification analysis. Of the remaining 20 individuals with one haplotype, 18 had M. galloprovincialis haplotypes and the other two carried the M. chilensis haplotype. These last two correspond to the same individuals classified as M. chilensis by COI (B12 and B42), as expected by the fact that the mitochondrial genome is considered one locus with 16S rRNA and COI variants inherited linked. This finding could indicate an asymmetric hybrid zone in the Arauco Gulf area, in which M. galloprovincialis being the predominant species, with a lower frequency of the native M. chilensis.
Current aquaculture practices in the Arauco Gulf zone involve production of Mediterranean and Chilean mussels in the same area. Interestingly, Westfall & Gardener64 also found introgressed individuals with nuclear M. galloprovincialis genotypes and mitochondrial M. chilensis haplotypes in Cocholgue, a location 12 km away from our sampling point in Dichato, supporting the concept of a hybrid zone.
PCR-RFLP Me15-16 follows a Mendelian inheritance pattern94 and is an extremely robust and reliable diagnostic marker for routine specimen identification53,95,96. Therefore, this method is the most common DNA-based technique for identifying mussel species. Of course, Me15-16 alone is not able to distinguish introgressed individuals40, and also, is not able to differentiate M. chilensis from the Southern Hemisphere lineage of M. galloprovincialis from New Zealand, because in both species, this fragment of genome is not cut by AciI due to the substitution of the allele “G” by “T” in the restriction site97.
The mono-locus approach offers some advantages: this method is fast, relatively easy to preform and simple to score with the fully diagnostic markers for the species analyzed here (Me15-16, ITS and COI). However, they show weakness, such as the fact that some markers have multiple alleles that are not fully fixed in each species (mac-1 and 16S rRNA), making them only semi-diagnostic. Also, the presence of multiple alleles sometimes hinders interpretation. Another problem with the mono-locus approach is that some markers cannot identify all of the species analyzed here when used alone, and when two or more are used simultaneously, they produce contradictory results32,35,61. Moreover, mitochondrial markers (COI and 16S rRNA) must be used simultaneously with a nuclear marker to detect introgression in hybrid zones.
The discrepancies observed among the PCR-RFLP markers were expected, as each Mytilus species diagnostic marker targets a single locus in distinct zones of the nuclear or mitochondrial genome, likely with different times to common ancestor or gene-genealogy98. Moreover, it is widely recognized that evolutionary forces act differently and in an uncoordinated way on nuclear and mitochondrial genomes, and even on different regions within the nuclear genome11,41, for example, in monocopy vs. multicopy genes or introns vs. exons. On the other hand, hybridization is also associated with conflicting results, as backcrossing with one or both parental taxa can lead to introgression of alleles from one taxon into the other99. In this case, analyses including different types of markers are preferable41.
Given that genome of smooth-shelled mussels is about 1.6 Gb100, species identification using a single locus or a very small number of loci is relatively straightforward. However, specimen identification based on simultaneous use of the information provided by each locus, known as the multi-locus approach, allows us to consider the evolutionary forces acting on different genomic regions. Therefore, this approach provides more coherent and reliable outcomes, especially when introgression has occurred58.
The development of genotyping-by-sequencing methods to affordably discover and genotype hundreds or even thousands of single-nucleotide polymorphism (SNP) markers makes it possible to apply a multi-locus approach to specimen identification40,101. New multi-locus SNP panels have been developed recently, allowing for species identification using only the most informative SNPs38,40,96,102–105. As an example of the multi-locus approach in mixed populations, hybrids and introgressed individuals were detected by a new twelve-SNP diagnostic panel in European Mytilus samples previously analyzed with PCR-RFLP Me15-1640. The multi-locus approach can also be applied to specimen identification, simultaneously analyzing the mitochondrial and genomic markers traditionally used in mono-locus analysis58.
In this work, we used a multi-locus strategy with two mtDNA (PCR-RFLP COI and PCR-RFLP 16S rRNA) and three nuclear (PCR-RFLP ITS, mac-1 and PCR-RFLP Me15-16) markers. The re-assignment analyses using the multi-locus panel, excluding the reference marker PCR-RFLP Me15-16, incorrectly classified 31.8% of M. galloprovincialis as M. edulis. This finding is not wholly unexpected, as these species are closely related due to a long history of hybridization and introgression in Europe36,44,76,106,107. Therefore, discriminating between these two species poses steep analytical challenges. With this panel, the same two M. galloprovincialis individuals (B12 and B42) from MG-2 were again classified as M. chilensis, likely due to the absence of the PCR-RFLP Me15-16 marker. On the other hand, the full multi-locus panel including all five markers was fully concordant with PCR-RFLP Me15-16, clearly separating the four species, similar to results for a 49-SNP panel analyzed by Larraín et al.38. These results indicate that a multi-locus approach, as described here, might improve the accuracy of PCR-RFLP marker-based specimen identification. Furthermore, using a single marker may result in poor performance, especially in mixed populations. The capabilities and limitations of each marker in a mono-locus and multi-locus approaches summarized in Table 2, can be useful when analyzing the results of previous studies in which these markers were applied.
Table 2.
Summary of marker performance in the specimen identification of Mytilus mussels (M. trossulus, M. edulis, M. chilensis and M. galloprovincialis).
| Marker | Diagnostic level by species | Capable to detect | |||
|---|---|---|---|---|---|
| Fully diagnostic | Semi diagnostic | Not diagnostic | F1 hybrids | introgressed individuals | |
| Me 15-16 | M. trossulus M. edulis M. chilensis M. galloprovincialis | — | — | Yes | No |
| mac-1 | — | M. trossulus M. edulis M. chilensis M. galloprovincialis | — | No | No |
| ITS | M. trossulus | — | M. edulis M. chilensis M. galloprovincialis | Yes | No |
| COI XbaI | M. chilensis | — | M. trossulus M. edulis M. galloprovincialis | No | No |
| 16s rRNA | M. trossulus M. chilensis | M. edulis M. galloprovincialis | — | Yes* | No |
| 4 markers: | |||||
| mac-1, ITS, COI XbaI, 16S rRNA | M. trossulus M. chilensis | M. edulis M. galloprovincialis | — | Yes | Yes |
| All markers: | |||||
| Me 15-16, mac-1, ITS, COI XbaI, 16s rRNA | M. trossulus M. edulis M. chilensis M. galloprovincialis | — | — | Yes | Yes |
Fully diagnostic: Characteristic allele or haplotype fixed in the species and absent in the others.
Semi diagnostic: Characteristic allele or haplotype not fixed in the species and present in the others.
Not diagnostic: No characteristic allele or haplotype allowed to differentiate the species.
*Only in males.
We conclude that the PCR-RFLP markers Me15-16, ITS and COI produce largely equivalent results when applied using a mono-locus approach; however, the latter two are useful only for separating M. trossulus and M. chilensis from the three other species (Table 2). Me15-16 distinguished among the four species tested but, as expected form single locus data, was not able to detect introgression. Mono-locus results for the nuclear mac-1 and mitochondrial 16S rRNA markers, due to their semi-diagnostic status, were difficult to interpret and showed low concordance with the results derived from the multi-locus approach. All five markers used simultaneously in a multi-locus approach produced more reliable and robust identifications, outperforming each of the markers when used separately, and comparable performance of SNPs panels. These findings support the use of a multi-locus approach when studying this important marine resource, with implications for research on food quality and safety, sustainable production, biodiversity and conservation.
Supplementary information
Acknowledgements
This research was funded by CONICYT through grants FONDECYT 1130302, FONDEF IDeA ID16I10013, REDES180143 and FONDECYT 1191765. The authors thank Dr. Santiago Aubourg, Mrs. Marcela Cárcamo, Dr. Zhongqi Chen, Dr. Alicia Rodriguez and Prof. Eduardo Tarifeño for their assistance with sample collection, also to Felipe Medina and Felipe Jilberto for assistance in plots design.
Author contributions
P.G. and C.P. contributed to the laboratory work. M.A.L. and C.A. contributed to obtaining funding, experimental design, data analysis and manuscript writing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pía González and Claudio Pérez.
Supplementary information
is available for this paper at 10.1038/s41598-019-55855-8.
References
- 1.Hilbish TJ, et al. Origin of the antitropical distribution pattern in marine mussels (Mytilus spp.): routes and timing of transequatorial migration. Mar. Biol. 2000;136:69–77. doi: 10.1007/s002270050010. [DOI] [Google Scholar]
- 2.Gérard K, Bierne N, Borsa P, Chenuil A, Féral J-P. Pleistocene separation of mitochondrial lineages of Mytilus spp. mussels from Northern and Southern Hemispheres and strong genetic differentiation among southern populations. Mol. Phylogenet. Evol. 2008;49:84–91. doi: 10.1016/j.ympev.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 3.ITIS. Integrated Taxonomic Information System. Available at: http://www.itis.gov (2019).
- 4.WoRMS. World Register of Marine Species, http://www.marinespecies.org at VLIZ. Accessed 2019-05-06. doi:10.14284/170. Available at: http://www.marinespecies.org (2019).
- 5.Crego-Prieto V, et al. Aquaculture and the spread of introduced mussel genes in British Columbia. Biol. Invasions. 2015;17:2011–2026. doi: 10.1007/s10530-015-0853-z. [DOI] [Google Scholar]
- 6.Inoue K, et al. A possible hybrid zone in the Mytilus edulis complex in Japan revealed by PCR markers. Mar. Biol. 1997;128:91–95. doi: 10.1007/s002270050072. [DOI] [Google Scholar]
- 7.Kartavtsev YP, Katolikova MV, Sharina SN, Chichvarkhina OV, Masalkova NA. A population genetic study of the hybrid zone of Mytilus trossulus Gould, 1850 and an introduced species, M. galloprovincialis Lamarck, 1819,(Bivalvia: Mytilidae) in Peter the Great Bay in the Sea of Japan. Russ. J. Mar. Biol. 2014;40:208–216. doi: 10.1134/S1063074014030055. [DOI] [Google Scholar]
- 8.Mathiesen SS, et al. Genetic diversity and connectivity within Mytilus spp. in the subarctic and Arctic. Evol. Appl. 2017;10:39–55. doi: 10.1111/eva.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oyarzún PA, Toro JE, Cañete JI, Gardner J. Bioinvasion threatens the genetic integrity of native diversity and a natural hybrid zone: smooth‐shelled blue mussels (Mytilus spp.) in the Strait of Magellan. Biol. J. Linn. Soc. 2016;117:574–585. doi: 10.1111/bij.12687. [DOI] [Google Scholar]
- 10.Rawson PD, Agrawal V, Hilbish TJ. Hybridization between the blue mussels Mytilus galloprovincialis and M. trossulus along the Pacific coast of North America: evidence for limited introgression. Mar. Biol. 1999;134:201–211. doi: 10.1007/s002270050538. [DOI] [Google Scholar]
- 11.Riginos C, Cunningham CW. Invited review: local adaptation and species segregation in two mussel (Mytilus edulis × Mytilus trossulus) hybrid zones. Mol. Ecol. 2005;14:381–400. doi: 10.1111/j.1365-294X.2004.02379.x. [DOI] [PubMed] [Google Scholar]
- 12.Väinölä R, Hvilsom MM. Genetic divergence and a hybrid zone between Baltic and North Sea Mytilus populations (Mytilidae: Mollusca) Biol. J. Linn. Soc. 1991;43:127–148. doi: 10.1111/j.1095-8312.1991.tb00589.x. [DOI] [Google Scholar]
- 13.Wilhelm R, Hilbish TJ. Assessment of natural selection in a hybrid population of mussels: evaluation of exogenous vs endogenous selection models. Mar. Biol. 1998;131:505–514. doi: 10.1007/s002270050342. [DOI] [Google Scholar]
- 14.Hoarau P, Damiens G, Roméo M, Gnassia-Barelli M, Bebianno MJ. Cloning and expression of a GST-pi gene in Mytilus galloprovincialis. Attempt to use the GST-pi transcript as a biomarker of pollution. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2006;143:196–203. doi: 10.1016/j.cbpc.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Lacroix, C. et al. Seasonal monitoring of blue mussel (Mytilus spp.) populations in a harbor area: A focus on responses to environmental factors and chronic contamination. Mar. Environ. Res. (2017). [DOI] [PubMed]
- 16.Sureda A, et al. Biochemical responses of Mytilus galloprovincialis as biomarkers of acute environmental pollution caused by the Don Pedro oil spill (Eivissa Island, Spain) Aquat. Toxicol. 2011;101:540–549. doi: 10.1016/j.aquatox.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Invasive Species Specialist Group ISSG. Species profile: Mytilus galloprovincialis. The Global Invasive Species Database (GISD) (2017).
- 18.Villaseñor-Parada C, Pauchard A, Macaya EC. Ecología de invasiones marinas en Chile continental:¿ Qué sabemos y que nos falta por saber? Rev. Biol. Mar. Oceanogr. 2017;52:1–17. doi: 10.4067/S0718-19572017000100001. [DOI] [Google Scholar]
- 19.FAO. Global Statistical Collections. http://www.fao.org/fishery/statistics/en Accesion date: 2019-04-19 Available at: http://www.fao.org/fishery/statistics/en (2019).
- 20.Fernández-Tajes J, et al. Alternative PCR–RFLP methods for mussel Mytilus species identification. Eur. Food Res. Technol. 2011;233:791–796. doi: 10.1007/s00217-011-1574-x. [DOI] [Google Scholar]
- 21.Krapivka S, et al. Shell-shape variation along the latitudinal range of the Chilean blue mussel Mytilus chilensis (Hupe 1854) Aquac. Res. 2007;38:1770–1777. doi: 10.1111/j.1365-2109.2007.01839.x. [DOI] [Google Scholar]
- 22.Groenenberg DSJ, et al. On the identity of broad-shelled mussels (Mollusca, Bivalvia, Mytilus) from the Dutch delta region. Contrib. to Zool. 2011;80:95–106. doi: 10.1163/18759866-08002001. [DOI] [Google Scholar]
- 23.Santaclara FJ, et al. Development of a method for the genetic identification of mussel species belonging to Mytilus, Perna, Aulacomya, and other genera. J. Agric. Food Chem. 2006;54:8461–8470. doi: 10.1021/jf061400u. [DOI] [PubMed] [Google Scholar]
- 24.Beaumont AR, Toro JE. Allozyme genetics of Mytilus edulis subjected to copper and nutritive stress. J. Mar. Biol. Assoc. United Kingdom. 1996;76:1061–1071. doi: 10.1017/S0025315400040959. [DOI] [Google Scholar]
- 25.Cárcamo C, Comesaña AS, Winkler FM, Sanjuan A. Allozyme identification of Mussels (Bivalvia: Mytilus) on the Pacific coast of South America. J. Shellfish Res. 2005;24:1101–1115. doi: 10.2983/0730-8000(2005)24[1101:AIOMBM]2.0.CO;2. [DOI] [Google Scholar]
- 26.McDonald JH, Seed R, Koehn RK. Allozymes and morphometric characters of three species of Mytilus in the northern and southern hemispheres. Mar. Biol. 1991;111:323–333. doi: 10.1007/BF01319403. [DOI] [Google Scholar]
- 27.Katolikova M, Khaitov V, Väinölä R, Gantsevich M, Strelkov P. Genetic, Ecological and Morphological Distinctness of the Blue Mussels Mytilus trossulus Gould and M. edulis L. in the White Sea. PLoS One. 2016;11:e0152963. doi: 10.1371/journal.pone.0152963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbadi M, et al. Species identification of bivalve molluscs by pyrosequencing. J. Sci. Food Agric. 2017;97:512–519. doi: 10.1002/jsfa.7754. [DOI] [PubMed] [Google Scholar]
- 29.Astorga MP, Cardenas L, Vargas J. Phylogenetic Approaches to Delimit Genetic Lineages of the Mytilus Complex of South America: How Many Species are there? J. Shellfish Res. 2015;34:919–930. doi: 10.2983/035.034.0322. [DOI] [Google Scholar]
- 30.Pickett T, David AA. Global connectivity patterns of the notoriously invasive mussel, Mytilus galloprovincialis Lmk using archived CO1 sequence data. BMC Res. Notes. 2018;11:231. doi: 10.1186/s13104-018-3328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rego I, et al. PCR technique for identification of mussel species. J. Agric. Food Chem. 2002;50:1780–1784. doi: 10.1021/jf0110957. [DOI] [PubMed] [Google Scholar]
- 32.Rawson PD, Joyner KL, Meetze K, Hilbish TJ. Evidence for intragenic recombination within a novel genetic marker that distinguishes mussels in the Mytilus edulis species complex. Heredity (Edinb). 1996;77:599–607. doi: 10.1038/hdy.1996.187. [DOI] [PubMed] [Google Scholar]
- 33.Heath DD, Rawson PD, Hilbish TJ. PCR-based nuclear markers identify alien blue mussel (Mytilus spp.) genotypes on the west coast of Canada. Can. J. Fish. Aquat. Sci. 1995;52:2621–2627. doi: 10.1139/f95-851. [DOI] [Google Scholar]
- 34.Rawson PD, Secor C, Hilbish TJ. The effects of natural hybridization on the regulation of doubly uniparental mtDNA Inheritance in blue mussels (Mytilus spp.) Genetics. 1996;144:241–248. doi: 10.1093/genetics/144.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zbawicka M, Drywa A, Śmietanka B, Wenne R. Identification and validation of novel SNP markers in European populations of marine Mytilus mussels. Mar. Biol. 2012;159:1347–1362. doi: 10.1007/s00227-012-1915-8. [DOI] [Google Scholar]
- 36.Zbawicka M, Sańko T, Strand J, Wenne R. New SNP markers reveal largely concordant clinal variation across the hybrid zone between Mytilus spp. in the Baltic Sea. Aquat. Biol. 2014;21:25–36. doi: 10.3354/ab00566. [DOI] [Google Scholar]
- 37.Saarman, N. P. & Pogson, G. H. Introgression between invasive and native blue mussels (genus Mytilus) in the central California hybrid zone. Mol. Ecol. (2015). [DOI] [PubMed]
- 38.Larraín MA, Zbawicka M, Araneda C, Gardner JPA, Wenne R. Native and invasive taxa on the Pacific coast of South America: Impacts on aquaculture, traceability and biodiversity of blue mussels (Mytilus spp.) Evol. Appl. 2018;11:298–311. doi: 10.1111/eva.12553. [DOI] [Google Scholar]
- 39.Chen, X. et al. An effective method for identification of three mussel species and their hybrids based on SNPs. Conserv. Genet. Resour. 10.1007/s12686-018-1051-y (2018).
- 40.Wilson J, Matejusova I, McIntosh RE, Carboni S, Bekaert M. New diagnostic SNP molecular markers for the Mytilus species complex. PLoS One. 2018;13:e0200654. doi: 10.1371/journal.pone.0200654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kartavtsev YP, Masalkova NA, Katolikova MV. Genetic and Morphometric Variability in Settlements of Two Mussel Species (Mytilus ex. gr. Edulis), Mytilus trossulus and Mytilus galloprovincialis, in the Northwestern Sea of Japan. J. Shellfish Res. 2018;37:103–119. doi: 10.2983/035.037.0109. [DOI] [Google Scholar]
- 42.Freitas R, et al. Effects of seawater acidification and salinity alterations on metabolic, osmoregulation and oxidative stress markers in Mytilus galloprovincialis. Ecol. Indic. 2017;79:54–62. doi: 10.1016/j.ecolind.2017.04.003. [DOI] [Google Scholar]
- 43.Stuckas H, et al. Combining hydrodynamic modelling with genetics: can passive larval drift shape the genetic structure of Baltic Mytilus populations? Mol. Ecol. 2017;26:2765–2782. doi: 10.1111/mec.14075. [DOI] [PubMed] [Google Scholar]
- 44.Casoli E, et al. A massive ingression of the alien species Mytilus edulis L.(Bivalvia: Mollusca) into the Mediterranean Sea following the Costa Concordia cruise-ship disaster. Mediterr. Mar. Sci. 2016;17:404–416. doi: 10.12681/mms.1619. [DOI] [Google Scholar]
- 45.Rawson PD, Hilbish TJ. Evolutionary relationships among the male and female mitochondrial DNA lineages in the Mytilus edulis species complex. Mol. Biol. Evol. 1995;12:893–901. doi: 10.1093/oxfordjournals.molbev.a040266. [DOI] [PubMed] [Google Scholar]
- 46.Westfall KM, Wimberger PH, Gardner JPA. An RFLP assay to determine if Mytilus galloprovincialis Lmk. (Mytilidae; Bivalvia) is of Northern or Southern hemisphere origin. Mol. Ecol. Resour. 2010;10:573–575. doi: 10.1111/j.1755-0998.2009.02779.x. [DOI] [PubMed] [Google Scholar]
- 47.Burzyński A, Zbawicka M, Skibinski DOF, Wenne R. Doubly uniparental inheritance is associated with high polymorphism for rearranged and recombinant control region haplotypes in Baltic Mytilus trossulus. Genetics. 2006;174:1081–1094. doi: 10.1534/genetics.106.063180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beynon CM, Skibinski DOF. The evolutionary relationships between three species of mussel (Mytilus) based on anonymous DNA polymorphisms. J. Exp. Mar. Bio. Ecol. 1996;203:1–10. doi: 10.1016/0022-0981(96)02565-8. [DOI] [Google Scholar]
- 49.Ohresser M, Borsa P, Delsert C. Intron-length polymorphism at the actin gene locus mac-1: A genetic marker for population studies in the marine mussels Mytilus galloprovincialis Lmk and M edulis L. Mol. Mar. Biol. Biotechnol. 1997;6:123–130. [PubMed] [Google Scholar]
- 50.Bierne N, David P, Boudry P, Bonhomme F. Asortative fertilization and selection at larval stage in the mussels Mytilus edulis and M. galloprovincialis. Evolution (N. Y). 2002;56:292–298. doi: 10.1111/j.0014-3820.2002.tb01339.x. [DOI] [PubMed] [Google Scholar]
- 51.Rawson PD, Hayhurst S, Vanscoyoc B. Species composition of blue mussel populations in the northeastern gulf of Maine. J. Schellfish Researc. 2001;20:31–38. [Google Scholar]
- 52.Riginos C, Wang D, Abrams AJ. Geographic Variation and Positive Selection on M7 Lysin, an Acrosomal Sperm Protein in Mussels (Mytilus spp.) Mol. Biol. Evol. 2006;23:1952–1965. doi: 10.1093/molbev/msl062. [DOI] [PubMed] [Google Scholar]
- 53.Kijewski T, Wijsman JWM, Hummel H, Wenne R. Genetic composition of cultured and wild mussels Mytilus from The Netherlands and transfers from Ireland and Great Britain. Aquaculture. 2009;287:292–296. doi: 10.1016/j.aquaculture.2008.10.048. [DOI] [Google Scholar]
- 54.Inoue K, Waite JH, Matsuoka M, Odo S, Harayama S. Interspecific variations in adhesive protein sequences of Mytilus edulis, M. galloprovincialis, and M. trossulus. Biol. Bull. 1995;189:370–375. doi: 10.2307/1542155. [DOI] [PubMed] [Google Scholar]
- 55.Jilberto F, Araneda C, Larraín MA. High resolution melting analysis for identification of commercially-important Mytilus species. Food Chem. 2017;229:716–720. doi: 10.1016/j.foodchem.2017.02.109. [DOI] [PubMed] [Google Scholar]
- 56.Kijewski T, Zbawicka M, Väinölä R, Wenne R. Introgression and mitochondrial DNA heteroplasmy in the Baltic populations of mussels Mytilus trossulus and M. edulis. Mar. Biol. 2006;149:1371–1385. doi: 10.1007/s00227-006-0316-2. [DOI] [Google Scholar]
- 57.Zbawicka M, Burzyński A, Skibinski D, Wenne R. Scottish Mytilus trossulus mussels retain ancestral mitochondrial DNA: Complete sequences of male and female mtDNA genomes. Gene. 2010;456:45–53. doi: 10.1016/j.gene.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 58.Kijewski T, et al. Distribution of Mytilus taxa in European coastal areas as inferred from molecular markers. J. Sea Res. 2011;65:224–234. doi: 10.1016/j.seares.2010.10.004. [DOI] [Google Scholar]
- 59.Riginos C, Sukhdeo K, Cunningham CW. Evidence for selection at multiple allozyme loci across a mussel hybrid zone. Mol. Biol. Evol. 2002;19:347–351. doi: 10.1093/oxfordjournals.molbev.a004088. [DOI] [PubMed] [Google Scholar]
- 60.Borsa P, Daguin C, Bierne N. Genomic reticulation indicates mixed ancestry in Southern-Hemisphere Mytilus spp. mussels. Biol. J. Linn. Soc. 2007;92:747–754. doi: 10.1111/j.1095-8312.2007.00917.x. [DOI] [Google Scholar]
- 61.Gérard K, et al. Does natural selection explain the fine scale genetic structure at the nuclear exon Glu-5′ in blue mussels from Kerguelen? Ecol. Evol. 2015;5:1456–1473. doi: 10.1002/ece3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson AS, Bilodeau AL, Gilg MR, Hilbish TJ. Routes of introduction of the Mediterranean mussel(Mytilus galloprovincialis) to Puget Sound and Hood Canal. J. Shellfish Res. 2002;21:75–79. [Google Scholar]
- 63.Larraín MA, Díaz NF, Lamas C, Vargas C, Araneda C. Genetic composition of Mytilus species in mussel populations from southern Chile. Lat. Am. J. Aquat. Res. 2012;40:1077–1084. doi: 10.3856/vol40-issue4-fulltext-23. [DOI] [Google Scholar]
- 64.Westfall KM, Gardner JPA. Genetic diversity of Southern hemisphere blue mussels (Bivalvia: Mytilidae) and identification of non-indigenous taxa. Biol. J. Linn. Soc. 2010;101:898–909. doi: 10.1111/j.1095-8312.2010.01549.x. [DOI] [Google Scholar]
- 65.Borsa P, Rolland V, Daguin-Thiébaut C. Genetics and taxonomy of Chilean smooth-shelled mussels Mytilus spp. (Bivalvia: Mytilidae) C. R. Biol. 2012;335:51–61. doi: 10.1016/j.crvi.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Bierne N, Daguin C, Bonhomme F, David P, Borsa P. Direct selection on allozymes is not required to explain heterogeneity among marker loci across a Mytilus hybrid zone. Mol. Ecol. 2003;12:2505–2510. doi: 10.1046/j.1365-294X.2003.01936.x. [DOI] [PubMed] [Google Scholar]
- 67.Toro JE. PCR-based nuclear and mtDNA markers and shell morphology as an approach to study the taxonomic status of the Chilean blue mussel, Mytilus chilensis (Bivalvia) Aquat. Living Resour. 1998;11:347–353. doi: 10.1016/S0990-7440(98)80006-5. [DOI] [Google Scholar]
- 68.Palumbi, S. et al. The simple fool’s guiden to PCR. (Laboratory, Department of Zoology and Kewalo Marine Laboratory 2002).
- 69.Zardi GI, et al. Re-assessing the origins of the invasive mussel Mytilus galloprovincialis in southern Africa. Mar. Freshw. Res. 2018;69:607–613. doi: 10.1071/MF17132. [DOI] [Google Scholar]
- 70.Garrido-Ramos MA, Stewart DT, Sutherland BW, Zouros E. The distribution of male-transmitted and female-transmitted mitochondrial DNA types in somatic tissues of blue mussels: implications for the operation of doubly uniparental inheritance of mitochondrial DNA. Genome. 1998;41:818–824. doi: 10.1139/g98-081. [DOI] [Google Scholar]
- 71.Narum SR, et al. Differentiating salmon populations at broad and fine geographical scales with microsatellites and single nucleotide polymorphisms. Mol. Ecol. 2008;17:3464–3477. doi: 10.1111/j.1365-294x.2008.03851.x. [DOI] [PubMed] [Google Scholar]
- 72.Archer FI, Adams PE, Schneiders B. B. stratag: An r package for manipulating, summarizing and analysing population genetic data. Mol. Ecol. Resour. 2017;17:5–11. doi: 10.1111/1755-0998.12559. [DOI] [PubMed] [Google Scholar]
- 73.Rannala B, Mountain JL. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA. 1997;94:9197–9201. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Piry S. GENECLASS2: A Software for Genetic Assignment and First-Generation Migrant Detection. J. Hered. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- 75.Daguin C, Borsa P. Genetic characterisation of Mytilus galloprovincialis Lmk. in North West Africa using nuclear DNA markers. J. Exp. Mar. Bio. Ecol. 1999;235:55–65. doi: 10.1016/S0022-0981(98)00163-4. [DOI] [Google Scholar]
- 76.Bierne N, et al. Introgression patterns in the mosaic hybrid zone between Mytilus edulis and M. galloprovincialis. Mol. Ecol. 2003;12:447–461. doi: 10.1046/j.1365-294X.2003.01730.x. [DOI] [PubMed] [Google Scholar]
- 77.Larraín MA, Díaz NF, Lamas C, Uribe C, Araneda C. Traceability of mussel (Mytilus chilensis) in southern Chile using microsatellite molecular markers and assignment algorithms. Exploratory survey. Food Res. Int. 2014;62:104–110. doi: 10.1016/j.foodres.2014.02.016. [DOI] [Google Scholar]
- 78.Altman D, Bland M. Statistics Notes: Diagnostic tests 1: sensitivity and specificity. BMC. 1994;308:1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Loong T-W. Understanding sensitivity and specificity with the right side of the brain. BMJ. 2003;327:716–719. doi: 10.1136/bmj.327.7417.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deeks, J. J. & Altman, D. G. Diagnostic tests 4: likelihood ratios. BMJ329 (2004). [DOI] [PMC free article] [PubMed]
- 81.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PMM. The diagnostic odds ratio: a single indicator of test performance. J. Clin. Epidemiol. 2003;56:1129–1135. doi: 10.1016/S0895-4356(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 82.Littenberg B, Moses LE. Estimating diagnostic-accuracy from multiple conflicting reports - A new metaanalitic method. Med. Decis. Mak. 1993;13:313–321. doi: 10.1177/0272989X9301300408. [DOI] [PubMed] [Google Scholar]
- 83.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 84.Collins RA, Cruickshank RH. The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 2013;13:969–975. doi: 10.1111/1755-0998.12046. [DOI] [PubMed] [Google Scholar]
- 85.Dellicour S, Flot J-F. The hitchhiker’s guide to single-locus species delimitation. Mol. Ecol. Resour. 2018;18:1234–1246. doi: 10.1111/1755-0998.12908. [DOI] [PubMed] [Google Scholar]
- 86.Blair, C. & Bryson, R. W. Cryptic diversity and discordance in single‐locus species delimitation methods within horned lizards (Phrynosomatidae: Phrynosoma). Mol. Ecol. Resour. 10.1111/1755-0998.12658 (2017). [DOI] [PubMed]
- 87.Dakin EE, Avise JC. Microsatellite null alleles in parentage analysis. Heredity (Edinb). 2004;93:504–509. doi: 10.1038/sj.hdy.6800545. [DOI] [PubMed] [Google Scholar]
- 88.Hillis DM, Dixon MT. Ribosomal DNA: Molecular Evolution and Phylogenetic Inference. Q. Rev. Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 89.Tarifeño E, et al. Erroneous identification of the mussel, Mytilus galloprovincialis (Lamarck 1819) as the specie, Mytilus chilensis (Hupe 1854) in the Bay of Concepcion, Chile. Gayana (Concepción) 2012;76:167–172. doi: 10.4067/S0717-65382012000300010. [DOI] [Google Scholar]
- 90.Daguin C, Borsa P. Genetic relationships of M galloprovinciallis Lmk populations worldwide. Evidence from nuclear-DNA markers. Bivalve Systematics and Evolution. 2000;177:389–397. [Google Scholar]
- 91.Steinert G, Huelsken T, Gerlach G, Bininda-Emonds OP. Species status and population structure of mussels (Mollusca: Bivalvia: Mytilus spp.) in the Wadden Sea of Lower Saxony (Germany) Org. Divers. Evol. 2012;12:387–402. doi: 10.1007/s13127-012-0075-5. [DOI] [Google Scholar]
- 92.Rawson PD, Hilbish TJ. Distribution of male and female mtDNA lineages in populations of blue mussels, Mytilus trossulus and M. galloprovincialis, along the Pacific coast of North America. Mar. Biol. 1995;124:245–250. doi: 10.1007/BF00347128. [DOI] [Google Scholar]
- 93.Zouros E. Biparental Inheritance Through Uniparental Transmission: The Doubly Uniparental Inheritance (DUI) of Mitochondrial DNA. Evol. Biol. 2013;40:1–31. doi: 10.1007/s11692-012-9195-2. [DOI] [Google Scholar]
- 94.Wood, A. R., Beaumont, A. R., Skibinski, D. O. F. & Turner, G. Analysis of a nuclear-DNA marker for species identification of adults and larvae in the Mytilus edulis complex. J. Molluscan Stud. (2003).
- 95.Coghlan B, Gosling E. Genetic structure of hybrid mussel populations in the west of Ireland: two hypotheses revisited. Mar. Biol. 2007;150:841–852. doi: 10.1007/s00227-006-0408-z. [DOI] [Google Scholar]
- 96.Beaumont AR, Hawkins MP, Doig FL, Davies IM, Snow M. Three species of Mytilus and their hybrids identified in a Scottish Loch: natives, relicts and invaders? J. Exp. Mar. Bio. Ecol. 2008;367:100–110. doi: 10.1016/j.jembe.2008.08.021. [DOI] [Google Scholar]
- 97.Gardner, J. P. A., Zbawicka, M., Westfall, K. M. & Wenne, R. Invasive blue mussels threaten regional scale genetic diversity in mainland and remote offshore locations: the need for baseline data and enhanced protection in the Southern Ocean. Glob. Chang. Biol. n/a-n/a 10.1111/gcb.13332 (2016). [DOI] [PubMed]
- 98.Wakeley, J. Coalescent Theory: An introduction. (Roberts and Company Publishers 2009).
- 99.Harrison RG. Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends Ecol. Evol. 1989;4:6–11. doi: 10.1016/0169-5347(89)90006-2. [DOI] [PubMed] [Google Scholar]
- 100.Murgarella M, et al. A First Insight into the Genome of the Filter-Feeder Mussel Mytilus galloprovincialis. PLoS One. 2016;11:e0151561. doi: 10.1371/journal.pone.0151561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Araneda C, Larraín MA, Hecht B, Narum S. Adaptive genetic variation distinguishes Chilean blue mussels (Mytilus chilensis) from different marine environments. Ecol. Evol. 2016;6:3632–3644. doi: 10.1002/ece3.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zbawicka M, Trucco MI, Wenne R. Single nucleotide polymorphisms in native South American Atlantic coast populations of smooth shelled mussels: hybridization with invasive European Mytilus galloprovincialis. Genet. Sel. Evol. 2018;50:5. doi: 10.1186/s12711-018-0376-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wenne, R., Drywa, A., Kent, M., Sundsaasen, K. K. & Lien, S. SNP Arrays for Species Identification in Salmonids. in Marine Genomics: Methods and Protocols (ed. Bourlat, J. S.) 97–111, 10.1007/978-1-4939-3774-5_6 (Springer New York 2016). [DOI] [PubMed]
- 104.Zbawicka M, Gardner JPA, Wenne R. Cryptic diversity in smooth-shelled mussels on Southern Ocean islands: connectivity, hybridisation and a marine invasion. Front. Zool. 2019;16:32. doi: 10.1186/s12983-019-0332-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Popovic, I., Matias, A. M. A., Bierne, N. & Riginos, C. Twin introductions by independent invader mussel lineages are both associated with recent admixture with a native congener in Australia. Evol. Appl. 0 (2019). [DOI] [PMC free article] [PubMed]
- 106.Gosling E, Doherty S, Howley N. Genetic characterization of hybrid mussel (Mytilus) populations on Irish coasts. J. Mar. Biol. Assoc. United Kingdom. 2008;88:341–346. doi: 10.1017/S0025315408000957. [DOI] [Google Scholar]
- 107.Rawson PD, Hilbish TJ. Asymmetric Introgression of Mitochondrial DNA Among European Populations of Blue Mussels (Mytilus Spp.) Evolution (N. Y). 1998;52:100–108. doi: 10.1111/j.1558-5646.1998.tb05142.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.