Abstract
The spatio-temporal reduction and oxidation of protein thiols is an essential mechanism in signal transduction in all kingdoms of life. Thioredoxin (Trx) family proteins efficiently catalyze thiol-disulfide exchange reactions and the proteins are widely recognized for their importance in the operation of thiol switches. Trx family proteins have a broad and at the same time very distinct substrate specificity – a prerequisite for redox switching. Despite of multiple efforts, the true nature for this specificity is still under debate. Here, we comprehensively compare the classification/clustering of various redoxins from all domains of life based on their similarity in amino acid sequence, tertiary structure, and their electrostatic properties. We correlate these similarities to the existence of common interaction partners, identified in various previous studies and suggested by proteomic screenings. These analyses confirm that primary and tertiary structure similarity, and thereby all common classification systems, do not correlate to the target specificity of the proteins as thiol-disulfide oxidoreductases. Instead, a number of examples clearly demonstrate the importance of electrostatic similarity for their target specificity, independent of their belonging to the Trx or glutaredoxin subfamilies.
Keywords: Mathematical biosciences, Biocomputational method, Biomolecules, Molecular docking, Gromov-Wasserstein distance, Protein-protein interaction, Redox signaling, Electrostatics, Thioredoxin, Glutaredoxin
Mathematical biosciences; Biocomputational method; Biomolecules; Molecular docking; Gromov-Wasserstein distance; Protein-protein interaction; Redox signaling; Electrostatics; Thioredoxin; Glutaredoxin
1. Introduction
Redox modifications of cysteinyl and also methionyl side chains are a vital part of numerous signal transduction pathways as well as the reaction cycle of essential metabolic enzymes [1, 2, 3, 4]. Many of these redox reactions are directly or indirectly catalyzed by members of the Trx family of proteins. This group of proteins share a common basic structural motif – the Trx fold. Their active sites, in most cases consisting of two cysteinyl residues separated by two amino acids (Cys-X-X-Cys), are the basis of their redox activity [5]. Proteins of this family are known to catalyze the reduction of disulfides in target proteins, the de- or glutathionylation of proteins, they catalyze the oxidative folding of proteins and are able to reduce redox modifications like sulfenic acids [4]. Trx family proteins are encoded in essentially all genomes and are localized in all compartments of eukaryotic cells, e.g. the cytosol, ER, mitochondria, nucleus, and plastids – often in multiple isoforms. Most members of the family have a broad, but distinct substrate specificity. The nature of this specificity is the focus of this work.
The Trx family divides into subfamilies, two of the major groups are the Trxs themselves and the glutaredoxins (Grxs). Trxs catalyze thiol-disulfide exchange reactions and the trans-nitrosylation of thiol groups [4, 6]. The disulfide formed in their consensus Cys-Gly-Pro-Cys active site during their reaction cycle is reduced by specific reductases named thioredoxin reductases (TrxRs) [7, 8]. The members of one of the Grx subfamilies (dithiol Grxs, with a consensus active site of Cys-Pro-Tyr-Cys) catalyze thiol-disulfide oxidoreductions as well, however, when oxidized these proteins are reduced by the tripeptide glutathione (GSH). The mechanisms of these reactions have been discussed in great detail before, see for instance [4, 9, 10, 11]. The members of a second subclass of the Grxs (monothiol Grxs, with a consensus Cys-Gly-Phe-Ser active site) do not catalyze thiol-disulfide exchange reactions at significant rates. Instead, they function in the regulation of iron metabolism or in the transfer of iron-sulfur centers [12, 13, 14].
Traditionally, Grxs and Trxs were named in each organism in order of their discovery, for instance in mammals, the firstly discovered cytosolic Trx1 [15] and the later discovered mitochondrial Trx2 [11]. An other example are the yeast Grxs 1–8, summarized in [16]. This historical naming, however, does not include any information on structural or functional differences with the Trx family of proteins. A more advanced classification and naming system based on the active site sequences was presented for plants Grxs and became widely accepted [17]. This nomenclature defines three classes, i.e. class I, that contains the largely redox-active dithiol Grxs, class II, including all monothiol Grxs, and class III, including the land plant-specific CC-type Grxs, also known as ROXYs [18].
In most species, including bacteria, fungi, mammals, and plants, multiple Trx family proteins are present in the same compartment, prompting questions on overlapping functions. Various proteomic studies, also summarized in this work, indicate a rather high degree of substrate specificity for each member of the family, with only some overlapping substrate/target proteins. Various attempts have been made to understand the substrate specificity and reactivity of the Trx family proteins. Previous suggestions primarily addressed the thermodynamics of the reaction, including the nucleophilicity of the more N-terminal active site thiol, the differences in redox potential, and entropic changes during the reaction [19, 20, 21]. Recently, based on the analysis of E. coli phosphoadenylyl sulfate (PAPS) reductase that can react with many, but not all Trxs and Grxs [22, 23], we proposed that different electrostatic properties of the redoxins govern their target specificity and reactivity [24].
In this work, we provide a detailed comparison on the coherence between the similarity of redoxins in (1st) primary structure, (2nd) tertiary structure, and (3rd) electrostatic properties. Where possible, we correlate these different methods of clustering/classification to known functions of the redoxins. We have focused on the redoxins encoded in the human genome and all redoxins from various species with experimentally determined structures deposited in the protein data base. Our results provide further evidence for the importance of the electrostatic properties of the proteins for their distinct target specificity. This clustering may allow a new functional classification of the redoxins and may enable the prediction of common functions and interactions partners.
2. Methods and procedures
2.1. Structures and molecular modeling
Structures, when available, were obtained from the protein data bank (https://www.rcsb.org); the PDB entries used are listed in the supplementary table. Molecular modeling was performed using the Swiss Model web server [25, 26, 27, 28, 29]. The final model was chosen from structures modeled with different templates that displayed the highest sequence identity with the target protein based on the quality assessment provided, i.e. the lowest QMEAN with no major outlier in the global or local quality estimates of Cβ, all atom, solvation, or torsion. The individual template structures used and the QMEAN values are summarized in the supplementary table (structures).
2.2. Sequence and structure comparison
Sequences of the proteins were obtained from the uniprot resources. In case of multi-domain proteins, the sequences encoding the redoxin domains only were mostly extracted as annotated in the respective uniprot entries, i.e. based on PROSITE-ProRule [30]. In some critical cases, for instance human nucleoredoxin, multiple sequence alignments were performed including sequences from various species. These typically share a higher degree of homology within the functional domains and a lower in the joining peptides. Primary structure alignments and the generation of the corresponding distance trees were performed with the CLC sequence viewer (Qiagen bioinformatics, Hilden, Germany) and Clustal omega [31]. The three dimensional structures were aligned using UCSF chimera [32] (MatchMaker) including structure-based multiple sequence alignments. From these primary structure and 3-D structure alignments, the corresponding distance trees were generated with the CLC sequence viewer applying the neighbor joining method and the Jukes-Cantor protein distance measure.
2.3. Electrostatic calculations
The structures in the PDB files were aligned in the desired orientation using UCSF chimera. The electrostatic properties of the proteins were computed from the pdb files as follows: the reconstruction of any missing atoms, the addition of hydrogens, the assignment of atomic charges and radii was performed using pdb2pqr with the amber force field [33]. The electrostatic parameters were calculated using the Adaptive Poisson-Boltzmann Solver (APBS) [34] within the vmd (visual molecular dynamics) software package [35]. The following parameters were used: 150 mM mobile ions, solvent dielectric constant: 78.54, temperature: 298.15 K. Images were rendered depicting the secondary structures of the proteins (with the N-terminal active site cysteinyl residues facing towards the camera perspective), the electrostatic potential mapped to the surface of the proteins (from -4 in red to 4 K T·e−1 in blue), and the isosurfaces of the electrostatic potential at -1 in red and 1 K T·e−1 in blue. These pictures were used to generate a summary picture using ImgageMagick. All steps following the 3D alignment of the structures were automatized with the help of scripts and a graphical interface. These can be obtained from: https://github.com/WillyBruhn/MutComp.
2.4. Electrostatic distances and clustering
Here, we compared the 3-dimensional isosurfaces of both the negative and positive electrostatic potential using the Gromov-Wasserstein-distance [36, 37]. Solving this problem, is NP-hard as the objective function is not convex. However, three lower bounds for the Gromov-Wasserstein-distance can be calculated in polynomial time. Our empirical tests demonstrated that not all points of the isosurface shall be calculated, instead we limited the sample to n points randomly distributed on the isosurfaces and calculate the lower bound for them. This was repeated m times, the obtained values were summarized in form of a histogram. This comparison was performed pairwise for all proteins. To get a measure of similarity between the histograms, the earth-mover's-distance was used [38]. For the hierarchical clustering, the unweighted pair group method with arithmetic mean (UPGMA) was used. This method yields the mean distance between all points from the new cluster to all points of another cluster. The result of this clustering were displayed in form of a dendrogram. Further details of this mathematical approach will be presented elsewhere. All code of the software produced here (C++, R, scripts) can be obtained free of charge and open source here: https://github.com/BerensF/ComparingProteins.
2.5. Interactome data and comparison
Interactome data were retrieved from the IntAct database [39] (as of Sept. 2018) as well as the BioGRID resources [40] (Vers. 3.4, as of Sept. 2018). ID mapping was performed using the uniprot resources (https://www.uniprot.org). All entries are listed with their unique UniprotKB ID. When available, additional resources for interactions – not yet listed in the upper mentioned databases – were included, for instance from some dedicated publications. All entries identified are listed in the supplementary tables (interactome). The matrix of common interactions as well as the Venn diagrams were computed using R (https://www.r-project.org) from RStudio (https://www.rstudio.com).
3. Results and discussion
All presently established classifications of Trx family proteins are based on the comparison of, and the clustering according to primary structures. We know, however, that these systems do not, or at best partially, reflect the various functions of the proteins. As examples, some only distantly related Trxs and Grxs share overlapping functions, while some closely related Grxs do not. The disulfide formed in E. coli PAPS reductase during its catalytic cycle can be reduced by the distantly related Trx1 and Grx1 (primary sequence identity: 19.6%), but not by Grx3 (sequence identity to Grx1: 41.7%) [23, 41]. The primary aim of this study is to provide a thorough comparison between the clustering and classification according to primary structure, 3-D structure, and electrostatic characteristics. Further more, we aim to evaluate the practicality of the comparison of electrostatic properties for the functional classification of Trx family proteins and the prediction of functions.
In the first part, we focus on the Trx family proteins from human (see Table 1). Functions of the proteins were deduced from the literature, identified interacting proteins were collected from the literature, our own data sets, and some major interaction databases, i.e. IntAct and BioGRID (see methods section). These sources summarize studies dedicated to single interactions as well as proteomic approaches. If available, 3-D structures were obtained from the protein data base (pdb). Missing structures were obtained by homology modeling. The electrostatic calculations were performed using pdb2pqr and APBS implemented in vmd. For the comparison and clustering of the electrostatic similarities, we have adopted new strategies based on Gromov-Wasserstein distances of lower boundaries and Earth-Movers distances. These computational strategies are outlined in the methods section.
Table 1.
Redoxins and redoxin domains encoded in the human genome. loc: localization, c: cytosol, e: endoplamic reticulum, g: golgi apparatus, l: lipid membrane, m: mitochondria, n: nucleus, o: outside of the cell, secreted; ‘.d’ marks individual domains. n.a.: not analyzed (as no information on the individual domains were available).
| Protein name | ID | Active site | pdb | Functions | loc. | Interactome (number) |
|---|---|---|---|---|---|---|
| Homo sapiens glutaredoxins | ||||||
| Grx1 | P35754 | CPYC | 1b4q, ... | (de)-glutathionylation | c/n | 24 |
| Grx2a/c | Q9NS18 | CSYC | 2fls, ... | Fe/S, redox sensor [52] | c/m/n | 64 [53] |
| Grx3.d2 | O76003 | CGFS | 3zyw | Fe/S, iron metabolism [54, 55] | c | 98 |
| Grx3.d3 | CGFS | 2yan | ||||
| Grx5 | Q86SX6 | CGFS | 2wul, ... | Fe/S biogenesis [56, 57] | m | 22 |
| TrxR1v3.d1 | Q16881-1 | CTRC | c | n.a. | ||
| TrxR3.d1 | Q86VQ6 | CPHS | 2h8q | c | n.a. | |
| GrxCR1.d2 | A8MXD5 | FERC | c | 9 | ||
| PTGES2.d1 | Q9H7Z7 | CPFC | 2pbj | prostaglandin synthesis [58] | c/l | 31 |
| SH3BGRL3 | Q9H299 | KSQQ | 1sj6 | cancer progression [59] | c/n | 10 |
| Homo sapiens thioredoxins | ||||||
| Trx1 | P10599 | CGPC | 1ert, ... | electron donor, redox signaling, ... [4] | c/n/o | 247 [60] |
| Trx2 | Q99757 | CGPC | 1wh4, ... | m | 102 | |
| Grx3.d1 | O76003 | APQC | 2diy, ... | Fe/S, iron metabolism [54, 55] | c | see above |
| Nrx.d2 | Q6DKJ4 | CPPC | redox signaling | c/n | 629 [61] | |
| Nxnl1 | Q96CM4 | CPQC | n/l | 4 | ||
| Nxnl2 | Q5VZ03 | CAPS | c | 2 | ||
| Txnl1 | O43396 | CGPC | 1gh2, ... | c/n | 62 | |
| Txnl4A | P83876 | DPTC | 1qgv, ... | n | 39 | |
| Txnl4B | Q9NX01 | DPVC | 3gix, ... | n | 21 | |
| Tmx1 | Q9H3N1 | CPAC | 1x5e | e/l/o | 79 | |
| Tmx2 | Q9Y320 | SNDC | 2dj0 | l/o | 103 | |
| Tmx4 | Q9H1E5 | CPSC | l/o | 25 | ||
| Txndc2 | Q86VQ3 | CGPC | spermatogenesis [62, 63] | c | 4 | |
| Txndc3 | Q8N427 | CGPC | spermatogenesis [64] | c | 8 | |
| Txndc6 | Q86XW9 | CGPC | microtuble dynamics, cancer progression [65, 66] | c | - | |
| Txndc8 | Q6A555 | CGPC | spermatogenesis [67] | c/g | - | |
| Txndc9 | O14530 | TFRC | protein complex assembly [68] | c/n | 91 | |
| Txndc11.d1 | Q6PKC3 | CGQS | e/l | 83 | ||
| Txndc11.d2 | CGFC | |||||
| Txndc12 | O95881 | CGAC | 1sen, ... | PDI | e | 30 |
| Txndc15 | Q96J42 | CRFS | ciliogenesis [69] | l/o | 44 | |
| Txndc16 | Q9P2K2 | QAVS | meningioma-associated antigen [70] | e/s | 29 | |
| Txndc17 | Q9BRA2 | CPDC | 1wou | disulfide and cystine reduction, denitrosylation [71, 72] | c | 39 |
| Qsox1 | O00391 | CGHC | 3q6q, ... | disulfide formation [73] | g/o | 17 |
| Qsox2 | Q6ZRP7 | CGHC | disulfide formation [73] | c/l/n/o | 28 | |
3.1. Human Grxs and Trxs
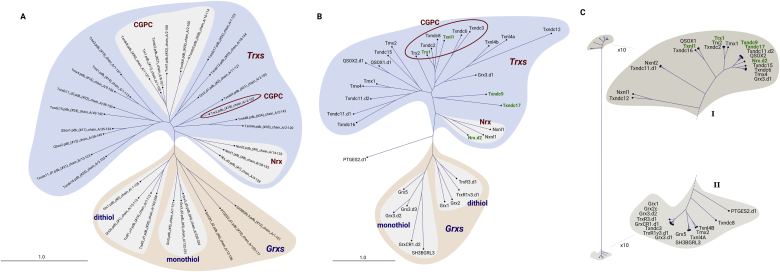
The human genome encodes about 9 Grxs or Grx domain- containing proteins and 24 Trxs or Trx domain-containing proteins (see Table 1). From the in total 35 Trx-fold domains in these 33 proteins, the 3-D structure of 20 had already been determined experimentally. The remaining 15 were predicted by homology modeling, see supplementary Table, sheet 1 – modeling. Similar to the comparison of the plant redoxins, the human redoxins divide into similar groups in the trees generated from primary and tertiary structure (Figure 1A and B). The subgroup of the Grxs both show the separate monothiol and dithiol Grx groups. The Trx group is quite diverse, however, the Nrxs and the Trxs with the consensus active site motif Cys-Gly-Pro-Cys (minus mitochondrial Trx2) clearly separate from the others in both analyses. The electrostatic characteristics (Figure 2) define two major groups (Figure 1C). Group ‘I’ contains Trxs and Trx domain-containing proteins exclusively. Group ‘II’ contains all of the Grxs and Grxs domains next to some Trx-related proteins and domains.
Figure 1.
Clustering of human redoxins. (A) Phylogram based on primary structure comparison, computed by Clustal Omega and CLC sequence viewer. (B) Similarity tree based on the similarity of the 3D structures extracted from the pdb and generated by homology modeling; the tree was computed using UCSF Chimera and the CLC sequence viewer. (C) The electrostatic similarity of the whole proteins was computed as outlined in the methods section; the tree was generated using ‘R’. The protein abbreviations highlighted in green are referred to in the main text. The Trx proteins with a Cys-Gly-Pro-Cys active site motif were highlighted with a red circle in B.
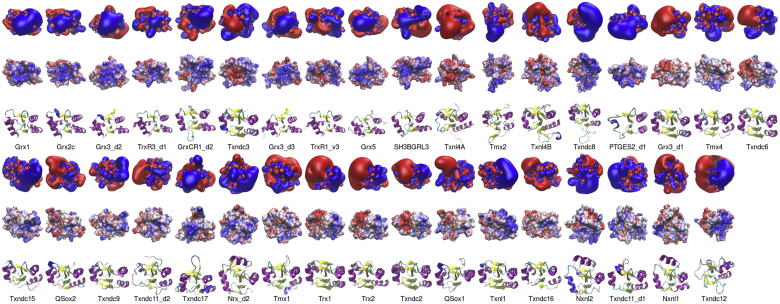
Figure 2.
Electrostatic features of the active site contact areas of the human redoxins. The first rows depict the electrostatic potential isosurfaces at +/- 1 K T·e−1. The second row depicts the electrostatic potential at +/- 4 K T·e−1 mapped to the water-accessible surface of the proteins. Blue: positive, red negative potential. The third row depicts the proteins in cartoon models, helices are colored in purple, sheets in yellow. The proteins were arranged with the N-terminal active site thiol in the middle of the models. The electrostatic similarity of the whole proteins was computed as outlined in the methods section.
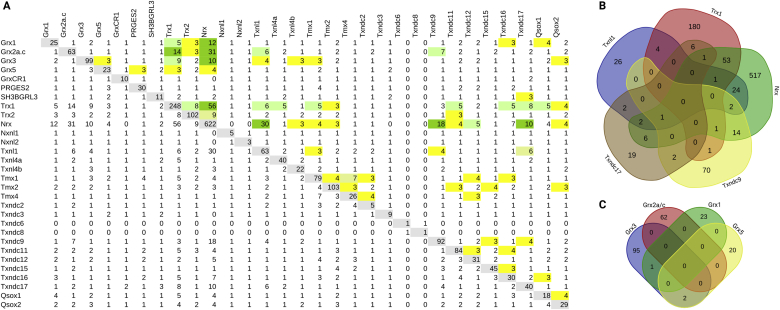
Using various sources, see Table 1, we collected almost 2000 potential interactions partners of these 33 proteins (all entries are available in the supplementary tables, sheet H.s. interactome). The pair-wise comparison of common targets in these sets is summarized in Figure 3A. The largest degree of overlapping interacting proteins was found between the thioredoxins Trx1, Txnl1, Txndc9, Txndc17, and Nrx, also summarized in the Venn diagram depicted in Figure 3B. This suggests some overlapping functions of various pairs of these five proteins. Triple and quadruple overlaps, on the other hand, are rare and no common potential interaction partner of all five of these redoxins was identified nor suggested so far. In the trees generated from the primary structure and tertiary structure comparisons, these five proteins of the Trx subfamily are localized in three different branches, separated by large distances (Figure 1A and B, green labels), only two of them contain the consensus Cys-Gly-Pro-Cys active site motif (Trx1 and Txnl1, see Table 1). In the tree based on their similarity in electrostatic properties, these five proteins are all localized close to each other in the terminal branches of cluster ‘I’ (Figure 1C, green labels). The glutaredoxins Grx1, Grx2, Grx3, and Grx5 share only three potential interaction partners, one between Grx1 and Grx2, two between Grx3 and Grx5, out of a total of 210 suggested target proteins (see Figure 3C). The primary reasons for this may be their different subcellular localization, dithiol Grx1 and monothiol Grx3 are cytosolic, dithiol Grx2 and monothiol Grx5 primarily mitochondrial.
Figure 3.
Common interaction partners between the human redoxins. (A) Pair-wise comparison between all human redoxins. The total numbers of potential interactions partners collected from various data sources is depicted with gray background in the diagonal; yellow background: 3–4 common interaction partners; light green background: 5–9 common interaction partners; green background: ≥ 10 common interaction partners. The full list of interaction partners can be found in the supplementary tables. (B–C) Venn diagrams of the overlapping potential interactions partners between Trx1, Nrx, Txndc9, Txndc17, and Txnl1 (B) as well as Grx1, Grx2, Grx3, and Grx5 (B).
This study primarily focused on redox-interactions of the redoxins with target proteins. The main reason for this was the availability of data. However, it has been suggested early on that the Trx-fold domains might act as platform for protein-protein interactions, e.g. as processivity factor of T7 DNA polymerase [42] or as the basis for their redox-independent chaperon activity, summarized in [43]. Our study implies that redox-inactive redoxins (mostly domains) will show a similar target specificity as electrostatically similar redox-active redoxins. The N-terminal Trx domain of human Grx3 (Grx3_d1), for instance, contains the ‘active’ site motif Ala-Pro-Gln-Cys, hence it cannot catalyze thiol-disulfide exchange reactions. Electrostatically, this domain is most similar to the redox-active Grx1 and Grx2, as well as the Grx-domains of TrxR1 transcript variant 3 [44] and TrxR3 [45], see Figure 1C (cluster II). It remains to be established whether these proteins and domains share a similar pattern of interacting proteins or other substrates as implied by our study.
3.2. All representative redoxin structures from the pdb
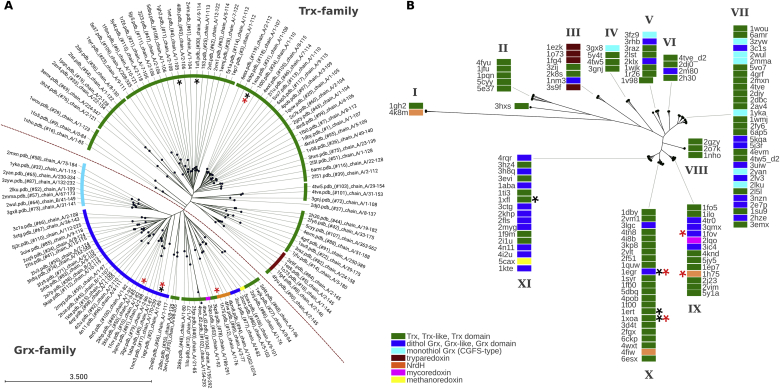
For a more comprehensive clustering of Trx-family proteins, we have selected all Trx- and Grx-fold structures from the pdb for analysis. For the electrostatic analysis, we selected only non-mutated proteins and, if required, extracted the Trx-/Grx-domains. Other molecules included in some of the structures, i.e. cofactors or water, were excluded. For the present analysis, we did not include other Trx-family members, such as DsbA/B/C proteins, protein disulfide isomerases, GSH peroxidases, or arsenate reductases. Our final collection included 119 structures (see supplementary Table, sheet 5 – pdb structures) from all domains of life, including some phage/virus-encoded proteins. For these structures, we generated both primary structure and electrostatic similarity trees, see Figure 4. The sequence-based tree clearly separates the structures into the Grx and Trx subfamilies (Figure 4A), independent of their sometimes confusing annotation in the pdb and sequence databases. Within the Grx branch, the genuine monothiol Grxs (Cys-Gly-Phe-Ser) form a well defined side branch, as well as the dithiol Grxs. The remaining structures include redoxins such as mycoredoxin, methanoredoxin, and NrdHs proteins. The Trx subfamily contains six distinct groups, two eukaryotic, three bacterial, and the tryparedoxin branches (Figure 4A). In contrast to sequence similarity, the electrostatic similarity tree separates into eleven branches, marked as I-XI in Figure 4B, see also Figure 5, of which most include structures from both the Trx and Grx subfamilies. None of these groups overlap with a branch of the sequence-based tree.
Figure 4.
Clustering of all representative redoxins in the pdb. (A) Phylogram based on primary structure comparison, computed by Clustal Omega and CLC sequence viewer. The dashed red line separates the Trx and Grx subfamilies. (B) The electrostatic similarity of the whole proteins was computed as outlined in the methods section; the tree was generated using ‘R’. The red asterisks mark proteins interacting with E. coli RNR, the black asterisks proteins interacting with E. coli PAPS reductase. The color code is included in the figure. Further information on the protein structures can be obtained from the supplementary Table, sheet 5.
Figure 5.
Electrostatic features of all representative redoxins in the pdb. The first rows depict the electrostatic potential isosurfaces at +/- 1 K T·e−1. The second row depicts the electrostatic potential at +/- 4 K T·e−1 mapped to the water-accessible surface of the proteins. Blue: positive, red negative potential. The third row depicts the proteins in cartoon models, helices are colored in purple, sheets in yellow. The pdb entry code of the structures is indicated in the fourth row. The proteins were arranged with the N-terminal active site thiol in the middle of the models. The electrostatic similarity of the whole proteins was computed as outlined in the methods section.
So what can be said about the correlation between the two clustering methods and the functional interaction of the different redoxins with target proteins? The two first discovered functions of Trxs and Grxs offer some insights. Trx1 (pdb: 1xoa) from E. coli was first discovered as electron donor for ribonucleotide reductase (RNR) [46]. Later studies demonstrated that this function could also be fulfilled by Grx1 (1egr) [47, 48], Grx3 (1fov) [49], and NrdH (1h75) [50] from E. coli. In the sequence based tree, these four proteins can be found in three distant branches (Figure 4A, red asterisks). In the electrostatic similarity tree, however, these four proteins can be found in the very close neighbouring branches IX and X (Figures 4B and 5, red asterisks). The requirement of Trxs for sulfate reduction was first described in yeast [51]. Subsequently, Trx1 was also demonstrated to be a cosubstrate of PAPS reductase in E. coli. Similar to RNR, Grx1 can replace Trx1 in vivo and in vitro in this function [23, 41], however, Grx3 and NrdH failed to do so [23, 24]. E. coli PAPS reductase can be reduced by a number of redoxins from various species, while some others cannot interact with the protein [22, 24]. Positive interactions partners are, for instance human and Arabidopsis thaliana Trx1 (1ert) and TrxH1 (1xfl), respectively. Again, these functionalities and non-functionalities cannot be predicted or explained by their similarities in primary sequence (Figure 4A, black asterisks). In the electrostatic similarity tree, three of the four proteins are located in cluster ‘X’, one (A.t. TrxH1) in cluster ‘XI’. It should also be mentioned that cluster ‘XI’ contains proteins that cannot interact productively with PAPS reductase, i.e. human Grx2 (2fls), and T4 Grx (1aba) [24]. It appears that – at least some – proteins included in the electrostatic characteristics cluster ‘X’ can interact with both PAPS reductase and RNR, proteins of cluster ‘IX’ also with RNR, but not PAPS reductase. These two examples clearly support our hypothesis that the electrostatic similarity between the redoxins correlates to their profile of interacting proteins or may even guide these specific interactions [24].
For this study, the points analyzed for electrostatic similarity were distributed randomly on the surface of the proteins. One might argue that more weight on the properties of the immediate contact surface surrounding the active site could even further improve the model. To test this, we have developed a strategy to extract the electrostatic isosurfaces of the areas surrounding the N-terminal active site residue, that is usually the cysteinyl residue that forms an intermediate mixed-disulfide with the (redox-) target proteins and analyzed them in the same way as the features of the whole proteins. This strategy and the results obtained when it was applied to the ‘representative structures from the pdb’ data set were summarized in the supplementary material (suppl. Figure 1 and text). In brief, the hierarchical clustering of the electrostatic similarities in the area close to the active site did not reflect the functions of some redoxins as electron donor of PAPS reductase or RNR anywhere near as good as the clustering of the electrostatic similarities of the whole proteins. The redoxins that were previously shown to donate electrons to PAPS reductase clustered all within the distance of 9.8% of the maximum distance of all redoxins, the ones donating electrons to RNR within 12.7% of the maximum distance (Figure 4B) when the features of the whole proteins were compared. When only the electrostatic features of the active site faces of the proteins were compared, these redoxins did not cluster in close proximity and the relative distances increased to 37.4% and 62.6% of the maximum distance for PAPS reductase and RNR, respectively (suppl. Figure 1, nodes a and b). These results indicate that the global properties of the proteins may play a more important role than previously assumed. Moreover, they favor a model of redoxin-target interaction in which the recognition of the two proteins is controlled by attractive and repulsive electrostatic forces that, presumably, take part in pre-orientation of the two proteins before they can form a productive encounter complex, rather than contact surface complementarity only. Further experimental studies will have to address this hypothesis.
4. Conclusions
Here, we evaluated the practicality of a mathematical model for the automated clustering of the electrostatic properties of proteins for the functional classification of Trx family proteins and the prediction of functions. The analysis of the human, and pdb-wide redoxin structures clearly demonstrate that primary and tertiary structure (backbone) similarity do not correlate to the target specificity of the proteins as thiol-disulfide oxidoreductases and neither does their redox potential, see [24]. Instead, the examples of the human and pdb-wide redoxins clearly demonstrate the importance of the electrostatic properties of the whole protein for target specificity and discrimination. The mathematical model evaluated here is the first step towards an automated analysis and comparison of electrostatic properties of a large number of protein structures.
Declarations
Author contribution statement
Manuela Gellert: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Md Faruq Hossain: Performed the experiments; Analyzed and interpreted the data.
Felix Jacob Ferdinand Berens, Lukas Willy Bruhn: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Claudia Urbainsky: Performed the experiments.
Volkmar Liebscher: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Christopher Horst Lillig: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by Deutsche Forschungsgemeinschaft, German Research Foundation (DFG): Li984/3-2, GRK1947-A1. We acknowledge support for the Article Processing Charge from the DFG (393148499) and the Open Access Publication Fund of the University of Greifswald.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at GitHub under the URLs: https://github.com/WillyBruhn/MutComp and https://github.com/BerensF/ComparingProteins.
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2019.e02943.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Ghezzi P., Bonetto V., Fratelli M. Thiol disulfide balance: from the concept of oxidative stress to that of redox regulation. Antioxidants Redox Signal. 2005;7:964–972. doi: 10.1089/ars.2005.7.964. [DOI] [PubMed] [Google Scholar]
- 2.Janssen-Heininger Y.M.W., Mossman B.T., Heintz N.H., Forman H.J., Kalyanaraman B., Finkel T., Stamler J.S., Rhee S.G., van der Vliet A. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drazic A., Winter J. The physiological role of reversible methionine oxidation. Biochim. Biophys. Acta. 2014;1844:1367–1382. doi: 10.1016/j.bbapap.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Hanschmann E.-M., Godoy J.R., Berndt C., Hudemann C., Lillig C.H. Thioredoxins, glutaredoxins, and peroxiredoxins--molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxidants Redox Signal. 2013;19:1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin J.L. Thioredoxin--a fold for all reasons. Struct. Lond. Engl. 1993. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 6.Sengupta R., Holmgren A. The role of thioredoxin in the regulation of cellular processes by S-nitrosylation. Biochim. Biophys. Acta. 2011 doi: 10.1016/j.bbagen.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Arnér E.S., Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. FEBS. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 8.Arnér E.S.J. Focus on mammalian thioredoxin reductases - important selenoproteins with versatile functions. Biochim. Biophys. Acta. 2009;1790:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Mieyal J.J., Gallogly M.M., Qanungo S., Sabens E.A., Shelton M.D. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxidants Redox Signal. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lillig C.H., Berndt C. Glutaredoxins in thiol/disulfide exchange. Antioxidants Redox Signal. 2013;18:1654–1665. doi: 10.1089/ars.2012.5007. [DOI] [PubMed] [Google Scholar]
- 11.Spyrou G., Enmark E., Miranda-Vizuete A., Gustafsson J. Cloning and expression of a novel mammalian thioredoxin. J. Biol. Chem. 1997;272:2936–2941. doi: 10.1074/jbc.272.5.2936. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Manzaneque M.T., Tamarit J., Bellí G., Ros J., Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bandyopadhyay S., Gama F., Molina-Navarro M.M., Gualberto J.M., Claxton R., Naik S.G., Huynh B.H., Herrero E., Jacquot J.P., Johnson M.K. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mühlenhoff U., Molik S., Godoy J.R., Uzarska M.A., Richter N., Seubert A., Zhang Y., Stubbe J., Pierrel F., Herrero E. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metabol. 2010;12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore E.C. A thioredoxin — thioredoxin reductase system from rat tumor. Biochem. Biophys. Res. Commun. 1967;29:264–268. doi: 10.1016/0006-291x(67)90446-9. [DOI] [PubMed] [Google Scholar]
- 16.Herrero E., Bellí G., Casa C. Structural and functional diversity of glutaredoxins in yeast. Curr. Protein Pept. Sci. 2010;11:659–668. doi: 10.2174/138920310794557637. [DOI] [PubMed] [Google Scholar]
- 17.Rouhier N., Gelhaye E., Jacquot J.-P. Plant glutaredoxins: still mysterious reducing systems. Cell. Mol. Life Sci. CMLS. 2004;61:1266–1277. doi: 10.1007/s00018-004-3410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutsche N., Thurow C., Zachgo S., Gatz C. Plant-specific CC-type glutaredoxins: functions in developmental processes and stress responses. Biol. Chem. 2015;396:495–509. doi: 10.1515/hsz-2014-0300. [DOI] [PubMed] [Google Scholar]
- 19.Roos G., Foloppe N., Messens J. Understanding the pK(a) of redox cysteines: the key role of hydrogen bonding. Antioxidants Redox Signal. 2013;18:94–127. doi: 10.1089/ars.2012.4521. [DOI] [PubMed] [Google Scholar]
- 20.Gallogly M.M., Starke D.W., Mieyal J.J. Mechanistic and kinetic details of catalysis of thiol-disulfide exchange by glutaredoxins and potential mechanisms of regulation. Antioxidants Redox Signal. 2009;11:1059–1081. doi: 10.1089/ars.2008.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palde P.B., Carroll K.S. A universal entropy-driven mechanism for thioredoxin-target recognition. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7960–7965. doi: 10.1073/pnas.1504376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwenn J.D., Schriek U. PAPS-reductase from Escherichia coli: characterization of the enzyme as probe for thioredoxins. Z. Für Naturforschung C J. Biosci. 1987;42:93–102. doi: 10.1515/znc-1987-1-216. [DOI] [PubMed] [Google Scholar]
- 23.Lillig C.H., Prior A., Schwenn J.D., Aslund F., Ritz D., Vlamis-Gardikas A., Holmgren A. New thioredoxins and glutaredoxins as electron donors of 3’-phosphoadenylylsulfate reductase. J. Biol. Chem. 1999;274:7695–7698. doi: 10.1074/jbc.274.12.7695. [DOI] [PubMed] [Google Scholar]
- 24.Berndt C., Schwenn J.-D., Lillig C.H. The specificity and efficiency of thioredoxins and glutaredoxins is determined by electrostatic and geometric complementarity. Chem. Sci. 2015 doi: 10.1039/c5sc01501d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinforma. Oxf. Engl. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertoni M., Kiefer F., Biasini M., Bordoli L., Schwede T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017;7:10480. doi: 10.1038/s41598-017-09654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bienert S., Waterhouse A., de Beer T.A.P., Tauriello G., Studer G., Bordoli L., Schwede T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45:D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigrist C.J.A., De Castro E., Langendijk-Genevaux P.S., Le Saux V., Bairoch A., Hulo N. ProRule: a new database containing functional and structural information on PROSITE profiles. Bioinforma. Oxf. Engl. 2005;21:4060–4066. doi: 10.1093/bioinformatics/bti614. [DOI] [PubMed] [Google Scholar]
- 31.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 33.Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., Baker N.A. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 36.Mémoli F. Gromov–wasserstein distances and the metric approach to object matching. Found. Comput. Math. 2011;11:417–487. [Google Scholar]
- 37.Mémoli F. The Gromov-Wasserstein distance: a brief overview. Axioms. 2014;3:335–341. [Google Scholar]
- 38.Ling H., Okada K. An efficient Earth Mover’s Distance algorithm for robust histogram comparison. IEEE Trans. Pattern Anal. Mach. Intell. 2007;29:840–853. doi: 10.1109/TPAMI.2007.1058. [DOI] [PubMed] [Google Scholar]
- 39.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stark C., Breitkreutz B.-J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russel M., Model P., Holmgren A. Thioredoxin or glutaredoxin in Escherichia coli is essential for sulfate reduction but not for deoxyribonucleotide synthesis. J. Bacteriol. 1990;172:1923–1929. doi: 10.1128/jb.172.4.1923-1929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mark D.F., Richardson C.C. Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 1976;73:780–784. doi: 10.1073/pnas.73.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berndt C., Lillig C.H., Holmgren A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim. Biophys. Acta BBA - Mol. Cell Res. 2008;1783:641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Rundlöf A.-K., Janard M., Miranda-Vizuete A., Arnér E.S.J. Evidence for intriguingly complex transcription of human thioredoxin reductase 1. Free Radic. Biol. Med. 2004;36:641–656. doi: 10.1016/j.freeradbiomed.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Su D., Novoselov S.V., Sun Q.-A., Moustafa M.E., Zhou Y., Oko R., Hatfield D.L., Gladyshev V.N. Mammalian selenoprotein thioredoxin-glutathione reductase. Roles in disulfide bond formation and sperm maturation. J. Biol. Chem. 2005;280:26491–26498. doi: 10.1074/jbc.M503638200. [DOI] [PubMed] [Google Scholar]
- 46.Laurent T.C., Moore E.C., Reichard P. Enzymatic synthesis of deoxyribonucleotides. iv. isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J. Biol. Chem. 1964;239:3436–3444. [PubMed] [Google Scholar]
- 47.Holmgren A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. U. S. A. 1976;73:2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holmgren A. Glutathione-dependent synthesis of deoxyribonucleotides. Purification and characterization of glutaredoxin from Escherichia coli. J. Biol. Chem. 1979;254:3664–3671. [PubMed] [Google Scholar]
- 49.Aslund F., Ehn B., Miranda-Vizuete A., Pueyo C., Holmgren A. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jordan A., Aslund F., Pontis E., Reichard P., Holmgren A. Characterization of Escherichia coli NrdH. A glutaredoxin-like protein with a thioredoxin-like activity profile. J. Biol. Chem. 1997;272:18044–18050. doi: 10.1074/jbc.272.29.18044. [DOI] [PubMed] [Google Scholar]
- 51.Gonzalez Porqué P., Baldesten A., Reichard P. The involvement of the thioredoxin system in the reduction of methionine sulfoxide and sulfate. J. Biol. Chem. 1970;245:2371–2374. [PubMed] [Google Scholar]
- 52.Lillig C.H., Berndt C., Vergnolle O., Lonn M.E., Hudemann C., Bill E., Holmgren A. Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc. Natl. Acad. Sci. 2005;102:8168–8173. doi: 10.1073/pnas.0500735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schütte L.D., Baumeister S., Weis B., Hudemann C., Hanschmann E.-M., Lillig C.H. Identification of potential protein dithiol-disulfide substrates of mammalian Grx2. Biochim. Biophys. Acta. 2013;1830:4999–5005. doi: 10.1016/j.bbagen.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 54.Haunhorst P., Berndt C., Eitner S., Godoy J.R., Lillig C.H. Characterization of the human monothiol glutaredoxin 3 (PICOT) as iron-sulfur protein. Biochem. Biophys. Res. Commun. 2010;394:372–376. doi: 10.1016/j.bbrc.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 55.Haunhorst P., Hanschmann E.-M., Bräutigam L., Stehling O., Hoffmann B., Mühlenhoff U., Lill R., Berndt C., Lillig C.H. Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol. Biol. Cell. 2013;24:1895–1903. doi: 10.1091/mbc.E12-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johansson C., Roos A.K., Montano S.J., Sengupta R., Filippakopoulos P., Guo K., von Delft F., Holmgren A., Oppermann U., Kavanagh K.L. The crystal structure of human GLRX5: iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem. J. 2011;433:303–311. doi: 10.1042/BJ20101286. [DOI] [PubMed] [Google Scholar]
- 57.Camaschella C., Campanella A., De Falco L., Boschetto L., Merlini R., Silvestri L., Levi S., Iolascon A. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110:1353–1358. doi: 10.1182/blood-2007-02-072520. [DOI] [PubMed] [Google Scholar]
- 58.Murakami M., Nakashima K., Kamei D., Masuda S., Ishikawa Y., Ishii T., Ohmiya Y., Watanabe K., Kudo I. Cellular prostaglandin E2 production by membrane-bound prostaglandin E synthase-2 via both cyclooxygenases-1 and -2. J. Biol. Chem. 2003;278:37937–37947. doi: 10.1074/jbc.M305108200. [DOI] [PubMed] [Google Scholar]
- 59.Chiang C.-Y., Pan C.-C., Chang H.-Y., Lai M.-D., Tzai T.-S., Tsai Y.-S., Ling P., Liu H.-S., Lee B.-F., Cheng H.-L. SH3BGRL3 protein as a potential prognostic biomarker for urothelial carcinoma: a novel binding partner of epidermal growth factor receptor. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015;21:5601–5611. doi: 10.1158/1078-0432.CCR-14-3308. [DOI] [PubMed] [Google Scholar]
- 60.Weingarten, L. Identification of novel cytosolic thioredoxin-1 target proteins in mammalian cells by mechanism-based kinetic trapping. Available online: https://archiv.ub.uni-heidelberg.de/volltextserver/8923/(accessed on Sep 5, 2018).
- 61.Urbainsky Claudia, Rolf Nölker, Imber Marcel, Adrian Lübken, Mostertz Jörg, Hochgräfe Falko, Godoy Jose. Eva-maria Hanschmann, and Christopher Horst Lillig nucleoredoxin-dependent targets and processes in neuronal cells. Oxid. Med. Cell. Longev. 2018 doi: 10.1155/2018/4829872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiménez A., Johansson C., Ljung J., Sagemark J., Berndt K.D., Ren B., Tibbelin G., Ladenstein R., Kieselbach T., Holmgren A. Human spermatid-specific thioredoxin-1 (Sptrx-1) is a two-domain protein with oxidizing activity. FEBS Lett. 2002;530:79–84. doi: 10.1016/s0014-5793(02)03417-8. [DOI] [PubMed] [Google Scholar]
- 63.Miranda-Vizuete A., Ljung J., Damdimopoulos A.E., Gustafsson J.A., Oko R., Pelto-Huikko M., Spyrou G. Characterization of Sptrx, a novel member of the thioredoxin family specifically expressed in human spermatozoa. J. Biol. Chem. 2001;276:31567–31574. doi: 10.1074/jbc.M101760200. [DOI] [PubMed] [Google Scholar]
- 64.Sadek C.M., Damdimopoulos A.E., Pelto-Huikko M., Gustafsson J.A., Spyrou G., Miranda-Vizuete A. Sptrx-2, a fusion protein composed of one thioredoxin and three tandemly repeated NDP-kinase domains is expressed in human testis germ cells. Genes Cells Devoted Mol. Cell. Mech. 2001;6:1077–1090. doi: 10.1046/j.1365-2443.2001.00484.x. [DOI] [PubMed] [Google Scholar]
- 65.Sadek C.M., Jiménez A., Damdimopoulos A.E., Kieselbach T., Nord M., Gustafsson J.-A., Spyrou G., Davis E.C., Oko R., van der Hoorn F.A. Characterization of human thioredoxin-like 2. A novel microtubule-binding thioredoxin expressed predominantly in the cilia of lung airway epithelium and spermatid manchette and axoneme. J. Biol. Chem. 2003;278:13133–13142. doi: 10.1074/jbc.M300369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu Y., Zhao X., Luo G., Shen G., Li K., Ren G., Pan Y., Wang X., Fan D. Thioredoxin-like protein 2b facilitates colon cancer cell proliferation and inhibits apoptosis via NF-κB pathway. Cancer Lett. 2015;363:119–126. doi: 10.1016/j.canlet.2014.12.048. [DOI] [PubMed] [Google Scholar]
- 67.Jiménez A., Zu W., Rawe V.Y., Pelto-Huikko M., Flickinger C.J., Sutovsky P., Gustafsson J.-A., Oko R., Miranda-Vizuete A. Spermatocyte/spermatid-specific thioredoxin-3, a novel Golgi apparatus-associated thioredoxin, is a specific marker of aberrant spermatogenesis. J. Biol. Chem. 2004;279:34971–34982. doi: 10.1074/jbc.M404192200. [DOI] [PubMed] [Google Scholar]
- 68.Wang D., De Deken X., Milenkovic M., Song Y., Pirson I., Dumont J.E., Miot F. Identification of a novel partner of duox: EFP1, a thioredoxin-related protein. J. Biol. Chem. 2005;280:3096–3103. doi: 10.1074/jbc.M407709200. [DOI] [PubMed] [Google Scholar]
- 69.Shaheen R., Szymanska K., Basu B., Patel N., Ewida N., Faqeih E., Al Hashem A., Derar N., Alsharif H., Aldahmesh M.A. Characterizing the morbid genome of ciliopathies. Genome Biol. 2016;17:242. doi: 10.1186/s13059-016-1099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harz C., Ludwig N., Lang S., Werner T.V., Galata V., Backes C., Schmitt K., Nickels R., Krause E., Jung M. Secretion and immunogenicity of the meningioma-associated antigen TXNDC16. J. Immunol. Baltim. Md 1950. 2014;193:3146–3154. doi: 10.4049/jimmunol.1303098. [DOI] [PubMed] [Google Scholar]
- 71.Jeong W., Jung Y., Kim H., Park S.J., Rhee S.G. Thioredoxin-related protein 14, a new member of the thioredoxin family with disulfide reductase activity: implication in the redox regulation of TNF-alpha signaling. Free Radic. Biol. Med. 2009;47:1294–1303. doi: 10.1016/j.freeradbiomed.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 72.Pader I., Sengupta R., Cebula M., Xu J., Lundberg J.O., Holmgren A., Johansson K., Arnér E.S.J. Thioredoxin-related protein of 14 kDa is an efficient L-cystine reductase and S-denitrosylase. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6964–6969. doi: 10.1073/pnas.1317320111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thorpe C., Hoober K.L., Raje S., Glynn N.M., Burnside J., Turi G.K., Coppock D.L. Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch. Biochem. Biophys. 2002;405:1–12. doi: 10.1016/s0003-9861(02)00337-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.