Abstract
BACKGROUND
Automated, accurate, objective, and quantitative medical image segmentation has remained a challenging goal in computer science since its inception. This study applies the technique of convolutional neural networks (CNNs) to the task of segmenting carotid arteries to aid in the assessment of pathology.
AIM
To investigate CNN’s utility as an ancillary tool for researchers who require accurate segmentation of carotid vessels.
METHODS
An expert reader delineated vessel wall boundaries on 4422 axial T2-weighted magnetic resonance images of bilateral carotid arteries from 189 subjects with clinically evident atherosclerotic disease. A portion of this dataset was used to train two CNNs (one to segment the vessel lumen and the other to segment the vessel wall) with the remaining portion used to test the algorithm’s efficacy by comparing CNN segmented images with those of an expert reader.
RESULTS
Overall quantitative assessment between automated and manual segmentations was determined by computing the DICE coefficient for each pair of segmented images in the test dataset for each CNN applied. The average DICE coefficient for the test dataset (CNN segmentations compared to expert’s segmentations) was 0.96 for the lumen and 0.87 for the vessel wall. Pearson correlation values and the intra-class correlation coefficient (ICC) were computed for the lumen (Pearson = 0.98, ICC = 0.98) and vessel wall (Pearson = 0.88, ICC = 0.86) segmentations. Bland-Altman plots of area measurements for the CNN and expert readers indicate good agreement with a mean bias of 1%-8%.
CONCLUSION
Although the technique produces reasonable results that are on par with expert human assessments, our application requires human supervision and monitoring to ensure consistent results. We intend to deploy this algorithm as part of a software platform to lessen researchers’ workload to more quickly obtain reliable results.
Keywords: Carotid arteries, Segmentation, Convolutional neural network, Magnetic resonance imaging, Vessel wall
Core tip: Accurate segmentation of carotid arteries is useful in assessing the degree of heart disease in general and vascular diseases (such as atherosclerosis) in particular. Until recently, obtaining accurate segmentation could only be accomplished through the work of an experienced researcher requiring a large investment of time and effort. Over the last several years, the method of convolutional neural networks has demonstrated its efficacy in a number of fields. In this study, we apply this method to magnetic resonance images acquired from subjects with clinically evident atherosclerotic disease and compare the resulting segmentations with those determined by experienced researchers.
INTRODUCTION
Carotid artery disease and stroke have been cited as the leading cause of death in the United States and worldwide[1]. Notably, atherosclerotic disease of the carotid vessels accounts for up to 20% of transient ischemic attacks or ischemic strokes[2]. Ischemic stroke arising from the development of vascular disease and associated plaque formation can cause vessel stenosis resulting in compromised hemodynamics[3]. The degree of the vessel stenosis has been shown to be related the risk of recurrent stroke[4].
Magnetic resonance imaging (MRI), can provide both structural and functional information, including lumen stenosis[5], vessel wall measurements, plaque composition, blood flow velocity, and flow rate[6]. Additionally, MRI has demonstrated the ability to characterize morphological aspects of the carotids such as lumen and wall area[7].
Despite MRI’s capabilities, automated, accurate, objective, and repeatable quantitative analysis of MRI data has proven a challenging goal in computer science for years. In the field of vessel wall imaging, virtually every image-processing algorithm has been applied to the task of accurately and objectively delineating vessel wall boundaries in order to segment the vessels. These techniques include deformable splines[8], active contours[9], active shapes[10], level sets[11], cluster analysis[12], and countless others.
Although many of these techniques worked well in a restricted regime, more complex analysis frequently required combining the approaches to achieve reasonable results. For example, gradient-based ellipse fitting has been combined with fuzzy clustering to delineate carotid artery walls in a semi-automated approach[13]. Similarly, segmentation of arterial vessel walls, both on MR and computed tomography data using a combination of techniques has been proposed. In another study, deformable models were used to segment the aortic lumen while a K nearest neighbor (KNN) classifier was employed to identify the associated thrombus in an aneurysm[14].
Recent advances in convolutional neural networks (CNN)[15,16] have dramatically improved efforts in both the classification of images as well as the segmentation of objects within images themselves. In the field of medical image analysis, the application of CNNs to the task of image segmentation has recently become ubiquitous[17]. Whether it is applied to facilitate the identification of brain regions[18] or to segment skeletal structures[19], the CNN approach has proven to be both general-purpose and remarkably effective. Recently CNNs have been employed in the segmentation of blood vessels in the retina[20], heart[21], as well as in the characterization of plaque in the carotid vessels[22,23].
This study applies the technique of CNNs to the task of segmenting carotid arteries in order to aid in the assessment of pathology. Accumulated plaque in carotid arteries has been established as a reliable marker of underlying cardiovascular disease. Intima media thickness (IMT) measured by ultrasound has been shown to be associated with future cardiovascular events[24]. Previous studies have also shown a strong correlative relationship between wall thickness and wall area measures using dark blood MRI and ultrasound based IMT[25]. Studies in the MESA cohort have also established relationships between wall morphology measurements by MRI and future cardiovascular events[26].
We anticipate incorporating CNN technology into a software platform designed to facilitate the analysis of carotid images thereby streamlining what was once an onerous time-consuming task into something more manageable.
MATERIALS AND METHODS
Images analyzed in this study were culled from a prior study[27] in which 189 subjects (ranging in age from 18 to 74 years) with clinically evident atherosclerotic disease underwent MRI of the carotid arteries. The subjects from that study presented with either type 2 diabetes mellitus or impaired glucose tolerance were randomized to receive placebo (n = 94) or canakinumab 150 mg monthly (n = 95) for 12 mo. Imaging was performed at multiple sites using a 3.0-T whole-body MRI scanner, including Trio, TIM Trio, or Verio (Siemens Medical SolutionsUSA, Inc., Malvern, PA, United States) or Achieva (Philips, Amsterdam, the Netherlands) platforms. The local Institutional Review Board approved both studies with all subjects providing informed consent.
A range of 6 to 13 high-resolution axial ECG-gated T2-weighted MR images of bilateral carotid arteries were acquired using a 4-channel carotid array (MachnetB.V., Roden, Netherlands) on Siemens scanners and an equivalent multi-channel (4 to 8) phased array carotid coil (Shanghai Medical, Shanghai, China) was used on the Philips scanners. In plane pixel resolution for the acquired images ranged from 0.52 mm to 0.7 mm with a slice thickness of 3 mm. These images were subsequently analyzed by an expert reader who manually segmented individual images of the carotid wall using VesselMass (LKEB, the Division of Image Processing, Department of Radiology, the Leiden University Medical Center, The Netherlands) resulting in a dataset of 4422 segmented images. Based on this manual segmentation, metrics describing the carotid vessel were generated and recorded thereby establishing a standard against which the efficacy of automated analysis could be judged.
In preparation for automated segmentation, the original dataset was divided into 3 groups: A “training dataset” (3581 images), a “validation dataset” (398 images), and a “test dataset” (443 images). Each dataset was composed of anatomical images and their associated manually segmented images. While the training dataset was used to dynamically adjust the CNN’s parameters during training, the validation dataset was used only to evaluate model performance during training and avoid over-fitting. These datasets were used to train two separate CNNs (one for carotid lumen segmentation and the other for carotid wall segmentation).
To prepare the CNNs for training, each manually segmented image was identified and the information describing that image’s segmented vessel was converted from VesselMass contour information to a binary mask image using a custom Matlab (Mathworks, Natick, MA, United States) script. Additionally, each image was cropped to a fixed size (64 × 64 pixel) to center on the carotid vessel with the luminance adjusted to maximize brightness and contrast. The order of images was randomly shuffled before they were submitted to the CNN for training.
For this study, we used a segmentation model developed for spinal cord gray matter segmentation by Perone[28]. The CNN trained to segment the carotid vessel wall was compiled to use an Adam optimizer, a learning rate of 0.001, with the DICE metric[29] selected as the loss function to be minimized. The network was trained with a batch size of 32 for 1000 epochs. The data was hosted on an AMD Ryzen7 3.6GHz computer although the actual neural network training ran on the computer’s Nvidia GeForce 1080 Ti GPU card. Training software was implemented using Tensorflow version 1.10 and Keras version 2.2.2. The CNN trained to segment the carotid vessel’s lumen was configured similarly but required far less training and converged in 100 epochs.
Once the CNNs had been trained, the images from the test dataset were processed to produce segmentations as binary images. The resulting binary images were then compared with expert segmentations to determine the overall efficacy of the approach. In order to compute traditional vessel metrics on the binary images, some minor post-processing (such as removing isolated pixels identified as noise) was necessary. Vessel wall metrics of the CNN’s test dataset were compared with those independently produced by expert reader to determine acceptability.
RESULTS
The DICE loss metric for the CNN used to segment the carotid’s wall decreased steadily during training with a final score of 0.85 for the training dataset and 0.87 for the validation dataset. The loss metric for the CNN used to segment the carotid’s lumen decreased very quickly with a final DICE score of 0.96 for both the training and the validation dataset. Following the training phase, the network used the computed weights to process 398 images of the test dataset.
Initial qualitative evaluation of the images produced by the CNN when applied to the test dataset was very good although technical issues stemming from some images with extremely narrow carotid walls affected the analysis. Visual inspection suggested that the majority of segmentations achieved by automated means closely approximated those that were segmented manually, with many appearing almost indistinguishable.
Overall quantitative assessment between automated and manual segmentations was determined by computing the DICE coefficient for each pair of segmented images in the test dataset (Figure 1). Applying the CNN trained to segment the carotid vessel wall to the test dataset resulted in segmented images with an average DICE coefficient of 0.87. Applying the CNN trained to segment the carotid lumen to the test dataset resulted in segmented images with an average DICE coefficient of 0.96.
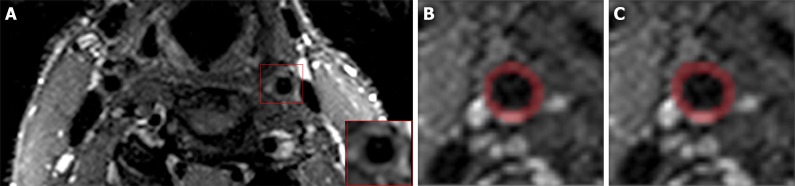
Figure 1.
Representative axial T2-weighted magnetic resonance image of bilateral carotid arteries with cropped vessel area highlighted (A). Carotid vessel wall segmentations using convolutional neural networks (B) and expert reader (C). DICE = 0.91.
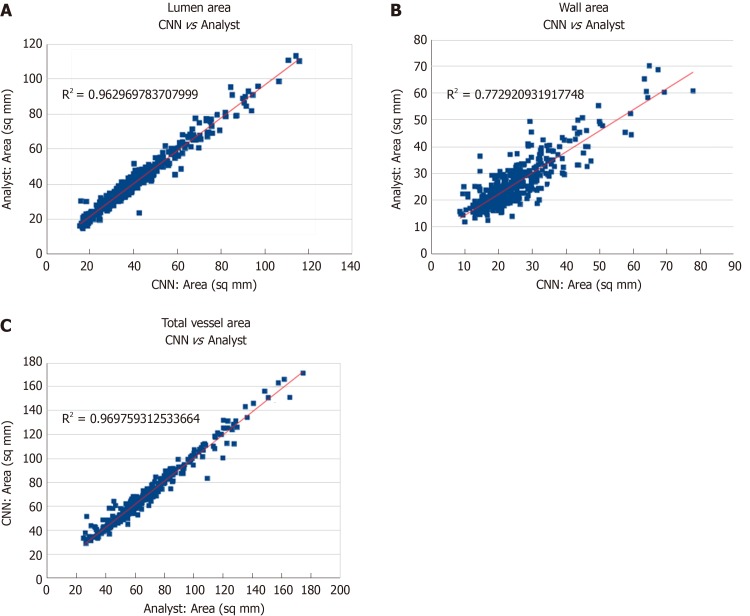
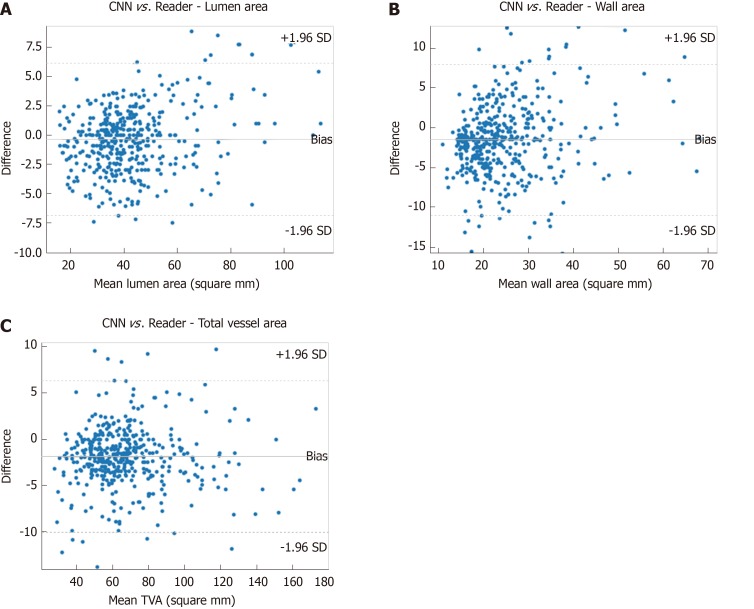
Pearson correlation values as well as intra-class correlation coefficients (ICC) were computed for vessel area metrics as determined for the expert reader and the CNN to assess the agreement of measurements. Excellent agreement was observed in the segmentation of lumen area (Pearson correlation = 0.98, ICC = 0.98) as well as in the segmentation of vessel wall area (Pearson correlation = 0.88, ICC = 0.86). Scatter plots of these areas comparing the expert reader’s measurements against the CNN measurements are shown in Figure 2. Additionally, Bland-Altman plots of these measurements (Figure 3) for the CNN and reader indicate good agreement and support earlier findings. Results from the CNN compared against the experienced reader using Bland Altman analysis and limits of agreement were comparable to the inter-observer variability observed in the manual approach[30]. Although measurements of vessel parameters can be problematic (and will be addressed in the Discussion section), the DICE coefficient is useful as an overall indicator of segmentation similarity.
Figure 2.
Comparison of measurements between convolutional neural network and an expert reader for lumen area (A), wall area (B), and total vessel area (C). CNN: Convolutional neural network.
Figure 3.
Bland-Altman plots demonstrating agreement of convolutional neural networks and expert reader in assessing lumen area (A), wall area (B), and total vessel area (C). CNN: Convolutional neural network.
DISCUSSION
CNNs have proven to be a remarkable advance in the field of image processing that has radically changed how we approach problems of image classification and object segmentation. CNN’s ability to derive salient features from existing training data has arguably obviated the need for the development of descriptors based on theoretical requirements.
In this study, a CNN was able to accurately identify the carotid vessel, its lumen, and make reasonable determinations of the vessel’s wall area. In this section, we will examine how data is prepared for CNN training, the training process itself, as well as the post-processing and subsequent assessment of efficacy.
The initial pre-processing step required that manually generated contour data be prepared for CNN training by converting X-Y data points to generate binary mask images of the carotid wall. This seemingly straightforward step may however become difficult when pixels on the edge of a region are only partially covered by the contours set by X-Y vertices. We made use of Matlab’s “poly2mask” function for this purpose. Although the function works well in most instances, it can sometimes generate problematic mask images such as non-toroidal carotid walls or vessels with unrealistically thin walls. With higher resolution images or with larger physiological structures this may not be an issue but in our case, it is a real concern.
While an analysis of the intricacies of the CNN training phase is beyond the scope of this discussion, it is important to note the plethora of CNN segmentation models currently available[31]. Furthermore, every model allows for the selection of many “hyper-parameters” such as epochs, batch size, dropout, and learning rate. Owing to the complexity of these models, setting these parameters is essentially an empirical process and cannot be determined a priori.
Originally, we trained a single model to segment the vessel wall and attempted to label the resulting images using conventional image processing techniques. This ostensibly simple task of identifying lumen, vessel wall, and background regions was potentially complicated as the segmented vessel wall border could narrow to a single pixel and make region identification based on pixel connectivity challenging. It quickly became apparent that training a separate CNN to segment the vessel lumen would result in a less ambiguous result. In addition, the typically high contrast between the lumen and surrounding tissue allowed the CNN to converge extremely quickly with very high accuracy.
In the interest of simplicity, we directed each CNN to segment the region of interest (either vessel wall or lumen). In that same spirit, we minimized experimentation to allow for reasonable processing time and adapt to memory constraints imposed by our hardware. Following the training phase, test data that had been set aside was processed by the CNN to generate segmented images. Post processing of the resultant images was limited to removing pixel “noise” that established isolated regions too small to be of significance for our purpose.
Our initial cursory side-by-side examination of manual and post-processed CNN image segmentations was encouraging. Our subjective impression was subsequently borne out by comparing traditional area measures of the vessel wall, lumen, and total vessel area for both the CNN and manual methods. As these measures may not convey morphological differences between the two segmentations, we also calculated the DICE coefficient for corresponding images from these datasets. It should be noted that although the DICE metric objectively describes the degree of overlap of two segmentations, a relatively low score could sometimes be misleading. Alternatively, calculation of traditional vessel metrics was complicated by the fact that the neural network approach is a pixel-based technique that may result in obvious erroneous results despite their high DICE score. For example, the neural network may produce an image with pixels sparsely distributed making identification of vessel structures problematic.
Ultimately, establishing the utility of CNNs must be assessed not in absolute terms, but in comparing its performance to that of manual analysis carried out by experts. In essence, a CNN must be shown to be accurate to within limits established by expert readers and be judged by that same criteria.
In some respects, the effectiveness of this technology depends on its intended use. For example, we initially applied the technique to images with a much larger field of view and a trained network was able to both identify and segment the vessel wall but with occasional failures. Most failures occurred when the CNN incorrectly identified the vessel of interest but largely succeeded in delineating vessel wall boundaries for that incorrect vessel. For our purposes, it was decided to focus less on vessel identification and concentrate on aspects of the vessel itself. As a practical matter, this shifts the burden of identifying the vessel to a user but potentially just for the initial image in a series. Consequently, we intend to implement this approach into a workflow whereby a researcher first identifies a vessel of interest in its initial presentation in an axial view. The researcher will then establish a bounding box of the vessel in that image and specify the number of axial images to process. Alternatively, these parameters could be established as a function of the subject’s physical dimensions and adjusted for established physiological landmarks. Once these parameters have been set, the CNN will proceed to process each image in turn with the segmentation of one image's vessel providing the bounding box for its neighboring slice in the series.
In conclusion, in this study, we have demonstrated the effectiveness of CNN technology in its application to the task of delineating carotid vessel walls thereby facilitating the detection of potential pathology. Although the technique produces reasonable results that are on par with expert human assessments, in our application it requires human supervision and monitoring to ensure consistent results. We intend to deploy this algorithm as part of a software platform to lessen researchers workload to more quickly obtain reliable results.
ARTICLE HIGHLIGHTS
Research background
Segmentation of arterial vessels is an important step is the assessment of vascular disease. For many years, the accepted method of producing segmentations was through manual approach performed by expert researchers. We apply the technique of convolutional neural networks (CNNs) to the task of segmentation of carotid arteries and compare the results to the manual method.
Research motivation
The accepted standard of manual segmentation by expert researchers is an onerous and time-consuming task that is inherently subjective. Consequently, constructing an algorithm from such an opaque process is problematic. Creation and adoption of a reliable segmentation algorithm could lead to significant savings through automation.
Research objectives
The objective in this study was to examine the feasibility of applying CNNs to the task of segmenting carotid arteries of subjects with vascular disease.
Research methods
Subsets of magnetic resonance images of the carotid arteries of 189 subjects with atherosclerotic disease were used to train and subsequently validate the CNN. Image segmentations used to train the CNN were produced by an expert reader who manually segmented individual images of the carotid wall using conventional means resulting in a dataset of 4422 segmented images. In preparation for automated segmentation, the original dataset was divided into 3 groups: A “training dataset” (3581 images), a “validation dataset” (398 images), and a “test dataset” (443 images). These datasets were used to train two separate segmentation CNNs (one for carotid lumen and the other for carotid wall). After training, images from the test dataset were processed to produce segmentations as binary images.
Research results
Overall quantitative assessment between manual and automated segmentations was determined by computing the DICE coefficient for each pair of segmented images in the test dataset. The average DICE coefficient between automated and manual segmentations was 0.87 for the carotid vessel wall and 0.96 for the carotid lumen. Intra-class correlation coefficients (ICC) as well as Pearson correlation values were computed for vessel area metrics as determined for the expert reader and the CNN to assess the agreement of measurements. Excellent agreement was observed in the segmentation of lumen area (Pearson correlation = 0.98, ICC = 0.98) as well as in the segmentation of vessel wall area (Pearson correlation = 0.88, ICC = 0.86). Additionally, Bland-Altman plots of these measurements for the CNN and reader indicate good agreement.
Research conclusions
In this study, we have demonstrated the effectiveness of CNN technology in its application to the task of delineating carotid vessel walls thereby facilitating the detection of potential pathology.
Research perspectives
Although the technique produces reasonable results that are on par with expert human assessments, in our application it requires human supervision and monitoring to ensure consistent results. We intend to deploy this algorithm as part of a software platform to lessen researchers workload to more quickly obtain reliable results.
Footnotes
Institutional review board statement: The study was approved by the Institutional Review Board of the Icahn School of Medicine at Mount Sinai.
Informed consent statement: Waiver of institutional review board (IRB) approval was obtained from the IRB as only deidentified data was used in this study. The images analyzed for this study were anonymized and devoid of any Protected Health Information.
Conflict-of-interest statement: No conflicts to disclose.
STROBE statement: The authors have read the STROBE Statement-checklist of items, and the manuscript was prepared and revised according to the STROBE Statement-checklist of items.
Manuscript source: Unsolicited manuscript
Peer-review started: July 21, 2019
First decision: September 21, 2019
Article in press: November 20, 2019
Specialty type: Radiology, nuclear medicine and medical imaging
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Qi XS, Tajiri K S-Editor: Yan JP L-Editor: A E-Editor: Li X
Contributor Information
Daniel D Samber, Translational and Molecular Imaging Institute (TMII), Icahn School of Medicine at Mount Sinai, New York, NY 10029, United States. daniel.samber@mssm.edu.
Sarayu Ramachandran, Translational and Molecular Imaging Institute (TMII), Icahn School of Medicine at Mount Sinai, New York, NY 10029, United States.
Anoop Sahota, Translational and Molecular Imaging Institute (TMII), Icahn School of Medicine at Mount Sinai, New York, NY 10029, United States.
Sonum Naidu, Translational and Molecular Imaging Institute (TMII), Icahn School of Medicine at Mount Sinai, New York, NY 10029, United States.
Alison Pruzan, Translational and Molecular Imaging Institute (TMII), Icahn School of Medicine at Mount Sinai, New York, NY 10029, United States.
Zahi A Fayad, Translational and Molecular Imaging Institute (TMII), Icahn School of Medicine at Mount Sinai, New York, NY 10029, United States.
Venkatesh Mani, Translational and Molecular Imaging Institute (TMII), Icahn School of Medicine at Mount Sinai, New York, NY 10029, United States.
Data sharing statement
Once published and after appropriate safeguard to ensure that the data is devoid of any identifiers, the data used for the analysis for this study will be shared on the Mount Sinai data sharing portal according to Institutional guidelines.
References
- 1.Writing Group Members, Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jiménez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB; American Heart Association Statistics Committee; Stroke Statistics Subcommittee. Executive Summary: Heart Disease and Stroke Statistics--2016 Update: A Report From the American Heart Association. Circulation. 2016;133:447–454. [Google Scholar]
- 2.Fairhead JF, Rothwell PM. The need for urgency in identification and treatment of symptomatic carotid stenosis is already established. Cerebrovasc Dis. 2005;19:355–358. doi: 10.1159/000085201. [DOI] [PubMed] [Google Scholar]
- 3.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 4.North American Symptomatic Carotid Endarterectomy Trial Collaborators, Barnett HJM, Taylor DW, Haynes RB, Sackett DL, Peerless SJ, Ferguson GG, Fox AJ, Rankin RN, Hachinski VC, Wiebers DO, Eliasziw M. Beneficial effect of carotid endarterectomy in symptomatic patients with high-grade carotid stenosis. N Engl J Med. 1991;325:445–453. doi: 10.1056/NEJM199108153250701. [DOI] [PubMed] [Google Scholar]
- 5.Nederkoorn PJ, van der Graaf Y, Hunink MG. Duplex ultrasound and magnetic resonance angiography compared with digital subtraction angiography in carotid artery stenosis: a systematic review. Stroke. 2003;34:1324–1332. doi: 10.1161/01.STR.0000068367.08991.A2. [DOI] [PubMed] [Google Scholar]
- 6.Markl M, Schnell S, Wu C, Bollache E, Jarvis K, Barker AJ, Robinson JD, Rigsby CK. Advanced flow MRI: emerging techniques and applications. Clin Radiol. 2016;71:779–795. doi: 10.1016/j.crad.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan C, Beach KW, Smith LH, Jr, Hatsukami TS. Measurement of atherosclerotic carotid plaque size in vivo using high resolution magnetic resonance imaging. Circulation. 1998;98:2666–2671. doi: 10.1161/01.cir.98.24.2666. [DOI] [PubMed] [Google Scholar]
- 8.Klein AK, Lee F, Amini AA. Quantitative coronary angiography with deformable spline models. IEEE Trans Med Imaging. 1997;16:468–482. doi: 10.1109/42.640737. [DOI] [PubMed] [Google Scholar]
- 9.Yan P, Kassim AA. Segmentation of volumetric MRA images by using capillary active contour. Med Image Anal. 2006;10:317–329. doi: 10.1016/j.media.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 10.de Bruijne M, van Ginneken B, Viergever MA, Niessen WJ. Interactive segmentation of abdominal aortic aneurysms in CTA images. Med Image Anal. 2004;8:127–138. doi: 10.1016/j.media.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Manniesing R, Velthuis BK, van Leeuwen MS, van der Schaaf IC, van Laar PJ, Niessen WJ. Level set based cerebral vasculature segmentation and diameter quantification in CT angiography. Med Image Anal. 2006;10:200–214. doi: 10.1016/j.media.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Itskovich VV, Samber DD, Mani V, Aguinaldo JG, Fallon JT, Tang CY, Fuster V, Fayad ZA. Quantification of human atherosclerotic plaques using spatially enhanced cluster analysis of multicontrast-weighted magnetic resonance images. Magn Reson Med. 2004;52:515–523. doi: 10.1002/mrm.20154. [DOI] [PubMed] [Google Scholar]
- 13.Adame IM, van der Geest RJ, Wasserman BA, Mohamed MA, Reiber JH, Lelieveldt BP. Automatic segmentation and plaque characterization in atherosclerotic carotid artery MR images. MAGMA. 2004;16:227–234. doi: 10.1007/s10334-003-0030-8. [DOI] [PubMed] [Google Scholar]
- 14.Olabarriaga SD, Rouet JM, Fradkin M, Breeuwer M, Niessen WJ. Segmentation of thrombus in abdominal aortic aneurysms from CTA with nonparametric statistical grey level appearance modeling. IEEE Trans Med Imaging. 2005;24:477–485. doi: 10.1109/tmi.2004.843260. [DOI] [PubMed] [Google Scholar]
- 15.Fukushima K. Neocognitron: a self organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern. 1980;36:193–202. doi: 10.1007/BF00344251. [DOI] [PubMed] [Google Scholar]
- 16.Lecun Y, Bengio Y. Convolutional Networks for Images, Speech, and Time Series. In: Arbib MA, editor. The handbook of brain theory and neural networks. 1st ed. In: Arbib MA, editor. Cambridge, MA: MIT Press; 1998. pp. 255–258. [Google Scholar]
- 17.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 18.de Brebisson A, Montana G. Deep neural networks for anatomical brain segmentation. 2015 Preprint. Available from: https://arxiv.org/abs/1502.02445. [Google Scholar]
- 19.Cernazanu-Glavan C, Holban S. Segmentation of bone structure in x-ray images using convolutional neural network. Adv Electr Comput En. 2013;13:87–94. [Google Scholar]
- 20.Liskowski P, Krawiec K. Segmenting Retinal Blood Vessels With Deep Neural Networks. IEEE Trans Med Imaging. 2016;35:2369–2380. doi: 10.1109/TMI.2016.2546227. [DOI] [PubMed] [Google Scholar]
- 21.López-Linares K, Aranjuelo N, Kabongo L, Maclair G, Lete N, Ceresa M, García-Familiar A, Macía I, González Ballester MA. Fully automatic detection and segmentation of abdominal aortic thrombus in post-operative CTA images using Deep Convolutional Neural Networks. Med Image Anal. 2018;46:202–214. doi: 10.1016/j.media.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Lekadir K, Galimzianova A, Betriu A, Del Mar Vila M, Igual L, Rubin DL, Fernandez E, Radeva P, Napel S. A Convolutional Neural Network for Automatic Characterization of Plaque Composition in Carotid Ultrasound. IEEE J Biomed Health Inform. 2017;21:48–55. doi: 10.1109/JBHI.2016.2631401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudha S, Jayanthi KB, Ramalingan C, Madian N, Sunder T. Convolutional Neural Network for Segmentation and Measurement of Intima Media Thickness. J Med Syst. 2018;42:154. doi: 10.1007/s10916-018-1001-y. [DOI] [PubMed] [Google Scholar]
- 24.Lorenz MW, Gao L, Ziegelbauer K, Norata GD, Empana JP, Schmidtmann I, Lin HJ, McLachlan S, Bokemark L, Ronkainen K, Amato M, Schminke U, Srinivasan SR, Lind L, Okazaki S, Stehouwer CDA, Willeit P, Polak JF, Steinmetz H, Sander D, Poppert H, Desvarieux M, Ikram MA, Johnsen SH, Staub D, Sirtori CR, Iglseder B, Beloqui O, Engström G, Friera A, Rozza F, Xie W, Parraga G, Grigore L, Plichart M, Blankenberg S, Su TC, Schmidt C, Tuomainen TP, Veglia F, Völzke H, Nijpels G, Willeit J, Sacco RL, Franco OH, Uthoff H, Hedblad B, Suarez C, Izzo R, Zhao D, Wannarong T, Catapano A, Ducimetiere P, Espinola-Klein C, Chien KL, Price JF, Bergström G, Kauhanen J, Tremoli E, Dörr M, Berenson G, Kitagawa K, Dekker JM, Kiechl S, Sitzer M, Bickel H, Rundek T, Hofman A, Mathiesen EB, Castelnuovo S, Landecho MF, Rosvall M, Gabriel R, de Luca N, Liu J, Baldassarre D, Kavousi M, de Groot E, Bots ML, Yanez DN, Thompson SG PROG-IMT study group. Predictive value for cardiovascular events of common carotid intima media thickness and its rate of change in individuals at high cardiovascular risk - Results from the PROG-IMT collaboration. PLoS One. 2018;13:e0191172. doi: 10.1371/journal.pone.0191172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mani V, Aguiar SH, Itskovich VV, Weinshelbaum KB, Postley JE, Wasenda EJ, Aguinaldo JG, Samber DD, Fayad ZA. Carotid black blood MRI burden of atherosclerotic disease assessment correlates with ultrasound intima-media thickness. J Cardiovasc Magn Reson. 2006;8:529–534. doi: 10.1080/10976640600675245. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Guallar E, Malhotra S, Astor BC, Polak JF, Qiao Y, Gomes AS, Herrington DM, Sharrett AR, Bluemke DA, Wasserman BA. Carotid Artery Wall Thickness and Incident Cardiovascular Events: A Comparison between US and MRI in the Multi-Ethnic Study of Atherosclerosis (MESA) Radiology. 2018;289:649–657. doi: 10.1148/radiol.2018173069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhury RP, Birks JS, Mani V, Biasiolli L, Robson MD, L'Allier PL, Gingras MA, Alie N, McLaughlin MA, Basson CT, Schecter AD, Svensson EC, Zhang Y, Yates D, Tardif JC, Fayad ZA. Arterial Effects of Canakinumab in Patients With Atherosclerosis and Type 2 Diabetes or Glucose Intolerance. J Am Coll Cardiol. 2016;68:1769–1780. doi: 10.1016/j.jacc.2016.07.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perone CS, Calabrese E, Cohen-Adad J. Spinal cord gray matter segmentation using deep dilated convolutions. Sci Rep. 2018;8:5966. doi: 10.1038/s41598-018-24304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 30.El Aidi H, Mani V, Weinshelbaum KB, Aguiar SH, Taniguchi H, Postley JE, Samber DD, Cohen EI, Stern J, van der Geest RJ, Reiber JH, Woodward M, Fuster V, Gidding SS, Fayad ZA. Cross-sectional, prospective study of MRI reproducibility in the assessment of plaque burden of the carotid arteries and aorta. Nat Clin Pract Cardiovasc Med. 2009;6:219–228. doi: 10.1038/ncpcardio1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Garcia A, Orts-Escolano S, Oprea S, Villena-Martinez V, Garcia-Rodriguez J. A Review on Deep Learning Techniques Applied to Semantic Segmentation. 2017 Preprint. Available from: https://arxiv.org/abs/1704.06857. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Once published and after appropriate safeguard to ensure that the data is devoid of any identifiers, the data used for the analysis for this study will be shared on the Mount Sinai data sharing portal according to Institutional guidelines.