Abstract
Background and Objectives
Metallic antimicrobial materials are of growing interest due to their potential to control pathogenic and multidrug-resistant bacteria. Yet we do not know if utilizing these materials can lead to genetic adaptations that produce even more dangerous bacterial varieties.
Methodology
Here we utilize experimental evolution to produce strains of Escherichia coli K-12 MG1655 resistant to, the iron analog, gallium nitrate (Ga(NO3)3). Whole genome sequencing was utilized to determine genomic changes associated with gallium resistance. Computational modeling was utilized to propose potential molecular mechanisms of resistance.
Results
By day 10 of evolution, increased gallium resistance was evident in populations cultured in medium containing a sublethal concentration of gallium. Furthermore, these populations showed increased resistance to ionic silver and iron (III), but not iron (II) and no increase in traditional antibiotic resistance compared with controls and the ancestral strain. In contrast, the control populations showed increased resistance to rifampicin relative to the gallium-resistant and ancestral population. Genomic analysis identified hard selective sweeps of mutations in several genes in the gallium (III)-resistant lines including: fecA (iron citrate outer membrane transporter), insl1 (IS30 tranposase) one intergenic mutations arsC →/→ yhiS; (arsenate reductase/pseudogene) and in one pseudogene yedN ←; (iapH/yopM family). Two additional significant intergenic polymorphisms were found at frequencies > 0.500 in fepD ←/→ entS (iron-enterobactin transporter subunit/enterobactin exporter, iron-regulated) and yfgF ←/→ yfgG (cyclic-di-GMP phosphodiesterase, anaerobic/uncharacterized protein). The control populations displayed mutations in the rpoB gene, a gene associated with rifampicin resistance.
Conclusions
This study corroborates recent results observed in experiments utilizing pathogenic Pseudomonas strains that also showed that Gram-negative bacteria can rapidly evolve resistance to an atom that mimics an essential micronutrient and shows the pleiotropic consequences associated with this adaptation.
Lay summary
We utilize experimental evolution to produce strains of Escherichia coli K-12 MG1655 resistant to, the iron analog, gallium nitrate (Ga(NO3)3). Whole genome sequencing was utilized to determine genomic changes associated with gallium resistance. Computational modeling was utilized to propose potential molecular mechanisms of resistance.
Keywords: experimental evolution, gallium, Escherichia coli, genomics
INTRODUCTION
The spread of resistance to traditional antibiotics has spurred the search for new antimicrobial substances [1–3]. Ionic and nanoparticle metals have been proposed, including silver, copper, excess iron (II, III) and gallium. Unfortunately, many of these studies were conducted by material scientists and engineers who had little understanding of the evolutionary dynamics of populations exposed to toxic materials [4, 5]. These studies claimed the fact that metals impacted so many different aspects of bacterial physiology, that they represented steep obstacles to the evolution of resistance [4].
Recently several studies have examined the impact of the iron analog gallium upon the growth of pathogenic bacteria [6]. The mechanism targeted by this approach was to use gallium (Ga3+) to saturate bacterial siderophores that normally take up ferric iron (Fe3+). Studies on the impact of gallium suggest that it may impede growth without killing cells (bacteriostatic) or eventually kill cells via prolonged iron starvation (bactericidal) [6]. Gallium’s (Ga3+) capacity to quench bacterial siderophores is due to this element’s similarity with ionic radius and binding properties of iron (III, Fe3+) [6]. Gallium is a group IIIA metal with atomic number 31 and shares certain properties with Fe3+, where the octahedral ionic radius of Ga3+ is 0.620 Å compared with 0.645 Å for Fe3+ and the tetrahedral ionic radius is 0.47 Å for Ga3+ compared with 0.49 Å for Fe3+ [7].
It is known that iron (as well as other micronutrient metals Mg, Mn, S and Zn) play key roles in the capacity for pathogenic bacteria to maintain their virulence. These metals often serve as enzymatic co-factors. Therefore, their intracellular concentration must be maintained and tightly regulated to maintain cell viability [8]. Iron is one of the most important micronutrients as it fulfills many biological roles [9]. Iron-containing proteins (heme-proteins, iron-sulfur cluster proteins and di-iron and mononuclear enzymes) play roles in nitrogen fixation and metabolism and serve as electron carriers for respiration [10]. Despite its critical role in bacterial metabolism, acquiring iron is one of the greatest challenges for bacterial growth [8, 11] and iron deficiency is one of the most common nutritional stresses [12]. Despite iron being an essential metal, it can also be extremely toxic under aerobic conditions [13].
In aerobic environments, iron predominantly occurs as ferric iron (Fe3+), however, Fe(OH)3 is poorly soluble in aqueous solution (as low as 10−18 M at pH 7.0). Under anaerobic conditions, the equilibrium shifts to ferrous iron (Fe2+) that is more readily bioavailable to microorganisms and availability is a key to pathogenesis for a variety of microbes, thus many innate immunity mechanisms utilize iron sequestration (e.g. serum albumin, calprotectin [14, 15]). As iron is essential, microbes have evolved means to take it up from their extracellular surroundings; for example, siderophores. The siderophore, enterobactin, in Escherichia coli is synthesized via genes such as entC and fep genes (in strain K-12: A, B, C, D, E and G). These siderophores are tightly controlled by the global iron homeostasis regulator, Fur [8]. Under aerobic conditions, excess iron has been shown to induce oxidative damage, whereas both Fe2+ and Fe3+ can interact with hydrogen peroxide and superoxides, respectively, which generate highly reactive hydroxyl radicals leading to cell damage and eventual cell death [13]. This is in addition to a variety of other effects summarized in Table 1. When oxygen free radicals form in their cells, bacteria have genes to defend themselves from the accrued stressors. This is aided by many proteins including OxyR that responds to the presence of hydrogen peroxide, SoxS and SoxR that respond to redox active compounds and RpoS that responds to general oxidative stress [17–19]. It is therefore important that intracellular iron be regulated to prevent toxicity by this essential metal [20, 21].
Table 1.
Mechanisms of excess iron and gallium toxicity
| Mechanism | Fe | Ga |
|---|---|---|
| Reactive oxygen species | + | + |
| Disruption of transcription/translation | + | − |
| Damage to cell wall/membrane | + | + |
| Interfering with respiration | + | + |
| Release of cellular components | + | + |
| Binding to thiol groups | ? | ? |
| Change in zinc homeostasis | − | + |
| Disruption of iron metabolism | − | + |
We have recently shown that E. coli can evolve resistance to excess iron (II and III) [22, M. D. Thomas, J. L. Graves, A. J. Ewunkem et al. in preparation]. Furthermore, an unexpected result of excess iron resistance in these strains was resistance to gallium which is an iron analog possessing the same nuclear radius as iron (Fe3+). For this reason, gallium has been shown to exchange for iron in siderophores such as in Pseudomonas aeruginosa where the siderophore pyochelin transports gallium into the cell [23], it also represses pvdS, a transcriptional regulator of the siderophore pyoverdine [6]. Once inside the cell gallium cannot be used for essential iron catalyzed reactions and is therefore inhibits growth (bacteriostatic) due to mismetallation. In addition, gallium has been shown to perturb the regulation of iron acquisition pathways and can eventually cause cell death [6].
Until recently, experiments showing the evolution of gallium resistance were limited to Pseudomonas [24–26]. However, a recent study utilized the Keio collection which consists of 3985 single, non-essential gene knockouts in E. coli BW25113 and examined the effect on gallium resistance [27, 28]. Each mutant was scored as to whether it increased or decreased gallium sensitivity on a scale of −0.6 to +0.6 normalized to colony size. The mutants were classified by their impact on cellular metabolism (transcription, translation, DNA metabolism, RNA metabolism, protein metabolism, protein folding and secretion, cell exterior functions, biosynthetic functions, degradation, energy, cellular processes, response to stimulus and other pathways). Of the 3945 mutations studied, they found a relatively equal distribution of mutations that raised or lowered gallium sensitivity (1761 raised and 1878 lowered). However, the vast majority of these effects on colony size were minor. These sorts of studies are limited in their capacity to predict how bacteria might actually evolve resistance to gallium or any stressing material, as the gene knockouts were of non-essential functions. Furthermore, knocking out a gene may produce unanticipated pleiotropic effects on fitness that are hard to interpret in any given environment.
Therefore, in this study we utilize experimental evolution on E. coli K-12 MG1655 to ask whether and how does E. coli evolve resistance to excess gallium. In addition, since we have already shown that this strain can evolve excess iron (II, III) resistance, we wanted to know whether gallium resistant E. coli would also have a correlated response that confers iron (II, III) resistance. Furthermore, we asked whether gallium resistance confers resistance to metals that may not have similar bacteriostatic/bactericidal effects to gallium such as silver (Table 1). We also examined the genomic changes, that allow E. coli to survive in environments containing excess gallium and compared those with the knockout results of Gugala et al. [28]. Finally, we modeled the structural changes in one of the proteins altered by selection to propose a mechanism by which this mutation conferred gallium resistance.
METHODOLOGY
Evolution experiment
Experimental evolution is defined as the study of populations over defined and reproducible conditions over multiple generations under laboratory or natural conditions [29]. It has been used to study adaptation in a variety of organisms and conditions, including the study of antimicrobial resistance via antibiotics (ampicillin, beta lactams, chloramphenicol and others), as well as metals [30–34]. Other methods have been used, such as step-wise selection to isolate metal resistant mutants in E. coli [35]. However, the underlying principle of artificial selection is the same as that employed in experimental evolution. However, our method allows the study of adaptation to increase antimicrobial metal concentration over time, which does not result from the step-wise selection protocol.
Escherichia coli K-12 MG1655 (ATCC #47076) was chosen for this study due to the paucity of known metal resistant loci in this bacterium. This is also the strain we have used in our previous studies evaluating the evolution of silver and iron resistance [21, 22, 33, 34]. Its chromosome has 4 641 652 nucleotides (GenBank: NC_000913.3) and contains no plasmids. We have sequenced this strain and polymorphisms not present in the published reference genome are given in in previous studies [33, 34].
In our experiments, E. coli was cultured in Davis Minimal Broth (DMB; Difco, Sparks, MD) with 1 g of dextrose per liter (Fisher Scientific, Fair Lawn, NJ) carbon source. The medium is enriched with thiamine hydrochloride with 10 µl in the final volume of 10 ml, for a concentration of 0.3 µM. This is kept in 50-ml Erlenmeyer flasks and placed in a shaking incubator at 115 rpm at 37°C. Each replicate was founded from a unique colony on an agar plate isolated via serial dilution from the ancestral stock culture. These samples were placed in separate flasks and allowed to grow for 24 hours before being transferred to fresh media. Five replicates of the controls (C1–C5) which are grown in standard DMB medium without the addition of gallium and five replicates of the Ga(NO3)3 selected (Ga3+_1–Ga3+_5) were founded and propagated by subculturing 0.1 ml into 9.9 ml of fresh sterile DMB daily. The population density increased from ∼107 to ∼109 cells per ml over the course of each daily transfer cycle. Replication of selection treatments is important because it allows for the observation of parallelism, that is, adaptations that are general responses to the selection regime.
A minimum inhibitory concentration (MIC) assay utilizing the ancestral E. coli K-12 MG1655 strain allowed the determination of the sublethal concentration of each gallium nitrate to be utilized for selection purposes. This was determined to be 100 mg/l of Ga(NO3)3 during 10 days of culture. The gallium selected and control populations were frozen at −80°C for future analysis. To determine the potential impact of the nitrate ion () on the growth of the controls and gallium nitrate selected populations, a MIC assay was conducted on both populations in sodium nitrate (NaNO3). The reduction in 24-hour growth as measured by optical density at 625 nm from 6.25 to 500 mg/l was 0.012 for the controls and 0.014 for the gallium-selected populations. From these results, we determined that resistance to the nitrate ion would not be a significant factor in the experiment.
Phenotypic assays: 24-hour growth
Measurements of 24-hour growth in excess metal (Ga3+, Fe2+, Fe3+ and Ag+) and traditional antibiotics for the ancestral, gallium-selected and control populations were determined at 10 days of evolution. These values were compared with the five samples of the E. coli K-12 MG1655 ancestor which was grown over night in standard DMB medium. The range used was 0–1000 mg/l for the Ga3+; 0–1750 mg/l for the Fe2+ and Fe3+ assays, 0–100 mg/l for Ag+ and finally 0–500 mg/l for the antibiotics (chloramphenicol, rifampicin, sulfanilamide and tetracycline). Previous studies of E. coli K-12 MG1655 have shown that this strain has no resistance to these antibiotics [36]. Statistical analysis of the effect of selection regime and concentration (and their interaction) for all 24-hour growth data was performed via General Linear Model utilizing SPSS version 23 (SPSS Inc, Armonk, NY, USA). All graphs in this paper were made via SigmaPlot version 14. Finally, the phenotypic data from these studies will be submitted into DRYAD (https://datadryad.org/) upon acceptance of this manuscript for publication.
Genomic analysis
DNA was extracted from each population after 10 days of culture using the EZNA Bacterial DNA extraction kit (Omega Bio-tek®) as per manufacturer instructions. DNA concentrations were normalized using the QuantiFluor® dsDNA system. Genomic libraries were prepared using the Illumina Nextera XT kit and samples were sequenced using the Illumina MiSeq sequencing platform. The depth of coverage of the sequencing runs ranged from ∼20× to ∼80×, with most exceeding 40× coverage. The SRA accession number for sequencing data from this study are PRJNA521749 (Ga3+-resistant and controls).
Sequence alignment and variant calling from the samples was achieved by use of the breseq 0.30.0 pipeline [37]. The breseq pipeline uses three types of evidence to predict mutations, read alignments, missing coverage and new junctions, and any reads that indicate a difference between the sample and the reference genome that cannot be resolved to describe precise genetic changes are listed as ‘unassigned’ [37]. These unassigned reads are not described or interpreted here.
The position at which the FecA mutations occurred were then identified in the published structure of FecA in complex with iron-citrate (PDB1PO3) [38] and visualized using PYMOL [39]. This was done in order to predict the functional consequences of the identified mutations.
RESULTS
Phenotypic changes in presence of metals
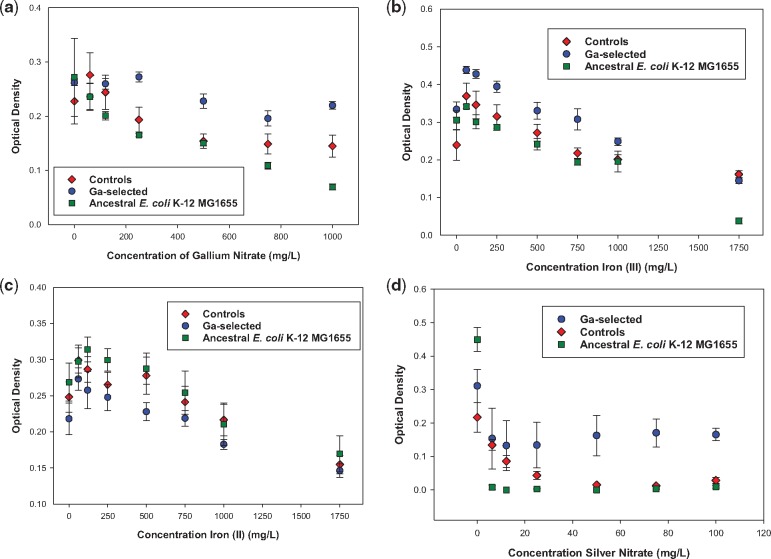
Figure 1a–d shows 24-hour growth across increasing concentrations of gallium, iron (II), iron (III) and silver nitrate. The Ga3+-selected populations showed superior growth relative to the controls and ancestral strain across concentration for gallium, iron (III) and silver nitrate. The controls and ancestral populations showed superior growth relative to the Ga3+-selected populations in iron (II). For all substances tested, the Ga3+-selected, controls and ancestral population decreased their 24-hour growth with increased concentration of the metal tested. There were two significant interactions between the population and concentration variables, in gallium and iron (II) resistance comparisons between the Ga3+-selected and ancestral population. The F-statistics and P-values for all comparisons are given in Table 2.
Figure 1.
(a) The mean and SE of 24-hour growth for populations in increasing concentration of gallium (Ga3+) to 1000 mg/l after 10 days of evolution. Ga3+ selected were significantly > controls = ancestors. (b) The mean and SE of 24-hour growth for populations in increasing concentration of iron III (Fe3+) to 1750 mg/l after 10 days of evolution. Ga3+ selected were significantly > controls = ancestors, the ancestors had virtually no ability to grow in iron (III) at the highest concentration measured. (c) The mean and SE of 24-hour growth for populations in increasing concentration of iron II (Fe2+) to 1750 mg/l after 10 days of evolution. The controls = ancestors were significantly > than the Ga3+-selected populations. (d) The mean and SE of 24-hour growth for populations in increasing concentration of silver (Ag+) to 100 mg/l after 10 days of evolution. The Ga3+-selected were significantly > controls > ancestors
Table 2.
F statistics and P-values for phenotypic tests
| Substance | Range tested | Population | Concentration | Interaction |
|---|---|---|---|---|
| Gallium resistant > Controls | ||||
| Gallium Ga(NO3)3 | 200–1000 mg/l | F = 11.42, P = 0.0001 | F = 5.00, P = 0.001 | F = 0.81, P = 0.99, NS |
| Iron (III) Fe2(SO4)3 | 6.2–1750 mg/l | F = 30.40, P = 0.0001 | F =27.41, P = 0.0001 | F = 1.22, P = 0.30, NS |
| Silver nitrate AgNO3 | 6.2–100 mg/l | F = 16.00, P = 0.0001 | F = 3.83, P = 0.003 | F= 0.06, P = 0.99, NS |
| Gallium resistant > Ancestor | ||||
| Gallium Ga(NO3)3 | 200–1000 mg/l | F= 110.64, P = 0.0001 | F = 18.09, P = 0.0001 | F = 7.18, P = 0.0001 |
| Iron (III) Fe2(SO4)3 | 6.2–1750 mg/l | F= 139.00, P = 0.0001 | F= 84.15, P = 0.0001 | F = 1.13, P = 0.357, NS |
| Silver nitrate AgNO3 | 6.2–100 mg/l | F = 33.10, P = 0.0001 | F = 0.08, P = 0.995, NS | F = 13.61, P = 0.001 |
| Rifampicin | 6.2–25 mg/l | F = 37.0, P = 0.0001 | F = 7.7, P = 0.0001 | F = 1.1, P = 0.356 |
| Sulfanilamide | 100–250 mg/l | F = 20.8, P = 0.0001 | F = 0.877, P = 0.431 | F = 0.238, P = 0.790 |
| Gallium resistant = controls | ||||
| Chloramphenicol | 6.2–250 mg/l | F = 1.81, P = 0.182 | F = 19.68, P = 0.0001 | F = 0.35, P = 0.92, NS |
| Sulfanilamide | 6.2–250 mg/l | F = 0.287, P = 0.532 | F =0.532, P = 0.807 | F = 0.829, P = 0.56, NS |
| Tetracycline | 6.2–250 mg/l | F = 0.135, P = 0.885 | F = 0.85, P = 0.526 | F = 155, P = 0.171, NS |
| Gallium resistant = ancestor | ||||
| Chloramphenicol | 6.2–250 mg/l | F = 0.355, P = 0.554 | F = 2.88, P = 0.01 | F = 0.170, P = 0.99 |
| Sulfanilamide | 6.2–75 mg/l | F = 0.01, P = 0.907 | F = 0.30, P = 0.870 | F = 1.44, P = 0.23 |
| Tetracycline | 6.2–250 mg/l | F = 0.504, P = 0.481 | F =1.20, P = 0.27 | F = 1.27, P = 0.28 |
| Rifampicin | 50–250 mg/l | F = 0.07, P = 0.79 | F = 1.3, P = 0.27 | F = 3.0, P = 0.03 |
| Controls > gallium resistant | ||||
| Iron (II) Fe2SO4 | 6.2–1750 mg/l | F = 8.98, P = 0.004 | F =11.44, P = 0.0001 | F = 0.22, P = 0.97, NS |
| Rifampicin | 6.2–250 mg/l | F = 33.51, P = 0.0001 | F =7.75, P = 0.0001 | F = 0.68, 0.60, NS |
| Controls > Ancestor | ||||
| Gallium | 200–1000 mg/l | F = 9.34, P = 0.003 | F = 13.55, P =0.0001 | F = 0.568, P = 0.724, NS |
| Iron (III) Fe2(SO4)3 | 6–1750 mg/l | F = 13.62, P = 0.001 | F = 36.16, P = 0.0001 | F = 1.72, P = 0.133, NS |
| Silver nitrate | 6–100 mg/l | F = 82.83, P = 0.0001 | F =13.92, P = 0.0001 | F = 13.6, P = 0.001 |
| Rifampicin | 6–250 mg/l | F = 309.5, P = 0.0001 | F = 7.7, P = 0.0001 | F = 2.6, P = 0.019 |
| Sulfanilamide | 100–250 mg/l | F = 19.5, P = 0.0001 | F = 0.07, P = 0.93 | F = 0.63, P = 0.54 |
| Controls = Ancestor | ||||
| Iron (II) Fe2SO4 | 6–1, 750 mg/l | F = 0.05, P = 0.813, | F = 30.13, P = 0.0001 | F = 3.72, P = 0.003 |
| Chloramphenicol | 6–250 mg/l | F = 2.83, P = 0.09 | F = 3.28, P = 0.005 | F= 0.559, P = 0.78 |
| Sulfanilamide | 6–75 mg/l | F = 0.87, P = 0.355 | F = 3.51, P = 0.015 | F = 0.436, P = 0.781 |
| Tetracycline | 6–250 mg/l | F = 0.004, P = 0.952 | F = 1.19, P = 0.323 | F = 0.886, P = 0.525 |
| Ancestor > gallium resistant | ||||
| Iron (II) Fe2SO4 | 6.2–1750 mg/l | F = 36.22, P = 0.0001 | F =30.13, P = 0.0001 | F = 3.72, P = 0.003 |
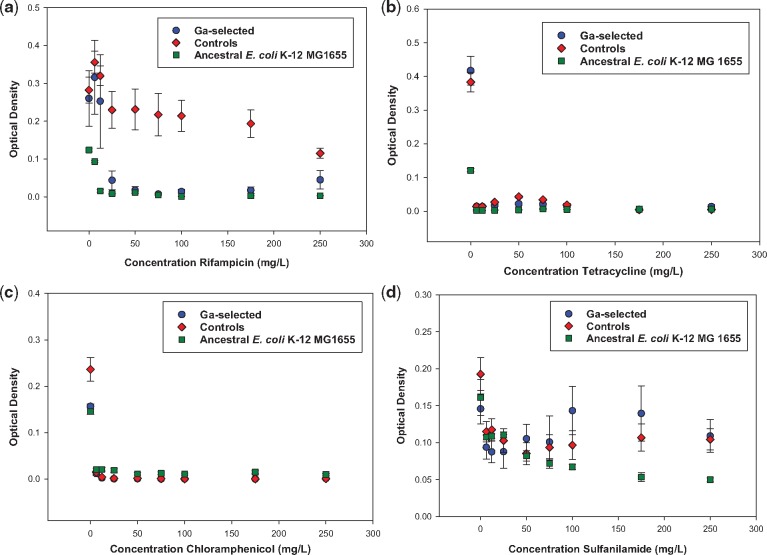
The 24-hour growth of the Ga3+-selected, control and ancestral populations from 0 to 250 mg/l in rifampicin, tetracycline, chloramphenicol and sulfanilamide are shown in Figure 2a–d. The control populations showed superior growth relative to the Ga3+-selected and ancestral population in increasing concentrations of rifampicin. There was no significant difference between the selected, control and ancestral populations in chloramphenicol and tetracycline (growth ceased at 6.2 mg/l). There was no difference between Ga3+-selected, control and ancestral populations from 6 to 75 mg/l of sulfanilamide. However, from 100 to 250 mg/l, both the Ga3+-selected and control populations showed equivalent growth which was statistically significantly greater than that of the ancestral population.
Figure 2.
(a) The mean and SE of 24-hour growth for populations in increasing concentration of rifampicin to 250 mg/l after 10 days of evolution. The controls from 25 to 250 mg/l were significantly > then the Ga3+-selected populations. The Ga3+-selected > ancestral from 6 to 25 mg/l and were equivalent from 50 to 250 mg/l. (b) The mean and SE of 24-hour growth for populations in increasing concentration of tetracycline to 250 mg/l after 10 days of evolution. The controls, Ga3+-selected and ancestral populations effectively showed no growth from 6 to 250 mg/l. (c) The mean and SE of 24-hour growth for populations in increasing concentration of chloramphenicol to 250 mg/l after 10 days of evolution. There was effectively no growth and no difference between the Ga3+-selected populations, controls and the ancestors from 6 to 250 mg/l. (d) The mean and SE of 24-hour growth for populations in increasing concentration of sulfanilamide to 250 mg/l after 10 days of evolution. There was no difference between the Ga3+-selected, controls and ancestral populations from 6 to 75 mg/l; however, the Ga3+-selected = controls and both were significantly > than the ancestral populations from 100 to 250 mg/l
Genomic results
Table 3 shows selective sweeps in the Ga3+-resistant populations at day 10. There were eight hard selective sweeps (0.000 ancestor–1.000 in the Ga3+-resistant populations); one hard sweep found in (Ga3+_1, Ga3+_2 and Ga3+_5), Ga3+_3 displayed two hard sweeps and Ga3+_4 showed four hard sweeps. Of the hard sweeps, all populations showed a mutation in fecA (UniProtKB-P77279). Ga3+_1, Ga3+_2, Ga3+_3 and Ga3+_5 showed the same mutation, and Ga3+_4 had a different mutation in this gene. Ga3+_3 showed a hard selective sweep in insl1 (UniProtKB-P0CF91), Ga3+_4 in yedN (UniProtKB-A0A023BD4). There was one hard sweep in an intergenic region (qmcA ←/→ fetA). The qmcA (UniProtKB-P0AA53) gene encodes a prohibitin homology domain-anchored (PHB) domain-anchored putative protease and fetA (UniProtKB-P77279) encodes an iron exporter, ATP binding subunit, ABC transporter FETAB subunit; peroxidase resistance protein. There were seven significant polymorphisms (0.351–0.39) distributed across the five populations. Ga3+_1 and Ga3+_3 show an identical mutation in fecE (dicitrate transport ATP binding protein, UniProtKB-P15031). Ga3+_4 shows a mutation in fecB (binds citrate-dependent Fe3+; part of the binding protein-dependent transport system for uptake of citrate-dependent Fe3+, UniProtKB-P15028). Ga3+_3 displays a mutation in ydfE (Qin prophage, pseudogene; phage or prophage related, UniProtKB-Q7138). The remaining significant polymorphisms are intergenic, Ga3+_1 (fepD ←/→ entS; fepD UniProtKB-23876 is part of the binding protein-dependent transport system for ferric enterobactin. Probably responsible for the translocation of the substrate across the membrane; entS UniProtKB-24077 has enterobactin transmembrane transporter activity, also involved in cellular response to DNA damage stimulus, and response to antibiotic). Ga3+_3 has an intergenic mutation [yfgF ←/→ yfgG; yfgF UniProtKB-P77172 is a phosphodiesterase (PDE) that catalyzes the hydrolysis of cyclic-di-GMP (c-di-GMP) to 5′-pGpG; yfgG UniProtKB-P64545 is an uncharacterized protein]. Finally, six low-level polymorphisms were observed. Ga3+_1 displayed a mutation in prlF UniProtKB-15373, antitoxin component of a type II toxin-antitoxin (TA) system, Ga3+_2 had a mutation in nhaB UniProtKB-P0AFA7, a Na+/H+ antiporter that extrudes sodium in exchange for external protons and Ga3+_5 had one in rpoS UniProtKB-P13445, RNA polymerase sigma factor S. The remaining low-level polymorphisms were intergenic with Ga3+_1 showing ypjC ←/← ileY; ypjC UniProtKB-76613 predicted protein and ileY tRNA isoleucine; agaI →/→ yraH, agaI UniProtKB-P42912, putative deaminase, galactosamine-6 phosphate isomerase; yraH UniProtKB-P42913, putative fimbrial-like adhesin protein; dcuB ←/← dcuR, dcuB, UniProtKB-P0ABN9, anaerobic C4-dicarboxylate transporter DcuB and DcuR are members of the two-component regulatory system DcuB/DcuR, involved in the C4-dicarboxylate-stimulated regulation of the genes encoding the anaerobic fumarate respiratory system. Table 4 and 5 report the annotation and functional description of these genes. Of the selective sweeps in these populations, six were non-synonymous single nucleotide polymorphisms, seven were intergenic, two were insertions in coding regions and two were insertions in pseudogenes.
Table 3.
Selective Sweeps in Ga3+-resistant populations at day 10
| Gene | Position | Mutation | Ga1 | Ga2 | Ga3 | Ga4 | Ga5 |
|---|---|---|---|---|---|---|---|
| qmcA ← / → fetA | 515, 859 | C→G | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 |
| fepD ← / → entS | 622, 244 | IS5 (–) +4 bp | 0.351 | 0.000 | 0.639 | 0.000 | 0.000 |
| nhaB ← | 1, 234, 632 | A→G | 0.000 | 0.190 | 0.000 | 0.000 | 0.000 |
| ydfE → | 1, 650, 461 | C→A | 0.000 | 0.000 | 0.452 | 0.000 | 0.000 |
| yedN ← | 2, 011, 665 | G→T | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 |
| yfgF ← / → yfgG | 2, 629, 042 | IS5 (+) +4 bp | 0.000 | 0.000 | 0.500 | 0.000 | 0.000 |
| ypjC ← / ← ileY | 2, 785, 563 | G→A | 0.193 | 0.000 | 0.000 | 0.000 | 0.000 |
| rpoS ← | 2, 866, 695 | IS1 (+) +9 bp | 0.000 | 0.000 | 0.000 | 0.000 | 0.273 |
| prlF → | 3, 277, 273 | (TTCAACA)2→3 | 0.184 | 0.000 | 0.000 | 0.000 | 0.000 |
| agaI → / → yraH | 3, 287, 319 | C→T | 0.161 | 0.000 | 0.000 | 0.000 | 0.000 |
| dcuB ← / ← dcuR | 4, 349, 066 | T→A | 0.147 | 0.000 | 0.000 | 0.000 | 0.000 |
| insI1 ← | 4, 507, 739 | G→T | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 |
| fecE ← | 4, 510, 928 | A→T | 0.514 | 0.000 | 0.574 | 0.000 | 0.000 |
| fecB ← | 4, 514, 119 | C→T | 0.000 | 0.000 | 0.000 | 0.479 | 1.000 |
| fecA ← | 4, 515, 480 | C→T | 1.000 | 1.000 | 1.000 | 0.000 | 1.000 |
| fecA ← | 4, 516, 171 | G→C | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 |
Color coding: yellow, fixation; green, major variant; blue, minor variant.
Table 4.
Annotation of genes in Table 2a
| Gene | Annotation |
|---|---|
| qmcA ← / → fetA | intergenic (-86/-60) |
| fepD ← / → entS | intergenic (-55/-53) |
| nhaB ← | L29S (TTA→TCA) |
| ydfE → | pseudogene (384/765 nt) |
| yedN ← | pseudogene (376/678 nt) |
| yfgF ← / → yfgG | intergenic (-104/-245) |
| ypjC ← / ← ileY | intergenic (-552/+199) |
| rpoS ← | coding (849-857/993 nt) |
| prlF → | coding (272/336 nt) |
| agaI → / → yraH | intergenic (+294/-107) |
| dcuB ← / ← dcuR | intergenic (-322/+249) |
| insI1 ← | D293E (GAC→GAA) |
| fecE ← | L177Q (CTG→CAG) |
| fecB ← | D64N (GAT→AAT) |
| fecA ← | G400S (GGC→AGC) |
| fecA ← | N169K (AAC→AAG) |
Color coding: blue, missense mutation; red, nucleotides changed.
Table 5.
Description of genes in Table 2a
| Gene | Description |
|---|---|
| qmcA ← / → fetA | PHB domain membrane-anchored putative protease/iron exportera |
| fepD ← / → entS | Iron-enterobactin transporter subunit/enterobactin exporterb |
| nhaB ← | Sodium: proton antiporter |
| ydfE → | Qin prophage; pseudogene; Phage or Prophage Related |
| yedN ← | Pseudogene, IpaH/YopM family |
| yfgF ← / → yfgG | Cyclic-di-GMP PDEc |
| ypjC ← / ← ileY | Pseudogene/tRNA-Ile |
| rpoS ← | RNA polymerase, sigma S (sigma 38) factor |
| prlF → | Antitoxin of the SohA(PrlF)-YhaV TA system |
| agaI → / → yraH | Galactosamine-6-phosphate isomerased |
| dcuB ← / ← dcuR | C4-dicarboxylate transporter, anaerobic; DcuS co-sensore |
| insI1 ← | IS30 transposase |
| fecE ← | Iron-dicitrate transporter subunit E |
| fecB ← | Iron-dicitrate transporter subunit B |
| fecA ← | Ferric citrate outer membrane transporter |
Genes that are directly related to iron metabolism are highlighted in red.
ATP-binding subunit, ABC transporter FetAB subunit; peroxide resistance protein.
Iron-regulated.
Anaerobic/uncharacterized protein.
Putative fimbrial-like adhesin protein.
Response regulator in two-component regulatory system with DcuS.
The most notable mutations observed in the control populations were in an intergenic mutation occurring between arsC →/→ yhiS in the C4 population; and a series of mutations in rpoB → in C1–C3, at position 4 183 378 ranging in frequency from 0.655 to 0.320; in C4 at position 4 182 820 in a frequency of 0.338 and in C4 at position 4 183 379 in frequency of 0.607. The arsC (UniProtKB-POAB96) gene is involved in reducing arsenate to arsenite allowing for efflux of this toxic material from the cell and yhiS is an uncharacterized protein. RNA polymerase subunit B (rpoB) is a DNA-dependent RNA polymerase that catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates; and resistance to the antibiotics salinamide A, salinamide B, rifampicin, streptolydigin, CBR703, myxopyronin and lipiarmycin can result from mutations in this protein (UniProtKB-POA8V2).
All polymorphisms classified by breseq in the Ga3+-selected populations and the controls are found in Supplementary Tables 1a–c and 2a–c. None of the suspected mutations associated with Ga3+ resistance were found in the controls, or have ever been observed in the control populations of our previous studies [21, 22, 33, 34].
DISCUSSION
These studies have clearly shown that E. coli K-12 MG1655 can rapidly evolve resistance to gallium. We predicted that this would be true based upon our previous work that demonstrated that this strain could evolve resistance to excess iron (II, III) and that in each case, increased resistance to gallium resulted [21, 22]. Our result is in direct opposition to previously published claims concerning gallium quenching as an evolutionarily stable antimicrobial treatment [24]. That study examined experimental evolution of gallium resistance and found no evidence of anti-gallium resistance over the course of 12 days for P. aeruginosa. On the face of it that result is counter intuitive, given that a 2013 study did find evidence of anti-gallium mutations in P. aeruginosa [26] and a recent 2018 study corroborated the 2013 result [25]. Thus, it seems clear that Gram-negative bacteria do have the capacity to evolve resistance to gallium quenching.
In addition to gallium resistance, correlated resistance to excess iron (III, Fe3+) and ionic silver (Ag+) was observed in the Ga3+-resistant populations. The latter result can be questioned, in that the genomic analysis showed no selection in genes that have previously been associated with silver resistance. Although selection for iron (II) and iron (III) resistance in our previous studies did confer a minor increase in silver resistance without evidence of selection in the genes cusS, ompR, that played a major role in conferring resistance [21, 22, 33, 34]. Therefore, it is possible that gallium selection is conferring a minor increase in silver resistance by mechanisms, we do not as of yet understand. Finally, our Fe2+- and Fe3+-resistant populations showed cross resistance to iron (II) and iron (III). The Ga3+-resistant populations are resistant to iron (III), but not to iron (II).
The Ga3+-selected populations also showed inferior 24-hour growth relative to the controls and the ancestors in the traditional antibiotic rifampicin. This can be linked to the selective sweeps in rpoB that were found in the control populations, but not in the Ga3+-resistant populations or ancestors. Mutations in rpoB are commonly observed in E. coli experimental evolution, including adaptation to media-containing minimal amounts of carbon source (glucose/lactose) and these are associated with increased resistance to rifampicin [40–42]. It seems that pleiotropic effects of such mutations may be a crucial means by which the evolution of antibiotic resistance proceeds in the absence of antibiotics [43, 44].
There was no significant difference between the Ga3+-resistant, control and ancestral populations in chloramphenicol and tetracycline. The Ga3+-resistant, control and ancestral populations showed equivalent growth at lower concentrations of sulfanilamide (6–75 mg/l). However, both the Ga3+-resistant and control populations performed better at higher concentrations (100–250 mg/l) of sulfanilamide compared with the ancestor. As neither of these populations were exposed to sulfanilamide, it is likely that this improvement in growth is due to some other aspect of the environment shared by both populations (such as minimum sugar content). ATCC cultures this strain using Luria broth which has a higher sugar content than DMB (https://www.atcc.org/products/all/47076.aspx#culturemethod).
The lack of correlated antibiotic resistance in the Ga3+-resistant populations is significant in that our iron-selected populations (Fe2+ and Fe3+) did show correlated resistance to antibiotics [21, 22]. The difference between these strains in correlated responses to selection is best explained by the differences in the genomic foundations of their respective resistances. All the Ga3+-resistant populations showed a mutation in the fecA gene (ferric citrate outer membrane transporter, the G400S mutation was observed in all populations except Ga3+_4 which displayed N169K). This gene encodes the protein FecA is responsible for Fe3+ transport in aerobic conditions [11]. We propose that these mutations reduce the function of FecA, thereby limiting Ga3+ import in these strains. For example, we have shown the FecA mutations (A559T and G243C) observed in our iron (II)-resistant populations express 128-fold less fecA than the controls in absence of iron indicating a likely preemptive mechanism of defense [22].
Several of the other genomic variants showing strong selection involved in conferring gallium resistance are in genes and intergenic regions associated with iron metabolism (fecE; fecB; qmcA/fetA; fepD/entS). The remaining polymorphisms showing evidence of selection have no clear relationship to iron metabolism, thus some could be responses to excess NO3− in the medium, or they could have yet undiscovered relationships to gallium resistance. It is also important to note the difference in our genomics results (via experimental evolution) and those obtained via knock out mutations in E. coli [9] in that none of our genes resulting from selection for gallium resistance were found in the previous study. This results from the fact that the Keio strains were produced by knockout of non-essential genes in E. coli K-12 [27] and our selective sweeps occurred in genes essential to iron metabolism in E. coli K-12 (e.g. fecA, fecE and fecB). In addition, the knock out method cannot identify intergenic regions that contribute to gallium adaptation, as no intergenic regions are knocked out in these strains. Thus, at best, knock out methods can identify genes that could possibly play a role in adaptation, but this technique is not necessarily going to tell you which genes will play a role in adaptation.
Our studies of experimental evolution of iron resistance showed that co-selection of metal and antibiotic resistance, and this can occur from linkage as well as pleiotropy. In the case of metals and antibiotics, pleiotropy is well known. For example, resistance mechanisms to metals such as reduction in cell wall permeability, substance alteration, efflux, alteration of cellular targets and sequestration are wide spread, and these traits also produce resistance to traditional antibiotics [45, 46].
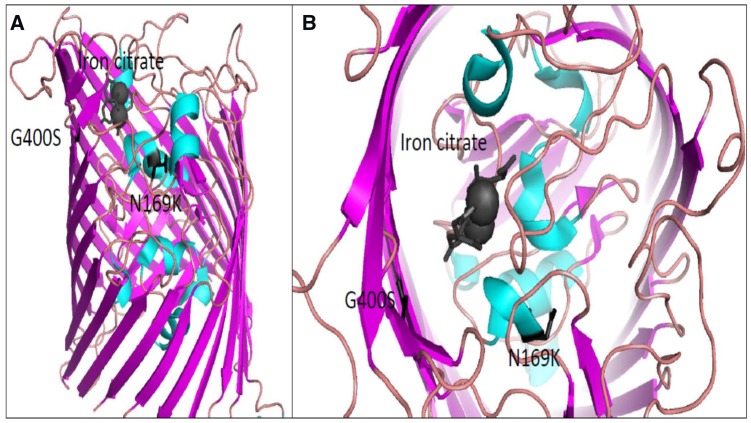
However, our Ga3+-resistant lines, do not display pleiotropy between gallium and antibiotic resistance whereas our iron (II, III) populations showed positive pleiotropy for five antibiotics: ampicillin, chloramphenicol, rifampicin, sulfanilamide and tetracycline [33, 34]. Genomic analysis illustrated that four of the five Fe2+-selected replicates showed at least one or more of these populations identified hard sweeps in genes such as murC, cueR, tolC, yeaG and ptsP. Several of these genes have been shown to be associated with antibiotic resistance, such as in the case of the murC which plays a role in cell wall biosynthesis as well as repairing oxidative damage [47]. However, the gallium-resistant populations do not display any selective sweeps in genes with an established relationship to antibiotic resistance, whereas the controls did display mutations in rpoB, that are consistently associated with antibiotic resistance [40–42]. We believe that the most functionally relevant mutations observed in this study is in FecA. As previously stated, FecA, through the siderophore citrate, is an iron (III) transporter under aerobic conditions. FecA is a transmembrane β-barrel and its structures has previously been solved both in its apo form and in complex with iron citration [48]. We used the complexed structure (PDB1PO3) as a guide to predict functional outcomes of the observed mutations. Both residues identified in this study (G400S and N169K) were found directly in the ligand binding pocket (Figure 3). The G400 amino acid sits directly adjacent to iron-citrate and the introduction of a polar group through mutation to a serine could easily hinder binding of the siderophore to the pocket. The second residue N169 sits on the backside of a helix which is also in contact with the iron citrate. The mutation to a large and positively charged amino acid such as lysine could potentially displace this helix into the ligand-binding pocket, also occluding entry of the siderophore. These mechanisms remain to be evaluated.
Figure 3.
(a) Depicts a full view of the FecA protein, iron-citrate is bound on the periplasmic side and colored dark grey. Both the G400 and N169 are colored black as their mutation to G400S and N169K were identified to play a role in conferring gallium resistance. (b) Close up view of the FecA protein from the top of the beta-barrel shows that G400 is directly adjacent to iron-citrate. N169 sits on a helix which is in direct contact also with iron-citrate. Figures were generated using PDB1P03 in PyMOL.
In summary, we have found that E. coli K-12 MG1655 can rapidly evolve resistance to gallium in opposition to claims of the evolutionary stability of gallium quenching. This trait also conferred resistance to ferric iron (iron III) and ionic silver, but no correlated responses to antibiotics were observed. The genomic results indicated that gallium resistance results from selection on mutations associated with iron metabolism. This is not surprising as gallium is an iron analog. We propose that these mutations are likely to be associated with the down regulation of genes involved in iron acquisition; as we observed in our iron (II)- and iron (III)-resistant populations [33, 34].
As in the evolution of silver, copper and iron resistance, a small number of genomic changes were required to produce the gallium-resistant phenotypes. All replicates showed at least one hard selective sweep (fecA). The parallelism of the mutations in fecA is a strong evidence that this gene played a major role in conferring gallium resistance. The Ga3+_4 replicate showed three additional hard sweeps, while the other populations had associated polymorphisms associated with iron metabolism not observed in the ancestor or control populations. We also suspect that these genomic changes must be associated with a profound difference in gene expression associated with iron and general metal metabolism. This was unmeasured in this study but is consistent with our previous results [33, 34]. Finally, our studies have important applications from the view of informing attempts to utilize metals such as gallium to control pathogenic and multidrug resistant bacteria. Despite optimism concerning the sustainability of metallic antimicrobials [4, 5, 24], bacteria can also rapidly evolve resistance to these materials as well.
Supplementary Material
Acknowledgements
This work was funded via support from the Joint School of Nanoscience & Nanoengineering, North Carolina A&T State University and UNC Greensboro, by a Research Experiences for Teachers (RET) award to Mr Jason Ward bia Biocomputational Evolution in Action (BEACON): An NSF Center for the Study of Evolution in Action (National Science Foundation Cooperative Agreement No. DBI-0939454), and by Characterizing the Evolutionary Behavior of Bacteria in the Presence of Iron Nanoparticles, NSF No. CBET-1602593. Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. We thank an anonymous reviewer for helpful comments on an earlier version of this manuscript.
Conflict of interest: None declared.
REFERENCES
- 1. Andersson DI. Persistence of antibiotic resistant bacteria. Curr Opin Microbiol 2003;6:452–6. [DOI] [PubMed] [Google Scholar]
- 2. Andersson DI, Hughes D.. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 2010;8:260–71. [DOI] [PubMed] [Google Scholar]
- 3. Wong A. Epistasis and the evolution of antimicrobial resistance. Front Microbiol 2017;8:246.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rai MK, Deshmukh SD, Ingle AP, Gade AK.. Silver nanoparticles: the powerful nanoweapon against multidrug-resistant bacteria. J Appl Microbiol 2012;112:841–52. [DOI] [PubMed] [Google Scholar]
- 5. He S, Feng Y, Gu N. et al. The effect of γ-Fe2O3 nanoparticles on Escherichia coli genome. Environ Pollut 2011;159:3468–73. [DOI] [PubMed] [Google Scholar]
- 6. Kaneko Y, Thoendel M, Olakanmi O. et al. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest 2007;117:877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chitambar CR. Gallium and its competing roles with iron in biological systems. Biochim Biophys Acta 2016;1863:2044–53. [DOI] [PubMed] [Google Scholar]
- 8. Fillat MF. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 2014;546:41–52. [DOI] [PubMed] [Google Scholar]
- 9. Escolar L, Perez-Martin J, Lorenzo V.. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriology 1999;181:6223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lill R. Function and biogenesis of iron-sulphur proteins. Nature 2009;460:831–8. [DOI] [PubMed] [Google Scholar]
- 11. Grass G. Iron transport in Escherichia coli: all has not been said and done. Biometals 2006;19:159–72. [DOI] [PubMed] [Google Scholar]
- 12. Coale KH, Johnson KS, Fitzwater SE. et al. A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean. Nature 1996;383: 495–501. [DOI] [PubMed] [Google Scholar]
- 13. Seo SW, Kim D, Latif H. et al. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun 2014;5:4910.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Konopka K, Neilands JB.. Effect of serum albumin on siderophore-mediated utilization of transferrin iron. Biochemistry 1984;23:2122–7. [DOI] [PubMed] [Google Scholar]
- 15. Nakashige TG, Zhang B, Krebs C, Nolan EM.. Human calprotectin is an iron-sequestering host-defense protein. Nat Chem Biol 2015;11:765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun HQ, Lu XM, Gao PJ. The exploration of the antibacterial mechanism of FE(3+) against bacteria. Braz J Microbiol 2011;42:410–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nunoshiba T, Hidalgo E, Amábile Cuevas CF, Demple B.. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol 1992;174:6054–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng M, Aslund F, Storz G.. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 1998;279:1718–21. [DOI] [PubMed] [Google Scholar]
- 19. Zheng M, Doan B, Schneider TD, Storz G.. OxyR and SoxRS regulation of fur. J Bacteriol 1999;181:4639–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Delany I, Rappuoli R, Scarlato V.. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol Microbiol 2004;52:1081–90. [DOI] [PubMed] [Google Scholar]
- 21. Shea CM, McIntosh MA.. Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli, 1991. Mol Microbiol 1991;5:1415–28. [DOI] [PubMed] [Google Scholar]
- 22. Graves JL, Ewunkem AJ, Thomas MD. et al. Experimental evolution of metal resistance in bacteria In Banzhaf W. (ed.) Evolution in Action—past, Present, and Future. Cham, Switzerland: Springer 2 International Publishing, AG; 2019. [Google Scholar]
- 23. Frangipani E, Bonchi C, Minandri F. et al. Pyochelin potentiates the inhibitory activity of gallium on Pseudomonas aeruginosa. Antimicrob Agents Chemother 2014;58:5572–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ross-Gillespie A, Weigert M, Brown SP, Kümmerli R.. Gallium-mediated siderophore quenching as an evolutionarily robust antibacterial treatment. Evol Med Public Health 2014;1: 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rezzoagli C, Wilson D, Weigert M. et al. Probing the evolutionary robustness of two repurposed drugs targeting iron uptake in Pseudomonas aeruginosa. Evol Med Public Health 2018;2018: 246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. García-Contreras R, Lira-Silva E, Jasso-Chávez R. et al. Isolation and characterization of gallium resistant Pseudomonas aeruginosa mutants. Int J Med Microbiol 2013;303:574–82. [DOI] [PubMed] [Google Scholar]
- 27. Baba T, Ara T, Hasegawa M. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006;2:2006.0008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gugala N, Chatfield-Reed K, Turner RJ, Chua G.. Using a chemical genetic screen to enhance our understanding of the antimicrobial properties of gallium against Escherichia coli. Genes 2019;10:pii: 34. DOI: 10.3390/genes10010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garland T, Rose MR.. Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments. Berkeley, CA: University of California Press, 2009. [Google Scholar]
- 30. Wong A, Kassen R.. Parallel evolution and local differentiation in quinolone resistance in Pseudomonas aeruginosa. Microbiology 2011;157(Pt 4): 937–44. [DOI] [PubMed] [Google Scholar]
- 31. Dettman JR, Rodrigue N, Aaron SD, Kassen R.. Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 2013;110:21065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vogwill T, Kojadinovic M, Furió V, MacLean RC.. Testing the role of genetic background in parallel evolution using the comparative experimental evolution of antibiotic resistance. Mol Biol Evol 2014;31:3314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Graves JL, Tajkarimi M, Cunningham Q. et al. Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front Genet 2015;6:DOI: 10.3389/fgene2015.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tajkarimi M, Rhinehardt K, Thomas M. et al. Selection for ionic confers silver nanoparticle resistance in Escherichia coli. Nanomed Nanotechnol 2017;5:1047. [Google Scholar]
- 35. Li XZ, Nikaido H, Williams KE.. Silver-resistant mutants of Escherichia coli display active efflux of Ag+ and are deficient in porins. J Bacteriol 1997;179:6127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Momtaz H, Safarpoor Dehkordi F, Taktaz T. et al. Shigatoxin-producing Escherichia coli isolated from bovine mastitic milk: serogroups, virulence factors, and antibiotic resistance properties. Sci World J 2012;2012:618709.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deatherage DE, Barrick JE.. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 2014;1151:165–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yue WW, Grizot S, Buchanan SK.. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J Mol Biol 2003;332:353–68. [DOI] [PubMed] [Google Scholar]
- 39. DeLano WL. Pymol: an open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography 2002;40:82–92. [Google Scholar]
- 40. Conrad TM, Joyce AR, Applebee MK. et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol 2009;10:R118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. LaCroix RA, Sandberg TE, O'Brien EJ. et al. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol 2015;81:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Campbell EA, Korzheva N, Mustaev A. et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell 2001;104:901–12. [DOI] [PubMed] [Google Scholar]
- 43. Knöppel A, Näsvall J (1), Andersson DI.. Evolution of antibiotic resistance without antibiotic exposure. Antimicrob Agents Chemother 2017;61:pii: e01495–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hershberg R. Antibiotic-independent adaptive effects of antibiotic resistance mutations. Trends Genet 2017;33:521–8. [DOI] [PubMed] [Google Scholar]
- 45. Seiler C, Berendonk TU.. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol 2012;3:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Di Cesare A, Eckert E, Corno G.. Co-selection of antibiotic and heavy metal resistance in freshwater bacteria. J Limnology 2015;75(S2):59–66. [Google Scholar]
- 47. Humnabadkar V, Prabhakar KR, Narayan A. et al. UDP-N-acetylmuramic acid l-alanine ligase (MurC) inhibition in a tolC mutant Escherichia coli strain leads to cell death. Antimicrob Agents Chemother 2014;58:6165–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Porcheron G, Garénaux A, Proulx J. et al. Iron, copper, zinc, and manganese transport and regulation in pathogenic enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front Cell Infect Microbiol 2013;3:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.