Abstract
Vitiligo is an autoimmune disease in which destruction of skin melanocytes results in patches of white skin and hair. Genome-wide linkage studies and GWAS in European ancestry cases identified over 50 vitiligo susceptibility loci, defining a model of melanocyte-directed autoimmunity. Vitiligo heritability is exceedingly high, ~2/3 coming from common and ~1/3 from rare genomic variants; ~20% of vitiligo risk is environmental. Vitiligo genetic risk is polygenic, with greater additive risk in multiplex vitiligo families than simplex cases. Vitiligo age-of-onset is bimodal, also involving a major genetic component; a MHC enhancer haplotype confers extreme risk for vitiligo (OR 8.1) and early disease onset, increasing expression of HLA-DQB1 mRNA and HLA-DQ protein and thus perhaps facilitating presentation of triggering antigens. Vitiligo triggering also involves a major environmental component; dramatic delay in vitiligo age-of-onset, especially from 1973–2004, suggests that exposure or response to a key vitiligo environmental trigger diminished during this period. Together, these findings provide deep understanding of vitiligo pathogenesis and genetic architecture, suggesting that vitiligo represents a tractable model for investigating complex disease genetic architecture and predictive aspects of personalized medicine.
Keywords: vitiligo, complex disease, genetic architecture, heritability, age-of-onset
Introduction
Vitiligo, because of its striking phenotype of patchy depigmentation of skin and hair, was among the first skin disorders to be recognized (Krauss, 2018) and described in a formal medical context (Le Cat, 1765). Nevertheless, consideration of the genetic basis of vitiligo came much later, when Stüttgen (1950) and Teindel (1950) simultaneously reported several families with multiple close relatives affected with vitiligo, so-called “multiplex families.” Furthermore, Stüttgen noted that several family members were additionally affected by autoimmune thyroid disease, with overall disease inheritance appearing to involve both dominant and recessive factors. Stüttgen’s description of the inheritance of autoimmunity in these families may represent one of the earliest descriptions of what would now be considered complex disease inheritance.
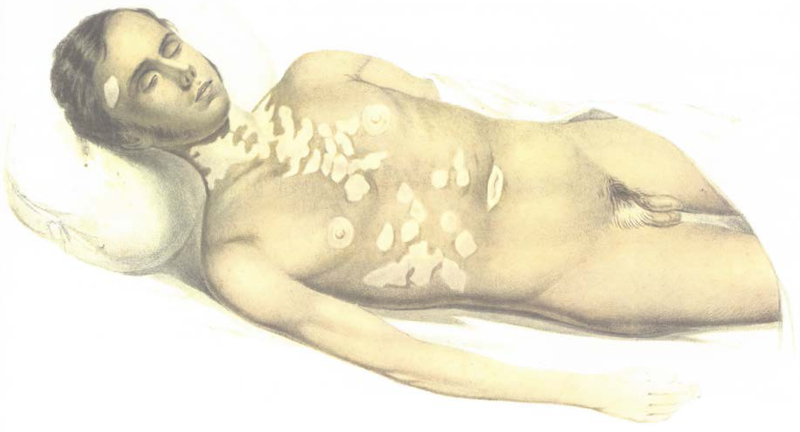
Though discussions of the specific genetic basis of vitiligo thus did not begin until the mid-20th century, epidemiological association of vitiligo with other autoimmune diseases had been observed far earlier. In 1855, Addison described vitiligo and pernicious anemia in two of his initial patients with Addison’s disease (Figure 1; Addison, 1855); all three of these diseases are now known to have an autoimmune basis. A century later, de Mowbray (1965) first applied the term “autoimmune” to vitiligo, reporting a patient with vitiligo, pernicious anemia, autoimmune thyroid disease, and type 1 diabetes, stating, “It is tempting to regard these multiple deficiencies in this patient as autoimmune disturbances having a familial incidence.” These and other such observations suggested that the epidemiologically-associated autoimmune diseases might share at least some of their genetic underpinnings.
Figure 1:
An illustration of one of Thomas Addison’s two patients with both Addison’s disease and vitiligo. Reprinted from Addison’s original case report (Addison, 1855).
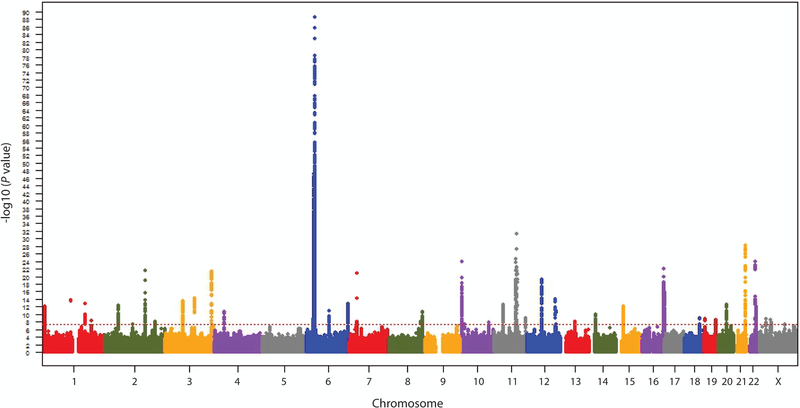
Beginning in the mid-1960s, researchers began to apply a series of evolving genetic technologies to elucidate the genetic basis of vitiligo and to identify specific underlying vitiligo susceptibility genes. These included heritability studies, candidate gene association studies, genetic linkage studies, and genome-wide association studies (GWAS). Today, at least 54 unique verified vitiligo susceptibility loci have been implicated in vitiligo cases of European ancestry (Spritz & Andersen, 2017; Roberts & Spritz, 2018), the most deeply studied human super-population. Of these loci, 50 were detected by GWAS, the most productive and reliable approach (Figure 2; Jin et al., 2016). For many of these vitiligo loci the corresponding underlying genes and causal genetic variants have now been identified, and as for many other genetically complex diseases, most of the causal variants appear to be regulatory in nature (Jin et al., 2016). Indeed, the variants with the greatest overall impact on disease risk genome-wide reside within transcriptional enhancers located in the class I and class II regions of the Major Histocompatibility Complex (MHC) on chromosome 6 (Cavalli et al., 2016; Hayashi et al., 2016; Jin et al., 2019a).
Figure 2:
Manhattan plot across the chromosomes of genome-wide meta-analysis results for 8,966,411 genotyped and imputed markers from three vitiligo GWAS (Jin et al., 2010, 2012, 2016) in a total 4,680 European-ancestry cases and 39,586 controls. Dotted line indicates the threshold for genome-wide significance (P < 5 × 10−8). Reprinted with from Jin et al. (2016) with permission.
Taken together, these genetic studies confirmed that vitiligo has an autoimmune basis, as about 85% of the identified vitiligo susceptibility genes encode proteins involved in immunity and apoptosis (Roberts et al., 2019b). Furthermore, about half of those genes have also been identified by independent genetic studies of other autoimmune diseases that are epidemiologically associated with vitiligo, supporting their role in shared autoimmune predisposition (Roberts & Spritz, 2018). The identified vitiligo susceptibility genes and corresponding pathways provided deep insights into the biological mechanisms that underlie vitiligo, a topic that has been addressed previously by several recent comprehensive reviews (Roberts & Spritz, 2018; Spritz & Andersen, 2017). Here, we instead address recent developments related to the overall genetic architecture of vitiligo as a complex disease, the genetic basis of clinical variation among vitiligo cases, and the relationship of vitiligo genetic susceptibility to environmental triggers. Discoveries in these areas provide remarkable new insights into the etiology of vitiligo and open new doors for future study.
Vitiligo Risk Involves both Genetic and Environmental Components
Various analytical approaches can be used to estimate the fraction of total vitiligo risk that is attributable to genetic variation, termed “heritability” (usually denoted h2), with the assumption that the rest is attributable to the environment. Different analytical approaches provide somewhat different estimates of heritability, and can be grouped generally as approaches based on analysis of disease recurrence in multiplex families or twins (h2FAM) (Tenesa & Haley, 2013) versus approaches that assess aggregate genetic similarity at hundreds of thousands to millions of genetic markers scored in unrelated, singleton (“simplex”) cases (h2SNP) (Lee et al., 2011). In the European-ancestry population, in which the prevalence of vitiligo is approximately 0.40 percent (Zhang et al., 2016), vitiligo h2FAM was recently estimated as approximately 0.75 – 0.84, depending on the types of relatives studied (Roberts et al, 2019b). In studies of other populations, estimates of vitiligo h2FAM have ranged from about 0.50 to 0.80 (Das et al., 1985; Hafez et al., 1983; Zhang et al., 2004; Arcos-Burgos et al., 2002). The differences are relatively minor, likely reflecting differing analytical approaches and somewhat differing vitiligo risk factors in different human super-populations. Most important, these estimates of vitiligo h2FAM are all quite high compared to many other complex diseases, for which h2FAM estimates are typically in the range 0.3 to 0.5 (Polderman et al., 2015).
Recent improvements in genetic resources and statistical methodology (Yang et. al., 2015) have allowed for improved estimation of h2SNP. Most important, very large European-ancestry genetic reference panels (McCarthy et al., 2016) have made possible “deep imputation” (Das et al., 2018), whereby millions of untyped common and rare variants can be imputed and used to compare the aggregate genetic similarity of vitiligo cases versus controls (Yang et al, 2015). By this approach, Roberts et al. (2019b) recently estimated vitiligo h2SNP in the European-ancestry population as 0.80, essentially matching the h2FAM estimates. It thus appears that essentially all vitiligo heritability estimated by family-based methods can now be captured by dense-array genotyping and imputation to large reference panels, and for vitiligo there is little or no remaining “missing heritability” (Manolio et al., 2009) (Figure 3).
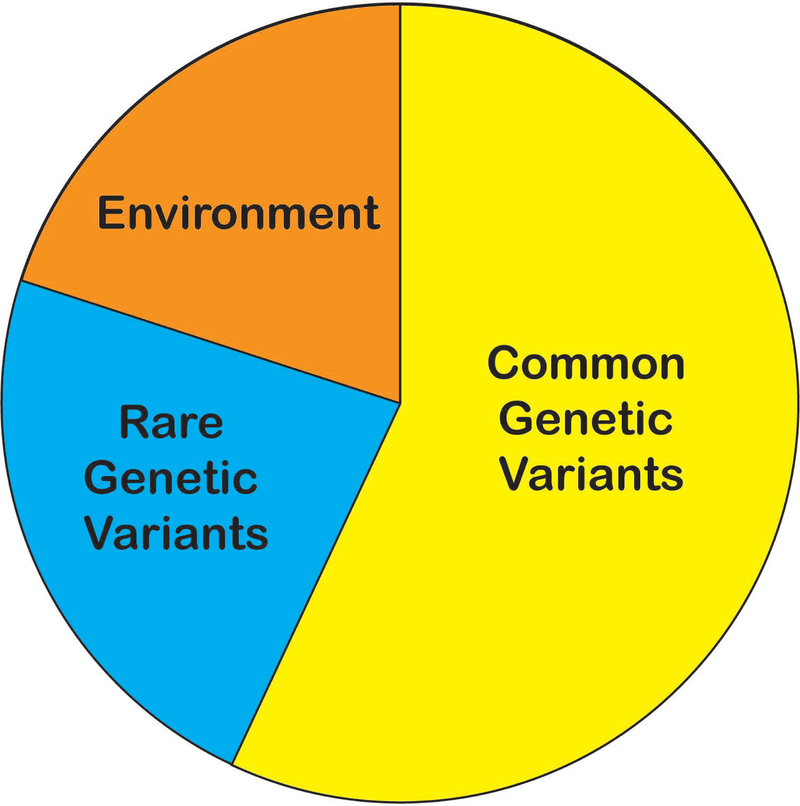
Figure 3.
Fraction of total vitiligo risk attributable to the environment (orange, 20%), common genetic variants (yellow, 57%), and rare genetic variants (blue, 23%).
Overall, then, in the European-ancestry vitiligo super-population, about 80% of vitiligo risk is attributable to genetic factors. This genetic component of vitiligo risk can be partitioned even farther. Common genetic variants (risk allele frequency > 0.01) account for about 71% of total vitiligo heritability and about 53% of total vitiligo risk, with a substantial fraction accounted for by the 50 genome-wide significant loci identified in GWAS (Jin et al., 2019a) (Figure 3; Jin et al., 2016). Furthermore, the effect sizes of many vitiligo-associated variants are strikingly large by comparison to other complex diseases, with odds ratios (ORs) above 1.5 for nearly 10% of the vitiligo loci identified by GWAS (Jin et al., 2016). Together, these findings explain why GWAS of vitiligo have been highly successful even with relatively modest sample sizes and further suggest that the genetic architecture of vitiligo may be less polygenic than that of other genetically complex diseases. Moreover, the relatively large fraction of vitiligo genetic risk attributable to common variants suggests that negative selection against these variants (and by extension, against individuals with vitiligo) is relatively low.
The remaining 29% of vitiligo heritability, and 23% of total vitiligo risk, is accounted for by rare variants (minor allele frequency 0.01– 0.0001) in aggregate. While most rare variants that contribute to vitiligo risk in the European-ancestry super-population can now be imputed, actual identification of the specific contributory rare variants as well as private variants will likely require either much larger GWAS sample sizes or the use of alternative analytical approaches such as family studies (Kosmicki et al., 2016).
Vitiligo Genetic Risk is Largely Polygenic
To date, 50 different vitiligo susceptibility loci have been identified by GWAS of cases of European ancestry. Genetic risk from these loci derives from relatively common variants, which can be additively combined to create a vitiligo “polygenic risk score”, with best performance achieved using only the most-associated variant at each locus (Roberts et al., 2019a). Altogether, this vitiligo polygenic risk score yields an OR of 8.79 comparing the highest 1% of the risk score distribution to the remaining 99%, substantially higher than polygenic risk score ORs for other genetically complex diseases such as coronary artery disease (OR = 4.83), atrial fibrillation (OR = 4.63), type II diabetes (OR = 3.30), inflammatory bowel disease (OR = 3.87), and breast cancer (OR = 3.36) (Khera et al., 2018). Indeed, the vitiligo polygenic risk score has a positive predictive value of approximately 71%, far better than polygenic risk scores for most other complex diseases (Wald & Old, 2019). Thus, while vitiligo risk is polygenic, it is apparently less polygenic than many other complex diseases, involving fewer total loci with relatively large effects from individual risk variants at these loci. This, together with the relatively high heritability, explains why the vitiligo polygenic risk score outperforms those for other complex diseases.
Vitiligo Genetic Risk is Polygenic in both Simplex and Multiplex Cases
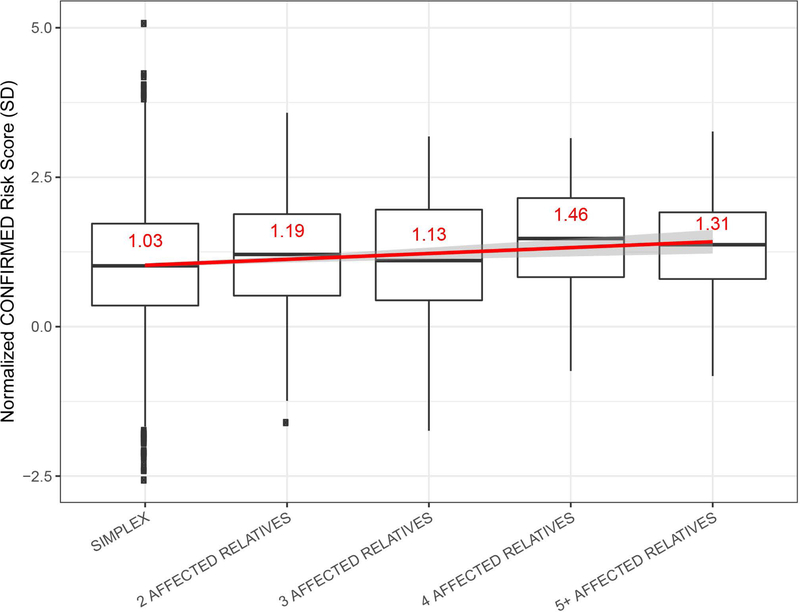
Large-scale epidemiological surveys (Alkhateeb et al., 2003; Roberts et al., 2019a) have shown that the vast majority of vitiligo cases (91%) are “simplex”, with no known close relatives affected by vitiligo, whereas about 9% belong to “multiplex” family clusters, reporting multiple close relatives with vitiligo. Roberts et al. (2019a) found that the vitiligo polygenic risk score is significantly higher in probands of such multiplex families than in simplex cases, indicating that the vitiligo polygenic burden is generally higher in multiplex cases. Furthermore, within vitiligo multiplex families, common vitiligo polygenic risk was over-transmitted from unaffected to affected relatives, and the polygenic risk score within multiplex families was roughly proportional to the total number of affected relatives (Figure 4; Roberts et al., 2019a). Even in a very large multiplex family that segregated a known, high-penetrance rare vitiligo susceptibility variant in FOXD3 (Alkhateeb et al., 2002, 2005), polygenic risk from common variants was also extraordinarily high.
Figure 4.
Vitiligo polygenic risk score categorized by the number of affected relatives reported by the proband. The Y-axis represents the normalized risk score; units represent standard deviations (SD) difference from the mean risk score in controls. Horizontal lines denote the median, and the value of the mean is in red. The red line shows the fit linear regression line and the corresponding 95% confidence interval (CI) surrounds the line in gray. Boxes denote first through third quartiles. Each vertical bar extends from the box to the largest or smallest value, no farther than 1.5 times the inter-quartile range. Data beyond the vertical bars are considered outliers and are plotted individually. Reprinted from Roberts et al. (2019) with permission.
Together, these findings indicate that vitiligo is typically polygenic in both simplex and multiplex cases, principally involving the same loci identified in GWAS of mostly simplex cases. The great majority of multiplex vitiligo families carry and transmit a relatively high burden of common vitiligo susceptibility alleles with modest effect sizes, although in at least some multiplex vitiligo families rare high-penetrance susceptibility variants also play a contributory role.
Vitiligo Triggering and Disease Onset Involves a Major Genetic Component
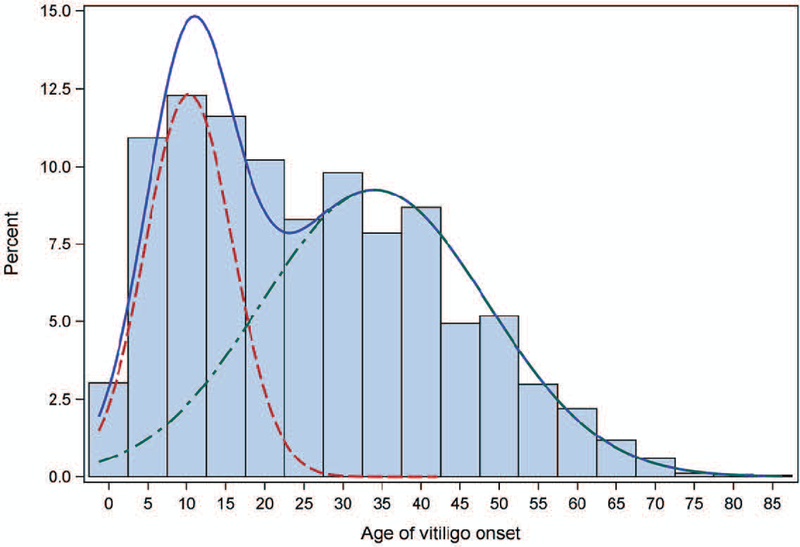
Epidemiological analysis of vitiligo cases of European ancestry revealed a bimodal distribution of vitiligo age-of-onset (Figure 5; Jin et al., 2019a), similar to several other autoimmune diseases such as type 1 diabetes mellitus (American Diabetes Association, 2017) and rheumatoid arthritis (Nigrovic et al., 2018). Overall, the mean age of vitiligo onset is 25.9 ± 16.6 years, apparently consisting of two overlapping age-of-onset subgroups. An early-onset vitiligo subgroup, comprising about 38% of cases, has a mean age-of-onset 10.3 ± 5.6 years, and a late-onset vitiligo subgroup, comprising about 62% of cases, has a mean age-of-onset 34.0 ± 14.5 years (Jin et al., 2019a).
Figure 5.
Distribution of age-of-onset and corresponding finite mixture model for 4,523 European-ancestry vitiligo cases. The blue line shows the full distribution, the red line the early-onset distribution (mean 10.3, SD 5.6 years; 38.4%), and the green line the late-onset distribution (mean 34.0, SD 14.5 years; 61.6%). Reprinted from Jin et al. (2019a) with permission.
Separate analyses of vitiligo cases from the early-onset and late-onset vitiligo subgroups indicated that the known GWAS loci contribute approximately equally to each group. However, stratified analysis of the early-onset subgroup revealed a novel, very strong specific association, with an uncommon insertion-deletion polymorphism in the class II region of the MHC on chromosome 6. This polymorphism was located within a transcriptional enhancer, in linkage disequilibrium with another MHC class II enhancer variant that is more generically associated with vitiligo risk in cases of European ancestry. The haplotype that contains both MHC enhancer variants is specifically associated with extreme vitiligo risk (OR 8.1) and early disease onset, occurring particularly at about ages 5 to 9 years. Functional studies showed that the extreme-risk, early-onset haplotype is associated with increased expression of both HLA-DQB1 mRNA and total HLA-DQ protein on peripheral blood monocytes and dendritic cells. Elevated HLA class II expression might facilitate presentation of triggering antigens on the surface of such professional antigen-presenting cells, thereby increasing the probability of autoreactive T-cell activation and loss of tolerance.
The extended extreme-risk, early-onset MHC enhancer haplotype also carries the classical HLA alleles HLA-DRB1*13:01—HLA-DRB3*01:01—HLA-DQA1*01:03—HLA-DQB1*06:03. Conditional genetic analysis indicated that these classical HLA alleles do not contribute to vitiligo risk; rather, the non-coding enhancer variation appears to drive vitiligo risk, consistent with elevated HLA-DQB1 gene expression in early-onset vitiligo. Nevertheless, the early-onset vitiligo subgroup had a significantly lower prevalence of other vitiligo-associated autoimmune diseases (17.0%) compared to the late-onset subgroup (22.4%). Interestingly, HLA-DRB1*13:01, which resides on the extreme-risk, early-onset vitiligo MHC class II haplotype, has been associated with protection from systemic lupus erythematosus, rheumatoid arthritis, and type 1 diabetes. Thus, this haplotype might simultaneously confer extreme genetic risk for vitiligo (with early onset) and at the same time confer relative protection from other autoimmune diseases that are often epidemiologically associated with vitiligo.
Vitiligo Triggering and Disease Onset Additionally Involves a Major Environmental Component
Vitiligo heritability of 0.80 in the European-ancestry population indicates that genetic factors account for about 80% of vitiligo risk, and non-genetic factors (presumably the environment) account for the remaining 20%. While no vitiligo environmental risk factors or triggers are known with certainty, the Koebner phenomenon is especially frequent in vitiligo patients (Sagi & Trau, 2011; van Geel et al., 2012) suggesting that skin damage, perhaps complicated by subclinical infection, might be important in vitiligo triggering. As yet, it is unknown whether vitiligo induction might involve one trigger versus multiple triggers, possibly with cumulative effects over time, ultimately resulting in loss of tolerance to melanocyte autoantigens.
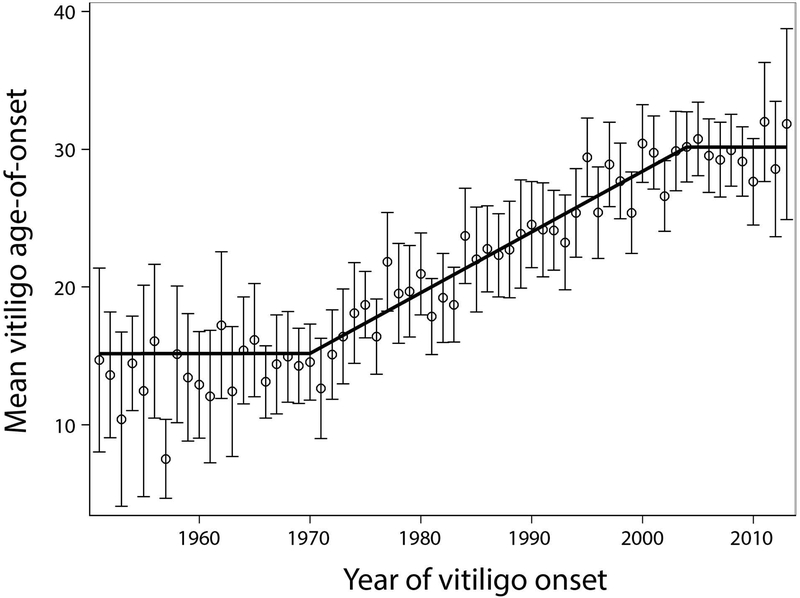
Very recently, an epidemiological study of vitiligo age-of-onset in patients of European ancestry showed there has been highly significant, progressive delay of vitiligo onset over the recent past (Jin et al., 2019b). These investigators were interested in whether vitiligo epidemiology has changed over time, perhaps in response to a changed repertoire of environmental triggers, as has been widely observed for type 1 diabetes mellitus (Patterson et al., 2009). As there are no longitudinal data on vitiligo prevalence over time, they analyzed retrospective vitiligo age-of-onset data from 1951 to 2013 for 4,406 incident European-ancestry vitiligo cases from North America and Europe. Remarkably, the mean age at vitiligo onset nearly doubled over the study period, most of the change having occurred during the period 1973 to 2000 (Figure 6; Jin et al., 2019b). Before 1970, vitiligo was principally a pediatric-onset disease with mean age-of-onset 14.6 ± 9.4 years. However, around 1973 vitiligo age-of-onset then began to increase by about 4 months per year, stabilizing about 2004 with vitiligo onset predominantly during adulthood with mean age-of-onset onset 30.2 ± 17.3 years. The overall pattern was almost identical in vitiligo cases from North America and in Europe, and is far in excess of increases in overall lifespan over this same period. This delay in vitiligo age-of-onset over time furthermore does not appear to involve the extreme-risk, early-onset MHC haplotype discussed above (Jin et al., 2019).
Figure 6.
Mean vitiligo age-of-onset in 4,406 unrelated European-ancestry cases by calendar year of onset 1951–2013. Circles indicate means and vertical bars 95% confidence intervals. Black segments indicate regression lines for time periods 1951–1969, 1970–2004, and 2005–2013. Reprinted from Jin et al. (2019b) with permission.
As the underlying genetics have not changed, this dramatic shift in vitiligo age-of-onset suggests strongly that either exposure to or biological response to a major vitiligo environmental trigger diminished between ~1973 and 2000. These authors speculated about various environmental changes during that period, of which perhaps the most intriguing are the introduction of “Sun Protection Factor” (SPF) sunscreens in 1974 and roughly contemporaneous improvements in skin wound care, both of which might have reduced skin damage and release of damage-associated molecular pattern (DAMP) and pathogen-associated molecular pattern (PAMP) motifs that initiate an immune response, thereby delaying vitiligo triggering (Jin et al., 2019b). At present, it is not known whether the temporal change in vitiligo epidemiology in North America and Europe is mirrored in other human super-populations; such comparison might highlight a corresponding temporal change in exposure to a specific potent environmental trigger or protective factor.
Conclusions and Future Directions
The last five years have seen remarkable advancement in understanding the biological basis of vitiligo. It is now possible to estimate the relative contributions of genes and the environment to vitiligo risk with reasonable accuracy. Overall, in the European-ancestry population, genes appear to account for about 80% and the environment about 20% of total vitiligo risk. Common variants account for about 71% of total genetic risk, with the remainder coming from rare variants. Genetic risk from both common and rare variants appears to be additive and polygenic in nature, and risk from the 50 common variants identified in GWAS can be combined in a polygenic risk score that is somewhat predictive of vitiligo risk. Predictive performance of the vitiligo polygenic risk score will undoubtedly improve with the discovery and inclusion of additional risk variants. Furthermore, studies to date have not identified genetically-based subgroups of vitiligo cases that have differing underlying polygenic pathobiology, and the ability to correctly assign vitiligo cases into genetically-defined polygenic subgroups could substantially improve performance of subgroup-specific polygenic risk scores that include only the loci relevant within each subgroup. Nevertheless, the overall vitiligo heritability of 0.80 establishes a ceiling on the ultimate ability to predict vitiligo risk based solely on genetics.
All of the over 50 vitiligo susceptibility loci found to date were detected by comparing affected European-derived cases to unaffected controls or to unaffected relatives. All of these loci are thus associated with case versus control status or, for the high-risk MHC haplotype, with early vitiligo onset. As yet, it remains entirely unknown whether any or all of these loci also influence the clinical course of vitiligo once immune tolerance to melanocyte antigens is broken. Additionally, it is important to note that a polygenic risk score based on the results of genetic studies of the European-derived super-population may not be applicable to other human super-populations, in which different risk loci and causal alleles may be predominant (Martin et al., 2017; Martin et al., 2019).
While vitiligo typically results from polygenic inheritance of multiple risk alleles that exert relatively small individual effects, that may not be universally true. Rare vitiligo cases might result from just one or a few rare, high-penetrance variants that have not yet been discovered, as is the case in rare Mendelian multiple autoimmunity syndromes that sometimes include vitiligo, such as APECED or IPEX. Accordingly, genomic analysis of multiplex vitiligo families that segregate low polygenic risk from known vitiligo susceptibility loci might thus enable discovery of rare or private variants that confer exceedingly high genetic risk. Conversely, some multiplex families with low polygenic risk might have shared exposure to an unusual and exceedingly strong non-genetic environmental trigger. In either case, exploration of the cause of family clusters with apparently low polygenic risk seems a promising next step in elucidating novel genetic and non-genetic causes of vitiligo.
Indeed, very little is known about environmental risk factors or triggers for vitiligo. Though the frequent occurrence of Koebner phenomenon in vitiligo suggests a role for skin damage, the number of potentially relevant environmental exposures is large and the ability to reliably track and measure such exposures is very limited, so it is hard to know where to start. The recent discovery that vitiligo age-of-onset changed substantially between about 1973 and 2000, transforming a disease that previously primarily affected children to one that now primarily affects adults, suggests that exposure to one or more important environmental triggers for vitiligo changed over approximately that period. Careful investigation of exposures that changed dramatically between approximately 1973 and 2000 might thus be one good starting point. Alternatively, the extreme-risk HLA haplotype that is specifically associated with vitiligo onset between 5 and 9 years of age suggests that investigation of exposures that especially pertain to that age range might be another good starting point. Similarly, investigation of children with very early vitiligo onset who do not carry the extreme-risk, early-onset MHC haplotype might also provide clues to important vitiligo environmental triggers.
Given that vitiligo heritability is quite high, that vitiligo risk is mostly polygenic involving the 50 current GWAS loci, and that a polygenic risk score additively combining these loci provides reasonably good assessment of genetic risk, vitiligo represents a near-ideal model for testing general hypotheses of complex disease genetic architecture and inheritance. As proof-of-concept, the vitiligo polygenic risk score was used as a tool to investigate a general question about the nature of complex disease inheritance in multiplex families versus simplex cases, showing that both types of cases involve polygenic risk from the same loci (Roberts et al., 2019a). Similar findings are emerging for other complex diseases such as testicular cancer (Loveday et al., 2018) and migraine (Gormley et al., 2018), suggesting that this aspect of complex disease biology may be generalizable. Vitiligo thus provides a particularly tractable complex disease model for investigations of both disease genetic architecture and predictive aspects of personalized medicine.
Acknowledgments
This work was supported by grants R01AR056292, R01AR057212, R01AR065951, and P30AR057212 from the National Institutes of Health. G.H.L.R. was supported by training grant T32AR007411 from the National Institutes of Health.
Footnotes
Competing Interests
None of the authors has any financial interest related to this work.
References
- Addison T (1855). On the constitutional and local effects of disease of the supra-renal capsules. Highley, London. [Google Scholar]
- Alkhateeb A, Stetler GL, Old W, Talbert J, Uhlhorn C, Taylor M, … & R.A. Spritz (2002). Mapping of an autoimmunity susceptibility locus (AIS1) to chromosome 1p31.3-p32.2. Hum Mol Genet 11, 661–667. [DOI] [PubMed] [Google Scholar]
- Alkhateeb A, Fain PR, Thody A, Bennett DC, & Spritz RA (2003). Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res 16, 208–214. [DOI] [PubMed] [Google Scholar]
- Alkhateeb A, Fain PR, & Spritz RA (2005). Candidate functional promoter variant in the FOXD3 melanoblast developmental regulator gene in autosomal dominant vitiligo. J Invest Dermatol 125, 388–391. [DOI] [PubMed] [Google Scholar]
- American Diabetes Association (2017). Classification and diagnosis of diabetes. Diabetes Care 40, S11–S24. [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Parodi E, Salgar M, Bedoya E, Builes J, Jaramillo D, … & Palacio G (2002). Vitiligo: complex segregation and linkage disequilibrium analyses with respect to microsatellite loci spanning the HLA. Hum Genet 110, 334–342. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Hayashi M, Jin Y, Yorgov D, Santorico SA, Holcomb C, … Dinarello CA (2016). MHC class II super-enhancer increases surface expression of HLA-DR and HLA-DQ and affects cytokine production in autoimmune vitiligo. Proc Natl Acad Sci USA 113, 1363–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Majumder PP, Majumdar TK, & Haldar B (1985). Studies on vitiligo. II. Familial aggregation and genetics. Genet Epidemiol 2, 255–262. [DOI] [PubMed] [Google Scholar]
- Das S, Abecasis GR, & Browning BL (2018). Genotype imputation from large reference panels. Annu Rev Genomics Hum Genet 19, 73–96. doi: 10.1146/annurev-genom-083117-021602. [DOI] [PubMed] [Google Scholar]
- De Mowbray R (1965). Addison’s disease with vitiligo, Addisonian anæmia, primary hypothyroidism and diabetes mellitus. Proc Royal Soc Med 58, 78–79. [PMC free article] [PubMed] [Google Scholar]
- Gormley P, Kurki MI, Hiekkala ME, Veerapen K, Häppölä P, Mitchell AA, … & Palotie A (2018). Common variant burden contributes to the familial aggregation of migraine in 1,589 families. Neuron 98, 743–753. doi: 10.1016/j.neuron.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez M, Sharaf L, & Abd el-Nabi SM (1983). The genetics of vitiligo. Acta Derm Venereol, 63, 249–251. [PubMed] [Google Scholar]
- Hayashi M, Jin Y, Yorgov D, Santorico SA, Hagman J, Ferrara T, … Spritz RA (2016). Autoimmune vitiligo is associated with gain of function by a transcriptional regulator that elevates expression of HLA-A*02:01 in vivo. Proc Natl Acad Sci USA 113, 1357–1162. doi: 10.1073/pnas.1525001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ, … & Spritz RA (2010). Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med 362, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL, … & Spritz RA (2012). Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet 44, 676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Andersen G, Yorgov D, Ferrara TM, Ben S, Brownson KM, … & Spritz RA (2016). Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat Genet 48, 1418–1424. doi: 10.1038/ng.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Roberts GHL, Ferrara TM, Ben S, van Geel N, Wolkerstorfer A, … & Spritz RA (2019a). Early-onset autoimmune vitiligo associated with an enhancer variant haplotype that upregulates class II HLA expression. Nat Comms 10, 391. doi: 10.1038/s41467-019-08337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Santorico SA, & Spritz RA (2019b). Pediatric to adult shift in vitiligo onset suggests altered environmental triggering. J Investig Dermatol 2019 Jun 28. pii: S0022-202X(19)31798-1. doi: 10.1016/j.jid.2019.06.131. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, … & Kathiresan S (2018). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmicki JA, Churchhouse CL, Rivas MA, & Neale BM (2016). Discovery of rare variants for complex phenotypes. Hum Genet 135, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, & Visscher PM (2011). Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet, 88, 294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, … & Visscher PM (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, … & Kenny EE (2017). Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet 100, 635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, & Daly MJ (2019). Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 51, 584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, … & Haplotype Reference Consortium. (2016). A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48, 1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigrovic PA, Raychaudhuri S, & Thompson SD (2018). Review: Genetics and the Classification of Arthritis in Adults and Children. Arthritis Rheumatol 70, 7–17. doi: 10.1002/art.40350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson CC, Dahlquist GG, Gyürüs E, Green A, & Soltész. G (2009). Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–20: a multicentre prospective registration study. Lancet 373, 2027–2033. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- Polderman TJ, Benyamin B, De Leeuw CA, Sullivan PF, Van Bochoven A, Visscher PM, & Posthuma D (2015). Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47, 702–709. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- Roberts GHL, Paul S, Yorgov D, Santorico SA, & Spritz RA (2019a). Family clustering of autoimmune vitiligo results principally from polygenic inheritance of common risk alleles. Am J Hum Genet 105, 364–372. doi: 10.1016/j.ajhg.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GHL, Santorico SA, & Spritz RA (2019b). Deep genotype imputation captures virtually all heritability of autoimmune vitiligo. Hum Molec Genet, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GHL & Spritz RA (2018). Genetics of Vitiligo. eLS, John Wiley & Sons, Ltd. doi: 10.1002/9780470015902.a0026934. [DOI] [Google Scholar]
- Sagi L & Trau H (2011). The Koebner phenomenon. Clinics Dermatol 29, 231–236. doi: 10.1016/j.clindermatol.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Spritz RA & Andersen GH (2017). Genetics of vitiligo. Dermatol Clin 35, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüttgen G (1950). Die Vitiligo in erbbiologischer Betrachtung [Hereditary aspects of vitiligo]. Zeitschr Haut- u. Geschlechtskrankheiten 9, 451–457. [PubMed] [Google Scholar]
- Tenesa A & Haley CS (2013). The heritability of human disease: estimation, uses and abuses. Nat Rev Genet 14, 139. doi: 10.1038/nrg3377. [DOI] [PubMed] [Google Scholar]
- Teindel H (1950). Familiäre Vitiligo [Familial vitiligo]. Zeitschr Haut- u. Geschlechtskrankheiten 9, 456–462. [PubMed] [Google Scholar]
- van Geel N, Speeckaert R, De Wolf J, Bracke S, Chevolet I, Brochez L, & Lambert J (2012). Clinical significance of Koebner phenomenon in vitiligo. Br J Dermatol 167, 1017–1024. doi: 10.1111/j.1365-2133.2012.11158.x. [DOI] [PubMed] [Google Scholar]
- Wald NJ & Old R (2019). The illusion of polygenic disease risk prediction. Genet Med 21, 1705–1707. [DOI] [PubMed] [Google Scholar]
- Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AA, Lee … & Visscher PM (2015). Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 47, 1114–1120. doi: 10.1038/ng.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Liu JB, Gui JP, Li M, Xiong QG, Wu HB, … & Yang Q (2004). Characteristics of genetic epidemiology and genetic models for vitiligo. J Am Acad Dermatol 51, 383–390. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cai Y, Shi M, Jiang S, Cui S, Wu Y, … & Chen H-D (2016). The prevalence of vitiligo: A meta-analysis. PLoS One 2016 Sep 27;11(9):e0163806 eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]