Abstract
Adenoviruses represent ubiquitous and clinically significant human pathogens, gene-delivery vectors, and oncolytic agents. The study of adenovirus-infected cells has long been used as an excellent model to investigate fundamental aspects of both DNA virus infection and cellular biology. While many key details supporting a well-established model of adenovirus replication have been elucidated over a period spanning several decades, more recent findings suggest that we have only started to appreciate the complex interplay between viral genome replication and cellular processes. Here we present a concise overview of adenovirus DNA replication, including the biochemical process of replication, the spatial organization of replication within the host cell nucleus, and insights into the complex plethora of virus-host interactions that influence viral genome replication. Finally, we identify emerging areas of research relating to the replication of HAdV genomes.
Keywords: Adenovirus, DNA, virus, viral replication compartment, liquid-liquid phase separation
Introduction:
Adenoviruses (family Adenoviridae) are medium-sized (90–100 nm), non-enveloped viruses with an icosahedral nucleocapsid containing a double-stranded DNA genome. The human adenoviruses (HAdV) comprise 7 species of the Mastadenovirus genus (A-G), including over 50 serotypes as defined by the absence of serological cross-neutralisation 1–3. Serotypes of the Mastadenovirus C species, in particular serotypes 2 and 5 (Ad2 and Ad5), are the best characterized in regard to their molecular biology, and are often utilized experimentally to study adenovirus DNA replication and other aspects of adenovirus biology. HAdV are prolific pathogens that represent a significant human disease burden 4. In addition to their clinical importance as pathogens, HAdV can also be utilized scientifically and clinically as gene expression vectors, oncolytic agents, and vaccine vectors 5–9. Furthermore, the study of HAdV as a prototype DNA virus has provided fundamental insights into important cellular processes, including DNA replication, the cellular DNA damage response (DDR), and RNA processing 10–15. HAdV therefore present excellent opportunities to investigate fundamental aspects of DNA virus and cellular biology.
Viruses are obligate intracellular pathogens, and must therefore infect a host cell and co-opt cellular processes in order to replicate viral genomes and ultimately produce infectious progeny. HAdV replicates in the host cell nucleus, utilizing components of the host nuclear machinery for viral processes, including expression of viral genes, and replication of viral DNA genomes. Following attachment and entry, viral particles undergo a complex process of disassembly to yield viral nucleocapsids, which are transported along microtubules to the nuclear pore complex, where viral genomes are imported into the nucleus 16–18. Replication of the HAdV genome generates de novo genomes that are required for several essential viral processes, including further amplification of viral genome copy number and expression of viral late genes necessary for production of viral particles. In addition, viral genomes are also packaged into particles to produce infectious progeny virions (Figure 1). Therefore, replication of HAdV genomes is essential for the production of viral progeny. Thus, our understanding of viral genome replication fundamentally underpins our understanding of HAdV infection. Decades of work have contributed to a detailed model of biochemical processes of HAdV DNA replication, defining its minimal requirements in vitro, characterizing the replication machinery, and elucidating the fundamental mechanism 12,13,19. However, viral genome replication within the host nuclear environment presents a number of challenges as well as opportunities. The virus must redirect cellular processes and co-opt cellular proteins, manipulate the nuclear architecture, and create its own nuclear sub-compartments to facilitate viral genome replication. Here we present an overview of adenovirus DNA replication, including the biochemical mechanism of replication, the spatial organization of genome replication within the host cell nucleus, and the recruitment of cellular proteins to replicating viral genomes. In doing so, we highlight the importance of understanding adenovirus replication in the context of the complex nuclear environment, and identify emerging areas of research relating to the replication of HAdV genomes.
Figure 1. Overview of HAdV replication cycle.
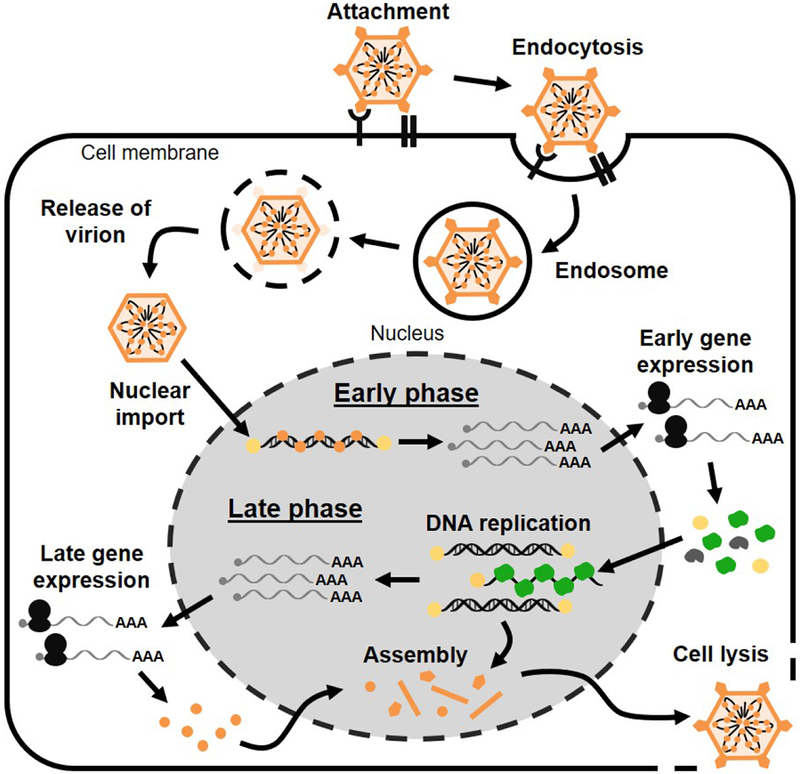
Virus entry and import of viral genomes into the nucleus leads to a program of early gene expression that includes the viral replication machinery. The onset of viral DNA replication marks progression from the early to the late phase of infection, and is a pre-requisite for both late gene expression and virion assembly.
Replication of the adenovirus genome:
Following entry into the nucleus, viral DNA genomes initiate a program of viral early gene expression that includes expression of genes encoding components of the viral DNA replication machinery. DNA replication machinery assembles on viral genomes and replication takes place. This enables productive infection, further genome amplification, and the full complement of viral gene expression. In this section we cover key viral components required for viral DNA replication and discuss how they function in concert to replicate viral genomes.
The adenovirus genome:
HAdV contain a linear, double-stranded DNA genome, typically 30-36 kb in length. By utilizing overlapping open reading frames and allowing transcription from both strands of the genome, as well as alternate splicing to maximize coding potential, this small viral genome encodes multiple genes (Figure 2). Viral early genes encode, amongst other proteins, the pre-terminal protein (pTP, encoded by E2B), DNA polymerase (Ad Pol, E2B), and DNA-binding protein (DBP, E2A), which together constitute the viral replication machinery. Late genes including the major late transcriptional unit (MLTU) are expressed following the onset of viral DNA replication, and encode the capsid and packaging proteins required for assembly of viral particles and encapsidation of the viral genome 20–22. The MLTU comprises 5 transcriptional units (L1-L5), which are typically expressed from the major late promoter (MLP) utilizing alternate splicing and differential polyadenylation 23–27. IX and IVa2, which are not encoded within the MLTU are often described as intermediate-late genes, since pIX and IVa2 facilitate the transcription of MLTU genes, in addition to their role in virion assembly 28–33. The central coding region of the genome is flanked by inverted terminal repeats (ITR) of approximately 100 bp at each of the 5’ and 3’ ends. The terminal ends of each ITR possess the origin of replication. These origins span approximately 50 bp and contain a minimal core origin at the extreme terminal ends, as well as an auxiliary origin. The core origin sequence contains a binding site for pTP and Ad Pol, while the auxiliary origin contains binding sites for the cellular transcription factors Nuclear Factor 1 (NF1) and the POU domain containing protein Oct-1 34,35. These proteins function with DBP to make up the pre-initiation complex that associates with the origin and initiates replication 13,35. In addition, one end of the genome also contains a packaging sequence (ψ) proximal to the ITR. This packaging sequence can be bound by viral packaging proteins, and is required for encapsulation (packaging) of the viral genome within the viral particle 36,37. Viral genomes present within capsids also have two copies of a mature form of the viral terminal protein (TP) covalently attached to the 5’ ends, resulting from the addition of pTP during replication and its subsequent proteolytic cleavage during virion maturation 38.
Figure 2. Schematic representation of the Ad5 genome.
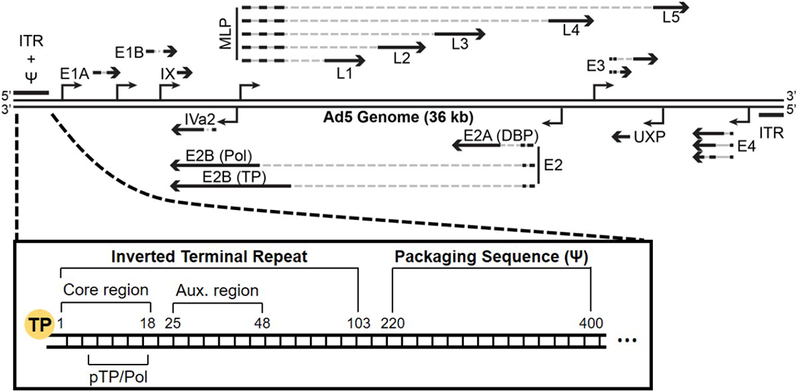
The organization of genes within the central coding region is shown, as are inverted terminal repeats (ITR) and packaging domain (ψ). The zoom-in provides further details of the terminal end that contains the packaging domain, including the core region, auxiliary (Aux.) region, and binding site for pre-terminal protein and the viral polymerase (pTP/Pol).
The viral replication machinery:
Three viral proteins - pTP, Ad Pol, and DBP - are essential for replication of the viral genome. Reconstitution of HAdV DNA replication in vitro has demonstrated that these viral proteins represent the minimal components necessary for replication, whilst the host proteins NF1 and Oct-1 can enhance replication 39–43. pTP is a 76.5 kDa protein that forms a heterodimer with Ad Pol, and along with Ad Pol and DBP forms part of the DNA replication pre-initiation complex where it functions in replication priming and is covalently coupled to the viral DNA as part of this process. Following encapsidation of the viral genome, pTP is cleaved by the adenovirus protease at two sites to yield the 37 kDa mature form of TP via a processing intermediate (iTP) 44. Thus, replicating viral DNA is associated with pTP, while mature TP is present on viral genomes within mature virions and on incoming viral genomes 38,45.
The Ad Pol is a 120 kDa protein that belongs to a subset of family B polymerases that utilize a protein primer for the initiation of replication 46,47. Sequence comparisons to polymerases of known structure, in particular the polymerase of the φ29 bacteriophage, have provided great insight into the likely structure of Ad Pol 48–50. In addition, many biochemical and mutagenesis studies have characterized its functional domains and demonstrated the requirement of these domains for its activity 12,51–55. The C-terminal half of Ad Pol contains the palm, finger, and thumb subdomains that are common to DNA polymerases and function in their 5’ to 3’ polymerase activity 12,56. In common with many other DNA polymerases, Ad Pol contains a 3’ to 5’ exonuclease domain within its N-terminal half that is necessary for its proofreading activity 12. An interesting feature of Ad Pol is the presence of two sub-domains, termed Terminal Protein Region 1 and 2 (TPR1 and TPR2), that are specific to family B polymerases that initiate replication by a protein-primed mechanism, and are thought to facilitate protein-primed replication 47,57–59.
DBP is a 70 kDa protein that likely performs several functions during HAdV genome replication. DBP was first isolated based on its ability to bind ssDNA 60, but can also bind dsDNA 61–63, and even RNA 64–66. A combination of biochemical assays, electron microscopy, and x-ray crystallography have demonstrated that DBP binds to ssDNA in a co-operative manner, and can oligomerize to form a protein chain associated with ssDNA 43,60,67–69. This binding along ssDNA is sequence-independent, with one DBP monomer every 10-15 nucleotides 70. Although DBP has been best characterized in relation to its function in the replication of viral DNA, it has been implicated in a number of different processes including transcription, mRNA stability, and even capsid assembly 29,71–76. DBP is essential for viral DNA replication recapitulated in vitro 77–79. Temperature-sensitive DBP mutant viruses fail to replicate in infected cells at non-permissive temperatures 73,77–80. DBP may contribute to viral DNA replication in multiple ways. Firstly, DBP is part of the replication pre-initiation complex and enhances the rate of initiation of replication 63,81–83. DBP also enhances Ad Pol processivity, and can facilitate template elongation and strand displacement by de-stabilizing the dsDNA helix 84–86. This ability to destabilize dsDNA is dependent on the multimerization of DBP on the displaced ssDNA during replication 87,88. In addition, DBP protects ssDNA intermediates generated during strand displacement from nuclease digestion, and regulates annealing of displaced ssDNA 89,90.
The mechanism of AdV genome replication:
HAdV replication occurs via a mechanism of strand-displacement similar to φ29-like dsDNA bacteriophages of the Podoviridae family 35,47,58,91,92 (Figure 3). Multiple protein-protein and protein-DNA interactions between the viral replication machinery, NF1, Oct-1 and the origin of replication result in the formation of the pre-initiation complex 35,93,94. NF1 and Oct-1 facilitate replication initiation by altering the origin conformation 35,95,96. The initiation of viral genome replication occurs by a protein priming mechanism in which pTP is covalently coupled to what will be the first nucleotide of the nascent chain. In the case of Ad5, the Ad Pol first covalently attaches a dCMP nucleotide to Ser580 of pTP to yield pTP-dCMP (pTP-C) 97,98. The generation of pTP-C acts as the first stage in the formation of the pTP-linked trinucleotide intermediate pTP-CAT. Although initiation occurs at position 4 of the template strand, a “jumping back” mechanism results in this pTP-CAT intermediate base pairing with positions 1-3 99. This is made possible by the presence of a short repeat sequence at the beginning of the AdV origin of replication 35. DBP facilitates initiation by enhancing binding of Ad Pol and NF1 to the origin of replication and enhancing coupling of the first nucleotide to pTP. This occurs on dsDNA, suggesting that DBP does not enhance initiation through origin unwinding, but instead may facilitate this process by modulating origin conformation 63. After replication priming, the Ad Pol dissociates from pTP, and synthesis of the nascent strand can proceed. The pre-initiation complex disassembles, with NF1 dissociating early during initiation and Oct-1 displaced following progression of the replication fork through the origin 94. Ad Pol elongates the nascent strand, facilitated by the dsDNA unwinding ability of DBP as it progressively oligomerizes along displaced ssDNA as the replication fork proceeds. This generates a new DNA duplex while displacing the non-template strand. Displaced ssDNA can subsequently be replicated to generate a new dsDNA genome. Interestingly, ssDNA replication intermediates do not appear to be short-lived, as they accumulate during infection 100–103. However, functional consequences of ssDNA accumulation are unclear. The ITRs of displaced single strands anneal together either through intramolecular or intermolecular interactions to form new dsDNA origins of replication 104,105. Thus, newly generated dsDNA genomes as well as ssDNA replication intermediates can enter into subsequent rounds of replication resulting in amplification of genome copy number that drives productive infection.
Figure 3. Replication of the HAdV genome by strand displacement.
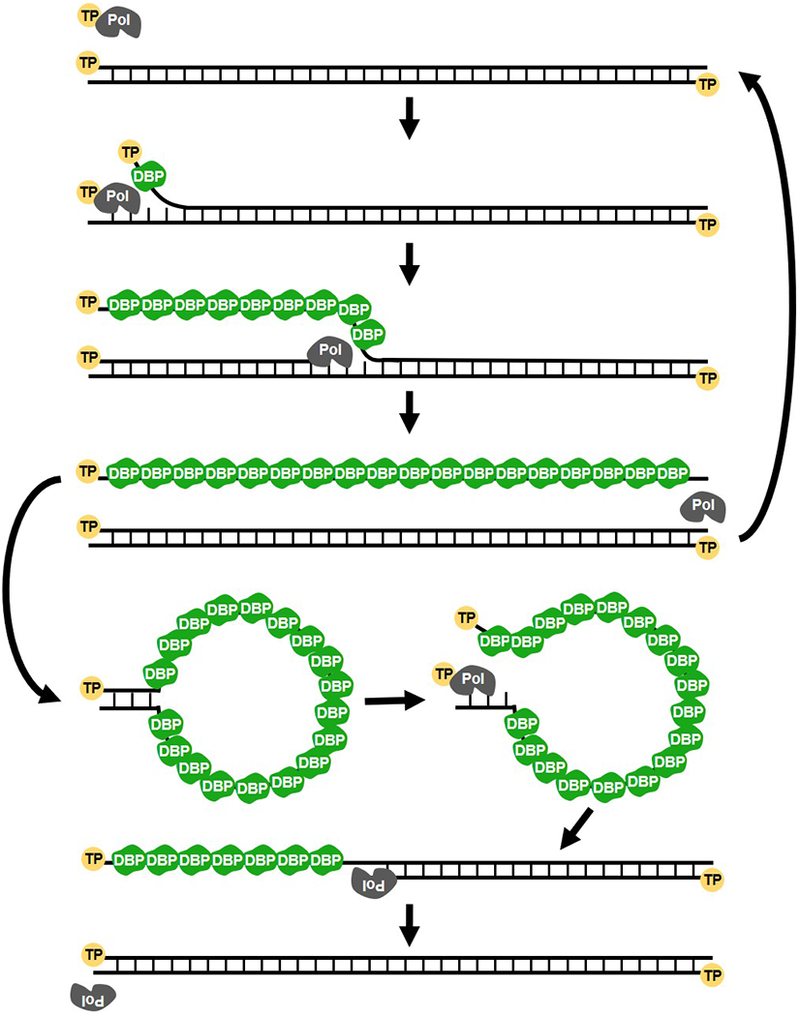
Following replication initiation, the viral polymerase (Pol) elongates the nascent strand displacing the existing strand and generating a new dsDNA template. This process is aided by the viral DNA-binding protein (DBP), which oligomerizes along displaced ssDNA. Both the newly generated dsDNA genome and displaced ssDNA intermediate can be used as templates for further replication.
Spatial organization of adenoviral genome replication and the role of genome replication in viral late processes:
The replication of HAdV genomes takes place within the host cell nucleus, which is a complex environment home to a vast repertoire of cellular processes. Accordingly, HAdV must co-opt or antagonize many cellular pathways, while reorganizing the existing nuclear environment. These architectural changes include the formation of virus-induced membrane-less nuclear compartments that harbor viral genome replication, termed viral replication compartments (VRCs). The formation of VRCs represents a strategy that is common to DNA viruses which replicate in the nucleus and is thought to provide the means to organize and concentrate viral and cellular factors beneficial to the virus, while excluding factors that are inhibitory 106,107. Despite their obvious importance, there is much we still do not understand about how VRCs form, how these compartments change as infection progresses, and how virus-host interactions influence the organization of viral processes at these sites. In this section we address the fate of incoming viral genomes in relation to initiation of VRCs at dedicated sites within the nucleus, discuss details of VRC formation, and review compartmentalization and spatial organization of HAdV genome replication at these sites. In addition, we discuss how viral genome replication is required for viral late processes, and the changes in VRC morphology that occur as infection progresses. In doing so, we highlight emerging questions regarding spatial organization of HAdV genome replication.
The fate of incoming viral genomes:
Following successful viral entry and import of the viral genome into the nucleus, several challenges must be overcome to begin replicating. Viral genomes must avoid cellular intrinsic antiviral defenses and homeostatic regulatory pathways such as the DDR that respond to the presence of foreign DNA and act to suppress viral gene expression and DNA replication 14,15,108–112(See also the review by P. Hearing in this issue). Incoming HAdV genomes that avoid restriction by cellular pathways undergo replication and initiate VRC formation in close proximity to nuclear bodies (NBs) marked by the promyelocytic leukemia (PML) protein 113,114. PML NBs regulate a number of important cellular processes including DNA repair, transcription regulation, cell senescence, apoptosis, and the interferon response, and have been proposed as major sites of the post-translational modification of proteins by small ubiquitin-like modifier (SUMO) 108,115–118. The initiation of VRCs at or in close proximity to PML NBs is common amongst DNA viruses that replicate in the nucleus 106,119. However, PML and components of PML NBs are recruited to sites associated with incoming genomes of DNA viruses and are known to restrict gene expression and replication of DNA viruses as part of an intrinsic antiviral defense. Indeed, many DNA viruses utilize strategies to antagonize this intrinsic defense 108,117–119. This has long raised the question whether the relationship between incoming viral genomes and PML NBs is the result of a viral strategy to utilize these nuclear domains, or a cellular strategy to resist infection. A study on HAdV demonstrated that incoming viral genomes do not localize to PML NBs at early times post-infection, suggesting instead that ongoing HAdV replication may be required to recruit PML NB proteins to VRCs 108. Thus, at least in the case of HAdV, initiation of VRCs at sites of PML NBs does not appear to result from a cellular response to target incoming viral genomes. PML NBs undergo a dramatic reorganization during HAdV infection, largely mediated by viral early proteins. For example, E4orf3 is responsible for a drastic redistribution of PML from spherical PML NBs into PML ‘tracks’ 120,121, while E1B-55K facilitates the proteasome-dependent degradation of DAXX to relieve DAXX-ATRX mediated suppression of viral gene expression 122,123. It has also been suggested that PML NB proteins may be utilized by HAdV, since both anti-viral and pro-viral roles for PML NB proteins have been reported 118–126. Interestingly, some PML NB proteins re-localize to VRCs during infection, including NDP55, specific isoforms of Sp100, and SUMO2/3 124–126. In the case of Sp100, the isoforms B, C, and HMG are reportedly sequestered at VRCs to inhibit their anti-viral function, while Sp100A is maintained in PML tracks so that it can be utilized to promote viral gene expression 126. Therefore, while some PML NB proteins are degraded or sequestered to limit their negative impact on virus infection, it is possible that others may be utilized by the virus to promote viral processes. It is likely that as our understanding of nuclear architecture and spatial and biophysical properties associated with sites of PML NBs advances, so too will our understanding of the role played by PML NBs during HAdV infection.
The formation of viral replication compartments:
Multiple VRCs can be observed in adenovirus infected nuclei, with the number of VRCs initiated dependent on MOI, up to a point of saturation where addition of more virus does not result in an increase in VRC number 114. This suggests that the number of VRCs formed is dependent on the number of incoming viral genomes, with each VRC likely initiated by a single genome. It also implies that the cell is only capable of supporting a limited number of VRCs. Evidence in support of this model can also be extrapolated from studies of other DNA viruses 127–131. The factors limiting the maximum number of VRCs initiated within each nucleus is unknown. However, given that VRCs form at PML NBs which reside within interchromosomal spaces, it is possible that VRC numbers may be limited by the number of sites viable for their initiation and growth. Interestingly, imaging of HAdV VRCs has revealed that host chromatin is excluded from these virus-induced domains, as they stain weakly for cellular histones and the DNA intercalator DAPI when compared to the rest of the nucleus 132–134 (see also the review by H. Wodrich in this issue). This suggests that VRC growth may require the reorganization of cellular chromatin. Exactly how cellular chromatin is excluded from VRCs is unknown, but viral proteins as well as chromatin remodeling factors and components of the cellular DDR have been implicated 132,135 (See also the review in this issue by D. Avgousti). The VRCs of HAdV and other DNA viruses also demonstrate the liquid-like ability to coalesce. Thus, as VRCs grow they may merge with nearby VRCs resulting in fewer, larger compartments that may contain progeny derived from more than one founder genome 131,136,137.
A particularly interesting question regarding the initiation and formation of viral VRCs is how the viral genomes and viral and cellular proteins are concentrated within these membrane-less compartments. An exciting emerging area of cell biology addresses the formation of biomolecular liquid-liquid condensates (LLCs) by the process of liquid-liquid phase separation (LLPS). It is now believed that many cellular membrane-less compartments exist as LLCs, including nuclear domains such as nuclear speckles, Cajal bodies and PML NBs 138,139. The formation of LLCs can be driven by frequent low-affinity interactions between biomolecules, and thus can be facilitated by interactions with proteins containing intrinsically disordered regions (IDRs) and the interaction of proteins with nucleic acids. Hallmarks of LLCs include liquid-like properties. For example, they typically exhibit spherical or rounded morphologies and the ability to coalesce with similar condensates. These features are also exhibited by viral VRCs 106,131. Furthermore, VRCs contain high concentrations of viral nucleic acids and viral proteins, and the presence of IDRs is a common feature in many viral proteins 140,141. Indeed, many HAdV proteins contain or are predicted to contain IDRs including E1A, Penton, E1B-55K, and EIB-93R 142–144. This has led to the suggestion that VRCs may be LLCs resulting from LLPS 107,145. However, a recent study found that although the VRCs of HSV-1 exhibit many of the microscopic hallmarks of LLCs, they were not dissolved by treatment with 1,6-hexanediol, a feature of other cellular LLCs. Furthermore, the authors report that the recruitment of RNA Pol II to VRCs and its movement within VRCs are best explained by a model other than LLPS 146. However, it is also necessary to consider that the role of LLPS in biological processes may be more subtle and complex than current prototype LLCs suggest 147. Nonetheless, it remains an interesting notion that LLPS may play a role in formation of VRCs, including those of HAdV. Alternatively, understanding how such seemingly liquid-like structure might come about independently of classical LLPS, and how biophysical properties of these compartments may influence viral processes is likely to be equally important. Thus, how the properties of resident biomolecules influence formation of VRCs, and how the biophysical properties of VRCs influence HAdV DNA replication, remains an exciting and interesting question.
Viral replication compartments as the sites of viral genome replication:
VRCs play a role in compartmentalizing the infected nucleus, concentrating factors beneficial to the virus and thus providing an environment conducive to viral processes. Consistent with VRCs as the site of adenovirus DNA genome replication, the three viral replication proteins (pTP, Ad Pol and DBP) and the co-opted cellular protein NF1 are known to localize to VRCs 101,103,113,148,149. Viral DNA can also be visualized at VRCs using fluorescent in-situ hybridization (FISH), or techniques that utilize the labelling of DNA through the incorporation of the nucleoside analogues such as 5-bromo-2’-deoxyuridine (BrdU), 5-ethynyl-2´-deoxyuridine (EdU) or 5-ethynyl-2’-deoxycytidine (EdC) during DNA replication 101–103,150–152. Since HAdV infection induces shutoff of host DNA replication, these nucleoside analogues are preferentially incorporated into viral DNA during infection 150,153. By labelling replicating viral DNA using short pulses of EdU, it is possible to mark replicating (nascent) DNA 150,154. Furthermore, DNA replication activity can be detected in VRCs isolated from HAdV-infected cells 155. Thus, VRCs have been confirmed as the bona fide sites of viral genome replication.
Adenovirus VRCs are also believed to be the sites of viral genome transcription and RNA processing, and may even act as sites of viral particle assembly and packaging 106,151. Investigations into the structure of VRCs and the localization of cellular and viral factors to these virus-induced domains suggest that VRCs may facilitate the spatial organization of viral DNA replication, as well as the aforementioned viral processes. Electron microscopy (EM) and immunofluorescence microscopy studies utilizing nucleoside-analogue labelling and FISH identified the accumulation of viral ssDNA replication intermediates within compact fibrillar structures termed single-stranded DNA accumulation sites (ssDAS) 101–103 (Figure 4A–B). Labelling and visualization of replicating DNA using tritiated thymidine identified ongoing viral genome replication within ssDAS in only a limited number of VRCs, suggesting that replication is intermittent within the ssDAS 101. In contrast, the same study found that viral genome replication was consistently detected in the surrounding fibrillo-granular areas of the nucleoplasm, termed the peripheral replicative zone (PRZ) (Figure 4A–B). Consistent with viral genome replication at the PRZ, Ad Pol and pTP predominantly label these sites, as does FISH of viral dsDNA 102,148. Similarly, detection of viral RNA by FISH and the labelling of nascent RNA using tritiated uridine suggest that the PRZ is the major site of viral transcription 156. However, another report suggest that DNA replicated at the PRZ subsequently moves out to the surrounding nucleoplasm, where both viral RNA and cellular RNA processing factors localize, suggesting that transcription of viral genomes and viral RNA biogenesis occurs in proximity to the PRZ 103. These somewhat conflicting interpretations may represent spatial differences in the sites of transcription vs. RNA processing. However, in light of more recent findings that HAdV VRCs undergo morphological changes during the late phase of infection 136,157,158, the possibility that reported differences may represent changes in spatial organization of viral processes at different stages in the viral replication cycle must also be considered. Indeed, the fate and function of viral genomes that are displaced from VRCs remains an interesting question, given that the viral processes of DNA replication, transcription, and perhaps even packaging are thought to be closely associated spatially with VRCs.
Figure 4. Viral replication compartments are reorganized during the late stage of infection.
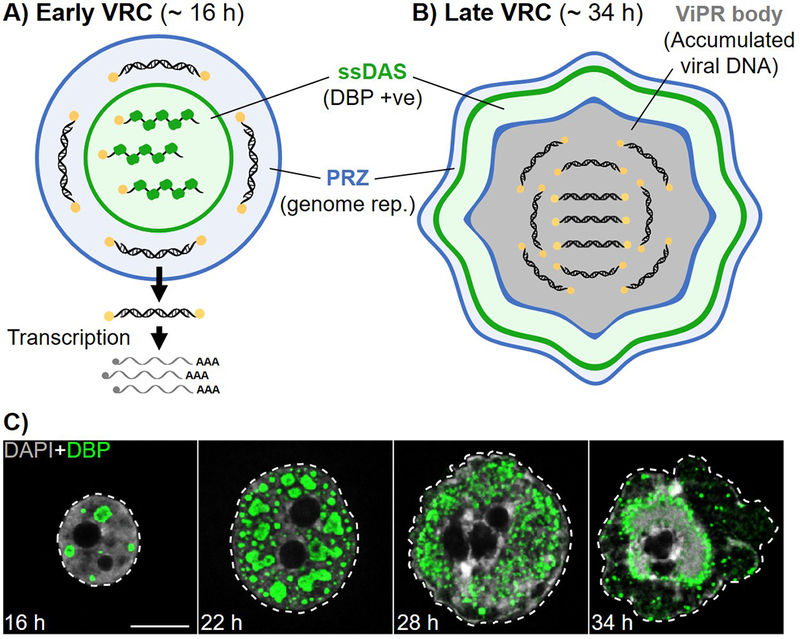
A. Schematic representation of an early viral replication compartment (VRC), showing the single-stranded DNA accumulation site (ssDAS) in which ssDNA replication intermediates bound by the viral DNA-binding protein (DBP) are present (green), and the peripheral replicative zone (PRZ) where viral DNA replication takes place (blue). Viral dsDNA genomes move away from VRCs where transcription and RNA processing take place. B. Schematic representation of a late VRC including virus-induced post-replicative (ViPR) body. C. Human bronchial epithelial cells infected with Ad5 showing VRC morphology at 16, 22, 28 or 34 hours post-infection (h). Nuclei were visualized by confocal microscopy with DBP immuno-labelled (green), and DNA labelled with DAPI (grey). Dashed lines outline nuclei. Scale bar = 10 μm. Note, at 34 h VRCs can be observed as a ring surrounding a single large ViPR body.
Viral genome replication and late gene expression:
Replication of HAdV genomes is a pre-requisite for expression of viral late genes. This dependency on genome replication for late gene expression has been demonstrated in multiple studies that prevented viral DNA replication using inhibitors of DNA replication, inhibitors of protein synthesis, or mutant viruses incapable of DNA replication upon shifting to a non-permissive temperature 80,159–161. Continuous replication does not appear to be essential for late gene expression, as ongoing late gene expression can be detected following inhibition of DNA replication provided that some viral genome replication has taken place prior to inhibition 80. Studies in which cells were superinfected with adenovirus demonstrated that progression of the initial infection into the late phase prior to superinfection was not sufficient to support late gene expression of superinfecting virus if replication of this virus was inhibited 159,162. This suggests that expression of late genes from the MLP may require genome replication in cis. Interestingly, inhibition of viral genome replication also results in an increase in expression of viral early genes 28,72,73,163–167. It is therefore likely that a modification of the viral genome or an interaction with a trans-acting factor that is dependent on replication in cis is required to switch from early to late transcriptional programs. It has been suggested that cellular chromatin organizing protein CTCF may play a role in this switch, since CTCF interacts with viral genomes in a replication-dependent manner, and knockdown of CTCF attenuates DNA replication and late gene expression, but not early gene expression 168. The authors of this study proposed that CTCF could promote HAdV genome replication and late gene expression by regulating viral chromatin, since CTCF is known to regulate cellular chromatin architecture and chromatin conformation of Kaposi’s sarcoma-associated Herpesvirus (KSHV) and Epstein-Barr virus (EBV) genomes 169–171. Others have suggested that viral core proteins or cellular histones associated with viral genomes during early stages of infection may need to be removed in a replication-dependent manner before late gene expression can proceed 135. This hypothesis is particularly attractive given recently reported findings that CTCF can displace nucleosomes from DNA 172. Ultimately, more work is required to understand mechanisms underlying the replication-dependent switch from early to late viral gene expression, which represents an essential feature of the adenovirus replication cycle.
Changes in spatial organization of viral replication compartments during late stages of infection:
VRCs form with the onset of viral DNA replication, following the early phase of infection. Although decades of work have contributed to our understanding of spatial organization of adenovirus genome replication at VRCs, it has only recently become apparent that changes in VRC morphology and nuclear organization during a final phase of infection may represent biologically significant diversions from the traditional view of VRCs organization 136,157,158. During the late phase of infection, DBP-marked VRCs transition from the more commonly described spherical and crescent shaped morphologies, into structure that first appear more diffuse and broken up before forming ring-like assemblies of smaller globular regions marked by DBP. This suggests that early VRCs transition from an early to late morphology as infection progresses (Figure 4C). These morphological changes coincide with the formation of structures recently referred to as virus-induced post-replicative (ViPR) bodies. These structures co-stain with DAPI, suggesting that they are enriched for DNA. In addition, ViPR bodies stained positive for viral core protein VII and packaging protein IVa2, but not capsid proteins pVI and IX 136,157,158. Visualization of viral genomes by EdU pulse-chase or an ANCHOR3/Par3 in vivo DNA-tagging system revealed that, although viral genome replication occurs at VRCs throughout the course of infection, viral genomes produced late in infection accumulate at ViPR bodies 136,157 (see also the review by H. Wodrich in this issue). This led to the suggestion that ViPR bodies may function as sites of DNA accumulation, playing a role in packaging of viral genomes into virions. The nucleolar proteins Mybbp1A, nucleophosmin (NPM1), UBTF, and nucleolin also localized to ViPR bodies 136,157,158. Depletion of NPM1 delayed ViPR body formation, increased resistance of viral genomes to nuclease digestion, and reduced production of infectious virions. Thus, the authors suggest that ViPR bodies contain viral genomes that have been remodelled in preparation for packaging into capsids, and that this remodelling is required for ViPR body formation 158. However, it is uncertain to what extent these phenotypes may also be influenced by the proposed role of nucleophosmin during viral genome replication 173. It is also noteworthy that although prevailing dogma suggests HAdV packages its genomes into pre-formed capsids as described by a sequential model of packaging 37, there is also evidence to support an alternate packaging model. A recent study that included immunofluorescence and electron microscopy identified viral DNA, the 52-55K packaging protein, and formed particles, as well as putative assembly intermediates, within the PRZ, leading to the suggestion that the PRZ is not only the site of viral DNA replication, but also the site of assembly and packaging 151. The model proposed by the authors suggests that packaging proteins interact with nascent viral genomes and facilitate concurrent assembly and packaging as the viral genome is replicated. Rationalizing these different findings will likely require future work to elucidate in detail the fate of viral genomes, and the spatial and temporal regulation of key interactions between viral genomes and cellular and viral proteins that are indicative of viral processes such as packaging.
Recruitment of cellular proteins to replicating viral genomes:
Although HAdV DNA replication can be recapitulated in vitro with minimal components, it is becoming increasingly clear that viral DNA replication within the infected host-cell nucleus is more complex. In addition to NF1 and Oct-1, which play a key role in replication initiation, many other cellular proteins can interact with replicating viral DNA. These include proteins that interact with viral DNA as part of cellular antiviral or homeostatic pathways, proteins that are recruited to viral genomes to facilitate replication, and proteins that are recruited to viral genomes to facilitate other viral processes. It is also evident that viral genomes exist within the nucleus not just as naked viral DNA bound by replication or transcription complexes, but as viral chromatin associated with viral core proteins and/or cellular histones. In this section we highlight recent advances in identification of cellular proteins associated with replicating HAdV DNA and review roles of recruited cellular proteins in viral processes.
Identification of factors recruited to or excluded from replicating viral genomes:
Recent advances allow for identification of proteins on nascent DNA (iPOND) by incorporating EdU into replicating DNA, covalently linking EdU to biotin in a click reaction, and purifying the labelled DNA in association with interacting proteins174–177. This approach has been used in conjunction with mass spectrometry by us and others to identify cellular proteins associated with replicating viral DNA during infection with DNA viruses 150,154. iPOND of HAdV identified cellular proteins involved in DNA replication, DNA repair, and chromatin remodelling, as well as proteins involved in transcription, RNA processing and nucleolar proteins. Identified cellular proteins representative of these processes were confirmed to localize to VRCs, validating their recruitment to the sites of viral genome replication 150. It is interesting to note that cellular proteins recruited to replicating HAdV DNA include not only proteins involved in DNA replication, but also those that interact with nascent genomes to influence subsequent genome functions. The identification of cellular proteins involved in transcription and RNA processing supports this notion and is consistent with the close spatial relationship between these processes (see earlier section: Viral replication compartments as the sites of viral genome replication). In addition, many viral proteins were found to be associated with replicating viral DNA. These included the viral replication machinery (pTP, Ad Pol and DBP), as well as viral proteins involved in RNA splicing, packaging, and even capsid proteins 150. This suggests that replication and transcription of viral genomes are intimately linked both spatially and temporally.
In addition to identification of cellular proteins recruited to replicating viral genomes, analysis of cellular proteins that were under-represented on replicating HAdV DNA compared to replicating host DNA also provided insight into host factors that may be actively excluded from viral genomes during replication. Amongst under-represented proteins were the components of the MRN complex - MRE11, RAD50, and NBS1 - consistent with known antagonism of the MRN complex by viral early proteins 11,178–183. Interestingly, Claspin was the most underrepresented protein on replicating HAdV DNA compared to host DNA and was excluded from VRCs 150. During replication stress and DNA damage, Claspin associates with the Chk1 kinase to facilitate Chk1-mediated signalling, slowing or stalling DNA replication as a result 184–186. It therefore seems likely that exclusion of Claspin from replicating viral genomes is the result of a viral strategy to antagonize the otherwise inhibitory effects of the DDR. Another such under-represented protein, TFII-I decreased in abundance during WT HAdV infection, and was re-localized to foci distinct from VRCs. In contrast, TFII-I localized to VRCs during infection with an E4-deleted virus lacking key antagonistic functions provided by E4orf3 and E4orf6, and was not reduced in abundance 150. TFII-I functions as a transcriptional repressor in many cellular processes 187, raising the possibility that TFII-I is prevented from interacting with replicating viral genomes to antagonize repression of viral gene expression. Interestingly, although many DDR proteins were excluded from replicating HAdV DNA, the structure-specific endonuclease subunit SLX4 was enriched 150. SLX4 promotes DNA repair as part of multiple DDR pathways, functioning as a SUMO E3 ligase and coordinating structure-specific endonucleases 188–193. Depletion of SLX4 demonstrated that SLX4 promotes viral genome accumulation and protein production, suggesting SLX4 is recruited to replicating viral genomes where it functions to promote viral processes 150. Thus, DDR proteins are not only antagonized by HAdV, but also exploited. In summary, comparing proteomes associated with replicating DNA in HAdV infected cells to uninfected cells presents an excellent tool to identify cellular factors that facilitate viral processes, as well as factors that are excluded from VRCs to prevent their interaction with replicating viral genomes.
Viral chromatin and the recruitment of nucleolar proteins:
Within the HAdV capsid, the viral genome exists as viral chromatin. Specifically, viral DNA in association with highly basic core proteins V, VII and Mu (μ) 17,37,194,195. During uncoating, μ and V dissociate from viral genomes, while VII and viral genomes are imported into the nucleus as a VII-DNA complex 16,17,135. As infection progresses, VII is lost from viral genomes, which become increasingly associated with cellular histones, in particular histone H3.3 135,196–198. The extent to which VII is lost, and cellular histones are added during the early phase of infection is still a matter of debate, but it is likely that a balance of both VII and cellular histones may be required for efficient viral early gene expression 135,196. However, the extent to which viral chromatin must be modified to promote replication during infection is less clear. In vitro replication assays in which viral genomes in association with viral core proteins are used as a template indicate that compacted core-associated genomes undergo only limited replication, suggesting that modification of viral chromatin may be required for efficient DNA replication. The cellular histone-chaperone proteins SET (TAF-Iβ) and nucleosome assembly protein 1 (Nap-1 aka TFA-II) have been shown to enhance replication in vitro when viral chromatin is used as template 173,199,200. The ability of these template-activating factors to promote replication is thought to be due to remodelling of viral chromatin, as has been suggested for SET 201–203.
It is also interesting to note that V, VII, and μ have been implicated in disruption of the nucleolus during HAdV infection, and redistribution of nucleolar proteins including nucleolin, nucleophosmin, nucleolar and coiled-body phosphoprotein 1 (NOLC1), and upstream-binding factor 1 (UBTF) 134,204–208. Similarly to SET and Nap-1, nucleophosmin has been shown to promote replication of viral chromatin in vitro, raising the possibility that nucleophosmin is also able to modify the structure of viral chromatin to facilitate replication 173. Furthermore, UBTF also appears to promote viral genome replication 208. It is therefore interesting that nucleolar proteins - including NOLC1, treacle protein (aka TCOF1), and components of the RNA polymerase I (POL I) complex and the small subunit (SSU) processome - were identified by iPOND to associate with replicating HAdV DNA, and localized to VRCs 150. NOLC1 and TCOF1 regulate ribosomal RNA biogenesis and processing via recruitment of POL I and SSU processome components 209–212. Depletion of TCOF1 resulted in failure of NOLC1, POL I, and SSU processome components to localize to VRCs in HAdV infected cells, and reduced viral protein production and genome copy number 150. This suggests that TCOF1 and TCOF1-mediated recruitment of POL I and SSU processome components promotes viral processes. Together, these findings suggest that redistribution of nucleolar proteins and their recruitment to VRCs represents a viral strategy to harness key nucleolar processes. Given the link between nucleolar proteins, viral core proteins, and genome replication, investigating how these nucleolar proteins function to promote viral processes may also shed more light on how the nature of viral chromatin influences replication of viral genomes.
While it is likely that the modification of viral chromatin influences viral DNA replication, how the presence of cellular histones on viral DNA may influence this process is unknown. Although cellular histones may be present on both incoming and de novo viral genomes 133,135,213, it is unclear whether these histones are removed by the process of replication, or indeed whether they must be removed or temporarily displaced for replication to take place. What is clear is that encapsidated viral genomes are devoid of cellular histones 37,135. This suggests either that cellular histones must be removed from viral genomes for encapsidation to occur, or that a proportion of viral genomes must avoid the addition of cellular histones altogether. Factor responsible for removal of cellular histones from viral genomes have not been identified. Thus, how adenovirus deals with the addition of cellular histones on its DNA to transcribe, replicate, and package genomes remains a fascinating question.
Conclusions and future perspectives:
Although many decades of research have contributed to a detailed model of HAdV DNA replication and to the characterization of viral replication machinery, we have only just begun to understand the complexity of HAdV genome replication in the context of the host-cell nucleus. Important questions regarding the formation and biophysical properties of VRCs remain. Furthermore, recent advances suggest that genome replication, transcription, and possibly even packaging are closely linked, both spatially and temporally. Thus, it will be interesting to determine how the vast repertoire of host proteins that interact with replicating viral DNA may impact not only viral genome replication, but also subsequent genome functions. Identification of morphological changes in VRCs raises further questions as to how viral late processes are organized spatially as infection progresses, and we can only begin to speculate as to how these late viral processes are coordinated in space and time to maintain concurrent DNA replication, transcription and packaging. Ultimately, future work aiming to elucidate in greater detail HAdV DNA replication in the context of the nuclear environment will be highly pertinent to our understanding of DNA viruses and nuclear processes.
Acknowledgments:
We thank members of the Weitzman lab for critical reading of the manuscript and helpful discussions. We apologize to those whose primary research papers could not be cited due to space constraints. Research on Adenovirus replication in the Weitzman laboratory is supported by grants to M.D.W. from the National Institutes of Health (CA097093 and AI21321).
Abbreviations
- Ad Pol
adenovirus DNA polymerase
- Ad2
adenovirus serotype 2
- Ad5
adenovirus serotype 5
- BrdU
5-bromo-2’-deoxyuridine
- DBP
DNA-binding protein
- DDR
DNA damage response
- EBV
Epstein-Barr virus
- EdC
5-ethynyl-2’-deoxycytidine
- EdU
5-ethynyl-2´-deoxyuridine
- EM
electron microscopy
- FISH
fluorescent in-situ hybridization
- HAdV
human adenovirus
- IDR
intrinsically disordered region
- iPOND
identification of proteins on nascent DNA
- iTP
intermediate terminal protein
- ITR
inverted terminal repeats
- KSHV
Kaposi’s sarcoma-associated Herpesvirus
- LLC
liquid-liquid condensate
- LLPS
liquid-liquid phase separation
- MLP
major late promoter
- MLTU
major late transcriptional unit
- NB
nuclear bodies
- NF1
nuclear Factor 1
- NOLC1
nucleolar and coiled-body phosphoprotein 1
- PML
promyelocytic leukemia
- POL I
RNA polymerase I
- PRZ
peripheral replicative zone
- pTP
pre-terminal protein
- ssDAS
single-stranded DNA accumulation sites
- SSU
small subunit
- SUMO
small ubiquitin-like modifier
- TP
terminal protein
- TPR1
terminal protein region 1
- TPR2
terminal protein region 1
- UBTF
upstream-binding factor 1
- ViPR
virus-induced post-replicative
- VRC
viral replication compartment
- WT
wild-type
- ψ
packaging sequence
References:
- 1.Ghebremedhin B Human adenovirus: Viral pathogen with increasing importance. Eur J Microbiol Immunol (Bp) 4, 26–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hage E, Gerd Liebert U, Bergs S, Ganzenmueller T & Heim A Human mastadenovirus type 70: a novel, multiple recombinant species D mastadenovirus isolated from diarrhoeal faeces of a haematopoietic stem cell transplantation recipient. J. Gen. Virol 96, 2734–2742 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Hage E et al. Three novel, multiple recombinant types of species of human mastadenovirus D (HAdV-D 73, 74 & 75) isolated from diarrhoeal faeces of immunocompromised patients. J. Gen. Virol 98, 3037–3045 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Khanal S, Ghimire P & Dhamoon AS The Repertoire of Adenovirus in Human Disease: The Innocuous to the Deadly. Biomedicines 6, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker AT, Aguirre-Hernández C, Halldén G & Parker AL Designer Oncolytic Adenovirus: Coming of Age. Cancers (Basel) 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee CS et al. Adenovirus-Mediated Gene Delivery: Potential Applications for Gene and Cell-Based Therapies in the New Era of Personalized Medicine. Genes Dis 4, 43–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wold WSM & Toth K Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther 13, 421–433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y et al. An adenovirus serotype 2-vectored ebolavirus vaccine generates robust antibody and cell-mediated immune responses in mice and rhesus macaques. Emerg Microbes Infect 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert SC Adenovirus-vectored Ebola vaccines. Expert Rev Vaccines 14, 1347–1357 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Berk AJ Discovery of RNA splicing and genes in pieces. Proc. Natl. Acad. Sci. U.S.A 113, 801–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pancholi NJ, Price AM & Weitzman MD Take your PIKK: tumour viruses and DNA damage response pathways. Philos. Trans. R. Soc. Lond., B, Biol. Sci 372, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Naismith JH & Hay RT Adenovirus DNA replication. Curr. Top. Microbiol. Immunol 272, 131–164 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Hoeben RC & Uil TG Adenovirus DNA replication. Cold Spring Harb Perspect Biol 5, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weitzman MD & Fradet-Turcotte A Virus DNA Replication and the Host DNA Damage Response. Annu Rev Virol 5, 141–164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luftig MA Viruses and the DNA Damage Response: Activation and Antagonism. Annu Rev Virol 1, 605–625 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Kremer EJ & Nemerow GR Adenovirus tales: from the cell surface to the nuclear pore complex. PLoS Pathog. 11, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee PK, Vayda ME & Flint SJ Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J. Mol. Biol 188, 23–37 (1986). [DOI] [PubMed] [Google Scholar]
- 18.Greber UF & Flatt JW Adenovirus Entry: From Infection to Immunity. Annu Rev Virol 6, 177–197 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Winnacker EL Adenovirus DNA: structure and function of a novel replicon. Cell 14, 761–773 (1978). [DOI] [PubMed] [Google Scholar]
- 20.Zhao H, Chen M & Pettersson U A new look at adenovirus splicing. Virology 456–457, 329–341 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Babich A, Nevins JR & Darnell JE Early capping of transcripts from the adenovirus major late transcription unit. Nature 287, 246–248 (1980). [DOI] [PubMed] [Google Scholar]
- 22.Fraser NW, Nevins JR, Ziff E & Darnell JE The major late adenovirus type-2 transcription unit: termination is downstream from the last poly(A) site. J. Mol. Biol 129, 643–656 (1979). [DOI] [PubMed] [Google Scholar]
- 23.Ramke M et al. The 5’UTR in human adenoviruses: leader diversity in late gene expression. Sci Rep 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chow LT, Gelinas RE, Broker TR & Roberts RJ An amazing sequence arrangement at the 5’ ends of adenovirus 2 messenger RNA. Cell 12, 1–8 (1977). [DOI] [PubMed] [Google Scholar]
- 25.Berget SM, Moore C & Sharp PA Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U.S.A 74, 3171–3175 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nevins JR & Darnell JE Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell 15, 1477–1493 (1978). [DOI] [PubMed] [Google Scholar]
- 27.Nevins JR & Chen-Kiang S Processing of adenovirus nuclear RNA to mRNA. Adv. Virus Res 26, 1–35 (1981). [DOI] [PubMed] [Google Scholar]
- 28.Binger MH & Flint SJ Accumulation of early and intermediate mRNA species during subgroup C adenovirus productive infections. Virology 136, 387–403 (1984). [DOI] [PubMed] [Google Scholar]
- 29.Christensen JB et al. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J. Virol 82, 9086–9093 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutz P & Kedinger C Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J. Virol 70, 1396–1405 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutz P, Rosa-Calatrava M & Kedinger C The product of the adenovirus intermediate gene IX is a transcriptional activator. J. Virol 71, 5102–5109 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks RJ Adenovirus protein IX: a new look at an old protein. Mol. Ther 11, 19–25 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Zhang W & Imperiale MJ Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol 77, 3586–3594 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hay RT The origin of adenovirus DNA replication: minimal DNA sequence requirement in vivo. EMBO J. 4, 421–426 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Jong RN, van der Vliet PC & Brenkman AB Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr. Top. Microbiol. Immunol 272, 187–211 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Ostapchuk P & Hearing P Regulation of adenovirus packaging. Curr. Top. Microbiol. Immunol 272, 165–185 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Ahi YS & Mittal SK Components of Adenovirus Genome Packaging. Front Microbiol 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangel WF & San Martín C Structure, function and dynamics in adenovirus maturation. Viruses 6, 4536–4570 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mul YM & Van der Vliet PC Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J. 11, 751–760 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mul YM, Verrijzer CP & van der Vliet PC Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J. Virol 64, 5510–5518 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coenjaerts FE, van Oosterhout JA & van der Vliet PC The Oct-1 POU domain stimulates adenovirus DNA replication by a direct interaction between the viral precursor terminal protein-DNA polymerase complex and the POU homeodomain. EMBO J. 13, 5401–5409 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Vries E, van Driel W, Tromp M, van Boom J & van der Vliet PC Adenovirus DNA replication in vitro: site-directed mutagenesis of the nuclear factor I binding site of the Ad2 origin. Nucleic Acids Res. 13, 4935–4952 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dekker J, van Oosterhout JA & van der Vliet PC Two regions within the DNA binding domain of nuclear factor I interact with DNA and stimulate adenovirus DNA replication independently. Mol. Cell. Biol 16, 4073–4080 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webster A, Leith IR, Nicholson J, Hounsell J & Hay RT Role of preterminal protein processing in adenovirus replication. J. Virol 71, 6381–6389 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell WC Adenoviruses: update on structure and function. J. Gen. Virol 90, 1–20 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Johansson E & Dixon N Replicative DNA polymerases. Cold Spring Harb Perspect Biol 5, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salas M Protein-priming of DNA replication. Annu. Rev. Biochem 60, 39–71 (1991). [DOI] [PubMed] [Google Scholar]
- 48.Kamtekar S et al. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell 16, 609–618 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Kamtekar S et al. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 25, 1335–1343 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berman AJ et al. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 26, 3494–3505 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joung I, Horwitz MS & Engler JA Mutagenesis of conserved region I in the DNA polymerase from human adenovirus serotype 2. Virology 184, 235–241 (1991). [DOI] [PubMed] [Google Scholar]
- 52.Joung I & Engler JA Mutations in two cysteine-histidine-rich clusters in adenovirus type 2 DNA polymerase affect DNA binding. J. Virol 66, 5788–5796 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Naismith JH & Hay RT Identification of conserved residues contributing to the activities of adenovirus DNA polymerase. J. Virol 74, 11681–11689 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brenkman AB, Heideman MR, Truniger V, Salas M & van der Vliet PC The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J. Biol. Chem 276, 29846–29853 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Brenkman AB, Breure EC & van der Vliet PC Molecular architecture of adenovirus DNA polymerase and location of the protein primer. J. Virol 76, 8200–8207 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steitz TA DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem 274, 17395–17398 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Rodríguez I et al. A specific subdomain in phi29 DNA polymerase confers both processivity and strand-displacement capacity. Proc. Natl. Acad. Sci. U.S.A 102, 6407–6412 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salas M, Holguera I, Redrejo-Rodríguez M & de Vega M DNA-Binding Proteins Essential for Protein-Primed Bacteriophage Φ29 DNA Replication. Front Mol Biosci 3, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dufour E et al. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol 304, 289–300 (2000). [DOI] [PubMed] [Google Scholar]
- 60.van der Vliet PC, Keegstra W & Jansz HS Complex formation between the adenovirus type 5 DNA-binding protein and single-stranded DNA. Eur. J. Biochem 86, 389–398 (1978). [DOI] [PubMed] [Google Scholar]
- 61.Fowlkes DM, Lord ST, Linné T, Pettersson U & Philipson L Interaction between the adenovirus DNA-binding protein and double-stranded DNA. J. Mol. Biol 132, 163–180 (1979). [DOI] [PubMed] [Google Scholar]
- 62.Schechter NM, Davies W & Anderson CW Adenovirus coded deoxyribonucleic acid binding protein. Isolation, physical properties, and effects of proteolytic digestion. Biochemistry 19, 2802–2810 (1980). [DOI] [PubMed] [Google Scholar]
- 63.van Breukelen B, Brenkman AB, Holthuizen PE & van der Vliet PC Adenovirus type 5 DNA binding protein stimulates binding of DNA polymerase to the replication origin. J. Virol 77, 915–922 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cleghon VG & Klessig DF Association of the adenovirus DNA-binding protein with RNA both in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A 83, 8947–8951 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seiberg M, Aloni Y & Levine AJ The adenovirus type 2 DNA-binding protein interacts with the major late promoter attenuated RNA. J. Virol 63, 1134–1141 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adam SA & Dreyfuss G Adenovirus proteins associated with mRNA and hnRNA in infected HeLa cells. J. Virol 61, 3276–3283 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanellopoulos PN, van der Zandt H, Tsernoglou D, van der Vliet PC & Tucker PA Crystallization and preliminary X-ray crystallographic studies on the adenovirus ssDNA binding protein in complex with ssDNA. J. Struct. Biol 115, 113–116 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Tucker PA et al. Crystal structure of the adenovirus DNA binding protein reveals a hook-on model for cooperative DNA binding. EMBO J. 13, 2994–3002 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuil ME, van Amerongen H, van der Vliet PC & van Grondelle R Complex formation between the adenovirus DNA-binding protein and single-stranded poly(rA). Cooperativity and salt dependence. Biochemistry 28, 9795–9800 (1989). [DOI] [PubMed] [Google Scholar]
- 70.van Amerongen H, van Grondelle R & van der Vliet PC Interaction between adenovirus DNA-binding protein and single-stranded polynucleotides studied by circular dichroism and ultraviolet absorption. Biochemistry 26, 4646–4652 (1987). [DOI] [PubMed] [Google Scholar]
- 71.Chang LS & Shenk T The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J. Virol 64, 2103–2109 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Babich A & Nevins JR The stability of early adenovirus mRNA is controlled by the viral 72 kd DNA-binding protein. Cell 26, 371–379 (1981). [DOI] [PubMed] [Google Scholar]
- 73.Carter TH & Blanton RA Possible role of the 72,000 dalton DNA-binding protein in regulation of adenovirus type 5 early gene expression. J. Virol 25, 664–674 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu X et al. Adenovirus DNA binding protein interacts with the SNF2-related CBP activator protein (SrCap) and inhibits SrCap-mediated transcription. J. Virol 75, 10033–10040 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahi YS, Vemula SV & Mittal SK Adenoviral E2 IVa2 protein interacts with L4 33K protein and E2 DNA-binding protein. J. Gen. Virol 94, 1325–1334 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Nicolas JC, Sarnow P, Girard M & Levine AJ Host range temperature-conditional mutants in the adenovirus DNA binding protein are defective in the assembly of infectious virus. Virology 126, 228–239 (1983). [DOI] [PubMed] [Google Scholar]
- 77.Dekker J et al. ATP-independent DNA unwinding by the adenovirus single-stranded DNA binding protein requires a flexible DNA binding loop. J. Mol. Biol 277, 825–838 (1998). [DOI] [PubMed] [Google Scholar]
- 78.Tsuji M, van der Vliet PC & Kitchingman GR Temperature-sensitive mutants of adenovirus single-stranded DNA-binding protein. Inability to support DNA replication is associated with an altered DNA-binding activity of the protein. J. Biol. Chem 266, 16178–16187 (1991). [PubMed] [Google Scholar]
- 79.Meyers ML et al. Purification and functional characterization of adenovirus ts111A DNA-binding protein. Fluorescence studies of protein-nucleic acid binding. J. Biol. Chem 265, 5875–5882 (1990). [PubMed] [Google Scholar]
- 80.Carter TH & Ginsberg HS Viral transcription in KB cells infected by temperature-sensitive ‘early’ mutants of adenovirus type 5. J. Virol 18, 156–166 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mul YM & van der Vliet PC The adenovirus DNA binding protein effects the kinetics of DNA replication by a mechanism distinct from NFI or Oct-1. Nucleic Acids Res. 21, 641–647 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cleat PH & Hay RT Co-operative interactions between NFI and the adenovirus DNA binding protein at the adenovirus origin of replication. EMBO J. 8, 1841–1848 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stuiver MH & van der Vliet PC Adenovirus DNA-binding protein forms a multimeric protein complex with double-stranded DNA and enhances binding of nuclear factor I. J. Virol 64, 379–386 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lindenbaum JO, Field J & Hurwitz J The adenovirus DNA binding protein and adenovirus DNA polymerase interact to catalyze elongation of primed DNA templates. J. Biol. Chem 261, 10218–10227 (1986). [PubMed] [Google Scholar]
- 85.Monaghan A, Webster A & Hay RT Adenovirus DNA binding protein: helix destabilising properties. Nucleic Acids Res. 22, 742–748 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zijderveld DC & van der Vliet PC Helix-destabilizing properties of the adenovirus DNA-binding protein. J. Virol 68, 1158–1164 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dekker J et al. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J. 16, 1455–1463 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Breukelen B, Kanellopoulos PN, Tucker PA & van der Vliet PC The formation of a flexible DNA-binding protein chain is required for efficient DNA unwinding and adenovirus DNA chain elongation. J. Biol. Chem 275, 40897–40903 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Mul YM, van Miltenburg RT, De Clercq E & van der Vliet PC Mechanism of inhibition of adenovirus DNA replication by the acyclic nucleoside triphosphate analogue (S)-HPMPApp: influence of the adenovirus DNA binding protein. Nucleic Acids Res. 17, 8917–8929 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zijderveld DC, Stuiver MH & van der Vliet PC The adenovirus DNA binding protein enhances intermolecular DNA renaturation but inhibits intramolecular DNA renaturation. Nucleic Acids Res. 21, 2591–2598 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kazlauskas D, Krupovic M & Venclovas Č The logic of DNA replication in double-stranded DNA viruses: insights from global analysis of viral genomes. Nucleic Acids Res. 44, 4551–4564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meijer WJ, Horcajadas JA & Salas M Phi29 family of phages. Microbiol. Mol. Biol. Rev 65, 261–287 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Jong RN & van der Vliet PC Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene 236, 1–12 (1999). [DOI] [PubMed] [Google Scholar]
- 94.van Leeuwen HC, Rensen M & van der Vliet PC The Oct-1 POU homeodomain stabilizes the adenovirus preinitiation complex via a direct interaction with the priming protein and is displaced when the replication fork passes. J. Biol. Chem 272, 3398–3405 (1997). [DOI] [PubMed] [Google Scholar]
- 95.Mysiak ME, Bleijenberg MH, Wyman C, Holthuizen PE & van der Vliet PC Bending of adenovirus origin DNA by nuclear factor I as shown by scanning force microscopy is required for optimal DNA replication. J. Virol 78, 1928–1935 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mysiak ME, Wyman C, Holthuizen PE & van der Vliet PC NFI and Oct-1 bend the Ad5 origin in the same direction leading to optimal DNA replication. Nucleic Acids Res. 32, 6218–6225 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smart JE & Stillman BW Adenovirus terminal protein precursor. Partial amino acid sequence and the site of covalent linkage to virus DNA. J. Biol. Chem 257, 13499–13506 (1982). [PubMed] [Google Scholar]
- 98.Desiderio SV & Kelly TJ Structure of the linkage between adenovirus DNA and the 55,000 molecular weight terminal protein. J. Mol. Biol 145, 319–337 (1981). [DOI] [PubMed] [Google Scholar]
- 99.King AJ & van der Vliet PC A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 13, 5786–5792 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Flint SJ, Berget SM & Sharp PA Characterization of single-stranded viral DNA sequences present during replication of adenovirus types 2 and 5. Cell 9, 559–571 (1976). [DOI] [PubMed] [Google Scholar]
- 101.Puvion-Dutilleul F & Puvion E Replicating single-stranded adenovirus type 5 DNA molecules accumulate within well-delimited intranuclear areas of lytically infected HeLa cells. Eur. J. Cell Biol 52, 379–388 (1990). [PubMed] [Google Scholar]
- 102.Puvion-Dutilleul F & Pichard E Segregation of viral double-stranded and single-stranded DNA molecules in nuclei of adenovirus infected cells as revealed by electron microscope in situ hybridization. Biol. Cell 76, 139–150 (1992). [DOI] [PubMed] [Google Scholar]
- 103.Pombo A, Ferreira J, Bridge E & Carmo-Fonseca M Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13, 5075–5085 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Graham FL Covalently closed circles of human adenovirus DNA are infectious. EMBO J. 3, 2917–2922 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Graham FL, Rudy J & Brinkley P Infectious circular DNA of human adenovirus type 5: regeneration of viral DNA termini from molecules lacking terminal sequences. EMBO J. 8, 2077–2085 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schmid M, Speiseder T, Dobner T & Gonzalez RA DNA virus replication compartments. J. Virol 88, 1404–1420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kobiler O & Weitzman MD Herpes simplex virus replication compartments: From naked release to recombining together. PLoS Pathog 15, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Komatsu T, Nagata K & Wodrich H The Role of Nuclear Antiviral Factors against Invading DNA Viruses: The Immediate Fate of Incoming Viral Genomes. Viruses 8, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Diner BA, Lum KK & Cristea IM The emerging role of nuclear viral DNA sensors. J. Biol. Chem 290, 26412–26421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma Z, Ni G & Damania B Innate Sensing of DNA Virus Genomes. Annu Rev Virol 5, 341–362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Everett RD Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol 8, 365–374 (2006). [DOI] [PubMed] [Google Scholar]
- 112.Weitzman MD & Ornelles DA Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24, 7686–7696 (2005). [DOI] [PubMed] [Google Scholar]
- 113.Ishov AM & Maul GG The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol 134, 815–826 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maul GG Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20, 660–667 (1998). [DOI] [PubMed] [Google Scholar]
- 115.Lallemand-Breitenbach V & de Thé H PML nuclear bodies: from architecture to function. Curr. Opin. Cell Biol 52, 154–161 (2018). [DOI] [PubMed] [Google Scholar]
- 116.Scherer M & Stamminger T Emerging Role of PML Nuclear Bodies in Innate Immune Signaling. J. Virol 90, 5850–5854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Everett RD & Chelbi-Alix MK PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89, 819–830 (2007). [DOI] [PubMed] [Google Scholar]
- 118.Everett RD, Boutell C & Hale BG Interplay between viruses and host sumoylation pathways. Nat. Rev. Microbiol 11, 400–411 (2013). [DOI] [PubMed] [Google Scholar]
- 119.Everett RD The spatial organization of DNA virus genomes in the nucleus. PLoS Pathog. 9, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Carvalho T et al. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol 131, 45–56 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ullman AJ & Hearing P Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol 82, 7325–7335 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schreiner S et al. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol 84, 7029–7038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schreiner S et al. Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res. 41, 3532–3550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Doucas V et al. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10, 196–207 (1996). [DOI] [PubMed] [Google Scholar]
- 125.Higginbotham JM & O’Shea CC Adenovirus E4-ORF3 Targets PIAS3 and Together with E1B-55K Remodels SUMO Interactions in the Nucleus and at Virus Genome Replication Domains. J. Virol 89, 10260–10272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Berscheminski J et al. Sp100 isoform-specific regulation of human adenovirus 5 gene expression. J. Virol 88, 6076–6092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sekine E, Schmidt N, Gaboriau D & O’Hare P Spatiotemporal dynamics of HSV genome nuclear entry and compaction state transitions using bioorthogonal chemistry and super-resolution microscopy. PLoS Pathog. 13, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dembowski JA & DeLuca NA Temporal Viral Genome-Protein Interactions Define Distinct Stages of Productive Herpesviral Infection. MBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kobiler O, Brodersen P, Taylor MP, Ludmir EB & Enquist LW Herpesvirus replication compartments originate with single incoming viral genomes. MBio 2, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tomer E et al. Coalescing replication compartments provide the opportunity for recombination between coinfecting herpesviruses. FASEB J. 33, 9388–9403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kobiler O & Weitzman MD Herpes simplex virus replication compartments: From naked release to recombining together. PLoS Pathog. 15, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Avgousti DC et al. A core viral protein binds host nucleosomes to sequester immune danger signals. Nature 535, 173–177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Komatsu T & Nagata K Replication-uncoupled histone deposition during adenovirus DNA replication. J. Virol 86, 6701–6711 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lam YW, Evans VC, Heesom KJ, Lamond AI & Matthews DA Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol. Cell Proteomics 9, 117–130 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Giberson AN, Davidson AR & Parks RJ Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res. 40, 2369–2376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Komatsu T et al. In Vivo Labelling of Adenovirus DNA Identifies Chromatin Anchoring and Biphasic Genome Replication. J. Virol 92, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chang L et al. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc. Natl. Acad. Sci. U.S.A 108, 8539–8540 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shin Y & Brangwynne CP Liquid phase condensation in cell physiology and disease. Science 357, (2017). [DOI] [PubMed] [Google Scholar]
- 139.Itakura AK, Futia RA & Jarosz DF It Pays To Be in Phase. Biochemistry 57, 2520–2529 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xue B et al. Structural disorder in viral proteins. Chem. Rev 114, 6880–6911 (2014). [DOI] [PubMed] [Google Scholar]
- 141.Tokuriki N, Oldfield CJ, Uversky VN, Berezovsky IN & Tawfik DS Do viral proteins possess unique biophysical features? Trends Biochem. Sci 34, 53–59 (2009). [DOI] [PubMed] [Google Scholar]
- 142.Pelka P, Ablack JNG, Fonseca GJ, Yousef AF & Mymryk JS Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol 82, 7252–7263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Flatt JW, Kim R, Smith JG, Nemerow GR & Stewart PL An intrinsically disordered region of the adenovirus capsid is implicated in neutralization by human alpha defensin 5. PLoS ONE 8, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sieber T et al. Intrinsic disorder in the common N-terminus of human adenovirus 5 E1B-55K and its related E1BN proteins indicated by studies on E1B-93R. Virology 418, 133–143 (2011). [DOI] [PubMed] [Google Scholar]
- 145.Heinrich BS, Maliga Z, Stein DA, Hyman AA & Whelan SPJ Phase Transitions Drive the Formation of Vesicular Stomatitis Virus Replication Compartments. MBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McSwiggen DT et al. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Alberti S, Gladfelter A & Mittag T Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Murti KG, Davis DS & Kitchingman GR Localization of adenovirus-encoded DNA replication proteins in the nucleus by immunogold electron microscopy. J. Gen. Virol 71, 2847–2857 (1990). [DOI] [PubMed] [Google Scholar]
- 149.Bosher J, Dawson A & Hay RT Nuclear factor I is specifically targeted to discrete subnuclear sites in adenovirus type 2-infected cells. J. Virol 66, 3140–3150 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Reyes ED et al. Identifying Host Factors Associated with DNA Replicated During Virus Infection. Mol. Cell Proteomics 16, 2079–2097 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Condezo GN & San Martín C Localization of adenovirus morphogenesis players, together with visualization of assembly intermediates and failed products, favor a model where assembly and packaging occur concurrently at the periphery of the replication center. PLoS Pathog. 13, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang I-H et al. Tracking viral genomes in host cells at single-molecule resolution. Cell Host Microbe 14, 468–480 (2013). [DOI] [PubMed] [Google Scholar]
- 153.Halbert DN, Cutt JR & Shenk T Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol 56, 250–257 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dembowski JA & Deluca NA Purification of Viral DNA for the Identification of Associated Viral and Cellular Proteins. J Vis Exp (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hidalgo P et al. Morphological, Biochemical, and Functional Study of Viral Replication Compartments Isolated from Adenovirus-Infected Cells. J. Virol 90, 3411–3427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Puvion-Dutilleul F & Puvion E Sites of transcription of adenovirus type 5 genomes in relation to early viral DNA replication in infected HeLa cells. A high resolution in situ hybridization and autoradiographical study. Biol. Cell 71, 135–147 (1991). [DOI] [PubMed] [Google Scholar]
- 157.Komatsu T et al. Tracking adenovirus genomes identifies morphologically distinct late DNA replication compartments. Traffic 17, 1168–1180 (2016). [DOI] [PubMed] [Google Scholar]
- 158.Genoveso MJ et al. Formation of adenovirus DNA replication compartments and viral DNA accumulation sites by host chromatin regulatory proteins including NPM1. FEBS J. (2019). [DOI] [PubMed] [Google Scholar]
- 159.Crossland LD & Raskas HJ Identification of adenovirus genes that require template replication for expression. J. Virol 46, 737–748 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shaw AR & Ziff EB Transcripts from the adenovirus-2 major late promoter yield a single early family of 3’ coterminal mRNAs and five late families. Cell 22, 905–916 (1980). [DOI] [PubMed] [Google Scholar]
- 161.Larsson S, Svensson C & Akusjärvi G Control of adenovirus major late gene expression at multiple levels. J. Mol. Biol 225, 287–298 (1992). [DOI] [PubMed] [Google Scholar]
- 162.Thomas GP & Mathews MB DNA replication and the early to late transition in adenovirus infection. Cell 22, 523–533 (1980). [DOI] [PubMed] [Google Scholar]
- 163.Craig EA & Raskas HJ Effect of cycloheximide on RNA metabolism early in productive infection with adenovirus 2. J. Virol 14, 26–32 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cross FR & Darnell JE Cycloheximide stimulates early adenovirus transcription if early gene expression is allowed before treatment. J. Virol 45, 683–692 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Katze MG, Persson H & Philipson L Control of adenovirus early gene expression: posttranscriptional control mediated by both viral and cellular gene products. Mol. Cell. Biol 1, 807–813 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lewis JB & Mathews MB Control of adenovirus early gene expression: a class of immediate early products. Cell 21, 303–313 (1980). [DOI] [PubMed] [Google Scholar]
- 167.Nevins JR, Ginsberg HS, Blanchard JM, Wilson MC & Darnell JE Regulation of the primary expression of the early adenovirus transcription units. J. Virol 32, 727–733 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Komatsu T, Sekiya T & Nagata K DNA replication-dependent binding of CTCF plays a critical role in adenovirus genome functions. Sci Rep 3, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Phillips JE & Corces VG CTCF: master weaver of the genome. Cell 137, 1194–1211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kang H, Wiedmer A, Yuan Y, Robertson E & Lieberman PM Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation. PLoS Pathog. 7, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tempera I, Klichinsky M & Lieberman PM EBV latency types adopt alternative chromatin conformations. PLoS Pathog. 7, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Owens N et al. CTCF confers local nucleosome resiliency after DNA replication and during mitosis. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Okuwaki M, Iwamatsu A, Tsujimoto M & Nagata K Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol 311, 41–55 (2001). [DOI] [PubMed] [Google Scholar]
- 174.Lopez-Contreras AJ et al. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep 3, 1105–1116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sirbu BM et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25, 1320–1327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sirbu BM, Couch FB & Cortez D Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc 7, 594–605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sirbu BM et al. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J. Biol. Chem 288, 31458–31467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Carson CT et al. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22, 6610–6620 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stracker TH, Carson CT & Weitzman MD Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418, 348–352 (2002). [DOI] [PubMed] [Google Scholar]
- 180.Evans JD & Hearing P Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol 79, 6207–6215 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mathew SS & Bridge E The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology 365, 346–355 (2007). [DOI] [PubMed] [Google Scholar]
- 182.Schwartz RA et al. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J. Virol 82, 9043–9055 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Brestovitsky A, Nebenzahl-Sharon K, Kechker P, Sharf R & Kleinberger T The Adenovirus E4orf4 Protein Provides a Novel Mechanism for Inhibition of the DNA Damage Response. PLoS Pathog. 12, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chini CCS & Chen J Human claspin is required for replication checkpoint control. J. Biol. Chem 278, 30057–30062 (2003). [DOI] [PubMed] [Google Scholar]
- 185.Lin S-Y, Li K, Stewart GS & Elledge SJ Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. U.S.A 101, 6484–6489 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang Y & Hunter T Roles of Chk1 in Cell Biology and Cancer Therapy. Int J Cancer 134, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Roy AL Biochemistry and biology of the inducible multifunctional transcription factor TFII-I. Gene 274, 1–13 (2001). [DOI] [PubMed] [Google Scholar]
- 188.Guervilly J-H et al. The SLX4 complex is a SUMO E3 ligase that impacts on replication stress outcome and genome stability. Mol. Cell 57, 123–137 (2015). [DOI] [PubMed] [Google Scholar]
- 189.Wei L & Zhao X Roles of SUMO in Replication Initiation, Progression, and Termination. Adv. Exp. Med. Biol 1042, 371–393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Andersen SL et al. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell 35, 128–135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fekairi S et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138, 78–89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Saito TT, Youds JL, Boulton SJ & Colaiácovo MP Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 5, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Muñoz IM et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell 35, 116–127 (2009). [DOI] [PubMed] [Google Scholar]
- 194.Maizel JV, White DO & Scharff MD The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology 36, 126–136 (1968). [DOI] [PubMed] [Google Scholar]
- 195.Russell WC, Laver WG & Sanderson PJ Internal components of adenovirus. Nature 219, 1127–1130 (1968). [DOI] [PubMed] [Google Scholar]
- 196.Komatsu T, Haruki H & Nagata K Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res. 39, 889–901 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ross PJ et al. Assembly of helper-dependent adenovirus DNA into chromatin promotes efficient gene expression. J. Virol 85, 3950–3958 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ross PJ, Kennedy MA & Parks RJ Host cell detection of noncoding stuffer DNA contained in helper-dependent adenovirus vectors leads to epigenetic repression of transgene expression. J. Virol 83, 8409–8417 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Matsumoto K, Nagata K, Ui M & Hanaoka F Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem 268, 10582–10587 (1993). [PubMed] [Google Scholar]
- 200.Kawase H et al. NAP-I is a functional homologue of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells 1, 1045–1056 (1996). [DOI] [PubMed] [Google Scholar]
- 201.Xue Y, Johnson JS, Ornelles DA, Lieberman J & Engel DA Adenovirus protein VII functions throughout early phase and interacts with cellular proteins SET and pp32. J. Virol 79, 2474–2483 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gyurcsik B, Haruki H, Takahashi T, Mihara H & Nagata K Binding modes of the precursor of adenovirus major core protein VII to DNA and template activating factor I: implication for the mechanism of remodeling of the adenovirus chromatin. Biochemistry 45, 303–313 (2006). [DOI] [PubMed] [Google Scholar]
- 203.Haruki H, Gyurcsik B, Okuwaki M & Nagata K Ternary complex formation between DNA-adenovirus core protein VII and TAF-Ibeta/SET, an acidic molecular chaperone. FEBS Lett. 555, 521–527 (2003). [DOI] [PubMed] [Google Scholar]
- 204.Puvion-Dutilleul F & Christensen ME Alterations of fibrillarin distribution and nucleolar ultrastructure induced by adenovirus infection. Eur. J. Cell Biol 61, 168–176 (1993). [PubMed] [Google Scholar]
- 205.Lee TWR, Blair GE & Matthews DA Adenovirus core protein VII contains distinct sequences that mediate targeting to the nucleus and nucleolus, and colocalization with human chromosomes. J. Gen. Virol 84, 3423–3428 (2003). [DOI] [PubMed] [Google Scholar]
- 206.Lee TWR et al. Precursor of human adenovirus core polypeptide Mu targets the nucleolus and modulates the expression of E2 proteins. J. Gen. Virol 85, 185–196 (2004). [DOI] [PubMed] [Google Scholar]
- 207.Matthews DA Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol 75, 1031–1038 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lawrence FJ, McStay B & Matthews DA Nucleolar protein upstream binding factor is sequestered into adenovirus DNA replication centres during infection without affecting RNA polymerase I location or ablating rRNA synthesis. J. Cell. Sci 119, 2621–2631 (2006). [DOI] [PubMed] [Google Scholar]
- 209.Hayano T et al. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J. Biol. Chem 278, 34309–34319 (2003). [DOI] [PubMed] [Google Scholar]
- 210.Lin C-I & Yeh N-H Treacle recruits RNA polymerase I complex to the nucleolus that is independent of UBF. Biochem. Biophys. Res. Commun 386, 396–401 (2009). [DOI] [PubMed] [Google Scholar]
- 211.Ciccia A et al. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc. Natl. Acad. Sci. U.S.A 111, 18631–18636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Werner A et al. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525, 523–527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Giberson AN et al. Human adenoviral DNA association with nucleosomes containing histone variant H3.3 during the early phase of infection is not dependent on viral transcription or replication. Biochem. Cell Biol 96, 797–807 (2018). [DOI] [PubMed] [Google Scholar]


