Abstract
Viruses alter host cell processes to optimize their replication cycle. Human Adenoviruses (Ad) encode proteins that promote viral macromolecular synthesis and counteract innate and adaptive responses to infection. The focus of this review is on how Ad evade innate cellular responses to infection, including an interferon (IFN) response and a DNA damage response (DDR). Ad blocks the IFN response by inhibiting cytoplasmic signaling pathways and the activation of IFN-stimulated genes (ISGs), as well as the functions of ISG products, such as PML. Ad also inhibits DDR sensors, for instance the Mre11-Rad50-Nbs1 complex, and DDR effectors like DNA ligase IV. These innate cellular responses impact many different viruses and studies on Ad have provided broad insight in these areas.
Keywords: Adenovirus, interferon, DNA damage response, innate cellular responses, E1A, E1B, E4, PML, SUMO
Viral Infection and the Induction of Innate Cellular Responses
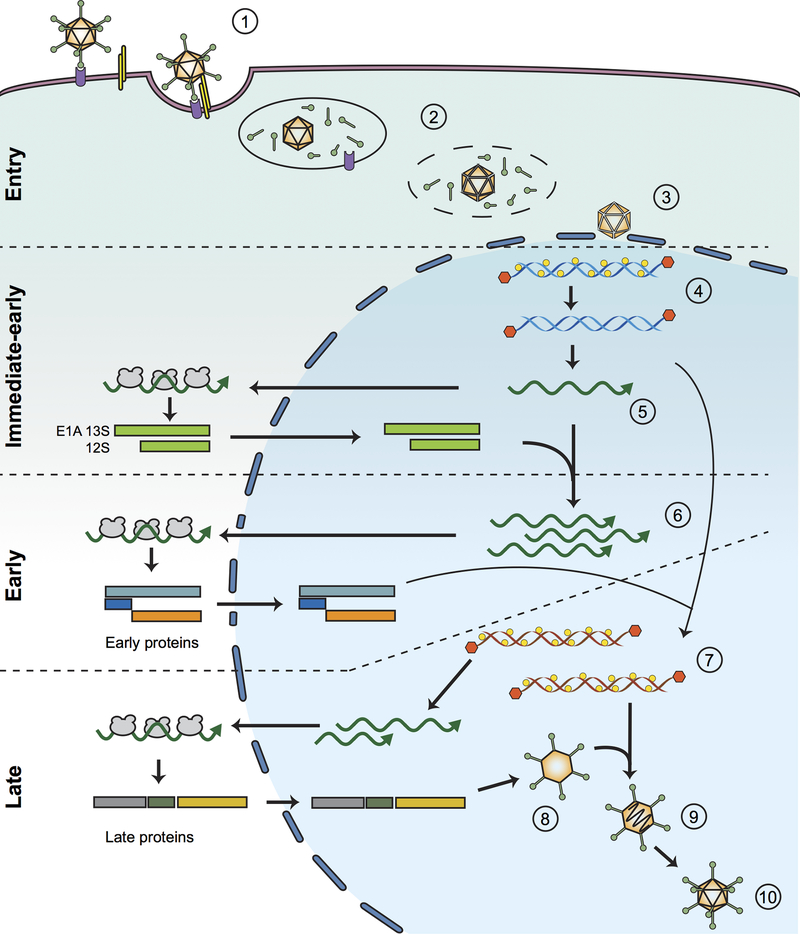
The Ad lytic replication cycle entails a complex suite of events (Figure 1). It begins with the Ad fiber protein of different Ad serotypes binding a primary receptor on the surface of cells (the Coxsackie-Adenovirus receptor (CAR) for most Ad subgroups, CD46 for subgroup C Ads, and desmoglein-2 for subgroup B Ads) followed by secondary interaction between the RGD peptide on the penton base and cellular integrins [1]. Species C Ads are internalized by receptor-mediated endocytosis via clathrin-coated pits, whereas some species B Ads enter by macropinocytosis. Ads evade degradation by escaping from early endosomes and, once liberated into the cytoplasm, the virion is transported to the nucleus along microtubules with the capsid undergoing slow disassembly en route. Upon reaching the nuclear pore complex, the protein VII-wrapped Ad DNA enters the nucleus [1] (Figure 1).
Figure 1. Ad lytic cycle.
Ad attachment to the host cell is initiated by the fiber protein interacting with the cellular CAR receptor or CD46, followed by interaction between the Ad penton base and cellular integrin (1). Ad enters the cell through the endocytic pathway and the viral particle is partially degraded inside the endosome before being released into the cytosol (2). At the nuclear-pore complex, the viral particle is disassembled (3). The Ad DNA-core protein VII complex is transported through the nuclear-pore complex and VII proteins are released from viral genome by early gene transcription (4). E1A is the first viral protein expressed (5) during Ad infection and induces the expression of other early genes that facilitate viral replication and counteract cellular responses to infection (6). After replication occurs (7), the major late promoter is activated to produce late proteins, which are required for the formation of empty procapsids (8). Following encapsidation of the viral genome (9), a viral encoded protease proteolyzes multiple viral proteins inside the virus particle to generate a mature virion that is released from the infected cell (10).
Depending on the cell type, human Ads are recognized by cytoplasmic and nuclear sensing mechanisms that trigger innate cellular responses to infection. In a number of different human cell lines, the Ad genome induces the activation of the cytoplasmic cGAS/STING (Cyclic GMP-AMP Synthase/Stimulator of Interferon Genes) pathway resulting in TBK1 (Tank Binding Kinase 1) and IRF3 (Interferon Regulatory Factor 3) activation and the induction of IFNβ gene expression [2]. In human THP1 monocyte cells, infection with subgroup B or C Ads triggers activation of the NLRP3 (Nucleotide-binding Oligomerization Domain-like Receptor Protein 3) inflammasome pathway to promote an innate response and caspase-dependent activation of IL-1β [3]. Ad-induced IL-1β release is dependent upon endosomal DNA sensor TLR9 sensing of the Ad genome [4]. Many studies in this area have been conducted using mice and mouse cells (see review by Dmitri Shayakhmetov in this issue). In mice, Ad infection induces high levels of type I interferons (IFNs) by plasmacytoid dendritic cells (pDCs), conventional DCs and macrophages. Recognition of Ad in pDCs is mediated by TLR9 (Toll-like Receptor 9), whereas Ad recognition in non-pDCs is TLR-independent and relies on cytosolic sensing of viral DNA [5]. In murine antigen-presenting cells (APCs) and primary lung fibroblasts, Ad infection stimulates IRF3-mediated IFN and proinflammatory responses through a TLR-independent DNA-sensing mechanism [6] dependent, in part, on the cGAS-STING pathway [2, 5, 7].
Ad triggers also nuclear innate cellular responses to infection. In fact, the Ad genome contains two features that promote a DNA damage response in the nucleus. First, the linear Ad DNA genome contains open ends that resemble double-stranded DNA breaks. These ends are recognized by the DDR sensor complex containing Mre11, Rad50, and Nbs1 (the MRN complex) [8]. The MRN complex activates a DDR that ligates the ends of the Ad genome into multimeric concatamers via non-homologous end-joining. Second, the Ad Terminal Protein (TP) is covalently attached to each 5’ end of the viral genome. This complex is equivalent to a DNA-protein adduct that would be expected to stimulate a repair mechanism that removes TP by proteolysis [9]. While this mechanism awaits formal demonstration, empirically, this would be expected to occur and inhibit viral DNA replication by removing TP, which primes the initiation of replication at each amplification cycle. If left unabated, DDR responses to the Ad genome can significantly inhibit viral DNA replication, and consequently, the completion of the viral life cycle [8]. During the early stages of Ad infection, the viral DNA is coated with core protein VII, which protects the genome from triggering a DDR [10]. Protein VII is displaced from the viral genome during the early phase of infection [11], but several viral proteins are expressed during this time that inhibit DDR pathways (see below).
Viral Early Gene Expression
Ad early gene expression is directed toward achieving three main goals. First, the infected cell is stimulated to enter S phase of the cell cycle, which provides an optimal intracellular environment for viral replication. Second, a number of Ad early proteins counteract cellular and host immune responses to infection and maintain a state of balance for persistent infection. Third, both viral gene products and cellular proteins must be produced and used in concert to carry out the replication cycle. We will focus only on Ad early proteins that function to overcome innate cellular responses to infection. Other reviews in this Issue describe Ad proteins that regulate different aspects of viral infection.
Early Region 1A (E1A)
E1A Functions
The first early region expressed after Ad infection is the immediate-early E1A gene. The E1A proteins in turn activate transcription from the other Ad early promoter regions. The E1A gene is comprised of two exons, and several E1A polypeptides are produced following alternative splicing of a primary RNA transcript. The most abundant E1A proteins are the E1A 243 amino acid (243aa, small E1a) and 289 amino acid (289aa, large E1a) products. The E1A 243aa and 289aa proteins act as regulators of viral early transcription, as well as important modulators of host cell gene expression and proliferation [12]. The E1A 243aa and 289aa proteins share two conserved regions within exon 1 (CR1 and CR2) and a conserved region at the C-terminus (CR4). The two proteins differ only in a 46-residue internal exon segment present in the 289aa protein, referred to as conserved region 3 (CR3). This region is important for transcriptional activation of the Ad early genes by the E1A 289aa protein [12].
The E1A proteins exert their effects by interacting with numerous cellular proteins, many of which are involved in transcriptional regulation [12, 13]. The best characterized of these include: 1) the retinoblastoma tumor suppressor family members pRb, p107, and p130 via CR1 and CR2; 2) transcriptional coactivators p300/CBP, PCAF, GCN5, TRRAP, and p400 via amino terminal sequences and CR1; 3) additional transcription factors, such as TATA-binding protein TBP, members of the ATF family, and the RNA polymerase II mediator complex protein MED23 via CR3; and 4) the transcriptional repressor CtBP via CR4. E1A activates gene expression and cellular proliferation via the E2F family of transcription factors [12, 13]. E2F transcription factors play a major role in the expression of cellular genes important for cell cycle progression. E1A proteins sequester Rb-family members and free E2Fs to activate viral and cellular gene expression. p300/CBP, PCAF, and GCN5 are all Histone Acetyl Transferases (HATs), while TRRAP and p400 serve as scaffolding proteins to bridge the interactions of HATs with other transcriptional regulators. E1A regulates viral and cellular gene expression profiles through these interactions to either recruit these activators to specific promoter regions or sequester them away from other promoter regions [12, 13]. E1A CR4 interacts with C-Terminal Binding Protein (CtBP), a transcriptional corepressor,, and cellular proteins like FOXK, DCAF7 and RuvBL1 [12, 13]. E1A also plays a role in the induction of apoptosis in infected cells. Sustained, unregulated E2F activity triggers cellular checkpoint signaling and causes an increase in the level of the tumor suppressor p53. p53 induces cell cycle arrest and cell death by inducing apoptosis [12]. The activation of p53 is deleterious to Ad replication, and therefore, Ad has evolved several proteins encoded by the E1B and E4 transcription units that repress p53 activity and inhibit apoptosis (see review by Thomas Dobner in this Issue). E1A proteins also interact with a plethora of other cellular binding partners [13] to regulate a wide range of other functions beyond the scope of this review.
E1A and Regulation of Innate Responses
IFNs are cytokines that have pleiotropic effects on cells and play important roles in both innate and adaptive immunity [14]. Type I, II and III IFNs are classified by their cell surface receptors. The binding of IFNs to receptors triggers the JAK-STAT signaling pathway resulting in the expression of IFN-stimulated genes (ISGs) [14]. Type I IFNs (α, β, ε, κ and ω) are produced by many cell types following pathogen recognition and function in both autocrine and paracrine manners. Type I IFNs bind to the IFNα receptor (IFNAR) complex, a heterodimer of IFNAR1 and IFNAR2, and induces autophosphorylation of receptor-associated protein kinases JAK1 (Janus Kinase 1) and TYK2 (Tyrosine Kinase 2). JAK1 and TYK2 phosphorylate and activate STAT (Signal Transducer and Activator of Transcription) proteins. A heterodimer of STAT1 and STAT2 associates with IRF9 (Interferon Regulatory Factor 9) to form the active transcription factor complex ISGF3 (Interferon-Stimulated Gene Factor 3), which translocates into the nucleus and activates ISG expression by binding to ISREs (IFN-Stimulated Response Elements) in cellular promoter regions [14]. Type II IFN (IFNγ) is produced by immune cells. IFNγ binds to its cognate receptor complex, IFNGR1 and IFNGR2, leading to autophosphorylation of JAK1 and homodimerization of STAT1 (termed GAF, IFNγ-Activating Factor). GAF binds to GAS elements (IFNγ-Activating Sequence) in promoter regions to induce ISG expression [14]. Type III IFNs (λs) play an important role in mucosal cell immunity. They bind to a distinct receptor, a dimer of IL-28 receptor α (IFNLR1) and IL-10 receptor 2 (IL-10Rβ) and trigger a type I-like IFN response [14].
The Ad E1A proteins are well characterized for their ability to block IFN signaling pathways and the induction of ISG expression by multiple mechanisms [15]. Early studies in cancer cell lines demonstrated that Ad infection, or E1A expression alone, blocks the induction of ISG expression by IFNα. Subsequently, it was shown that this E1A activity was the result of reduced formation of the ISGF3 complex due to decreased levels of its protein constituents [15]. Inhibition of IFN-induced ISG expression correlated with the ability of E1A to bind p300/CBP-ISGF3 and GAF transcriptional coactivators [15, 16]. In addition, the E1A protein was found to block the interaction of unphosphorylated STAT1 with IRF1 (Interferon Regulatory Factor 1) and to down-regulate the expression of LMP2, a subunit of the immunoproteasome [17]. Recently, E1A was found to inhibit ISG expression via the cellular protein RuvBL1. RuvBL1 affects type I IFN signaling. E1A was shown to bind to RuvBL1 and prevent the activation of RuvBL1-regulated promoters in an IFN-dependent manner [18]. The E1A 289aa protein also interacts with the ISG PML-II to enhance E1A transcriptional activation [19]. The cellular protein DREF (ZBED1) colocalizes with PML-NB and interacts with the C-terminus of E1A. E1A relocalizes DREF to the periphery of PML-NB during Ad infection, which promotes viral gene expression [20]. It is unknown if the binding of E1A to PML-II is related to DREF relocalization and DREF activation on viral promoter regions.
In primary human cells, a slightly different picture emerged. E1A blocked IFN-induced ISG expression in primary human airway epithelial cells through a direct interaction with STAT1 mediated by the E1A N-terminus [21]. Further, E1A expression resulted in down-regulation of STAT1 phosphorylation at later times during Ad infection [21]. This is in contrast to other observations, as Ad infection of A549 and HeLa cancer cells lines prolongs STAT1 phosphorylation, as well as the recruitment of STAT1 to Ad viral DNA replication centers, presumably to sequester its activity [22]. Thus, the cell context is very important is discerning the different activities of E1A on innate signaling pathways. This is not really surprising given that cancer cell lines have significantly altered signaling pathways, and consequently, may respond to viral infection and viral proteins in different ways.
Two interesting mechanisms were described by which E1A proteins inhibit IFN induction of ISG expression. As described above, E1A proteins bind to several HATs. The Berk laboratory showed that the E1A 243aa protein (small E1a) decreased Histone 3 lysine 18 acetylation and that p300/CBP are normally responsible for this modification [23]. Histone ubiquitination marks transcriptionally active chromatin [24]. The Mymryk laboratory analyzed global Histone 2B-ubiquitin levels and found that an Ad5 E1A mutant virus strongly induced H2B-ubiquitin lysine 120 levels in primary and transformed human cells, but that wild-type Ad5 did not. A similar induction of H2B-ubiquitin levels was observed following treatment with type I IFNs; E1A expression alone blocked this effect [25]. Human Bre1/RNF20 (hBre1/Ring Finger Protein 20) is an E3 ubiquitin ligase that monoubiquitinates H2B lysine 120 [26]. Foneseca et al. demonstrated that the N-terminal region of E1A binds to hBre1 and blocks the interaction between hBre1 and the catalytic subunit of the ubiquitin conjugating enzyme Ube2b [25]. This results in inhibition of H2B monoubiquitylation at the ISG loci and inhibition of the induction of ISG expression. Subsequent studies by this laboratory showed that E1A utilizes hBre1 to recruit the RNA polymerase II transcription elongation factor hPaf1 (RNA Polymerase-Associated Factor 1) to Ad promoters to stimulate viral early gene expression [27]. These elegant studies demonstrated how the multifunctional E1A proteins coopt cellular transcriptional regulators to promote viral gene expression and inhibit the expression of cellular genes involved in innate responses to infection, in this specific case type I IFN signaling.
In separate studies, Zemke and Berk showed the role of E1A CR4-binding partners in the regulation of gene expression [28]. FOXK1 and FOXK2 (members of the Forkhead Box family of transcription factors) and DCAF7 (DNA Damage Binding Protein 1, DDB1, and CUL4-Associated Factor 7) bind E1A CR4 and disruption of either interaction increases the cell-transformation activity of E1A in conjunction with activated H-Ras [29]. CtBP also binds to E1A CR4 and functions as a transcriptional repressor [30]. Growth-arrested human primary epithelial cells were infected with E1A CR4 mutant viruses that cannot bind CtBP and RNA-seq conducted to analyze changes in cellular gene expression [28]. Although only a small number of genes showed altered expression comparing infection of wild-type Ad5 with Ad5 CR4 mutants, a large overlap in overexpressed genes was observed with the mutant viruses. Several of these genes express proteins that have antiviral functions and their promoter regions were enriched for ISREs. Further analyses showed that the E1A CR4 mutant viruses activate transcription of a subset of ISGs through an increase in IRF3 levels by protein stabilization. A CR4 mutant bearing all mutations in combination displayed the same phenotype as individual mutants indicating that all three CR4 cellular proteins function as a complex and this idea was supported by biochemical experiments [28]. Collectively, these results show that E1A CR4 mediates down-regulation of ISG expression by influencing the stabilities of key transcription factors, particularly IRF3.
While the E1A proteins were originally described as transactivators of viral gene expression, it is clear that a major role of these effectors is to down-regulate innate cellular responses to infection, particularly with respect to different aspects of IFN signaling. E1A proteins block cytoplasmic IFN signaling pathways to dampen the cellular response to infection and E1A proteins down-regulate mechanisms that are used by the cell to promote ISG expression in the nucleus.
Early Region 1B (E1B)
E1B encodes the E1B-19K and E1B-55K proteins. The major roles of these proteins during Ad infection are to inhibit apoptosis and further modify the intracellular environment in order to make the cell more hospitable to viral protein production and viral DNA replication. Viruses with mutations in either E1B protein are significantly reduced in virus yield, due to cell death by apoptosis prior to the completion of the replication cycle [12]. The E1B-55K protein is essential for a variety of important functions in the viral life cycle, including the inhibition of p53-induced apoptosis [31]. The E1B-55K protein binds to the N-terminal transactivation domain of p53 and inhibits p53-induced transcription. E1B-55K also disrupts the interaction of p53 with the HAT PCAF (p300/CBP-Associated Factor) and interferes with p53 acetylation [31]. The E1B-55K protein is modified by the small, ubiquitin-like protein SUMO-1 [32], and indeed, the E1B-55K protein itself is a SUMO E3 ligase that mediates p53 sumoylation for maximal inhibition of p53 activity [33]. Finally, E1B-55K acts in a complex with another Ad early protein, E4-ORF6, to promote the proteasome-dependent degradation of p53 and other cellular protein targets [34, 35]. The E1B-19K protein also is involved in the inhibition of apoptosis. E1B-19K acts to block apoptotic pathways that do not rely on p53, such as the TNFα (Tumor Necrosis Factor Alpha) and Fas ligand cell-death pathways [36]. Interestingly, E1B-19K is a functional homologue of a cellular suppressor of apoptosis, BCL2 (B-Cell CLL/Lymphoma 2). E1B-19K acts in the same manner as BCL2 and predominantly inhibits apoptosis by binding pro-apoptotic activities BAX and BAK (BCL2-Associated X Protein and BCL2 Antagonist/Killer 1) [36]. E1B-19K also plays a role in the inhibition of TNFα-induced apoptosis by blocking the oligomerization of death-inducing complexes involving FADD (Fas-Associated Via Death Domain) [36].
E1B-55K and Regulation of Innate Responses to Ad Infection
The E1B-55K protein associates with other Ad early proteins, specifically the E4-ORF3 and E4-ORF6 proteins. Together with E4-ORF6, Ad5 E1B-55K binds to an E3 ubiquitin-ligase complex containing cellular proteins Elongins B and C, Cullin 5 (CUL5), and RBX1 (Ring Box Protein 1) [34, 35]. The E1B-55K/E4-ORF6 complex tethers this CUL5 complex to different cellular proteins in order to target them for ubiquitin conjugation and proteasomal degradation. E1B-55K serves as a substrate adaptor protein and E4-ORF6 recruits the ubiquitin ligase. This viral E3 ubiquitin-ligase complex targets multiple proteins related to the DDR including Mre11-Rad50-Nbs1 (MRN) complex components, DNA ligase IV, Bloom helicase (BLM), and Tankyrase 1 Binding Protein 1 (TNKS1BP1)[37–40]. By inducing the degradation of MRN proteins and DNA ligase IV, E1B-55K/E4-ORF6 block the induction and activity of the DDR induced by the Ad genome to allow efficient viral DNA replication. Interestingly, the E1B-55K/E4-ORF6 proteins of other Ad serotypes form ubiquitin-ligase complexes with either CUL2 or CUL5, which alters their substrate specificity, but all target one of more components of the DDR to ensure efficient Ad DNA replication [41–43]. Additional targets of the E1B-55K/E4-ORF6 ubiquitin-ligase complex include integrin α3, Tip60, SPOC1, ATRX, SMARCAL1, ALCAM, EPHA2, and PTPRF [44–49]. Ad5 E1B-55K also targets the chromatin-remodeling factor Daxx (death-associated protein) for proteasomal degradation by an E4-ORF6-independent mechanism [50] and Ad12 E4-ORF6 targets TOPBP1 (Topoisomerase-IIβ-Binding Protein 1) for CUL2-based degradation independent of E1B-55K [51]. While it is not clear why all of these substrates are targeted by Ad for degradation, several of them, for instance Tip60, SPOC1, and Daxx/ATRX, limit viral early gene expression and replication.
In the absence of E4-ORF6, E1B-55K colocalizes with E4-ORF3 and PML [52]. E1B-55K binds to PML isoforms IV and V and PML-IV promotes the association of E1B-55K with PML-NBs [53]. The role of interaction between E1B-55K and PML during Ad infection is not clear. E1B-55K counteracts the inhibition of Ad replication by type I IFNs in normal human cells by inhibiting ISG induction [54]. This function, however, is not related to the reorganization of PML-NB during Ad infection.
Early Region 4 (E4)
A common theme among the Ad early and late transcription units is that they encode multiple proteins of related functions. However, Ad early region 4 (E4) is the only transcription unit that produces proteins of relatively disparate activities. E4 encodes at least seven proteins according to analysis of open reading frames (ORF) and alternatively spliced mRNAs. Proteins expressed from the E4 region exhibit a wide range of activities and have been shown to be important for transcriptional regulation, viral DNA replication, viral mRNA transport and splicing, shutoff of host-cell protein synthesis, oncogenic transformation, and the regulation of apoptosis. Two E4 proteins play prominent roles in counteracting innate responses to infection. These are the E4-ORF3 and E4-ORF6 gene products, with the latter being discussed above in the context of E1B-55K.
E4-ORF3 and Regulation of Innate Responses
The E4-ORF3 protein is highly conserved among different Ads and is multifunctional. E4-ORF3 reorganizes nuclear structures alternatively known as PML nuclear bodies (PML-NB), PML oncogenic domains (PODs), or ND10. PML-NB are multiprotein complexes that exhibit a discreet, punctate appearance in the nucleus of a cell [55, 56]. E4-ORF3 is necessary and sufficient for PML-NB regulation. The reorganization of PML-NB by E4-ORF3 requires higher-order multimerization by this viral protein [57, 58]. E4-ORF3 targets PML isoform II during the process of PML-NB disruption and PML-II promotes the innate response to Ad infection [59–61]. PML-NB have been implicated in a number of cellular processes including transcriptional regulation, the regulation of apoptosis, DNA damage repair, protein modification, and antiviral responses [62]. PML-NB have also been shown to react to stresses, such as heat shock and heavy metals as well as interferon, suggesting a role in cellular defense mechanisms. PML-NB display antiviral properties, particularly with respect to nuclear DNA viruses, both intrinsically and following induction by IFNs [63], and E4-ORF3 counteracts these activities [64, 65]. Herpesvirus genomes either localize in proximity to PML-NB or PML-NB assemble de novo in proximity to Herpesvirus genomes, rapidly after nuclear entry and this promotes repression of viral gene expression through the formation of repressive chromatin [63]. It is not clear if this paradigm pertains to Ad infection since incoming Ad genomes neither localize at PML-NB nor recruit PML-NB proteins to viral genomes [66].
The E4-ORF3 proteins of subgroup C Ads (e.g., Ad2 and Ad5) inhibit a DDR. E4-ORF3 directs the reorganization of the MRN complex into PML-containing tracks to sequester and block their activity in a DDR [39, 67]. Interestingly, this function of E4-ORF3 is only conserved among group C Ads [68]. Furthermore, E4-ORF3 blocks p53 signaling by inducing heterochromatin formation at p53-induced promoters, which thereby blocks p53 transactivation and DNA binding [69]. E4-ORF3 also recruits a number of other host proteins to nuclear tracks, including transcription factors like TRIM24 (TIF1α), TRIM33 (TIF1γ), and TFII-I [70–72]. Since the relocalization of MRN and p53 into E4-ORF3 nuclear tracks inhibits their activities, it seems reasonable that TRIM24, TRIM33, and TFII-I may also have antiviral activities, although it is not yet clear.
E4-ORF3 and Regulation of Protein Sumoylation
The Ad5 E4-ORF3 protein was originally shown to induce sumoylation of Mre11 and Nbs1 in virus-infected cells [73]. Since then, increasing evidence supports the key role of E4-ORF3 in regulating the host SUMO pathway during Ad infection. The sumoylation system, which is important to maintain cell homeostasis, conjugates Small Ubiquitin-like MOdifiers (SUMOs) to lysine residues in substrate proteins [74]. SUMOs predominantly localize in the nucleus, where replication of most DNA viruses takes place. Five mammalian SUMO genes, SUMO-1 to SUMO-5, have been identified. Three major mammalian SUMO proteins (SUMO-1, SUMO-2, and SUMO-3) have been extensively studied. SUMO-2 shares 97% amino acid homology with SUMO-3 (often referred as SUMO-2/3) and 50% homology with SUMO1. SUMO-2/3 can form polymeric SUMO chains via conjugation at SUMO-2/3 lysine residue 11. The abundant pool of free SUMO-2/3 in cells may be involved in immediate responses to various cellular stresses. The effects of protein sumoylation are diverse and include the regulation of protein-protein interactions, subcellular localization, protein stability, and enzymatic activities [74].
Ad infection, or E4-ORF3 expression alone, causes a drastic change in subcellular localization of SUMO1 and SUMO2/SUMO3 [68, 73]. In uninfected cells, SUMO shows a diffuse nuclear pattern with some colocalization with PML-NBs. Following E4-ORF3 expression, SUMO is redistributed into E4-ORF3 nuclear tracks. A comparative SUMO proteomic study revealed that SUMO conjugation of multiple host proteins increased upon Ad5 E4-ORF3 expression [70]. Most of the identified E4-ORF3 SUMO targets are involved in DNA damage and repair processes [70], highlighting pivotal roles of E4-ORF3 and the SUMO pathway in modulating DNA damage responses during Ad infection. Like ubiquitylation, sumoylation is carried out by a cascade of enzymatic steps [74]. SUMO precursors are cleaved by SUMO-specific proteases (SENPs) at their carboxy-termini to expose a diglycine motif. The processed forms of SUMO are covalently linked at the C-terminal glycine residue to a catalytic cysteine in the heterodimeric E1 SUMO-activating enzyme SAE1/SAE2 by an ATP-dependent reaction. SUMOs are subsequently transferred to a catalytic cysteine residue of the E2 SUMO-conjugating enzyme Ubc9. SUMOs may then be transferred to a lysine residue(s) in a substrate protein directly by Ubc9 or using a SUMO E3 ligase. Biochemical studies using an in vitro SUMO-conjugation assay demonstrated that Ad5 E4-ORF3 functions as a SUMO E3 ligase [75].
The E4-ORF3 SUMO target proteins have two distinct fates. One group (e.g., TFII-I and TIF-1γ) is further degraded by the ubiquitin-proteasome pathway in the absence of any other viral proteins [75–77]. In contrast, degradation of the other group (e.g., Mre11 and Nbs1) is mediated by E1B-55K and E4-ORF6, but not by E4-ORF3. Conjugation of poly-SUMO chains onto substrates is known to serve as a signal for STUbL (SUMO-Targeted Ubiquitin Ligase)-mediated proteasomal degradation [78]. SUMO2 and SUMO3, but not SUMO1, are able to form polymeric SUMO chains. It remains unclear whether Mre11 and Nbs1 are modified by only mono-SUMO, but not by poly-SUMO chains, although E4-ORF3 has been shown to induce their conjugation to both SUMO1 and SUMO2 [73]. Since an E4-ORF3-deficient Ad5 is still able to degrade the MRN complex, E4-ORF3-mediated SUMO conjugation is dispensable for E1B-55K/E4-ORF6-mediated degradation. In yeast, Mre11 and Nbs1 proteins are modified by SUMO and also play an essential role in a SUMO-conjugation wave during the DDR [79]. However, in mammalian cells, their conjugation to SUMO has been observed only during Ad infection and the functional consequences thereof remain unknown. The two groups of E4-ORF3 targets also show distinct SUMO-conjugation patterns in the in vitro SUMOylation assay. E4-ORF3 leads to a significant increase in SUMO conjugation to TFII-I and TIF-1γ, whereas just a slight increase in Mre11 and Nbs1 modification [75, 80]. This opens up the possibility that additional cellular factors may be involved in SUMO conjugation to certain E4-ORF3 substrates. For instance, a study reported that one of the cellular SUMO E3 ligases, PIAS3 (Protein Inhibitor of Activated STAT3), colocalizes with E4-ORF3 [81], but a role of PIAS3 in E4-ORF3-mediated sumoylation is unknown. E4-ORF3 SUMO substrates are not limited to cellular proteins. Recently, it was found that E4-ORF3 enhances SUMO1 conjugation to E1B-55K during infection with an E4-ORF6-deficient virus and also using the in vitro SUMO-conjugation assay [80]. These latter experiments showed that E4-ORF3 SUMO ligase activities are conserved across the human Ad species, even though Ad5 E4-ORF3 shares only 50% or less amino acid-sequence identity with E4-ORF3 from other Ad species [80]. In cultured cells, E4-ORF3-mediated SUMO conjugation occurs only when target proteins are recruited into E4-ORF3 nuclear tracks generated by each of the Ad species. E4-ORF3 proteins from Ad12, 3, 5, 9 and 4, belonging to species A, B, C, D and E, respectively, have been shown to form nuclear tracks and recruit SUMO [73]. Among the characterized E4-ORF3 SUMO targets, TIF-1γ is sequestered and its SUMO conjugation is enhanced by E4-ORF3 from all five Ad genotypes [80]. However, only Ad5 E4-ORF3 is able to relocalize TFII-I and MRN components and to promote their SUMO modification [73, 76]. This emphasizes that colocalization between E4-ORF3 and target proteins is critical for E4-ORF3-mediated SUMOylation. However, SUMO conjugation does not play a role in E4-ORF3-mediated relocalization of target proteins. Inhibition of sumoylation by overexpression of SUMO-specific protease 1 (SENP1) did not impact either E4-ORF3 nuclear track formation or relocalization of the MRN complex [73]. Also, sumoylation-deficient TIF-1γ is still relocalized into E4-ORF3 nuclear tracks [80], implying that target proteins are recruited by SUMO-independent mechanisms and relocalization is a prerequisite for SUMO conjugation to occur.
Ad E4-ORF3 appears to be a very unique SUMO E3 ligase. All known SUMO ligases have RING (really interesting new gene) finger-type domains, SIMs (SUMO-interacting motifs), or both [74]. This enables ligases to interact with SUMO, UBC9, SUMO-bound UBC9, and SUMO-conjugated substrates. However, E4-ORF3 does not possess any of the typical domains. Although Ad5 E4-ORF3 is predicted to have SIMs at residues 70–73 (VCLL) and residues 103–106 (LIDL), based on the SIM consensus sequence, these residues are not conserved across Ad genotypes and not required for E4-ORF3 ligase activity [75]. This activity is impaired only when the E4-ORF3 protein lacks self-assembling ability, such as E4-ORF3 mutations in residues N82 or L103. Unlike other SUMO ligases, E4-ORF3 is unable to directly bind either SUMO or UBC9. However, E4-ORF3 can interact with non-covalently SUMO-bound UBC9, which implicated in poly-SUMO chain formation [80]. E4-ORF3 polymerization is crucial for this ternary binding. Collectively, these findings suggest two distinct modes of action for E4-ORF3 as a SUMO ligase. First, the E4-ORF3 nuclear tracks serve as a platform for SUMO conjugation in the nucleus of infected cells by mediating colocalization of the SUMOylation machinery and target proteins. Second, the interface generated by E4-ORF3 polymerization enables its interaction with SUMO-bound UBC9. This optimizes the conformation of UBC9 and thereby accellerates the SUMO-conjugation reaction.
IFN Signaling Promotes Persistent Ad Infection
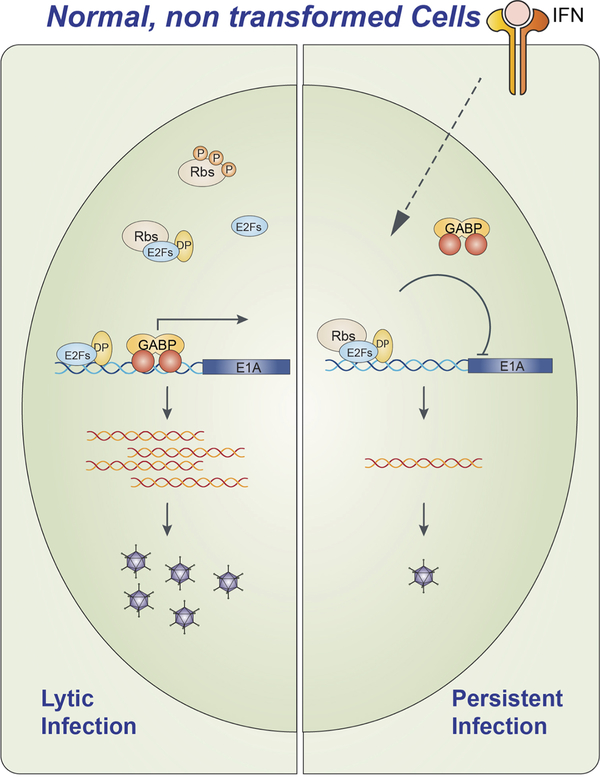
Following acute infection, Ads may establish persistent infections in the host (see review by Thomas Lion in this Issue). The molecular underpinnings of persistent Ad infection are not well characterized, but a recent report demonstrates that Ad may hijack IFN signaling to suppress the lytic cycle and promote persistent infection [82]. Both IFNα and IFNγ inhibit Ad5 replication in normal human cells (fibroblasts and epithelial cells). Type I and II IFNs inhibit the replication of divergent Ads in these cells. IFNα and IFNγ repress E1A transcription to block the Ad replication cycle. IFNα and IFNγ reduce the association of GA Binding Protein (GABP), the cellular activator of E1A transcription, with the E1A enhancer region during the early phase of infection. The repression of E1A expression by IFNs requires a conserved E2F-binding site in the E1A enhancer, and IFNs increase the enrichment of the E2F-associated repressor proteins, Rb and p107, at the E1A enhancer in vivo (Fig. 2). Consistent with this result, expression of the E1A 243aa protein, which disrupts the association between E2F and Rb-family proteins, rescues Ad replication in the presence of IFNs. Finally, IFNγ suppresses productive Ad replication over an extended period of time in primary cells dependent on the E2F-binding site in the E1A enhancer. This study reveals a novel mechanism by which Ads utilize IFN signaling to suppress lytic virus replication and thereby promote persistent infection (Figure 2)[82].
Figure 2. Ad infection and the IFN response in normal human cells.
During lytic infection in the absence of IFN (left), the cellular transcription factor GABP binds the E1A enhancer region and activates transcription of the E1A gene. This promotes the viral replication cycle and viral progeny are actively produced. During an IFN response (right), GABP is displaced and repressor E2F-Rb complexes bind to the E1A enhancer and thereby reduce E1A transcription and protein levels. Virus production is greatly diminished and a persistent infection may ensue.
Conclusions and Perspectives
Over 60 years of research on human and animal Ads have revealed unique insights into multiple cellular processes, spanning from oncogenesis to innate and adaptive immune responses. Innate cellular signaling pathways hinder productive Ad infection, and as such, Ad has evolved numerous mechanisms to counteract these host responses. Yet at the same time, Ad may utilize innate responses to promote persistent infection in the host. The impact that persistent Ad infections have on hosts over months, or perhaps years, is not clear and represents an important new avenue of research. The study of Ad infections continues to provide a wealth of knowledge on cellular and viral functions.
Acknowledgments
Research was supported by a grant from the National Institutes of Health, USA. We are very grateful to Dr. Yueting Zheng who created the figure.
Abbreviations
- Ad
Adenoviruses
- IFN
Interferon
- DDR
DNA damage response
- ISG
Interferon-stimulated gene
- CtBP
C-terminal binding protein
- SUMO
small ubiquitin-like modifier
References
- 1.Greber UF & Flatt JW (2019) Adenovirus Entry: From Infection to Immunity, Annu Rev Virol. [DOI] [PubMed] [Google Scholar]
- 2.Lam E, Stein S & Falck-Pedersen E (2014) Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade, J Virol. 88, 974–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ & Tschopp J (2008) The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response, Nature. 452, 103–7. [DOI] [PubMed] [Google Scholar]
- 4.Barlan AU, Griffin TM, McGuire KA & Wiethoff CM (2011) Adenovirus membrane penetration activates the NLRP3 inflammasome, J Virol. 85, 146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Huang X & Yang Y (2007) Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and -independent pathways, J Virol. 81, 3170–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nociari M, Ocheretina O, Murphy M & Falck-Pedersen E (2009) Adenovirus induction of IRF3 occurs through a binary trigger targeting Jun N-terminal kinase and TBK1 kinase cascades and type I interferon autocrine signaling, J Virol. 83, 4081–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maler MD, Nielsen PJ, Stichling N, Cohen I, Ruzsics Z, Wood C, Engelhard P, Suomalainen M, Gyory I, Huber M, Muller-Quernheim J, Schamel WWA, Gordon S, Jakob T, Martin SF, Jahnen-Dechent W, Greber UF, Freudenberg MA & Fejer G (2017) Key Role of the Scavenger Receptor MARCO in Mediating Adenovirus Infection and Subsequent Innate Responses of Macrophages, MBio. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weitzman MD & Ornelles DA (2005) Inactivating intracellular antiviral responses during adenovirus infection, Oncogene. 24, 7686–96. [DOI] [PubMed] [Google Scholar]
- 9.Stingele J, Bellelli R & Boulton SJ (2017) Mechanisms of DNA-protein crosslink repair, Nat Rev Mol Cell Biol. 18, 563–573. [DOI] [PubMed] [Google Scholar]
- 10.Karen KA & Hearing P (2011) Adenovirus Core Protein VII Protects the Viral Genome from a DNA Damage Response at Early Times after Infection, J Virol. 85, 4135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Morral N & Engel DA (2007) Transcription releases protein VII from adenovirus chromatin, Virology. 369, 411–22. [DOI] [PubMed] [Google Scholar]
- 12.Berk AJ (2005) Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus, Oncogene. 24, 7673–85. [DOI] [PubMed] [Google Scholar]
- 13.King CR, Zhang A, Tessier TM, Gameiro SF & Mymryk JS (2018) Hacking the Cell: Network Intrusion and Exploitation by Adenovirus E1A, MBio. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider WM, Chevillotte MD & Rice CM (2014) Interferon-stimulated genes: a complex web of host defenses, Annu Rev Immunol. 32, 513–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrickx R, Stichling N, Koelen J, Kuryk L, Lipiec A & Greber UF (2014) Innate immunity to adenovirus, Hum Gene Ther. 25, 265–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Routes JM, Li H, Bayley ST, Ryan S & Klemm DJ (1996) Inhibition of IFN-stimulated gene expression and IFN induction of cytolytic resistance to natural killer cell lysis correlate with E1A-p300 binding, J Immunol. 156, 1055–61. [PubMed] [Google Scholar]
- 17.Chatterjee-Kishore M, van Den Akker F & Stark GR (2000) Adenovirus E1A down-regulates LMP2 transcription by interfering with the binding of stat1 to IRF1, J Biol Chem. 275, 20406–11. [DOI] [PubMed] [Google Scholar]
- 18.Olanubi O, Frost JR, Radko S & Pelka P (2017) Suppression of Type I Interferon Signaling by E1A via RuvBL1/Pontin, J Virol. 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berscheminski J, Groitl P, Dobner T, Wimmer P & Schreiner S (2013) The adenoviral oncogene E1A-13S interacts with a specific isoform of the tumor suppressor PML to enhance viral transcription, J Virol. 87, 965–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radko S, Koleva M, James KM, Jung R, Mymryk JS & Pelka P (2014) Adenovirus E1A targets the DREF nuclear factor to regulate virus gene expression, DNA replication, and growth, J Virol. 88, 13469–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Look DC, Roswit WT, Frick AG, Gris-Alevy Y, Dickhaus DM, Walter MJ & Holtzman MJ (1998) Direct suppression of Stat1 function during adenoviral infection, Immunity. 9, 871–80. [DOI] [PubMed] [Google Scholar]
- 22.Sohn SY & Hearing P (2011) Adenovirus sequesters phosphorylated STAT1 at viral replication centers and inhibits STAT dephosphorylation, J Virol. 85, 7555–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK & Berk AJ (2008) Adenovirus small e1a alters global patterns of histone modification, Science. 321, 1084–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA & Strahl BD (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II, Mol Cell Biol. 25, 637–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fonseca GJ, Thillainadesan G, Yousef AF, Ablack JN, Mossman KL, Torchia J & Mymryk JS (2012) Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification, Cell Host Microbe. 11, 597–606. [DOI] [PubMed] [Google Scholar]
- 26.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C & Madhani HD (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control, Mol Cell. 11, 261–6. [DOI] [PubMed] [Google Scholar]
- 27.Fonseca GJ, Cohen MJ, Nichols AC, Barrett JW & Mymryk JS (2013) Viral retasking of hBre1/RNF20 to recruit hPaf1 for transcriptional activation, PLoS Pathog. 9, e1003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zemke NR & Berk AJ (2017) The Adenovirus E1A C Terminus Suppresses a Delayed Antiviral Response and Modulates RAS Signaling, Cell Host Microbe. 22, 789–800 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komorek J, Kuppuswamy M, Subramanian T, Vijayalingam S, Lomonosova E, Zhao LJ, Mymryk JS, Schmitt K & Chinnadurai G (2010) Adenovirus type 5 E1A and E6 proteins of low-risk cutaneous beta-human papillomaviruses suppress cell transformation through interaction with FOXK1/K2 transcription factors, J Virol. 84, 2719–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chinnadurai G (2009) The transcriptional corepressor CtBP: a foe of multiple tumor suppressors, Cancer Res. 69, 731–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackford AN & Grand RJ (2009) Adenovirus E1B 55-kilodalton protein: multiple roles in viral infection and cell transformation, J Virol. 83, 4000–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wimmer P, Schreiner S & Dobner T (2012) Human pathogens and the host cell SUMOylation system, J Virol. 86, 642–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pennella MA, Liu Y, Woo JL, Kim CA & Berk AJ (2010) Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies, J Virol. 84, 12210–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harada JN, Shevchenko A, Shevchenko A, Pallas DC & Berk AJ (2002) Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery, J Virol. 76, 9194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW & Branton PE (2001) Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex, Genes Dev. 15, 3104–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aarbiou J, Tjabringa GS, Verhoosel RM, Ninaber DK, White SR, Peltenburg LT, Rabe KF & Hiemstra PS (2006) Mechanisms of cell death induced by the neutrophil antimicrobial peptides alpha-defensins and LL-37, Inflamm Res. 55, 119–27. [DOI] [PubMed] [Google Scholar]
- 37.Baker A, Rohleder KJ, Hanakahi LA & Ketner G (2007) Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation, J Virol. 81, 7034–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orazio NI, Naeger CM, Karlseder J & Weitzman MD (2011) The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection, J Virol. 85, 1887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stracker TH, Carson CT & Weitzman MD (2002) Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex, Nature. 418, 348–52. [DOI] [PubMed] [Google Scholar]
- 40.Chalabi Hagkarim N, Ryan EL, Byrd PJ, Hollingworth R, Shimwell NJ, Agathanggelou A, Vavasseur M, Kolbe V, Speiseder T, Dobner T, Stewart GS & Grand RJ (2018) Degradation of a Novel DNA Damage Response Protein, Tankyrase 1 Binding Protein 1, following Adenovirus Infection, J Virol. 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng CY, Gilson T, Dallaire F, Ketner G, Branton PE & Blanchette P (2011) The E4orf6/E1B55K E3 ubiquitin ligase complexes of human adenoviruses exhibit heterogeneity in composition and substrate specificity, J Virol. 85, 765–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng CY, Gilson T, Wimmer P, Schreiner S, Ketner G, Dobner T, Branton PE & Blanchette P (2013) Role of E1B55K in E4orf6/E1B55K E3 ligase complexes formed by different human adenovirus serotypes, J Virol. 87, 6232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forrester NA, Sedgwick GG, Thomas A, Blackford AN, Speiseder T, Dobner T, Byrd PJ, Stewart GS, Turnell AS & Grand RJ (2011) Serotype-specific inactivation of the cellular DNA damage response during adenovirus infection, J Virol. 85, 2201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dallaire F, Blanchette P, Groitl P, Dobner T & Branton PE (2009) Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex, J Virol. 83, 5329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta A, Jha S, Engel DA, Ornelles DA & Dutta A (2013) Tip60 degradation by adenovirus relieves transcriptional repression of viral transcriptional activator EIA, Oncogene. 32, 5017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreiner S, Kinkley S, Burck C, Mund A, Wimmer P, Schubert T, Groitl P, Will H & Dobner T (2013) SPOC1-mediated antiviral host cell response is antagonized early in human adenovirus type 5 infection, PLoS Pathog. 9, e1003775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nazeer R, Qashqari FSI, Albalawi AS, Piberger AL, Tilotta MT, Read ML, Hu S, Davis S, McCabe CJ, Petermann E & Turnell AS (2019) Adenovirus E1B 55-Kilodalton Protein Targets SMARCAL1 for Degradation during Infection and Modulates Cellular DNA Replication, J Virol. 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schreiner S, Burck C, Glass M, Groitl P, Wimmer P, Kinkley S, Mund A, Everett RD & Dobner T (2013) Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes, Nucleic Acids Res. 41, 3532–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu YR, Turnell AS, Davis S, Heesom KJ, Evans VC & Matthews DA (2017) Comparison of protein expression during wild-type, and E1B-55k-deletion, adenovirus infection using quantitative time-course proteomics, J Gen Virol. 98, 1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schreiner S, Wimmer P, Sirma H, Everett RD, Blanchette P, Groitl P & Dobner T (2010) Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells, J Virol. 84, 7029–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blackford AN, Patel RN, Forrester NA, Theil K, Groitl P, Stewart GS, Taylor AM, Morgan IM, Dobner T, Grand RJ & Turnell AS (2010) Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation, Proc Natl Acad Sci U S A. 107, 12251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leppard KN & Everett RD (1999) The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components, J Gen Virol. 80 (Pt 4), 997–1008. [DOI] [PubMed] [Google Scholar]
- 53.Wimmer P, Schreiner S, Everett RD, Sirma H, Groitl P & Dobner T (2010) SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML, Oncogene. 29, 5511–22. [DOI] [PubMed] [Google Scholar]
- 54.Chahal JS, Qi J & Flint SJ (2012) The human adenovirus type 5 E1B 55 kDa protein obstructs inhibition of viral replication by type I interferon in normal human cells, PLoS Pathog. 8, e1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carvalho T, Seeler JS, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M & Dejean A (1995) Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies, J Cell Biol. 131, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM & Maul GG (1996) Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure, Genes Dev. 10, 196–207. [DOI] [PubMed] [Google Scholar]
- 57.Ou HD, Kwiatkowski W, Deerinck TJ, Noske A, Blain KY, Land HS, Soria C, Powers CJ, May AP, Shu X, Tsien RY, Fitzpatrick JA, Long JA, Ellisman MH, Choe S & O’Shea CC (2012) A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors, Cell. 151, 304–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patsalo V, Yondola MA, Luan B, Shoshani I, Kisker C, Green DF, Raleigh DP & Hearing P (2012) Biophysical and functional analyses suggest that adenovirus E4-ORF3 protein requires higher-order multimerization to function against promyelocytic leukemia protein nuclear bodies, J Biol Chem. 287, 22573–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atwan Z, Wright J, Woodman A & Leppard KN (2016) Promyelocytic leukemia protein isoform II inhibits infection by human adenovirus type 5 through effects on HSP70 and the interferon response, J Gen Virol. 97, 1955–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoppe A, Beech SJ, Dimmock J & Leppard KN (2006) Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption, J Virol. 80, 3042–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leppard KN, Emmott E, Cortese MS & Rich T (2009) Adenovirus type 5 E4 Orf3 protein targets promyelocytic leukaemia (PML) protein nuclear domains for disruption via a sequence in PML isoform II that is predicted as a protein interaction site by bioinformatic analysis, J Gen Virol. 90, 95–104. [DOI] [PubMed] [Google Scholar]
- 62.Lallemand-Breitenbach V & de The H (2018) PML nuclear bodies: from architecture to function, Curr Opin Cell Biol. 52, 154–161. [DOI] [PubMed] [Google Scholar]
- 63.Scherer M & Stamminger T (2016) Emerging Role of PML Nuclear Bodies in Innate Immune Signaling, J Virol. 90, 5850–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ullman AJ & Hearing P (2008) Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein, J Virol. 82, 7325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ullman AJ, Reich NC & Hearing P (2007) Adenovirus E4 ORF3 protein inhibits the interferon-mediated antiviral response, J Virol. 81, 4744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komatsu T, Nagata K & Wodrich H (2016) An Adenovirus DNA Replication Factor, but Not Incoming Genome Complexes, Targets PML Nuclear Bodies, J Virol. 90, 1657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evans JD & Hearing P (2005) Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication, J Virol. 79, 6207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stracker TH, Lee DV, Carson CT, Araujo FD, Ornelles DA & Weitzman MD (2005) Serotype-Specific Reorganization of the Mre11 Complex by Adenoviral E4orf3 Proteins, Journal of Virology. 79, 6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soria C, Estermann FE, Espantman KC & O’Shea CC (2010) Heterochromatin silencing of p53 target genes by a small viral protein, Nature. 466, 1076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sohn S-Y, Bridges RG & Hearing P (2015) Proteomic Analysis of Ubiquitin-Like Posttranslational Modifications Induced by the Adenovirus E4-ORF3 Protein, Journal of Virology. 89, 1744–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vink EI, Yondola MA, Wu K & Hearing P (2012) Adenovirus E4-ORF3-dependent relocalization of TIF1alpha and TIF1gamma relies on access to the Coiled-Coil motif, Virology. 422, 317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yondola MA & Hearing P (2007) The adenovirus E4 ORF3 protein binds and reorganizes the TRIM family member transcriptional intermediary factor 1 alpha, J Virol. 81, 4264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sohn S-Y & Hearing P (2012) Adenovirus Regulates Sumoylation of Mre11-Rad50-Nbs1 Components through a Paralog-Specific Mechanism, Journal of Virology. 86, 9656–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao X (2018) SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes, Mol Cell. 71, 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sohn S-Y & Hearing P (2016) The adenovirus E4-ORF3 protein functions as a SUMO E3 ligase for TIF-1γ sumoylation and poly-SUMO chain elongation, Proceedings of the National Academy of Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bridges RG, Sohn S-Y, Wright J, Leppard KN & Hearing P (2016) The Adenovirus E4-ORF3 Protein Stimulates SUMOylation of General Transcription Factor TFII-I to Direct Proteasomal Degradation, mBio. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Forrester NA, Patel RN, Speiseder T, Groitl P, Sedgwick GG, Shimwell NJ, Seed RI, Catnaigh PO, McCabe CJ, Stewart GS, Dobner T, Grand RJ, Martin A & Turnell AS (2012) Adenovirus E4orf3 targets transcriptional intermediary factor 1gamma for proteasome-dependent degradation during infection, J Virol. 86, 3167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abed M, Bitman-Lotan E & Orian A (2018) The Biology of SUMO-Targeted Ubiquitin Ligases in Drosophila Development, Immunity, and Cancer, J Dev Biol. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S & Zhao X (2012) Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint, Mol Cell. 45, 422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sohn S-Y & Hearing P (2019) Mechanism of Adenovirus E4-ORF3-Mediated SUMO Modifications, mBio. 10, e00022–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Higginbotham JM & O’Shea CC (2015) Adenovirus E4-ORF3 Targets PIAS3 and Together with E1B-55K Remodels SUMO Interactions in the Nucleus and at Virus Genome Replication Domains, J Virol. 89, 10260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng Y, Stamminger T & Hearing P (2016) E2F/Rb Family Proteins Mediate Interferon Induced Repression of Adenovirus Immediate Early Transcription to Promote Persistent Viral Infection, PLoS Pathog. 12, e1005415. [DOI] [PMC free article] [PubMed] [Google Scholar]