Abstract
Neuroblastoma (NB) is the most common extracranial solid tumor in childhood. Outcome for children with high-risk NB remains unsatisfactory. Accumulating evidence suggests that tripartite motif (TRIM) family proteins express diversely in various human cancers and act as regulators of oncoproteins or tumor suppressor proteins. This review summarizes the TRIM proteins involving in NB and the underlying molecular mechanisms. We expect these new insights will provide important implications for the treatment of NB by targeting TRIM proteins.
Keywords: E3 ubiquitin ligase, neuroblastoma, TRIM proteins, ubiquitination
Introduction
Neuroblastoma (NB) is the most common extracranial solid tumor in childhood, accounting for 8–10% of all children’s malignancies and about 15% of cancer-related deaths [1,2]. NB onset has a strong genetic component, with gene mutations commonly seen in MYCN, ALK and PHOX2B [3]. In addition, clinical outcomes of NB patients are highly heterogeneous, with differentiation status [4], disease stage [5], age at diagnosis [6] and MYCN amplification [7] all influencing prognosis. What’s more, genome sequencing studies revealed that the genetic variations of NB tumor are markedly lower than that found in many other adult solid tumors [8], indicating the importance of post-translational modification in the occurrence and development of NB. As a vital post-translational modification, ubiquitination is involved in the regulation of many eukaryotic signaling pathways and aberrant ubiquitin signaling is currently an active area in NB research. For example, recent studies have indicated ubiquitination modification functions as an important regulator for the stability of MYCN protein, a marker of poor NB prognosis [9]. Ubiquitin-specific protease 7 (USP7) promotes the growth of MYCN-amplified NB cell lines by inducing the deubiquitination and subsequent stabilization of MYCN [10], while the ubiquitin-ligase FBW7 promotes the proteasome-mediated MYCN degradation and leads to the opposite result [11].
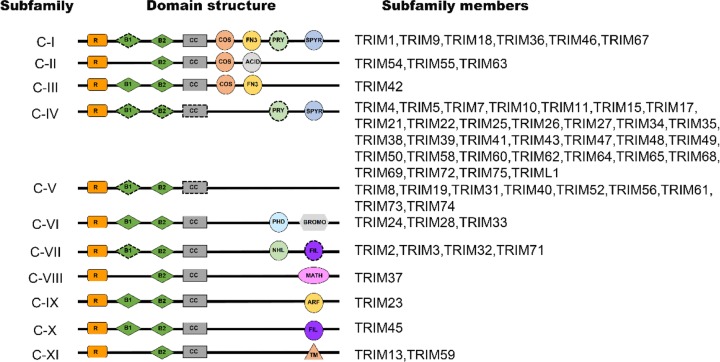
The tripartite motif (TRIM) family proteins, also known as RBBC proteins, are characterized by the common presence of RBBC motif comprises a RING domain, either one or two B-boxes (B1 and B2), and a coiled-coil (CC) domain in their amino terminal, followed by a highly variable carboxyl-terminal domain, which categorizes them into 11 subgroups (C-I – C-XI, Figure 1) [12,13]. In recent years, accumulating studies have found that TRIM proteins regulate a wide variety of biological processes and their dysregulation are associated with various diseases, including cancer [14–16]. The functions of TRIM proteins in tumor development and progression are complex and diverse. There are several established mechanisms of carcinogenesis involving the TRIM family, including: (1) Acquiring oncogenic potential through chromosomal translocations. One best-known example is TRIM19, which is encoded by the promyelocytic leukaemia (PML) gene and involves in the balanced translocation with retinoic acid receptor-α (RARα) to produce the PML-RARα fusion protein. This kind of fusion protein specifically occurs in patients with acute promyelocytic leukaemia (APL) [17], acting as a RARα transcriptional suppressor and thus inducing a block in the differentiation of promyelocytes, which is the underlying etiology of this blood cancer [18,19]. (2) Regulating the activation of nuclear receptors, such as androgen receptors (AR) and estrogen receptors (ER). For example, both TRIM24 and TRIM68 can interact with the AR and increase its transcriptional activity in the presence of dihydrotestosterone (DHT) in prostate cancer cells [20,21]. Several findings also indicate that TRIM proteins regulate transcription by modifying chromatin. For instance, TRIM16 has been shown to increase histone acetylation and reactivate the transcription of retinoic acid receptor β2 (RARβ2) in retinoid-resistant breast and lung cancer cells [22]. (3) Functioning as E3 ubiquitin ligases to regulate the degradation of oncoproteins and tumor suppressor proteins. For example, TRIM11 exerts its oncogenic effect in hepatocellular carcinoma through the inhibition of p53 [23], while TRIM67 suppresses colorectal cancer initiation and progression via the activation of p53 [24]. TRIM65 enhances the invasiveness of bladder urothelial carcinoma (BUC) cells by inducing epithelial–mesenchymal transition (EMT) and promoting the ubiquitination and degradation of Annexin A2 (ANXA2) [25].
Figure 1. TRIM proteins domain structure and classification of TRIMs (C-I to C-XI).
TRIMs have an N-terminal RING finger (R), one or two B-boxes (B1 and B2), and coiled coil (CC) domain. C-terminal domains comprise various domains including: ARF, ADP-ribosylation factor family domain; BROMO, bromodomain; COS, cos-box; FIL, filamin-type Ig domain; FN3, fibronectin type III repeat; MATH, meprin and TRAF-homology domain; NHL, NCL1, HT2A and LIN41 domain; PHD, PHD domain; PRY, PRY domain; SPRY, SPRY domain; TM, transmembrane region. Dashed-outline domains are those which are differentially present among subfamily members.
The involvement of TRIM protein family in various oncogenic processes makes it a current focus in cancer research. While the functions of TRIM proteins in multiple cancers have been described, their roles in NB have only recently begun to emerge and poorly understood. In this review, we summarize the current research progress of TRIM proteins in NB (Table 1).
Table 1. TRIM proteins and their roles in regulating neuroblastoma.
| TRIM proteins | Functions | References |
|---|---|---|
| TRIM11 | Interacting with PHOX2B and increasing the expression of DBH | [33] |
| TRIM16 | Enhancing retinoic acid receptor β (RAR-β) transcription | [23,43] |
| Down-regulating cytoplasmic vimentin and nuclear E2F1 | [45] | |
| Regulating G1/S cell cycle progression | [46] | |
| Modulating activity of caspase-2 and promoting apoptosis | [47] | |
| TRIM17 | Repressing apoptosis by stabilizing anti-apoptotic proteins Mcl-1 | [51,52] |
| TRIM32 | Enhancing transcriptional activity of RARα | [63] |
| Inducing asymmetric cell division (ACD) | [64] | |
| Facilitating c-MYC and n-MYC proteasomal degradation | [62,64] | |
| TRIM36 | Being hypermethylated in neuroblastoma tumors | [72] |
| TRIM59 | Inducing the expression of ANXA2 | [78] |
| Regulating Wnt/β-catenin signaling pathway | [80] |
TRIM11
TRIM11, also known as ring finger protein 92 (RNF92), belongs to the C-IV subfamily, containing a RING finger, one B-box domain, a coiled-coil domain and a C-terminal PRY domain and SPRY domain. As an E3 ubiquitin ligase, it is reported to be up-regulated in several human tumors. In lung cancer, TRIM11 is up-regulated and correlates with poor prognosis [26]. In lung adenocarcinoma, TRIM11 promotes tumor angiogenesis through activating transcription 3 (STAT3)/vascular endothelial growth factor A (VEGFA) pathway [27]. In hepatocellular carcinoma (HCC), enforced expression of TRIM11 promotes cell proliferation, invasion, and migration by activating of the PI3K/Akt signaling pathway [28]. A recent study also revealed that TRIM11 regulates the proliferation and apoptosis of breast cancer cells by regulating ERK1/2 and JNK1/2 signaling pathways [29]. These results indicate that TRIM11 is responsible for different oncogenic processes in various cancers.
Of interest, TRIM11 has recently been shown to involve in the process of neurogenesis. For example, it has been shown to mediate the degradation of PAX6 (a member of the paired-box family of transcription factors which play key roles in development) via the ubiquitin–proteasome system, thereby modulating cortical neurogenesis [30]. TRIM11 has also been reported to be strongly correlated with the differentiation status of malignant glioma cells [31]. More interestingly, TRIM11 can interact with Paired-Like Homeobox 2B (PHOX2B), a homeodomain transcription factor [32], to increase the expression of dopamine β hydroxylase (DBH) gene and participate in the development of noradrenergic (NA) neurons [32].
Universally known, NB originates from sympathoadrenal precursor cells of the sympathetic nervous system and the tumor cells have embryonic features, presumably as a consequence of an impaired capacity to respond to signals operating during normal differentiation [33]. Moreover, PHOX2B, whose coding gene is located on chromosome 4p13, is originally found in a NB cell line. It is expressed specifically in the nervous system and has a vital effect on the formation and differentiation of sympathetic neurons and chromaffin cells [34]. In addition, it has been reported that PHOX2B germline mutations, which account for 6% hereditary NBs, are involved in the initiation of NB tumorigenesis [35]. Thus, these findings provide the potential role of TRIM11 in NB by regulating PHOX2B.
TRIM16
TRIM16, also named as the estrogen responsive B box protein (EBBP), has been shown to play a negative role in the development and progression of several cancers. For example, the expression of TRIM16 is significantly down-regulated in HCC and knockdown of TRIM16 enhances HCC cell migration and invasion via the promotion of EMT process [36]. TRIM16 is also at low levels in prostate tumors and enforced expression of TRIM16 inhibits prostate cancer cell migration and invasion in a manner associated with the inhibition of Snail signaling pathway and EMT process [37]. The anti-tumor function of TRIM16 has also been elucidated in many other cancers, such as ovarian cancer [38], breast cancer [39], non-small cell lung cancer [40] and melanoma [41]. Together, these studies indicate a tumor-suppressing function for TRIM16 in human cancers.
Recent studies have demonstrated that TRIM16 acts as a tumor suppressor in NB. First, TRIM16 can enhance the transcription of retinoic acid receptor β (RARβ), and overexpression of TRIM16 significantly reduces the proliferation of RA-sensitive NB cells as well as RA-resistant lung and breast cancer cells [42]. These findings suggest an important role of TRIM16 in the response of these tumor cells to differentiating agents, such as RA, an effective inducer of NB cell differentiation that has been used in the clinic for the treatment of high-risk NB [43]. Further investigation showed that TRIM16 can reactivate the transcription of RARβ2 by increasing histone H3 acetylation, with the de-acetylation of histone H3 considered to be the most common mechanism of RARβ2 transcriptional repression in retinoid-resistant cancer cells [22]. These studies suggest that TRIM16 may modulate the transcriptional activity of RA-related receptors that required for NB cell differentiation and could be a novel therapeutic target for retinoid-resistant NB cancer. Second, TRIM16 can affect the migration and differentiation of NB cell through down-regulating vimentin and nuclear E2F1 protein (required for cell replication) [44]. TRIM16 can also influence NB proliferation in vitro and tumorigenicity in vivo through the regulation of cell cycle [45]. Third, TRIM16 can promote the apoptosis of BE(2)-C NB cells by directly interacting with caspase-2 and modulating its activity [46]. In conclusion, results above have suggested that TRIM16 serves as a tumor suppressor in NB, and defining an effective method to increase TRIM16 protein expression in NB might provide a novel strategy of the cancer therapy.
TRIM17
TRIM17, also known as Testis RING finger protein (TERF), is expressed not only in the testis but also in the brain during embryonic development [12], suggesting it may have a role in neuronal development. In a previous study, TRIM17 has been identified as a crucial E3 ubiquitin ligase that is necessary for neuronal apoptosis [47]. Unlike majority of TRIM proteins, such as TRIM5α, TRIM6, TRIM20 and TRIM21, which have been demonstrated to promote autophagy, TRIM17 is notable because it can inhibit autophagy [48]. Since neuronal apoptosis is crucial for normal development of the nervous system [49] and NB originates from precursor cells of the sympathetic nervous system, this suggests that TRIM17 may has some effects on NB.
Indeed, several studies in vivo in NB cells have shown the function of TRIM17 in promoting neuronal apoptosis. For instance, in NB Neuro2A cells, TRIM17 mediates the ubiquitination and degradation of Mcl-1, an anti-apoptotic Bcl-2 family protein that is necessary for initiating neuronal apoptosis [50]. TRIM17 has also been reported to stabilize BCL2A1, another anti-apoptotic Bcl-2 family protein that contributes to chemo-resistance in a subset of tumors [51]. In summary, these results suggest that TRIM17 is an important factor in neuronal apoptosis. However, studies on the function of TRIM17 in human cancers are still lacking. Whether the function of TRIM17 contributes to NB tumor onset and progression is not yet clear.
TRIM32
TRIM32 is known as Limb Girdle Muscular Dystrophy 2H (LGMD2H), based on the finding that TRIM32 deficiency in mice results in phenotypes characteristic of the human disease [52]. A role for TRIM32 in the regulation of skeletal muscle stem cell differentiation and normal adult muscle regeneration has been reported [53]. Moreover, TRIM32 is reported to exhibit tumor suppressor functions in many human cancers, including breast cancer, gastric cancer, lung cancer and skin cancer [54–57]. Mechanistically, TRIM32 is an E3 ubiquitin-ligase for some tumor suppressors, such as p53 and Abi2 (Abl-interactor 2) and can regulate their degradation [58,59]. In addition, TRIM32 can promote the proliferation and invasion of gastric cancer cell lines by activating β-catenin signaling pathway [55]. All these findings suggest that TRIM32 is involved in tumor formation and development.
Previously, several studies have shown the importance of TRIM32 in neuronal differentiation by several different mechanism as follows: First, TRIM32 can ubiquitylate and result in the proteasomal degradation of c-Myc, one of the key stem cell transcription factors (SCTFs) crucial for neuronal stem cell (NSC) renewal and maintaining an undifferentiated phenotype [60,61]. Second, TRIM32 can interact with RARα and enhance its transcriptional activity, bringing about enhancement of neural differentiation [62]. These findings suggest that TRIM32 may play an important role in the process of neural differentiation and can be a tumor-suppressor candidate of NB. Indeed, TRIM32 concentrates in one of the daughter cells during mitosis, where it interacts with MYCN and facilitates its proteasomal degradation, thus inducing asymmetric cell division (ACD) of human NB cells, resulting in one cell with neural progenitor cell activity and the other with potential for differentiation [63]. Furthermore, N-myc downstream regulated gene 2 (NDRG2), a new tumor suppressor gene that has recently been reported to suppress the growth and aggressiveness of NB cells [64], is identified as a novel target for TRIM32 [65]. Together, the above-mentioned studies provide some support for us to consider TRIM32 as a tumor suppressor in NB, offering an alternative direction for NB prognosis and/or treatment.
TRIM36
TRIM36, also known as ring finger protein 98 (RNF98), belongs to the C-I subfamily and contains a RING finger, one B-box domain, a coiled-coil domain and C-terminal domains (COS domain, FN3 domain and PYR domain). It is located in the tumor suppressor gene region at chromosome 5q22.3 [66], and has been reported to be an E3 ubiquitin-ligase for p53, one of the best-known tumor suppressors [67]. Overexpression of TRIM36 decelerates the cell cycle and limits cell growth [68]. TRIM36 can also act as a putative tumor suppressor by attenuating MAPK/ERK signaling pathways and regulating apoptosis-related pathways in prostate cancer [69,70]. These findings indicate that TRIM36 may have suppressive effects on tumors, specifically in prostate cancer.
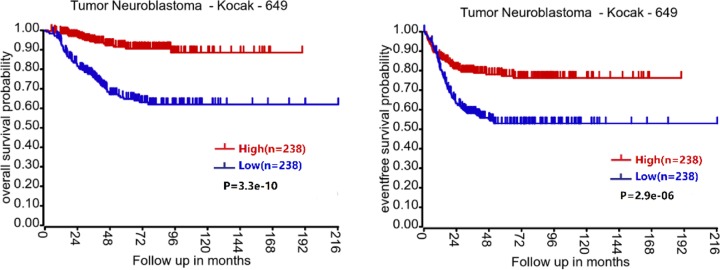
In agreement with above hypothesis, genome-wide methylation analysis on 60 NB tumors using Illumina 450K methylation arrays revealed that TRIM36 is hypermethylated and has lower expression in aggressive NB tumors compared with less aggressive ones [71], indicating that down-regulated TRIM36 has a significant correlation with NB tumor pathogenesis and progression. Furthermore, there is an observed trend for better survival in NB patients with high TRIM36 expression indicated by bioinformatic analysis of public available Kocak datasets, which were obtained from the R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl) (Figure 2). The same trend is seen in other NB datasets, such as SEQC and Oberthuer. The data presented above suggests that TRIM36 probably plays a negative role in NB development and progression. Unfortunately, information in regards to the specific roles of TRIM36 in NB is currently limited, and additional research is required in this area.
Figure 2. Kaplan–Meier analysis of overall survival (left side) and event-free survival (right side) for the Kocak datasets based on TRIM36 expression with the log-rank test P value indicated (n = 649, with 173 samples lack survival data and being omitted from the analysis).
TRIM59
TRIM59, also known as mouse ring finger protein 1 (MRF1), belongs to the C-XI TRIM subfamily with a RING finger, one B-box domain, a coiled-coil domain and a C-terminal trans-membrane region [72]. TRIM59 is found, among all the TRIM genes, to be the only member displaying marked up-regulation across all 12 cancer types in The Cancer Genome Atlas (TCGA) database [73], suggesting a probably oncogenic function of it in human cancers. Indeed, there is expanding evidence that TRIM59 is an important regulator of tumorigenesis. For instance, TRIM59 is highly expressed and acts as an early signal transducer of Ras signaling pathway in prostate cancer (CaP) mouse models, whereas the silencing of TRIM59 results in inhibition of cell growth and S-phase arrest in CaP cells [74]. Immunohistochemical analysis shows that TRIM59 expression is significantly up-regulated in multiple human cancers and overexpressed TRIM59 is associated with tumorigenesis and progression [75]. A study in glioblastoma (GBM) cells suggests that TRIM59 promotes GBM tumorigenesis through interaction with nuclear STAT3 and maintains its transcriptional activation by preventing its dephosphorylation [76].
Intriguingly, TRIM59 can induce the expression of ANXA2 [77], a protein that has been reported to enhance multidrug resistance in NB from work in my laboratory [78]. More interestingly, TRIM59 knockdown inhibits cell proliferation by downregulating the Wnt/β-catenin signaling pathway in human NB cells [79]. Collectively, these data imply a role of TRIM59 in NB progression. However, there is currently limited study on the role of TRIM59 in NB. Thus, further investigations are needed to understand its precise function and importance in this pediatric cancer.
Conclusion and perspectives
As an important post-translational modification, ubiquitination has received much attention for its wide and complex roles in many cellular processes, including protein degradation, cell survival and differentiation, innate and adaptive immunity, and signal transduction [80]. In particular, there has been some success in the use of drugs that act on ubiquitination pathway, such as proteasome inhibitors bortezomib and carfilzomib, which are approved by US FDA to be used for the treatment of multiple myeloma [81]. In parallel, targeting E3 ligases or deubiquitinating enzymes has also been a research focus in anti-tumor drug discovery. For example, the successful use of arsenic trioxide (ATO) to treat APL with PML (TRIM19)–RARα fusion protein. Compared with bortezomib treatment, which often leads to multiple side effects and drug resistance, drugs that specifically target E3 ubiquitin ligases, are likely to be more effective in the treatment of of different cancers due to their high substrate specificity. TRIM proteins, the largest subfamily of RING E3 ligases, are potentially strong candidates for therapeutic targeting as they are reported to be associated with cancer by mechanism including involvement in p53 pathway, activation of nuclear receptors and regulation of oncoproteins and tumor suppressors with their E3 ligase activity [16].
As noted above, recent genome sequencing studies revealed relatively low level of gene mutations in NB, suggesting that epigenetic regulation of gene expression may play an important role in the occurrence and development of NB. Thus, there are substantial biomedical research focusing on the contributions of post-translation modification to the origin and progression of NB. As we have discussed in this review, there are six TRIM proteins known to positively or negatively regulate the initiation or progression of NB, suggesting these TRIM proteins would be attractive drug targets for the disease. However, other TRIM proteins which are known to involve in tumorgenesis but not studied explicitly in NB, may play roles in the development of NB as well, such as TRIM protein regulating nuclear receptors (including TRIM19, TRIM24, TRIM25 and TRIM68) and the p53 pathway (including TRIM13, TRIM19, TRIM24, TRIM28 and TRIM29). Moreover, while other TRIM proteins are associated with the prognosis of some cancers, including NB, their precise mechanisms of action and contributions to cancer remain largely unknown. In addition, there are no current drugs targeting TRIM proteins at the laboratory or clinical level for tumor therapy. Thus, further detailed analysis of TRIM proteins is important to understand how to control TRIM proteins as seeds for effective therapy of NB in aim to improve the curative effect and prognosis of the disease.
Abbreviations
- ACD
asymmetric cell division
- ANXA2
Annexin A2
- APL
acute promyelocytic leukaemia
- AR
androgen receptor
- ATO
arsenic trioxide
- BUC
bladder urothelial carcinoma
- CaP
prostate cancer
- DBH
dopamine β hydroxylase
- DHT
dihydrotestosterone
- EBBP
estrogen responsive B box protein
- EMT
epithelial–mesenchymal transition
- ER
estrogen receptor
- GBM
glioblastoma
- HCC
hepatocellular carcinoma
- LGMD2H
limb girdle muscular dystrophy 2H
- MRF1
ring finger protein 1
- NB
neuroblastoma
- NDRG2
N-myc downstream regulated gene 2
- NSC
neuronal stem cell
- PHOX2B
paired-like homebox 2B
- PML
promyelocytic leukaemia
- RARα
retinoic acid receptor-α
- RARβ2
retinoic acid receptor β2
- RNF92
ring finger protein 92
- RNF98
ring finger protein 98
- SCTF
stem cell transcription factor
- TERF
testis RING finger protein
- TRIM
tripartite motif
- USP7
ubiquitin-specific protease 7
Author Contribution
Y.X. and Z.Z. wrote the manuscript. G.X. reviewed and participated in revision of the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 81672488]; and the Science and Technology Commission of Shanghai Municipality [grant number 19140905400].
References
- 1.Maris J.M., Hogarty M.D., Bagatell R. and Cohn S.L. (2007) Neuroblastoma. Lancet (London, England) 369, 2106–2120 10.1016/S0140-6736(07)60983-0 [DOI] [PubMed] [Google Scholar]
- 2.Park J.R., Eggert A. and Caron H. (2008) Neuroblastoma: biology, prognosis, and treatment. Pediatr. Clin. North Am. 55, 97–120, x. 10.1016/j.pcl.2007.10.014 [DOI] [PubMed] [Google Scholar]
- 3.Pugh T.J., Morozova O., Attiyeh E.F., Asgharzadeh S., Wei J.S., Auclair D. et al. (2013) The genetic landscape of high-risk neuroblastoma. Nat. Genet. 45, 279–284 10.1038/ng.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros I.M., Hata J., Joshi V.V., Roald B., Dehner L.P., Tuchler H. et al. (2002) Morphologic features of neuroblastoma (Schwannian stroma-poor tumors) in clinically favorable and unfavorable groups. Cancer 94, 1574–1583 10.1002/cncr.10359 [DOI] [PubMed] [Google Scholar]
- 5.Cohn S.L., Pearson A.D., London W.B., Monclair T., Ambros P.F., Brodeur G.M. et al. (2009) The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J. Clin. Oncol. 27, 289–297 10.1200/JCO.2008.16.6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.London W.B., Castleberry R.P., Matthay K.K., Look A.T., Seeger R.C., Shimada H. et al. (2005) Evidence for an age cutoff greater than 365 days for neuroblastoma risk group stratification in the Children’s Oncology Group. J. Clin. Oncol. 23, 6459–6465 10.1200/JCO.2005.05.571 [DOI] [PubMed] [Google Scholar]
- 7.Zimmerman M.W., Liu Y., He S., Durbin A.D., Abraham B.J., Easton J. et al. (2018) MYC Drives a Subset of High-Risk Pediatric Neuroblastomas and Is Activated through Mechanisms Including Enhancer Hijacking and Focal Enhancer Amplification. Cancer Discov. 8, 320–335 10.1158/2159-8290.CD-17-0993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molenaar J.J., Koster J., Zwijnenburg D.A., van Sluis P., Valentijn L.J., van der Ploeg I. et al. (2012) Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483, 589–593 10.1038/nature10910 [DOI] [PubMed] [Google Scholar]
- 9.Bonvini P., Nguyen P., Trepel J. and Neckers L.M. (1998) In vivo degradation of N-myc in neuroblastoma cells is mediated by the 26S proteasome. Oncogene 16, 1131–1139 10.1038/sj.onc.1201625 [DOI] [PubMed] [Google Scholar]
- 10.Tavana O., Li D., Dai C., Lopez G., Banerjee D., Kon N. et al. (2016) HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat. Med. 22, 1180–1186 10.1038/nm.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto T., Horn S., Brockmann M., Eilers U., Schuttrumpf L., Popov N. et al. (2009) Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell 15, 67–78 10.1016/j.ccr.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 12.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L. et al. (2001) The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatakeyama S. (2017) TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 42, 297–311 10.1016/j.tibs.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 14.Meroni G. and Diez-Roux G. (2005) TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. BioEssays 27, 1147–1157 10.1002/bies.20304 [DOI] [PubMed] [Google Scholar]
- 15.Ozato K., Shin D.M., Chang T.H. and Morse H.C. III (2008) TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8, 849–860 10.1038/nri2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatakeyama S. (2011) TRIM proteins and cancer. Nat. Rev. Cancer 11, 792–804 10.1038/nrc3139 [DOI] [PubMed] [Google Scholar]
- 17.de The H., Lavau C., Marchio A., Chomienne C., Degos L. and Dejean A. (1991) The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66, 675–684 10.1016/0092-8674(91)90113-D [DOI] [PubMed] [Google Scholar]
- 18.Kakizuka A., Miller W.H. Jr, Umesono K., Warrell R.P. Jr, Frankel S.R., Murty V.V. et al. (1991) Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 66, 663–674 10.1016/0092-8674(91)90112-C [DOI] [PubMed] [Google Scholar]
- 19.Chen Z. and Chen S.J. (1992) RARA and PML genes in acute promyelocytic leukemia. Leuk. Lymphoma 8, 253–260 [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi M., Okumura F., Tsukiyama T., Watanabe M., Miyajima N., Tanaka J. et al. (2009) TRIM24 mediates ligand-dependent activation of androgen receptor and is repressed by a bromodomain-containing protein, BRD7, in prostate cancer cells. Biochim. et Biophys. Acta 1793, 1828–1836 10.1016/j.bbamcr.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 21.Miyajima N., Maruyama S., Bohgaki M., Kano S., Shigemura M., Shinohara N. et al. (2008) TRIM68 regulates ligand-dependent transcription of androgen receptor in prostate cancer cells. Cancer Res. 68, 3486–3494 10.1158/0008-5472.CAN-07-6059 [DOI] [PubMed] [Google Scholar]
- 22.Raif A., Marshall G.M., Bell J.L., Koach J., Tan O., D’Andreti C. et al. (2009) The estrogen-responsive B box protein (EBBP) restores retinoid sensitivity in retinoid-resistant cancer cells via effects on histone acetylation. Cancer Lett. 277, 82–90 10.1016/j.canlet.2008.11.030 [DOI] [PubMed] [Google Scholar]
- 23.Liu J., Rao J., Lou X., Zhai J., Ni Z. and Wang X. (2017) Upregulated TRIM11 Exerts its Oncogenic Effects in Hepatocellular Carcinoma Through Inhibition of P53. Cell. Physiol. Biochem. 44, 255–266 10.1159/000484678 [DOI] [PubMed] [Google Scholar]
- 24.Wang S., Zhang Y., Huang J., Wong C.C., Zhai J., Li C. et al. (2019) TRIM67 Activates p53 to Suppress Colorectal Cancer Initiation and Progression. Cancer Res. 79, 4086–4098 [DOI] [PubMed] [Google Scholar]
- 25.Wei W.S., Chen X., Guo L.Y., Li X.D., Deng M.H., Yuan G.J. et al. (2018) TRIM65 supports bladder urothelial carcinoma cell aggressiveness by promoting ANXA2 ubiquitination and degradation. Cancer Lett. 435, 10–22 10.1016/j.canlet.2018.07.036 [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Shi W., Shi H., Lu S., Wang K., Sun C. et al. (2016) TRIM11 overexpression promotes proliferation, migration and invasion of lung cancer cells. J. Exp. Clin. Cancer Res. 35, 100. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Huang J., Tang L., Zhao Y. and Ding W. (2019) TRIM11 promotes tumor angiogenesis via activation of STAT3/VEGFA signaling in lung adenocarcinoma. Am. J. Cancer Res. 9, 2019–2027 [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z., Xu C., Zhang X., Huang L., Zheng C., Chen H. et al. (2017) TRIM11 Upregulation Contributes to Proliferation, Invasion, and EMT of Hepatocellular Carcinoma Cells. Oncol. Res. 25, 691–699 10.3727/096504016X14774897404770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai X., Geng F., Li M. and Liu M. (2019) Tripartite motifcontaining 11 regulates the proliferation and apoptosis of breast cancer cells. Oncol. Rep. 41, 2567–2574 [DOI] [PubMed] [Google Scholar]
- 30.Tuoc T.C. and Stoykova A. (2008) Trim11 modulates the function of neurogenic transcription factor Pax6 through ubiquitin-proteosome system. Genes Dev. 22, 1972–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di K., Linskey M.E. and Bota D.A. (2013) TRIM11 is overexpressed in high-grade gliomas and promotes proliferation, invasion, migration and glial tumor growth. Oncogene 32, 5038–5047 10.1038/onc.2012.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong S.J., Chae H., Lardaro T., Hong S. and Kim K.S. (2008) Trim11 increases expression of dopamine beta-hydroxylase gene by interacting with Phox2b. Biochem. Biophys. Res. Commun. 368, 650–655 10.1016/j.bbrc.2008.01.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maris J.M. (2010) Recent advances in neuroblastoma. N. Engl. J. Med. 362, 2202–2211 10.1056/NEJMra0804577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bielle F., Freneaux P., Jeanne-Pasquier C., Maran-Gonzalez A., Rousseau A., Lamant L. et al. (2012) PHOX2B immunolabeling: a novel tool for the diagnosis of undifferentiated neuroblastomas among childhood small round blue-cell tumors. Am. J. Surg. Pathol. 36, 1141–1149 10.1097/PAS.0b013e31825a6895 [DOI] [PubMed] [Google Scholar]
- 35.Mosse Y.P., Laudenslager M., Khazi D., Carlisle A.J., Winter C.L., Rappaport E. et al. (2004) Germline PHOX2B mutation in hereditary neuroblastoma. Am. J. Hum. Genet. 75, 727–730 10.1086/424530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L., Dong L., Qu X., Jin S., Lv X. and Tan G. (2016) Tripartite motif 16 inhibits hepatocellular carcinoma cell migration and invasion. Int. J. Oncol. 48, 1639–1649 10.3892/ijo.2016.3398 [DOI] [PubMed] [Google Scholar]
- 37.Qi L., Lu Z., Sun Y.H., Song H.T. and Xu W.K. (2016) TRIM16 suppresses the progression of prostate tumors by inhibiting the Snail signaling pathway. Int. J. Mol. Med. 38, 1734–1742 10.3892/ijmm.2016.2774 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Tan H., Qi J., Chu G. and Liu Z. (2017) Tripartite Motif 16 Inhibits the Migration and Invasion in Ovarian Cancer Cells. Oncol. Res. 25, 551–558 10.3727/096504016X14758370595285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim P.Y., Tan O., Liu B., Trahair T., Liu T., Haber M. et al. (2016) High TDP43 expression is required for TRIM16-induced inhibition of cancer cell growth and correlated with good prognosis of neuroblastoma and breast cancer patients. Cancer Lett. 374, 315–323 10.1016/j.canlet.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 40.Huo X., Li S., Shi T., Suo A., Ruan Z. and Yao Y. (2015) Tripartite motif 16 inhibits epithelial-mesenchymal transition and metastasis by down-regulating sonic hedgehog pathway in non-small cell lung cancer cells. Biochem. Biophys. Res. Commun. 460, 1021–1028 10.1016/j.bbrc.2015.03.144 [DOI] [PubMed] [Google Scholar]
- 41.Sutton S.K., Koach J., Tan O., Liu B., Carter D.R., Wilmott J.S. et al. (2014) TRIM16 inhibits proliferation and migration through regulation of interferon beta 1 in melanoma cells. Oncotarget 5, 10127–10139 10.18632/oncotarget.2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung B.B., Bell J., Raif A., Bohlken A., Yan J., Roediger B. et al. (2006) The estrogen-responsive B box protein is a novel regulator of the retinoid signal. J. Biol. Chem. 281, 18246–18256 10.1074/jbc.M600879200 [DOI] [PubMed] [Google Scholar]
- 43.Volchenboum S.L. and Cohn S.L. (2009) Progress in defining and treating high-risk neuroblastoma: lessons from the bench and bedside. J. Clin. Oncol. 27, 1003–1004 10.1200/JCO.2008.20.2739 [DOI] [PubMed] [Google Scholar]
- 44.Marshall G.M., Bell J.L., Koach J., Tan O., Kim P., Malyukova A. et al. (2010) TRIM16 acts as a tumour suppressor by inhibitory effects on cytoplasmic vimentin and nuclear E2F1 in neuroblastoma cells. Oncogene 29, 6172–6183 10.1038/onc.2010.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bell J.L., Malyukova A., Kavallaris M., Marshall G.M. and Cheung B.B. (2013) TRIM16 inhibits neuroblastoma cell proliferation through cell cycle regulation and dynamic nuclear localization. Cell Cycle (Georgetown, Tex) 12, 889–898 10.4161/cc.23825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim P.Y., Rahmanto A.S., Tan O., Norris M.D., Haber M., Marshall G.M. et al. (2013) TRIM16 overexpression induces apoptosis through activation of caspase-2 in cancer cells. Apoptosis 18, 639–651 10.1007/s10495-013-0813-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lassot I., Robbins I., Kristiansen M., Rahmeh R., Jaudon F., Magiera M.M. et al. (2010) Trim17, a novel E3 ubiquitin-ligase, initiates neuronal apoptosis. Cell Death Differ. 17, 1928–1941 10.1038/cdd.2010.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandell M.A., Jain A., Arko-Mensah J., Chauhan S., Kimura T., Dinkins C. et al. (2014) TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell 30, 394–409 10.1016/j.devcel.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan J. and Yankner B.A. (2000) Apoptosis in the nervous system. Nature 407, 802–809 10.1038/35037739 [DOI] [PubMed] [Google Scholar]
- 50.Magiera M.M., Mora S., Mojsa B., Robbins I., Lassot I. and Desagher S. (2013) Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons. Cell Death Differ. 20, 281–292 10.1038/cdd.2012.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lionnard L., Duc P., Brennan M.S., Kueh A.J., Pal M., Guardia F. et al. (2019) TRIM17 and TRIM28 antagonistically regulate the ubiquitination and anti-apoptotic activity of BCL2A1. Cell Death Differ. 26, 902–917 10.1038/s41418-018-0169-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kudryashova E., Wu J., Havton L.A. and Spencer M.J. (2009) Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum. Mol. Genet. 18, 1353–1367 10.1093/hmg/ddp036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicklas S., Otto A., Wu X., Miller P., Stelzer S., Wen Y. et al. (2012) TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PLoS One 7, e30445 10.1371/journal.pone.0030445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao T.T., Jin F., Li J.G., Xu Y.Y., Dong H.T., Liu Q. et al. (2018) TRIM32 promotes proliferation and confers chemoresistance to breast cancer cells through activation of the NF-kappaB pathway. J. Cancer 9, 1349–1356 10.7150/jca.22390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang C., Xu J., Fu H., Zhang Y., Zhang X., Yang D. et al. (2018) TRIM32 promotes cell proliferation and invasion by activating beta-catenin signalling in gastric cancer. J. Cell. Mol. Med. 22, 5020–5028 10.1111/jcmm.13784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin H., Li Z., Chen J. and Hu X. (2019) Expression and the potential functions of TRIM32 in lung cancer tumorigenesis. J. Cell. Biochem. 120, 5232–5243 10.1002/jcb.27798 [DOI] [PubMed] [Google Scholar]
- 57.Horn E.J., Albor A., Liu Y., El-Hizawi S., Vanderbeek G.E., Babcock M. et al. (2004) RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis 25, 157–167 10.1093/carcin/bgh003 [DOI] [PubMed] [Google Scholar]
- 58.Liu J., Zhang C., Wang X.L., Ly P., Belyi V., Xu-Monette Z.Y. et al. (2014) E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ. 21, 1792–1804 10.1038/cdd.2014.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kano S., Miyajima N., Fukuda S. and Hatakeyama S. (2008) Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 68, 5572–5580 10.1158/0008-5472.CAN-07-6231 [DOI] [PubMed] [Google Scholar]
- 60.Schwamborn J.C., Berezikov E. and Knoblich J.A. (2009) The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925 10.1016/j.cell.2008.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fults D., Pedone C., Dai C. and Holland E.C. (2002) MYC expression promotes the proliferation of neural progenitor cells in culture and in vivo. Neoplasia (New York, NY) 4, 32–39 10.1038/sj.neo.7900200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato T., Okumura F., Kano S., Kondo T., Ariga T. and Hatakeyama S. (2011) TRIM32 promotes neural differentiation through retinoic acid receptor-mediated transcription. J. Cell Sci. 124, 3492–3502 10.1242/jcs.088799 [DOI] [PubMed] [Google Scholar]
- 63.Izumi H. and Kaneko Y. (2014) Trim32 facilitates degradation of MYCN on spindle poles and induces asymmetric cell division in human neuroblastoma cells. Cancer Res. 74, 5620–5630 10.1158/0008-5472.CAN-14-0169 [DOI] [PubMed] [Google Scholar]
- 64.Li D., Mei H., Pu J., Xiang X., Zhao X., Qu H. et al. (2015) Intelectin 1 suppresses the growth, invasion and metastasis of neuroblastoma cells through up-regulation of N-myc downstream regulated gene 2. Mol. Cancer 14, 47 10.1186/s12943-015-0320-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mokhonova E.I., Avliyakulov N.K., Kramerova I., Kudryashova E., Haykinson M.J. and Spencer M.J. (2015) The E3 ubiquitin ligase TRIM32 regulates myoblast proliferation by controlling turnover of NDRG2. Hum. Mol. Genet. 24, 2873–2883 10.1093/hmg/ddv049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balint I., Muller A., Nagy A. and Kovacs G. (2004) Cloning and characterisation of the RBCC728/TRIM36 zinc-binding protein from the tumor suppressor gene region at chromosome 5q22.3. Gene 332, 45–50 10.1016/j.gene.2004.02.045 [DOI] [PubMed] [Google Scholar]
- 67.Gushchina L.V., Kwiatkowski T.A., Bhattacharya S. and Weisleder N.L. (2018) Conserved structural and functional aspects of the tripartite motif gene family point towards therapeutic applications in multiple diseases. Pharmacol. Ther. 185, 12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyajima N., Maruyama S., Nonomura K. and Hatakeyama S. (2009) TRIM36 interacts with the kinetochore protein CENP-H and delays cell cycle progression. Biochem. Biophys. Res. Commun. 381, 383–387 10.1016/j.bbrc.2009.02.059 [DOI] [PubMed] [Google Scholar]
- 69.Liang C., Wang S., Qin C., Bao M., Cheng G., Liu B. et al. (2018) TRIM36, a novel androgen-responsive gene, enhances anti-androgen efficacy against prostate cancer by inhibiting MAPK/ERK signaling pathways. Cell Death. Dis. 9, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kimura N., Yamada Y., Takayama K.I., Fujimura T., Takahashi S., Kume H. et al. (2018) Androgen-responsive tripartite motif 36 enhances tumor-suppressive effect by regulating apoptosis-related pathway in prostate cancer. Cancer Sci. 109, 3840–3852 10.1111/cas.13803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olsson M., Beck S., Kogner P., Martinsson T. and Caren H. (2016) Genome-wide methylation profiling identifies novel methylated genes in neuroblastoma tumors. Epigenetics 11, 74–84 10.1080/15592294.2016.1138195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y. and Yang W.B. (2017) Down-regulation of tripartite motif protein 59 inhibits proliferation, migration and invasion in breast cancer cells. Biomed. Pharmacother. 89, 462–467 [DOI] [PubMed] [Google Scholar]
- 73.Tan P., Ye Y., He L., Xie J., Jing J., Ma G. et al. (2018) TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PLoS Biol. 16, e3000051 10.1371/journal.pbio.3000051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valiyeva F., Jiang F., Elmaadawi A., Moussa M., Yee S.P., Raptis L. et al. (2011) Characterization of the oncogenic activity of the novel TRIM59 gene in mouse cancer models. Mol. Cancer Ther. 10, 1229–1240 10.1158/1535-7163.MCT-11-0077 [DOI] [PubMed] [Google Scholar]
- 75.Khatamianfar V., Valiyeva F., Rennie P.S., Lu W.Y., Yang B.B., Bauman G.S. et al. (2012) TRIM59, a novel multiple cancer biomarker for immunohistochemical detection of tumorigenesis. BMJ Open 2, e001410 10.1136/bmjopen-2012-001410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sang Y., Li Y., Song L., Alvarez A.A., Zhang W., Lv D. et al. (2018) TRIM59 Promotes Gliomagenesis by Inhibiting TC45 Dephosphorylation of STAT3. Cancer Res. 78, 1792–1804 10.1158/0008-5472.CAN-17-2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y., Zhou Z., Wang X., Zhang X., Chen Y., Bai J. et al. (2018) TRIM59 Is a Novel Marker of Poor Prognosis and Promotes Malignant Progression of Ovarian Cancer by Inducing Annexin A2 Expression. Int. J. Biol. Sci. 14, 2073–2082 10.7150/ijbs.28757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y., Chen K., Cai Y., Cai Y., Yuan X., Wang L. et al. (2017) Annexin A2 could enhance multidrug resistance by regulating NF-kappaB signaling pathway in pediatric neuroblastoma. J. Exp. Clin. Cancer Res. 36, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen G., Chen W., Ye M., Tan W. and Jia B. (2019) TRIM59 knockdown inhibits cell proliferation by down-regulating the Wnt/beta-catenin signaling pathway in neuroblastoma. Biosci. Rep. 39, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Popovic D., Vucic D. and Dikic I. (2014) Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- 81.Landgren O. and Iskander K. (2017) Modern multiple myeloma therapy: deep, sustained treatment response and good clinical outcomes. J. Intern. Med. 281, 365–382 10.1111/joim.12590 [DOI] [PubMed] [Google Scholar]