Abstract
Regulation of abscission is an important agricultural concern since precocious abscission can reduce crop yield. INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) peptide and its receptors the HAESA (HAE) and HAESA-like2 (HSL2) kinases have been revealed to be core components controlling floral organ abscission in the model plant Arabidopsis. However, it is still unclear whether the homologs of IDA-HAE/HSL2 in non-model plants are correlated to abscission. Previously, we found LcIDL1, a homolog of IDA from litchi, has a similar role to AtIDA in control of floral organ abscission in Arabidopsis. Here, we further isolated an HAESA-like homolog, LcHSL2, which is likely involved in the fruitlet abscission in litchi. Ectopic expression of LcHSL2 in wild type Arabidopsis has no effect on the floral organ abscission. However, its presence in the hae hsl2 mutant background completely rescued the floral organ abscission deficiency. LcHSL2 is localized in the cell membrane and the LcHSL2 gene is expressed at the pedicel abscission zone (AZ) of litchi and floral AZ of Arabidopsis. Real-time PCR analysis showed that the expression level of LcHSL2 was increased during ethephon-induced fruitlet abscission in litchi. Taken together, our findings suggest that HSL2 homologs have functional conservation in Arabidopsis and litchi, and LcHSL2 might play a critical role in regulation of fruitlet abscission in litchi.
Keywords: litchi, fruitlet abscission, LcHSL2, floral organ abscission, AZ
1. Introduction
Abscission in general is an important process in plants to shed unwanted or infected organs in response to internal or external cues [1]. However, untimely abscission of flowers or fruits is a challenge for farmers since it can cause severe losses in crop yield. Therefore, it is of great significance to understand the mechanism underlying the regulation of abscission. The abscission process in plants takes place at the cell files in the specialized abscission zone (AZ) between the organ to be shed and the main plant body. Generally, the AZ will go through three sequential developmental stages when the abscission process is activated: (i) Acquisition of competence to respond to abscission signals; (ii) cell wall loosening and expansion followed by organ separation; and (iii) transdifferentiation of the retained portion of the AZ to generate a protective layer [2,3,4]. For years, a core signaling pathway has been defined to regulate the floral organ abscission in model plant Arabidopsis [5,6,7,8,9].
In Arabidopsis, INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), a small secreted peptide, plays an essential role in the regulation of abscission. Mutation in IDA causes the failure of floral organ abscission to take place, whereas overexpression of IDA results in obvious precocious abscission events [10,11]. HAESA (HAE) and HAESA-like2 (HSL2), a pair of closely related leucine-rich repeat receptor-like kinases (LRR-RLKs), function redundantly to positively regulate the floral organ abscission process since the hae hsl2 double mutants were, like ida, totally deficient in abscission [5,12]. Further genetic investigation demonstrates that the role of IDA in the regulation of floral organ abscission is dependent on its receptors HAE/HSL2 [13]. Upon activation of the HAE/HSL2 by IDA, a mitogen-activated protein (MAP) kinase cascade consisting of the MAPK kinase 4 (MKK4)/MKK5 and the MAPK 3 (MPK3)/MPK6, is turned on [5], leading to the suppression of KNOTTED1-LIKE HOMEOBOX (KNOX) transcription factors, ultimately resulting in the induction of genes encoding cell wall remodeling enzymes [6]. Recently, it was shown that four somatic embryogenesis receptor kinase (SERK) family RLKs serve as co-receptors of HAE/HSL2 and form a complex with HAE/HSL2 upon binding of IDA to the receptor kinases, thus regulating the initiation of the floral organ abscission [9,14]. In contrast to many other species, Arabidopsis usually does not abscise cauline leaves, whole flowers, fruit, or leaves [15]. Orthologs of IDA-HAE/HSL2 have been identified and found to be expressed in the AZs of flowers, leaves, and fruits in all orders of flowering plants, suggesting that the IDA-HAE/HSL2 signaling module can induce the abscission processes in species besides Arabidopsis [16,17]. To date, a few cases validated the hypothesized conserved function of IDA-HAE/HSL2 across organ and species by complementation of the Arabidopsis ida mutant. For example, CitIDA3, a closest homolog of AtIDA in citrus, is suggested to be correlated to the process of fruit abscission since ectopic expression of CitIDA3 caused precocious abscission in wild type Arabidopsis, and was sufficient to rescue the abscission defect when expressed in the ida mutant background [18]. In addition, IDA peptides enhanced the leaf abscission in Populus and ripe fruit abscission in oil palm, which provides additional evidence for the conservation of the IDA–HAE–HSL2 pathway in widely different abscission contexts [17]. However, the proof of conserved function of IDA-HAE/HSL2 in regulation of abscission is still lacking, particularly in fruit crops.
Litchi (Litchi chinensis Sonn.) is an important tropical and subtropical fruit crop that is widespread and cultivated in Southeast Asia. There are three to four waves of physiological fruit abscission throughout litchi fruit development, causing low yield and heavy economic loss [19,20]. Thus, it is of interest to identify key components that are involved in regulation of the fruit abscission. Previously, we characterized and identified LcIDL1, an IDA homolog from litchi, which functioned similarly to AtIDA in the control of floral abscission in Arabidopsis [21]. Here, we further isolated LcHSL2—a homolog of HAE/HSL2 in litchi—and found that it was involved in the fruitlet abscission in litchi and played a role in regulating the floral organ abscission in Arabidopsis.
2. Results
2.1. Identification of HAE/HSL2 Homologs in Litchi
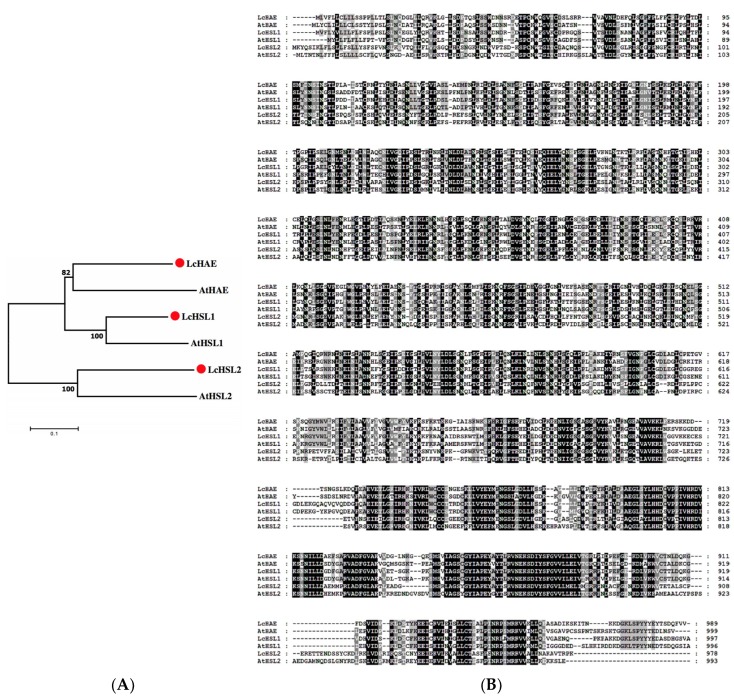
To identify the litchi HAE/HSL2 homologs, TBLASTN searches against the litchi genome (http://111.230.180.7:81/index.php) were performed at TBtools [22] using the amino acid sequences of the Arabidopsis HAE (AT4G28490), HSL1 (AT1G28440), and HSL2 (AT5G65710). As a result, three HAE/HSL2-like genes in the litchi genome were identified (File S1). They were named LcHAE, LcHSL1, and LcHSL2 based on their phylogenetic relationship with Arabidopsis HAE/HSL2 (Figure 1A). Protein sequence alignment showed that the amino acid similarity was 60.56% between LcHAE and AtHAE, 72.46% between LcHSL1 and AtHSL1, and 58.16% between LcHSL2 and AtHSL2 (Figure 1B). We thus hypothesized that LcHAE, LcHSL1, and LcHSL2 might have similar functions to Arabidopsis HAE/HSL2 in the regulation of abscission.
Figure 1.
Sequence and phylogenetic analysis of LcHSLs protein. (A) Phylogenetic relationships of LcHSLs with Arabidopsis AtHAE, AtHSL1, and AtHSL2. LcHAE (ID number: LITCHI029130.m1), LcHSL1 (ID number: LITCHI029130.m1), and LcHSL2 (ID number: LITCHI007137.m1) are indicated by red circles. The maximum likelihood phylogenetic tree was created using MEGA program (version 5.0). (B) Multiple alignment of LcHAE, LcHSL1, and LcHSL2 with AtHAE, AtHSL1, and AtHSL2.
2.2. Ectopic Expression of LcHSL2 Complements the Abscission Deficiency of the hae hsl2 Mutant
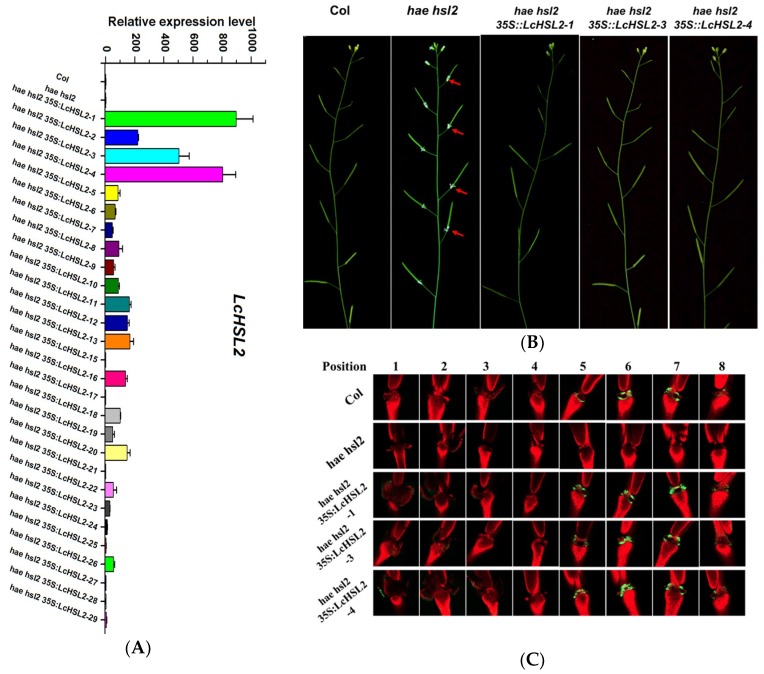
To test whether LcHAE, LcHSL1, and LcHSL2 have functions in the regulation of abscission, wild type Arabidopsis plants were transformed with a construct driving LcHAE, LcHSL1, and LcHSL2, respectively, by the strong constitutive cauliflower mosaic virus 35S promoter. More than 20 transgenic lines expressing each gene were generated. However, there were no visible phenotypic changes observed when compared to wild type Arabidopsis. Next, we investigated whether LcHAE, LcHSL1, and LcHSL2 had a conserved function to HAE/HSL2 in regulation of floral abscission in Arabidopsis. The hae hsl2 mutant plants were transformed with each construct as mentioned above. We also generated more than 20 transgenic lines expressing each gene. For all the transgenic lines expressing LcHAE or LcHSL1 in hae hsl2 background, the floral organs remained attached, which was the same as that in the hae hsl2 mutants. Interestingly, we found that 20 out of 29 T1 transgenic lines expressing LcHSL2 showed wild type abscission patterns (three representative transgenic lines were shown here, Figure 2A,B). Previous studies have demonstrated that organ abscission is closely associated with an increase in cytosolic pH in AZ cells, which can be easily detected by BCECF-AM staining [23]. Therefore, we performed BCECF assays using three representative lines, hae hsl2 35S:LcHSL2-1, hae hsl2 35S:LcHSL2-3, and hae hsl2 35S:LcHSL2-4, which displayed a relatively higher expression level of LcHSL2 in hae hsl2 background (Figure 2A). In wild-type Col, the BCECF signals in the floral AZ appeared starting from position five and went through position eight, while the BCECF signals disappeared in the floral AZ of hae hsl2 mutants, which is consistent with floral organ abscission deficiency of hae hsl2. When LcHSL2 was expressed in hae hsl2 mutants, the BCECF signals appeared again and showed a similar pattern to that in Col (Figure 2C). These findings indicate that the presence of LcHSL2 was sufficient to induce the floral organ abscission in hae hsl2 mutant plants. Thus, LcHSL2 was selected for further analysis.
Figure 2.
Ectopic expression of LcHSL2 in Arabidopsis hae hsl2 mutants rescues the floral organ abscission deficiency. (A) Expression levels of LcHSL2 in transgenic lines. (B) Comparison of the inflorescence of hae hsl2 and hse hsl2 35S:LcHSL2-1, hse hsl2 35S:LcHSL2-3, and hse hsl2 35S:LcHSL2-4. Red arrow heads indicate attached floral organs. (C) BCECF fluorescence micrographs of floral organ AZ of Col, hae hsl2, and hse hsl2 35S:LcHSL2 transgenic lines. Inflorescences were sampled separately, incubated in BCECF solution, and examined by a confocal laser scanning microscope. The microscopic fluorescence images represent merged images of BCECF fluorescence (green) with chlorophyll auto fluorescence (red) images. The images presented for each plant and positions are representative images out of three to four replicates.
2.3. LcHSL2 Is Localized in the Plasma Membrane and Expressed at the Floral AZ of Arabidopsis
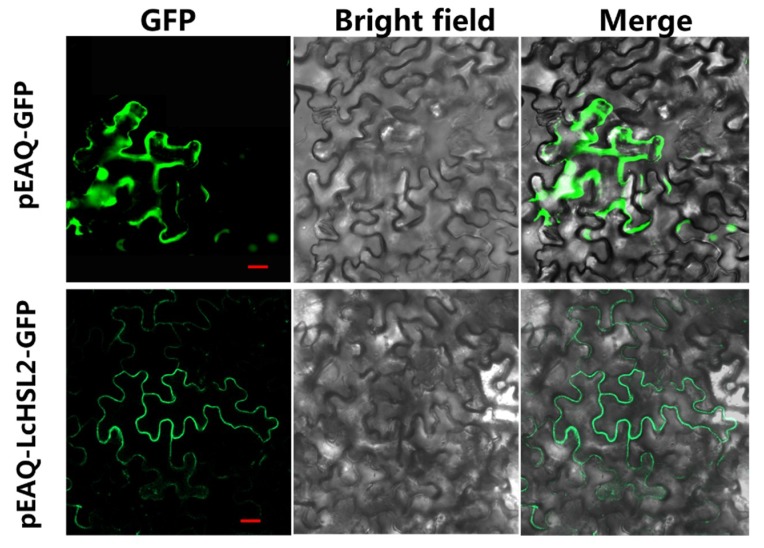
To investigate the subcellular localization of LcHSL2, the full-length coding sequence of LcHSL2 fused with green fluorescent protein (GFP) was transiently expressed in tobacco leaf epidermal cells. Fluorescence of fused GFP was predominantly observed in the cell membrane, and GFP signals of free control (pEAQ-GFP) were uniformly distributed throughout the whole cell (Figure 3). This indicated that LcHSL2 functions in the plasma membrane, which is consistent with its role as a receptor-like kinase.
Figure 3.
Subcellular localization assay of LcHSL2 in epidermal cells of tobacco leaves. GFP signals were observed with a fluorescence microscope after 48 h of infiltration. Bars = 25 μm.
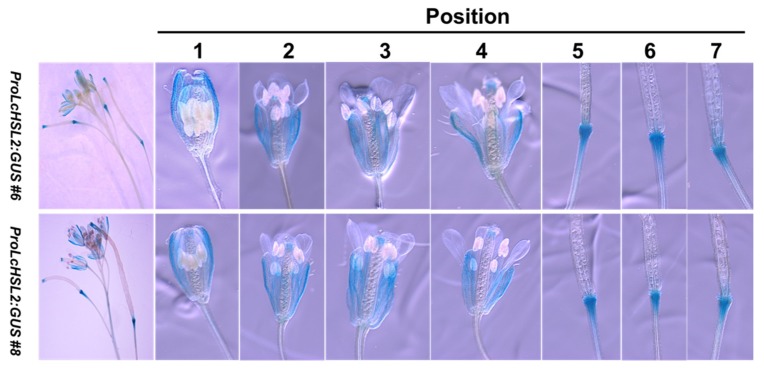
In addition, we investigated the spatial and temporal activity of the LcHSL2 promoter in Arabidopsis. A construct carrying a GUS reporter driven by the LcHSL2 promoter was transformed into Arabidopsis. We had two transgenic lines with different expression levels of GUS (Figure 4). The GUS expression driven by the LcHSL2 promoter was predominately localized at the floral AZ, and the GUS signals in the floral AZ appeared starting from position five (Figure 4), which is consistent with its function in the control of floral organ abscission in Arabidopsis.
Figure 4.
ProLcHSL2:GUS expression in Arabidopsis floral AZ. The numbers indicate flower position along the inflorescence. Position numbers were counted from the first flower with visible white petals at the top of the inflorescence.
2.4. LcHSL2 Is Expressed at the Fruit AZ and Its Expression Level Is Increased During the Fruitlet Abscission in Litchi
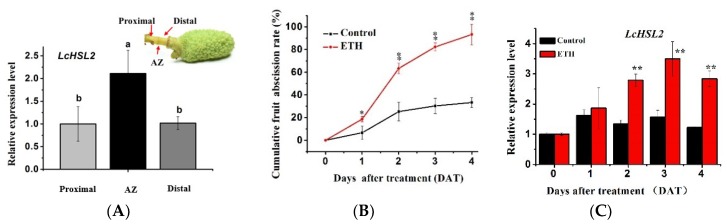
To test the involvement of LcHSL2 in the fruitlet abscission of litchi, we examined the expression profile of LcHSL2 in three regions of the pedicel: The fruit AZ, the distal region (between the fruitlet and the fruit AZ), and the proximal region (Figure 5A). LcHSL2 was expressed significantly higher in the fruit AZ than those in the adjacent tissues (Figure 5A).
Figure 5.
LcHSL2 is expressed in the fruit AZ and is increased during the ETH-induced fruitlet abscission in litchi. (A) LcHSL2 is predominantly expressed in the fruit AZ of litchi. Top right corner shows the fruit abscission zone (AZ), distal, and proximal regions of the litchi fruit peduncle. Different letters indicate significant differences as determined using Ducan’s multiple range test (p < 0.05). (B) Ethephon (ETH)-induced fruitlet abscission in litchi. The results are the means of three biological replicates. Error bars represent ± SE. Asterisks indicate a significant difference (Student’s t-test: p < 0.05 indicated by *; p < 0.01 indicated by **). (C) LcHSL2 expression is increased following the ETH-induced fruitlet abscission. The y-axis represents fold-change in expression relative to the control at 0 day, which was set to 1. Data represent the average of three biological replicates with three technical replicates each. Error bars represent ± SE. Asterisks indicate a significant difference (Student’s t-test: p < 0.01 indicated by **).
To gain a better understanding of the involvement of LcHSL2 in fruitlet abscission in litchi, we examined the expression pattern of LcHSL2 during the litchi fruitlet abscission induced by ethephon application (ETH). As shown in Figure 5B, fruitlets dropped starting from the third day after ETH treatments, the cumulative fruit abscission rate was up to 93.33% at the fifth day after ETH treatment, whereas only 33.33% of fruitlets dropped at fifth day in control (Figure 5B). qRT-PCR analysis showed that the expression level of LcHSL2 in the pedicel AZ was significantly induced by ETH treatment, with about 2.07-fold, 2.22-fold, and 2.31-fold higher in ETH-treated AZ tissues than that in control at third, fourth, and fifth day, respectively (Figure 5C). This suggests that LcHSL2 is associated with ethephon-induced fruitlet abscission in litchi.
3. Discussion
Unexpected fruit abscission is a severe problem for the litchi industry. In general, litchi trees will produce hundreds of female flowers per inflorescence, but less than 5% of the initial female flowers can develop into mature fruits [20,24]. The excessive abscission of flowers/fruitlets is one of the main factors causing universal low productivity in litchi [19,20]. Therefore, new litchi genotypes with a low rate of fruit abscission behavior derived from genetic engineering (transgenic or CRISPR gene editing technologies) would be favorable. For this, identifying the key components that are involved in regulation of fruit abscission in litchi is our priority.
In Arabidopsis, the IDA-HAE/HSL2 module was primarily found to initiate a signaling pathway in control of floral organ abscission [5,6,10,11,12]. This signaling module was also revealed to play an essential role in control of cell separation during lateral root emergence and root cap sloughing [25,26,27]. GmIDL2a and GmIDL4a, two homologs of IDA in soybean, could promote the cell separation during lateral root emergence through regulating the cell wall remodeling gene expression [28]. These findings suggest that the IDA-HAE/HSL2 signaling module can control cell separation events other than just floral organ abscission. Previously, we identified a homolog of IDA, LcIDL1, which is associated with the fruitlet abscission in litchi and has a similar function to Arabidopsis IDA in regulation of floral organ abscission [21]. In this study, we further revealed that homologs of HAE/HSL2 also exist in the litchi genome, and one member LcHSL2 is involved in the fruitlet abscission. In addition, LcHSL2 was demonstrated to play a role in abscission since ectopic expression of LcHSL2 in Arabidopsis hae hsl2 mutants background could rescue the floral organ abscission deficiency (Figure 2). However, whether LcIDL1 and LcHSL2 could form a ligand-receptor module in litchi to control the fruit abscission requires further study. Recent studies have showed that IDA-HAE/HSL2 signaling components were found across the plant kingdom [16,29]. Based on these findings, we propose that different cell separation processes might share the IDA-HAE/HSL2 signaling module that is conserved across plant species.
The interaction between the IDA-HAE/HSL2 module and ethylene signaling that functions in the abscission needs further clarification. In the model plant Arabidopsis, ida mutants share similar ethylene sensitivity to wild type Arabidopsis, and the deficiency in floral organ abscission of ida is unaffected by the exposure of exogenous ethylene [10]. In addition, the AtHAE promoter is expressed specifically in floral AZ and its expression does not differ between the ethylene-insensitive mutant etr1-1 and wild type [12], leading to the conclusion that the IDA-HAE/HSL2 signaling module acts independently of ethylene in regulation of abscission. Recently, a review which reevaluates the relation between ethylene and the IDA–HAE–HSL2 pathway, proposes that the IDA–HAE–HSL2 pathway is essential for the final stages of organ abscission, while ethylene plays a critical role in its initiation and progression [30]. In cultivated crops, ethylene activated the abscission-specific expression of soybean and tomato IDA-like genes, and application of ethylene inhibitor repressed the soybean IDA-like gene expression [31]. Additionally, recent reports revealed that ethylene activated the expression of HSL and IDA genes in the AZ of oil palm fruits [16], as well as in the AZ of lupine flowers [32]. In all the above studies, the IDA or HSL in the AZ were induced prior to the onset of abscission, indicating that the IDA-HAE/HSL2 signaling module is promoted by ethylene, and probably controls the abscission processes downstream of ethylene. In our study, we also found that LcHSL2 was increased during the ethephon (ETH)-induced fruitlet abscission in litchi (Figure 5). Our findings in this study, and in combination with a previous report for LcIDL1 [21], further support that the IDA-HAE/HSL2 module acts downstream of ethylene signaling in control of abscission. Therefore, it is of great interest in the future to determine which key components involved in the ethylene signaling pathway interact directly with the IDA-HAE/HSL2 module.
4. Materials and Methods
4.1. Plant Materials and Treatments
For litchi, three 16-year-old litchi trees (Litchi chinensis Sonn. cv. Feizixiao) grown in an orchard located in South China Agricultural University (Guangzhou, China) were selected randomly. Similar diameter shoots (about 5–8 mm) bearing 30 fruits and growing in different directions from each tree were tagged. ETH treatments, calculation of cumulative fruit abscission rate (CFAR), and collection of AZ tissues is previously described in [21].
For Arabidopsis to express LcHSL2 in hae hsl2 mutants (which are totally deficient in floral organ abscission [5]), the full-length open reading frame of LcHSL2 was subcloned into the vector pCAMBIA1302 under the control of the 35S promoter to generate 35S:LcHSL2 constructs using ClonExpress®II One Step Cloning Kit (Vazyme, Nanjing, China). Then, 35S:LcHSL2 constructs were transformed into hae hsl2 plants following the floral dip method [33]. T1 transgenic lines were used for phenotypic analysis. All the Arabidopsis plants were grown at 22 °C under long day (16 h light/8 h dark) conditions. To reduce variation, all genotypes tested in each experiment were grown together. Primers used here are listed in Table S1.
4.2. Quantitative RT-PCR Analysis
Total RNA was isolated from litchi AZ tissues or Arabidopsis leaves (20 days old) using 1 mL Trizol reagent (Invitrogen, Carlsbad, CA, USA). The first strand cDNA synthesis was generated using 2 μg total RNA according to the manual of the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen, Beijing, China). Quantitative RT-PCR analysis was performed as previously described [21]. Primers used here are listed in Table S1.
4.3. Subcellular Localization Analysis
The coding sequence of LcHSL2 was fused into a pBI121 vector that was tagged with GFP to generate 35S:LcHSL2-GFP constructs. Then, 35S:LcHSL2-GFP constructs were delivered into Agrobacterium tumefaciens strain EHA105, and were transformed into tobacco (Nicotiana benthamiana) leaves as previously described [34]. YFP and GFP fluorescence were observed with a confocal laser scanning microscope (LSM 7 DUO, ZEISS, Oberkochen, Germany). Primers used here are listed in Table S1.
4.4. BCECF Fluorescence Assay
BCECF fluorescence analysis was conducted as described previously [21]. In brief, inflorescences were cut from the plant body and immersed in 10 μM BCECF-AM (B1150, Invitrogen, Carlsbad, CA, USA) solution under darkness for 20 min. The inflorescences were then rinsed four times with phosphate-buffered saline (PBS, pH 7.4) to remove excess BCECE-AM. Images were snapped with a confocal laser scanning microscope (LSM 7 DUO, ZEISS, Germany). Samples were excited by both 488 nm and 633 nm light, then BCECF fluorescence and chlorophyll autofluorescence were detected through 494–598 and 647–721 filters, respectively.
4.5. Histochemical GUS Assays
The LcHSL2 promoter region (−1 to −2000 bp) was subcloned into the vector pCAMBIA1391 to generate the LcHSL2pro:GUS constructs. The LcHSL2pro:GUS constructs were delivered into Arabidopsis as mentioned above. T1 transgenic plants were used for GUS assays. Transgenic flowers were stained in GUS solution (10 mM EDTA, 0.1% Triton X-100, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, 100 μg mL−1 chloramphenicol, and 1 mg mL−1 X-Gluc in a 50 mM sodium phosphate buffer, pH 7.0) overnight at 37 °C and cleared in a 20% lactic acid/20% glycerol solution for 6 h at 37 °C and then cleared in 70% ethanol. GUS expression was visualized using a Zeiss SV11 stereoscope. Primers used here are listed in Table S1.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31601725), the Innovation Team Project of the Department of Education of Guangdong Province (grant no. 2016KCXTD011), and the Science and Technology Program of Guangzhou, China (grant no. 201804020063).
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/23/5945/s1.
Author Contributions
M.Z. and J.L. conceived and designed the experiments; F.W. and Z.Z. performed most of the experiments; Y.Y. provided assistance; M.Z. and J.L. wrote the paper.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31601725), the Innovation Team Project of the Department of Education of Guangdong Province (grant no. 2016KCXTD011), and the Science and Technology Program of Guangzhou, China (grant no. 201804020063).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bleecker A.B., Patterson S.E. Last exit: Senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell. 1997;9:1169–1179. doi: 10.1105/tpc.9.7.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patterson S.E. Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 2001;126:494–500. doi: 10.1104/pp.126.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aalen R.B., Wildhagen M., Sto I.M., Butenko M.A. IDA: A peptide ligand regulating cell separation processes in Arabidopsis. J. Exp. Bot. 2013;64:5253–5261. doi: 10.1093/jxb/ert338. [DOI] [PubMed] [Google Scholar]
- 4.Gubert C.M., Christy M.E., Ward D.L., Groner W.D., Liljegren S.J. ASYMMETRIC LEAVES1 regulates abscission zone placement in Arabidopsis flowers. BMC Plant Biol. 2014;14:195. doi: 10.1186/s12870-014-0195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho S.K., Larue C.T., Chevalier D., Wang H., Jinn T.L., Zhang S., Walker J.C. Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2008;105:15629–15634. doi: 10.1073/pnas.0805539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi C.L., Stenvik G.E., Vie A.K., Bones A.M., Pautot V., Proveniers M., Aalen R.B., Butenko M.A. Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell. 2011;23:2553–2567. doi: 10.1105/tpc.111.084608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liljegren S.J. Organ abscission: Exit strategies require signals and moving traffic. Curr. Opin. Plant Biol. 2012;15:670–676. doi: 10.1016/j.pbi.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Niederhuth C.E., Cho S.K., Seitz K., Walker J.C. Letting go is never easy: Abscission and receptor-like protein kinases. J. Integr. Plant Biol. 2013;55:1251–1263. doi: 10.1111/jipb.12116. [DOI] [PubMed] [Google Scholar]
- 9.Meng X., Zhou J., Tang J., Li B., de Oliveira M., Chai J., He P., Shan L. Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 2016;14:1330–1338. doi: 10.1016/j.celrep.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butenko M.A., Patterson S.E., Grini P.E., Stenvik G.E., Amundsen S.S., Mandal A., Aalen R.B. Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell. 2003;15:2296–2307. doi: 10.1105/tpc.014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenvik G.E., Butenko M.A., Urbanowicz B.R., Rose J.K., Aalen R.B. Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell. 2006;18:1467–1476. doi: 10.1105/tpc.106.042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinn T.L., Stone J.M., Walker J.C. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000;14:108–117. [PMC free article] [PubMed] [Google Scholar]
- 13.Stenvik G.E., Tandstad N.M., Guo Y., Shi C.L., Kristiansen W., Holmgren A., Clark S.E., Aalen R.B., Butenko M.A. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell. 2008;20:1805–1817. doi: 10.1105/tpc.108.059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santiago J., Brandt B., Wildhagen M., Hohmann U., Hothorn L.A., Butenko M.A., Hothorn M. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. Elife. 2016;5:e15075. doi: 10.7554/eLife.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patharkar O.R., Walker J.C. Core mechanisms regulating developmentally timed and environmentally triggered abscission. Plant Physiol. 2016;172:510–520. doi: 10.1104/pp.16.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sto I.M., Orr R.J., Fooyontphanich K., Jin X., Knutsen J.M., Fischer U., Tranbarger T.J., Nordal I., Aalen R.B. Conservation of the abscission signaling peptide IDA during Angiosperm evolution: Withstanding genome duplications and gain and loss of the receptors HAE/HSL2. Front. Plant Sci. 2015;6:931. doi: 10.3389/fpls.2015.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tranbarger T.J., Domonhedo H., Cazemajor M., Dubreuil C., Fischer U., Morcillo F. The PIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION enhances Populus leaf and Elaeis guineensis fruit abscission. Plants (Basel) 2019;8:143. doi: 10.3390/plants8060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estornell L.H., Wildhagen M., Perez-Amador M.A., Talon M., Tadeo F.R., Butenko M.A. The IDA peptide controls abscission in Arabidopsis and citrus. Front. Plant Sci. 2015;6:1003. doi: 10.3389/fpls.2015.01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan R., Huang H. Litchi fruit abscission: Its patterns, effect of shading and relation to endogenous abscisic acid. Sci. Hortic. 1988;36:281–292. doi: 10.1016/0304-4238(88)90063-5. [DOI] [Google Scholar]
- 20.Mitra S.K., Pereira L.S., Pathak P.K., Majumdar D. Fruit abscission pattern of lychee cultivars. Acta Hortic. 2005;665:215–218. doi: 10.17660/ActaHortic.2005.665.24. [DOI] [Google Scholar]
- 21.Ying P., Li C., Liu X., Xia R., Zhao M., Li J. Identification and molecular characterization of an IDA-like gene from litchi, LcIDL1, whose ectopic expression promotes floral organ abscission in Arabidopsis. Sci. Rep. 2016;6:37135. doi: 10.1038/srep37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C., Chen H., He Y., Xia R. TBtools, a Toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv. 2018:289660. doi: 10.1101/289660. [DOI] [Google Scholar]
- 23.Sundaresan S., Philosoph-Hadas S., Riov J., Belausov E., Kochanek B., Tucker M.L., Meir S. Abscission of flowers and floral organs is closely associated with alkalization of the cytosol in abscission zone cells. J. Exp. Bot. 2015;66:1355–1368. doi: 10.1093/jxb/eru483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stern R.A., Kigel J., Tomer E., Gazit S. ‘Mauritius’ lychee fruit development and reduced abscission after treatment with the auxin 2,4,5-TP. Am. Soc. Hortic. Sci. 1995;120:65–70. doi: 10.21273/JASHS.120.1.65. [DOI] [Google Scholar]
- 25.Kumpf R.P., Shi C.L., Larrieu A., Sto I.M., Butenko M.A., Peret B., Riiser E.S., Bennett M.J., Aalen R.B. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA. 2013;110:5235–5240. doi: 10.1073/pnas.1210835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi C.L., von Wangenheim D., Herrmann U., Wildhagen M., Kulik I., Kopf A., Ishida T., Olsson V., Anker M.K., Albert M., et al. The dynamics of root cap sloughing in Arabidopsis is regulated by peptide signalling. Nat. Plants. 2018;4:596–604. doi: 10.1038/s41477-018-0212-z. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Q., Shao Y., Ge S., Zhang M., Zhang T., Hu X., Liu Y., Walker J., Zhang S., Xu J. A MAPK cascade downstream of IDA-HAE/HSL2 ligand-receptor pair in lateral root emergence. Nat. Plants. 2019;5:414–423. doi: 10.1038/s41477-019-0396-x. [DOI] [PubMed] [Google Scholar]
- 28.Liu C., Zhang C., Fan M., Ma W., Chen M., Cai F., Liu K., Lin F. GmIDL2a and GmIDL4a, encoding the inflorescence deficient in abscission-like protein, are involved in soybean cell wall degradation during lateral root emergence. Int. J. Mol. Sci. 2018;19:2262. doi: 10.3390/ijms19082262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi C.L., Alling R.M., Hammerstad M., Aalen R.B. Control of organ abscission and other cell separation processes by evolutionary conserved peptide signaling. Plants (Basel) 2019;8:225. doi: 10.3390/plants8070225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meir S., Philosoph-Hadas S., Riov J., Tucker M.L., Patterson S.E., Roberts J.A. Re-evaluation of the ethylene-dependent and -independent pathways in the regulation of floral and organ abscission. J. Exp. Bot. 2019;70:1461–1467. doi: 10.1093/jxb/erz038. [DOI] [PubMed] [Google Scholar]
- 31.Tucker M.L., Yang R. IDA-like gene expression in soybean and tomato leaf abscission and requirement for a diffusible stelar abscission signal. AoB Plants. 2012;2012:pls035. doi: 10.1093/aobpla/pls035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilmowicz E., Kućko A., Ostrowski M., Panek K. INFLORESCENCE DEFICIENT IN ABSCISSION-like is an abscission-associated and phytohormone-regulated gene in flower separation of Lupinus luteus. Plant Growth Regul. 2018;85:1–10. doi: 10.1007/s10725-018-0375-7. [DOI] [Google Scholar]
- 33.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 2010;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 34.Fan Z.Q., Tan X.L., Chen J.W., Liu Z.L., Kuang J.F., Lu W.J., Shan W., Chen J.Y. BrNAC055, a novel transcriptional activator, regulates leaf senescence in Chinese flowering cabbage by modulating reactive oxygen species production and chlorophyll degradation. J. Agric. Food Chem. 2018;66:9399–9408. doi: 10.1021/acs.jafc.8b02309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.