Abstract
The serotonin receptor 4b (5-HTR4b) is expressed throughout the gastrointestinal tract, and its agonists are used in the treatment of irritable bowel syndrome with constipation (IBS-C). Today, there are no rapid assays for the identification of 5-HTR4b agonists. Here, we developed a luciferase-based 5-HTR4b assay capable of assessing one compound per second with a 38-fold dynamic range and nM limit of detection for serotonin. We used the assay to screen more than 1000 natural products and anti-infection agents and identified five new 5-HTR4b ligands: hordenine, halofuginone, proflavine, ethacridine, and revaprazan. We demonstrate that hordenine (antibiofilm), halofuginone (antiparasitic), and revaprazan (gastric acid reducer) activate 5-HTR4b in human colon epithelial cells, leading to increased cell motility or wound healing. The 5-HTR4b assay can be used to screen larger pharmaceutical libraries to identify novel treatments for IBS-C. This work shows that antimicrobials interact not only with the gut microbiota, but also with the human host.
Keywords: serotonin receptor, high-throughput screen, hordenine, halofuginone, revaprazan
In humans, 95% of serotonin (5-HT) is found in the gastrointestinal (GI) tract,1 where release and reception of 5-HT transmits information from the gut lumen to gut nerve cells and smooth muscles. Of the seven 5-HT receptor families, 5-HTR4 is broadly expressed in the gut: on nitrergic neurons that control smooth muscle relaxation, cholinergic and nitrergic neurons that control muscle contraction and relaxation, enterocytes that control chemical transport, and enteroendocrine cells that control the secretion of gastrointestinal hormones.2,3 Specifically, 5-HTR4b is highly expressed in the jejunum, ileum, and colon.4 5-HTR4 has been implicated in irritable bowel syndrome (IBS), which affects 15% of the world population.4,5 Agonists of 5-HTR4 are used for the treatment of irritable bowel syndrome with constipation (IBS-C), relieving constipation, abdominal pain, and bloating.
One of the major challenges in identifying novel 5-HTR4 agonists is the dearth of 5-HTR high-throughput assays to rapidly assess large libraries of chemicals. The two-day culture time required to test cell motility using colon cells, which naturally express 5-HTR4, would be time-prohibitive for a primary screening tool. Commercial G-protein coupled receptor (GPCR)-based assays, such as SelectScreen (Thermo Fisher) and gpcrMAXSM (Eurofins) lack screens for any member of the 5-HTR4 family. To our knowledge, only one large-scale 5-HTR4 screen has been performed to date, against 976 ToxCast chemicals using guinea pig brain tissue and radio-labeled ligand displacement.6
Previously, we engineered a fluorescence-based 5-HTR4b assay in yeast by linking expression of human 5-HTR4b on the yeast surface to green fluorescent protein (GFP) expression.7 The fluorescent reporter, however, posed some practical limitations. Fluorescent readout requires a 4 h chemical incubation step for robust GFP expression, and a high-throughput flow cytometer for signal readout, thus limiting the assay throughput to one 96-well plate per hour. Additionally, the GFP-based 5-HTR4 assay has only a 3-fold dynamic range and one order of magnitude linear range after activation with serotonin.
Here, we developed a luciferase-based 5-HTR4b assay with an overall assay time of 2.5 h, enabling the use of a luminescent plate reader, and achieving a screening throughput of one compound per second. Next, we used the assay to screen 1206 compounds coming from two commercial chemical libraries, a natural products library and an anti-infection library, and discovered five previously unidentified 5-HTR4b ligands. We validated three of the five ligands, hordenine, halofuginone, and revaprazan, as 5-HTR4b agonists in mammalian cells as they increase motility or wound healing in human colon epithelial cells. (Figure 1A).
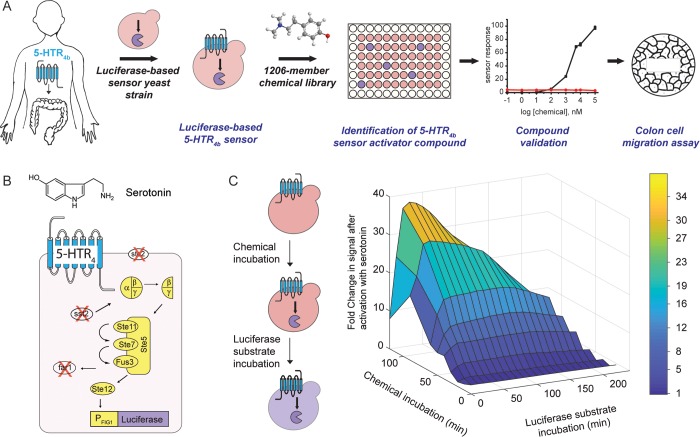
Figure 1.
Luciferase-based 5-HTR4 assay development. (A) Workflow for the identification and validation of HTR4b agonists. (B) Luciferase-based 5-HTR4 assay: human 5-HTR4b was expressed on the cell surface of a yeast engineered to link receptor activation to reporter to luciferase gene expression via the yeast mating pathway. (C) 5-HTR4-assay optimization at a pH = 7 and 100 mg/L serotonin.
This work has three significant outcomes. First, the same compound discovery workflow can be used, in the future, to screen pharmaceutical libraries for the identification of novel 5-HTR4b agonists for the treatment of IBS-C. Second, the increased assay signal provided by the luciferase reporter, when compared to the GFP reporter, should enable the generation of other high-throughput GPCR-based assays by simply swapping the receptor from the cell surface. Third, as antibiofilm (hordenine) and antiparasitic (halofuginone) agents affect colon cell motility and/or wound healing, antimicrobials may interact not only with the gut microbiota, but also with the human host leading to potential changes in gut movement and secretion. Finally, with a 5-HTR4b high-throughput assay in hand, we can now screen gut microbiota metabolites to further understand the link between host and gut microbiome.
Results
Luciferase-Based 5-HTR4b Assay Development
We replaced the GFP reporter from our GFP-based 5-HTR4b assay7 with NanoLuc luciferase,8 which we optimized for yeast expression (Figure 1B). As the pH of the GI tract hovers between 5.7 and 7.4, we optimized the assay at a pH of 7. The 5-HTR4b assay is composed of two steps, (i) ligand incubation leading to luciferase expression, and (ii) luciferase substrate incubation leading to luminescence. By co-optimizing both incubation steps using serotonin, we concluded that a 2 h ligand incubation followed by a 30 min luciferase substrate incubation results in the fastest overall assay conditions (2.5 h) with the highest signal increase after activation (38-fold) (Figure 1C). Of note, multiple 96-well plates can be incubated simultaneously, as reading the plate for luminescence takes 2 min.
Luciferase-Based 5-HTR4b Assay Validation
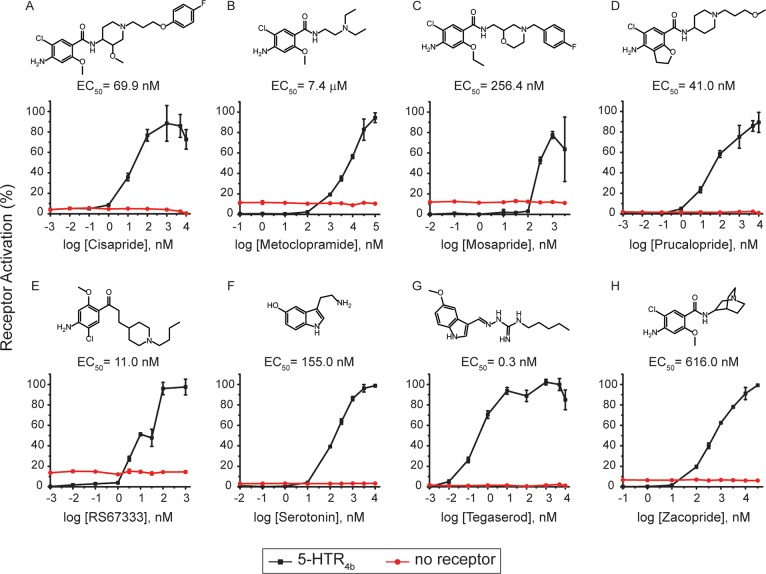
We demonstrate that the assay detects three known 5-HTR4 agonists for the treatment of IBS-C: tegaserod, prucalopride, and mosapride,9,10 and four other agonists used to treat gastroesophageal reflux (cisapride),11 depression (RS67333),12 anxiety (zacopride),13 and nausea (metoclopramide)10,14 (Figure 2A–H). To verify that the agonists led to cell luminescence due to 5-HTR4b activation and not via an alternative mechanism, we performed dose response curves with the agonists using a control strain carrying the luciferase reporter plasmid and a blank plasmid in lieu of 5-HTR4b. On the basis of EC50s, the potency of the agonists toward 5-HTR4b are tegaserod (0.3 nM) > RS67333 (11.0 nM) > prucalopride (41.0 nM) > cisapride (69.9 nM) > serotonin (155.0 nM) > mosapride (256.4nM) > zacopride (616.0 nM) > metoclopramide (7.4 μM). These results agree with previous studies that identified tegaserod and RS67333 to be more potent than serotonin,15,16 and zacopride and metoclopramide to be less potent.17−19 Taken together, the luciferase-based HTR4b assay is capable of identifying drugs with EC50s ranging from the low nM to the μM level.
Figure 2.
Validation of the 5-HTR4 assay. 5-HTR4 assay dose response curves with known 5-HTR4 agonists: (A) cisapride, (B) metoclopramide, (C) mosapride, (D) prucalopride, (E) RS-67333, (F) serotonin, (G) tegaserod, (H) zacopride. Data was collected in triplicate. Shown are means ± s.d.
5-HTR4b Assay High-Throughput Screening Validation
We validated the assay for 96-well plate high-throughput screening using a 3-day plate uniformity experiment20 (Figure S1). The assay had an average Z factor of 0.74 and an average coefficient of variation of 7.7%, meeting the two statistical parameters for high-throughput assay acceptance, i.e., a Z factor of >0.521 and a CV < 10%.22
Identifying Novel 5-HTR4b Ligands
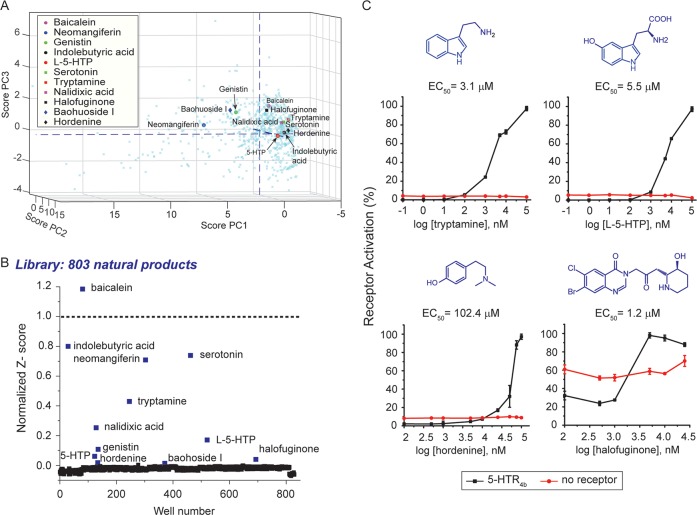
GPCRs expressed in the GI tract tend to bind gut microbial metabolites. For example, GPR41, GPR43, and GPR109 bind microbiota produced short chain fatty acids.23−25 We hypothesize that 5-HTR4b may bind microbial natural products. To explore the range of biological compounds that 5-HTR4b may bind, we used the assay to screen a commercial 803-member chemical natural products library. First, to understand the chemical diversity of the library, we broke down each library member into 23 chemical descriptors to perform a principal component analysis (Figure 3A, Table S4). The principal components (PC axes) reflect the common and unique variances of the chemical descriptors with the top three PCs accounting for 64% of the cumulative variances, with PC1, PC2, and PC3 capturing 45%, 11%, and 8% of the total variance, respectively. On the basis of their PC scores, chemicals can be separated into eight chemical spaces (CS) (Table S5). All chemical spaces are populated by compounds in the natural products library, indicating its chemical diversity. CS3, composed of small heteroaromatics with O-containing functionality, is the highest populated CS, containing 22% of all chemicals in the library. The least populated CSs are CS5 and CS7, each containing 6% of all the chemicals tested. In the future, the chemical diversity of the library could be improved by introducing more compounds with chiral centers and cyclic rings.
Figure 3.
5-HTR4 assay identifies natural products as novel ligands. (A) Principal component analysis of natural products library with 5-HTR4b screening hits highlighted. (B) 803-member natural product library screening results. The compounds were screened using the 5-HTR4b assay in singlets. Z-scores were normalized to the serotonin positive control, which was set to 1 (dotted line). (C) Dose response curves of the 4 validated natural product hits. Data was collected in triplicate. Shown are means ± s.d.
The screening of the natural products library resulted in 12 hits: serotonin, tryptamine, 3-indolebutyric acid, l-5-hydroxytryptophan (l-5-HTP), d/l-5-hydroxytryptophan (5-HTP), hordenine, genistin, neomangiferin, baohuoside I, baicalein, halofuginone, and nalidixic acid (Figure 3B). Previously, tryptamine26 and 5-HTP27 have been shown to bind 5-HTR4, and baicalein28 has been shown to bind 5-HTR7. Hordenine, nalidixic acid, halofuginone, 3-indolebutyric acid, genistin neomangiferin, and baohuoside I have not been previously shown to bind 5-HTR4.29
To confirm the natural products library hits, we ran dose response curves using the 5-HTR4b assay strain, and a control strain carrying the luciferase reporter and a blank plasmid in lieu of 5-HTR4b. We confirmed tryptamine and 5-HTP to be ligands of 5-HTR4. We could not confirm 3-indolebutyric acid, nalidixic acid, baicalein, genistin, baohuoside I, and neomangiferin as ligands of 5-HTR4b (Figure S2). We find, for the first time, that hordenine (EC50 102.4 μM) and halofuginone (EC50 1.2 μM) are HTR4b ligands (Figure 3C). Except for l-5-HTP, all natural products hits, i.e., hordenine, tryptamine, serotonin, and halofuginone, belong to CS8.
High-Throughput Screening of a 403-Member Anti-infection Library
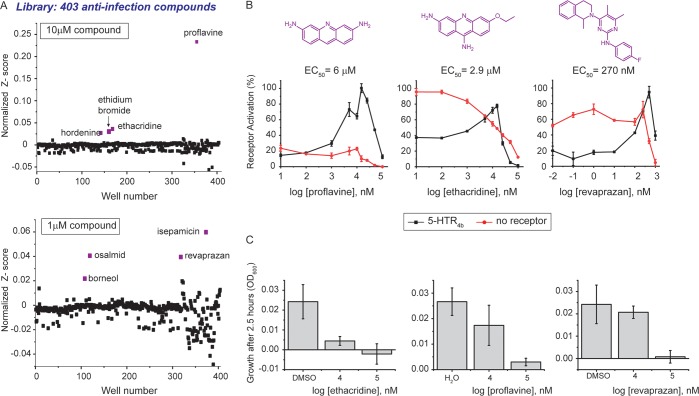
Hordenine has antibiofilm activity against Pseudomonas aeruginosa,30 and halofuginone is used for the treatment and prophylaxis of cryptosporidiosis in ruminants.31 Intrigued by the fact that antimicrobial agents activate 5-HTR4b, we explored whether other antimicrobials activate 5-HTR4b. To do this, we screened a commercial 403-member anti-infection library at two chemical concentrations: (i) 10 μM, the same concentration used to screen the natural products library, and (ii) 1 μM due to potential toxicity effects of the anti-infection compounds on yeast. On the basis of Z-scores, the 10 μM screen resulted in four hits: proflavine, ethidium bromide, ethacridine, and hordenine, while the 1 μM screen also resulted in four hits: isepamicin, osalmid, revaprazan, and borneol (Figure 4A). We attribute the absence of 1 μM hits in the 10 μM screen to toxicity issues, given that the 5-HTR4b assay is in cells. Neither proflavine, ethacridine, nor revaprazan have been previously shown to activate any serotonin receptor.29
Figure 4.
Screening of a 403 member anti-infective library. (A) Anti-infective compound screening results at 10 μM (top) and 1 μM (bottom). The compounds were screened using the 5-HTR4b assay in singlets. Z-scores were normalized to the serotonin positive control, which was set to 1 (dotted line). (B) Dose response curves of the 3 validated anti-infective hits. (C) Toxicity results after yeast was incubated for 2.5 h with varying concentration of the anti-infective hits. Data was collected in triplicate. Shown are means ± s.d.
To confirm the anti-infection library hits, we ran dose response curves using the 5-HTR4b assay strain, and a control strain carrying the luciferase reporter and a blank plasmid in lieu of 5-HTR4b. We confirmed proflavine (EC50 6 μM), ethacridine (EC50 2.9 μM), and revaprazan (EC50 270 nM) as HTR4b ligands (Figure 4B). We noticed a sharp decline in receptor activation at high compound concentrations. A toxicity assessment of these compounds shows they are toxic at high concentrations (Figure 4C). We could not confirm ethidium bromide, isepamicin, osalmid, or borneol as 5-HTR4b ligands (Figure S3).
Validating 5-HTR4b Ligands via Wound Healing and Motility Assays in Colon Epithelial Cells
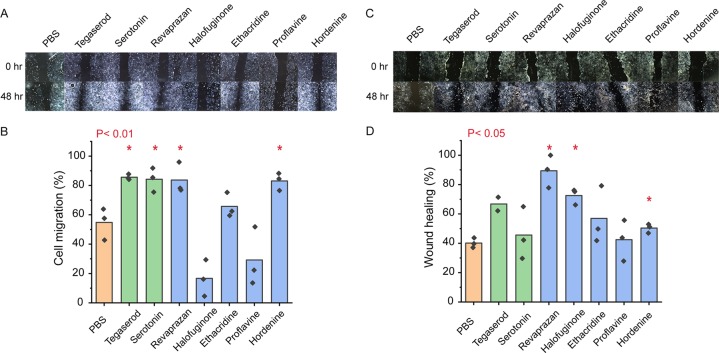
To assess the biological relevance of the five 5-HTR4b identified ligands, we tested their ability to activate 5-HTR4b in mammalian cells by inducing cell motility and wound healing in human colon epithelial cells (Caco-2). Caco-2 cells endogenously express 5-HTR432 and activation of 5-HTR4 using the agonist tegaserod leads to increased cell motility, wound healing, and cell proliferation.32 Tegaserod (Zelmac) is used for the treatment of IBS-C. To test cell motility, we used culture inserts to create a cell free zone to avoid cell death and damage.33 Revaprazan and hordenine significantly (P < 0.01) increase cell motility (Figure 5A,B). Wound healing was tested by creating a scratch in the cell monolayer using a pipet tip. This more closely mimics in vivo wound healing by creating cell damage, such as increasing reactive oxygen species at the wound boundary.34 Incubation of Caco-2 cells with hordenine, halofuginone, and revaprazan resulted in a statistically significant increase in wound healing (P < 0.05) when compared to the buffer control (Figure 5C,D). Proflavine and ethacridine did not result in a significant increase in either colon cell motility or wound healing. Taken together, revaprazan and hordenine results in both increased cell motility and wound healing, while halofuginone results only in increased wound healing.
Figure 5.
Antimicrobials enhanced colon epithelial cell motility and wound healing. (A) Representative photomicrographs of the colon cell (Caco-2) migration assay after 48 h with phosphate buffer control (PBS), tegaserod (5-HTR4B agonist, 1 μM), serotonin (1 μM), and the five 5-HTR4B ligands (10 μM). For all photomicrographs for migration after 24 and 48 h, see Figure S4. (B) Quantification of the colon cell migration assay after 48 h. The cell migration elicited by revaprazan and hordenine is similar to that observed by serotonin and tegaserod, and it is statistically significantly different than the PBS control (P < 0.01). (C) Representative photomicrographs of the Caco-2 wound healing scratch assay after 48 h with control (PBS), tegaserod (1 μM), serotonin (10 μM), and the 5 identified 5-HTR4B ligands (10 μM). For all photomicrographs of the wound healing scratch assay after 24 and 48 h, see Figure S5. (D) Quantification of the colon cell wound healing scratch assay. The wound healing elicited by revaprazan, halofuginone, and hordenine is statistically significantly different than the PBS control (P < 0.05).
Relevance of the Newly Identified 5-HTR4b Agonists in the Gut
Hordenine is present in malted barley and beer (strong beer: 5.16 mg/L; Pilsner: 2.7 mg/L35,36). Assuming all hordenine consumed reaches the colon, a pint of Pilsner beer would need to be ingested to reach concentrations that were observed to increase colon cell motility and wound healing. Other sources of hordenine are athletic performance and weight loss supplements.37
Halofuginone is used for the treatment and prophylaxis of cryptosporidiosis in ruminants31 with a maximum residue limit of 10–30 μg in bovine muscle, fat, kidney, and liver.38 Chickens treated with halofuginone can expect to lay eggs with up to 60 μg/kg halofuginone.39 One may only need to eat half a jumbo egg to reach the levels seen to increase colon cell wound healing, assuming all halofuginone reaches the colon.
Revaprazan is a proton pump inhibitor that reduces gastric acid secretion and reduces inflammation caused by Helicobacter pylori.40 At a daily dose of 200 mg/day and assuming it all reaches the colon, the concentration of revaprazan in the gut would be 983.6 μM, which is more than 3000 times higher than the EC50 of 270 nM.
Discussion
We have developed a rapid and robust luciferase-based 5-HTR4b assay in yeast with a 38-fold dynamic range and a limit of detection in the nM level for serotonin. We applied this assay to screen more than 1000 chemicals and identified five new 5-HTR4b ligands. Three of these ligands, hordenine, halofuginone, and revaprazan, activate 5-HTR4b in mammalian cells, eliciting colon cell motility and/or wound healing. The 5-HTR4b assay can now be used to screen large pharmaceutical libraries to identify 5-HTR4b agonists for the treatment of IBS-C. The increased sensitivity of the 5-HTR4b luciferase assay over our previous GFP-based assay should enable the generation of assays for other 5-HTRs, including 5-HTR1, the pharmaceutical target of antidepressant drugs.41
A current limitation of the luciferase-based 5-HTR4b assay is the relative high background in the no receptor control. In the absence of receptor, there is free floating Gαβλ, which does not need to be dissociated to activate the kinase cascade leading to increased gene expression.42 Sequestering Gαβλ by expression of the yeast endogenous GPCR (Ste2) may reduce basal luciferase levels in the control strain. Additionally, mixtures of heterologous GPCR/Ste2 in the sensor strains could achieve lower basal expression, but the system would have to be tuned.
The identification of antimicrobials binding 5-HTR4 is of profound impact to understanding conditions mediated by serotonin receptors. As shown in this work, antimicrobials may not only interact with the gut microbiota, but also with the human host leading to potential changes in gut movement and secretion. Changes in bowel movements and gastrointestinal hormone secretion may at least be partially caused by activation of serotonin receptors. Thus, our findings have implications for the treatment of IBS-C, which can be treated using antibiotics,43 and more generally to conditions related to serotonin receptor activation, such as depression. Finally, although antimicrobials have not been extensively considered as activating GPCRs, the availability of this high-throughput GPCR assay will enable the analysis of other GPCRs for their binding of antimicrobials.
Methods
Luciferase-Based 5-HTR4b Assay Construction
NanoLuc was codon optimized for Saccharomyces cerevisiae, commercially synthesized and cloned into pKM58644 between NcoI and NheI to generate pRS415-Leu2-pFIG1-NanoLuc (pEY15). pEY15 was sequenced verified using primer EY248. To construct the 5-HTR4b assay, pEY15, and pESC-His3-pTEF1-HTR4 (pTMC187) were cotransformed into the PPY14044 (W303 Δfar1 Δste2, Δsst2) to generate PPY1808. To construct the controls strain lacking 5-HTR4b, PPY140 was cotransformed with pEY15 and an empty vector (PPY111) to generate PPY1809.
Luciferase-Based 5-HTR4b Assay
Serotonin Detection
An overnight culture of PPY1808 was used to inoculate 20 mL of synthetic complete medium with 2% glucose lacking histidine and leucine (SD(HL–) to an OD600 = 0.06. After 18 h at 15 °C (150 rpm), PPY1808 was centrifuged (3500 rpm, 10 min), and resuspended to an OD600 = 1. In a white flat bottom 96-well plate, 190 μL of pH = 7 SD(HL–), 8 μL of PPY1808, and 2 μL of serotonin (final concentrations 0–4.3 mM) were added. After the chemical incubation step (2 h, plates covered with Breathe Easy Sealing Membrane, 30 °C, 250 rpm), 20 μL of 1:100 mixture of NanoLuc substrate to NanoLuc buffer was added45 for the luciferase substrate incubation step well (30 min, plates covered with Breathe Easy Sealing Membrane, 30 °C, 250 rpm). Luminescence was read immediately after luciferase substrate incubation in a Biotek Synergy 2 plate reader using default settings. For the time course assay, the cells were incubated at medium shaking speed in a Biotek Synergy 2.
Screening of Known 5-HTR4 Agonists
The same protocol for serotonin detection was followed. Instead of 2 μL of serotonin the same volume of RS67333, zacopride, and metoclopramide in water or cisapride, mosapride, tegaserod, and prucalopride in DMSO were used to reach a final concentrations between 0 and 10 000 nM. Dose response curves were fitted using the dose–response equation in Origin Pro 2016. EC50s were derived from this function by Origin.
High-Throughput Chemical Compound Screening
High-Throughput Assay Validation
A three-day assay validation was performed according to Iversen, et al.(20) The same serotonin detection protocol was followed. High signal wells have 5 μM of serotonin, mid signal wells have 60 nM of serotonin, and low signal wells have 0 nM of serotonin. Data were analyzed in the Excel template provided by Iversen, et al.(20) using activation assay settings with statistics calculated for single replicate screens.
Chemical Compound Screening
The same protocol used for serotonin detection was followed. Instead of 2 μL of serotonin, and 2 μL of chemical in DMSO or water from the natural product library (Selleck Chemicals L1400, 803 chemicals) or the anti-infection compound library (Selleck Chemicals L3100, 403 chemicals) was used to a final concentration of 10 μM (and 1 μM in the case of the anti-infection library). Each plate contained three negative control wells with DMSO or water and three positive controls with serotonin (5 μM) in DMSO. Natural product hits were identified using a Z-score as calculated below. The Z-scores were normalized to the serotonin positive control in each 96-well plates.
Principal Component Analysis
Data Gathering
Simplified molecular-input line-entry system (SMILES) for 801 of the 803 chemicals in the natural product library (lanolin and tea polyphenols are mixtures) were retrieved from Selleck Chemicals and PubChem to obtain data on 23 chemical descriptors with 8 descriptors representing chemical functional groups, and 15 descriptors according to Wenderski, et al.(46) The SMILES were inputted into Instant JChem (ChemAxon).
Data Analysis
PCA was run using Solo software (Eigenvector Research). Evaluation of the three PCs was conducted using Solo based on Eigenvalues. The three PC scores data from Solo were used to create 3-D scatterplots in MATLAB. The scatterplots were divided into eight distinct chemical spaces based on their 3-D coordinates.
Colon Cell Wound Healing Assay
Caco-2 cells (ATCC HTB-37) were grown at 37 °C with 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) with high glucose, sodium pyruvate and GlutaMAX or l-glutamine (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Media was changed every 2–3 days. Once 70–80% confluence was reached in T25 flasks, cells were detached using 0.05% trypsin–EDTA and diluted to a final volume of 15 mL with growth media. Cells were seeded into a 48 well plate and allowed to grow until they reached 90–100% confluence. Wounds were made with a P200 pipet tip and cells were washed with PBS. Cells were incubated in fresh growth media with either PBS, 10 μM of chemical hit or 1 μM of serotonin or tegaserod. Images were taken on a Leica inverted microscope at 0, 24, and 48 h. ImageJ MRI Wound Healing Tool [http://dev.mri.cnrs.fr/projects/imagej-macros/wiki/Wound_Healing_Tool] was used to measure wound size.
Cell Migration Assay
Caco-2 cells were maintained as stated above. Ibidi culture inserts were placed in a 24 well plate, and 70 μL of cells ((1–2) × 105 cells/mL) were placed in both insert wells. Cells were grown overnight to allow for attachment. Inserts were removed with sterile tweezers. Cells were washed twice with PBS then incubated with either PBS, 10 μM of chemical hit or serotonin, or 1 μM of tegaserod. Images were taken on a Leica inverted microscope at 0, 24, and 48 h. ImageJ MRI Wound Healing Tool was used to measure the cell free zone.
Acknowledgments
This work was funded by an NIH MIRA Award (R35GM124871) to P.P.-Y.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.9b00310.
Materials and methods; Sequences; Table S1: Table of plasmids; Table S2: Table of primers; Table S3: Table of strains; Table S4: Table of chemicals descriptors used in the PCA analysis; Table S5: Table of chemicals space composition of the natural product library; Figure S1: Three-day 96-well plate validation assay; Figure S2: Dose response curves of the 6 natural product hits that could not be confirmed; Figure S3: Dose response curves of the 4 anti-infection hits that could not be confirmed; Figure S4: Photomicrographs of colon cell migration assay after 0, 24, and 48 h; Figure S5: Photomicrographs of colon cell scratch wound healing assay after 0, 24, and 48 h. (PDF)
Author Contributions
E.A.Y. and P.P.-Y. conceived the assay, designed the experiments, and wrote the manuscript. E.A.Y. and A.A.A. carried out the experiments. W.S. carried out the principal component analysis. All authors approved the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Gershon M. D.; Tack J. (2007) The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology 132, 397–414. 10.1053/j.gastro.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Hoffman J. M.; Tyler K.; MacEachern S. J.; Balemba O. B.; Johnson A. C.; Brooks E. M.; Zhao H.; Swain G. M.; Moses P. L.; Galligan J. J.; Sharkey K. A.; Greenwood-Van Meerveld B.; Mawe G. M. (2012) Activation of colonic mucosal 5-HT(4) receptors accelerates propulsive motility and inhibits visceral hypersensitivity. Gastroenterology 142, 844–854. 10.1053/j.gastro.2011.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonini M. (2005) 5-Hydroxytryptamine effects in the gut: the 3, 4, and 7 receptors. Neurogastroenterol. Motil. 17, 637–642. 10.1111/j.1365-2982.2005.00716.x. [DOI] [PubMed] [Google Scholar]
- Wohlfarth C.; Schmitteckert S.; Härtle J. D.; Houghton L. A.; Dweep H.; Fortea M.; Assadi G.; Braun A.; Mederer T.; Pöhner S.; Becker P. P.; Fischer C.; Granzow M.; Mönnikes H.; Mayer E. A.; Sayuk G.; Boeckxstaens G.; Wouters M. M.; Simrén M.; Lindberg G.; Ohlsson B.; Schmidt P. T.; Dlugosz A.; Agreus L.; Andreasson A.; D’Amato M.; Burwinkel B.; Bermejo J. L.; Röth R.; Lasitschka F.; Vicario M.; Metzger M.; Santos J.; Rappold G. A.; Martinez C.; Niesler B. (2017) miR-16 and miR-103 impact 5-HT4 receptor signalling and correlate with symptom profile in irritable bowel syndrome. Sci. Rep. 7, 14680. 10.1038/s41598-017-13982-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikander A.; Rana S. V.; Prasad K. K. (2009) Role of serotonin in gastrointestinal motility and irritable bowel syndrome. Clin. Chim. Acta 403, 47–55. 10.1016/j.cca.2009.01.028. [DOI] [PubMed] [Google Scholar]
- Sipes N. S.; Martin M. T.; Kothiya P.; Reif D. M.; Judson R. S.; Richard A. M.; Houck K. A.; Dix D. J.; Kavlock R. J.; Knudsen T. B. (2013) Profiling 976 ToxCast Chemicals across 331 Enzymatic and Receptor Signaling Assays. Chem. Res. Toxicol. 26, 878–895. 10.1021/tx400021f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenworth A. M.; Claiborne T.; Peralta-Yahya P. (2017) Medium-Throughput Screen of Microbially Produced Serotonin via a G-Protein-Coupled Receptor-Based Sensor. Biochemistry 56, 5471–5475. 10.1021/acs.biochem.7b00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. P.; Unch J.; Binkowski B. F.; Valley M. P.; Butler B. L.; Wood M. G.; Otto P.; Zimmerman K.; Vidugiris G.; Machleidt T.; Robers M. B.; Benink H. A.; Eggers C. T.; Slater M. R.; Meisenheimer P. L.; Klaubert D. H.; Fan F.; Encell L. P.; Wood K. V. (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7, 1848–1857. 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y.; Min Y. S.; Sohn U. D. (2018) Recent advances in pharmacological research on the management of irritable bowel syndrome. Arch. Pharmacal Res. 41, 955–966. 10.1007/s12272-018-1068-5. [DOI] [PubMed] [Google Scholar]
- Manabe N.; Wong B. S.; Camilleri M. (2010) New-generation 5-HT4 receptor agonists: potential for treatment of gastrointestinal motility disorders. Expert Opin. Invest. Drugs 19, 765–775. 10.1517/13543784.2010.482927. [DOI] [PubMed] [Google Scholar]
- Champion M. C. (1997) Prokinetic therapy in gastroesophageal reflux disease. Can. J. Gastroenterol. 11, 55B–65B. [PubMed] [Google Scholar]
- Pascual-Brazo J.; Castro E.; Díaz A.; Valdizán E. M.; Pilar-Cuéllar F.; Vidal R.; Treceño B.; Pazos A. (2012) Modulation of neuroplasticity pathways and antidepressant-like behavioural responses following the short-term (3 and 7 days) administration of the 5-HT(4) receptor agonist RS67333. Int. J. Neuropsychopharmacol. 15, 631–643. 10.1017/S1461145711000782. [DOI] [PubMed] [Google Scholar]
- Costall B.; Domeney A. M.; Gerrard P. A.; Kelly M. E.; Naylor R. J. (1988) Zacopride - Anxiolytic Profile in Rodent and Primate Models of Anxiety. J. Pharm. Pharmacol. 40, 302–305. 10.1111/j.2042-7158.1988.tb05254.x. [DOI] [PubMed] [Google Scholar]
- Gralla R. J.; Itri L. M.; Pisko S. E.; Squillante A. E.; Kelsen D. P.; Braun D. W. Jr; Bordin L. A.; Braun T. J.; Young C. W. (1981) Antiemetic efficacy of high-dose metoclopramide: randomized trials with placebo and prochlorperazine in patients with chemotherapy-induced nausea and vomiting. N. Engl. J. Med. 305, 905–909. 10.1056/NEJM198110153051601. [DOI] [PubMed] [Google Scholar]
- Bender E.; Pindon A.; van Oers I.; Zhang Y. B.; Gommeren W.; Verhasselt P.; Jurzak M.; Leysen J.; Luyten W. (2000) Structure of the human serotonin 5-HT4 receptor gene and cloning of a novel 5-HT4 splice variant. J. Neurochem. 74, 478–489. 10.1046/j.1471-4159.2000.740478.x. [DOI] [PubMed] [Google Scholar]
- Mialet J.; Berque-Bestel I.; Sicsic S.; Langlois L.; Fischmeister R.; Lezoualc’h F. (2000) Pharmacological characterization of the human 5-HT(4(d)) receptor splice variant stably expressed in Chinese hamster ovary cells. Br. J. Pharmacol. 131, 827–835. 10.1038/sj.bjp.0703641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald C.; Adham N.; Kao H. T.; Olsen M. A.; Laz T. M.; Schechter L. E.; Bard J. A.; Vaysse P. J.; Hartig P. R.; Branchek T. A. (1995) The 5-HT4 receptor: molecular cloning and pharmacological characterization of two splice variants. EMBO J. 14, 2806–2815. 10.1002/j.1460-2075.1995.tb07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel O.; Gastineau M.; Dahmoune Y.; Langlois M.; Fischmeister R. (1998) Cloning, expression, and pharmacology of four human 5-hydroxytryptamine 4 receptor isoforms produced by alternative splicing in the carboxyl terminus. J. Neurochem. 70, 2252–2261. 10.1046/j.1471-4159.1998.70062252.x. [DOI] [PubMed] [Google Scholar]
- Claeysen S.; Sebben M.; Journot L.; Bockaert J.; Dumuis A. (1996) Cloning, expression and pharmacology of the mouse 5-HT(4L) receptor. FEBS Lett. 398, 19–25. 10.1016/S0014-5793(96)01132-5. [DOI] [PubMed] [Google Scholar]
- Iversen P. W., Benoit B., Chen Y.-F., Dere W., Devanarayan V., Eastwood B. J., Farmen M. W., Iturria S. J., Montrose C., Moore R. A., Weidner J. R., and Sittampalam G. S. (2012) In Assay Guidance Manual (Sittampalam G. S., Coussens N. P., Brimacombe K., Grossman A., Arkin M., Auld D., Austin C., Baell J., Bejcek B., Chung T. D. Y., Dahlin J. L., Devanaryan V., Foley T. L., Glicksman M., Hall M. D., Hass J. V., Inglese J., Iversen P. W., Kahl S. D., Kales S. C., Lal-Nag M., Li Z., McGee J., McManus O., Riss T., Trask O. J. Jr., Weidner J. R., Xia M., and Xu X., Eds.) pp 937–968, Eli Lilly and Company and the National Center for Advancing Translational Sciences, Bethesda, MD. [Google Scholar]
- Zhang J. H.; Chung T. D. Y.; Oldenburg K. R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screening 4, 67–73. 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Mishra A.; Dobritsa S. V.; Crouch M. L.; Rabenstein J.; Lee J. X.; Dhakshinamoorthy S. (2015) Establishment and validation of a 384-well antibacterial assay amenable for high-throughput screening and combination testing. J. Microbiol. Methods 118, 173–175. 10.1016/j.mimet.2015.09.019. [DOI] [PubMed] [Google Scholar]
- Samuel B. S.; Shaito A.; Motoike T.; Rey F. E.; Backhed F.; Manchester J. K.; Hammer R. E.; Williams S. C.; Crowley J.; Yanagisawa M.; Gordon J. I. (2008) Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. U. S. A. 105, 16767–16772. 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski K. M.; Vieira A. T.; Ng A.; Kranich J.; Sierro F.; Yu D.; Schilter H. C.; Rolph M. S.; Mackay F.; Artis D.; Xavier R. J.; Teixeira M. M.; Mackay C. R. (2009) Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286. 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.; Gurav A.; Sivaprakasam S.; Brady E.; Padia R.; Shi H.; Thangaraju M.; Prasad P. D.; Manicassamy S.; Munn D. H.; Lee J. R.; Offermanns S.; Ganapathy V. (2014) Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139. 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai Y.; Williams B. B.; Battaglioli E. J.; Whitaker W. R.; Till L.; Grover M.; Linden D. R.; Akiba Y.; Kandimalla K. K.; Zachos N. C.; Kaunitz J. D.; Sonnenburg J. L.; Fischbach M. A.; Farrugia G.; Kashyap P. C. (2018) Gut Microbiota-Produced Tryptamine Activates an Epithelial G-Protein-Coupled Receptor to Increase Colonic Secretion. Cell Host Microbe 23, 775–785. 10.1016/j.chom.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Martinez V.; Kimura H.; Tache Y. (2007) 5-Hydroxytryptophan activates colonic myenteric neurons and propulsive motor function through 5-HT4 receptors in conscious mice. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G419–428. 10.1152/ajpgi.00289.2006. [DOI] [PubMed] [Google Scholar]
- Gafner S.; Bergeron C.; Batcha L. L.; Reich J.; Arnason J. T.; Burdette J. E.; Pezzuto J. M.; Angerhofer C. K. (2003) Inhibition of [3H]-LSD binding to 5-HT7 receptors by flavonoids from Scutellaria lateriflora. J. Nat. Prod. 66, 535–537. 10.1021/np0205102. [DOI] [PubMed] [Google Scholar]
- Gaulton A.; Hersey A.; Nowotka M.; Bento A. P.; Chambers J.; Mendez D.; Mutowo P.; Atkinson F.; Bellis L. J.; Cibrián-Uhalte E.; Davies M.; Dedman N.; Karlsson A.; Magariños M. P.; Overington J. P.; Papadatos G.; Smit I.; Leach A. R. (2017) The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–D954. 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. W.; Luo H. Z.; Jiang H.; Jian T. K.; Chen Z. Q.; Jia A. Q. (2018) Hordenine: A Novel Quorum Sensing Inhibitor and Antibiofilm Agent against Pseudomonas aeruginosa. J. Agric. Food Chem. 66, 1620–1628. 10.1021/acs.jafc.7b05035. [DOI] [PubMed] [Google Scholar]
- de Graaf D. C.; Vanopdenbosch E.; Ortega-Mora L. M.; Abbassi H.; Peeters J. E. (1999) A review of the importance of cryptosporidiosis in farm animals. Int. J. Parasitol. 29, 1269–1287. 10.1016/S0020-7519(99)00076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spohn S. N.; Bianco F.; Scott R. B.; Keenan C. M.; Linton A. A.; O’Neill C. H.; Bonora E.; Dicay M.; Lavoie B.; Wilcox R. L.; MacNaughton W. K.; De Giorgio R.; Sharkey K. A.; Mawe G. M. (2016) Protective Actions of Epithelial 5-Hydroxytryptamine 4 Receptors in Normal and Inflamed Colon. Gastroenterology 151, 933–944. 10.1053/j.gastro.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poujade M.; Grasland-Mongrain E.; Hertzog A.; Jouanneau J.; Chavrier P.; Ladoux B.; Buguin A.; Silberzan P. (2007) Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl. Acad. Sci. U. S. A. 104, 15988–15993. 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic D. L.; Boettiger A. N.; Bar-Sagi D.; Carbeck J. D.; Shvartsman S. Y. (2006) Role of boundary conditions in an experimental model of epithelial wound healing. Am. J. Physiol. Cell Physiol. 291, C68–C75. 10.1152/ajpcell.00411.2005. [DOI] [PubMed] [Google Scholar]
- Brauers G.; Steiner I.; Daldrup T. (2013) Quantification of the biogenic phenethylamine alkaloid hordenine by LC-MS/MS in beer. Toxichem. Krimtech. 80, 323–326. [Google Scholar]
- Sommer T.; Dlugash G.; Hübner H.; Gmeiner P.; Pischetsrieder M. (2019) Monitoring of the dopamine D2 receptor agonists hordenine and N-methyltyramine during the brewing process and in commercial beer samples. Food Chem. 276, 745–753. 10.1016/j.foodchem.2018.10.067. [DOI] [PubMed] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Marcu A.; Guo A. C.; Liang K.; Vázquez-Fresno R.; Sajed T.; Johnson D.; Li C.; Karu N.; Sayeeda Z.; Lo E.; Assempour N.; Berjanskii M.; Singhal S.; Arndt D.; Liang Y.; Badran H.; Grant J.; Serra-Cayuela A.; Liu Y.; Mandal R.; Neveu V.; Pon A.; Knox C.; Wilson M.; Manach C.; Scalbert A. (2018) HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 46, D608–D617. 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagren V., Peippo P., and Lovgren T. (2005) Detecting and Controlling Veterinary Drug Residues in Poultry in Food Safety Control in the Poultry Industry (Mead G. C., Ed) 1st ed., pp 44–82, Woodhead Publishing. [Google Scholar]
- Alexander J.; Auđunsson G. A.; Benford D.; Cockburn A.; Cravedi J.-P.; Dogliotti E.; Di Domenico A.; Férnandez-Cruz M. L.; Fürst P.; Fink-Gremmels J.; Galli C. L.; Grandjean P.; Gzyl J.; Heinemeyer G.; Johansson N.; Mutti A.; Schlatter J.; van Leeuwen R.; Van Peteghem C.; Verger P. (2008) Opinion of the Scientific Panel on Contaminants in the Food chain on a request from the European Commission on cross-contamination of non-target feedingstuffs by halofuginone hydrobromide authorised for use as a feed additive. EFSA J. 657, 1–31. [Google Scholar]
- Lee J. S.; Cho J. Y.; Song H.; Kim E. H.; Hahm K. B. (2012) Revaprazan, a novel acid pump antagonist, exerts anti-inflammatory action against Helicobacter pylori-induced COX-2 expression by inactivating Akt signaling. J. Clin. Biochem. Nutr. 51, 77–83. 10.3164/jcbn.11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artigas F. (2013) Serotonin receptors involved in antidepressant effects. Pharmacol. Ther. 137, 119–131. 10.1016/j.pharmthera.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Klein S.; Reuveni H.; Levitzki A. (2000) Signal transduction by a nondissociable heterotrimeric yeast G protein. Proc. Natl. Acad. Sci. U. S. A. 97, 3219–3223. 10.1073/pnas.97.7.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino-Janeiro B. K.; Vicario M.; Alonso-Cotoner C.; Pascua-Garcia R.; Santos J. (2018) A Review of Microbiota and Irritable Bowel Syndrome: Future in Therapies. Adv. Ther. 35, 289–310. 10.1007/s12325-018-0673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K.; Bhattacharyya S.; Peralta-Yahya P. (2015) GPCR-Based Chemical Biosensors for Medium-Chain Fatty Acids. ACS Synth. Biol. 4, 1261–1269. 10.1021/sb500365m. [DOI] [PubMed] [Google Scholar]
- Masser A. E.; Kandasamy G.; Kaimal J. M.; Andreasson C. (2016) Luciferase NanoLuc as a reporter for gene expression and protein levels in Saccharomyces cerevisiae. Yeast 33, 191–200. 10.1002/yea.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenderski T. A.; Stratton C. F.; Bauer R. A.; Kopp F.; Tan D. S. (2015) Principal Component Analysis as a Tool for Library Design: A Case Study Investigating Natural Products, Brand-Name Drugs, Natural Product-Like Libraries, and Drug-Like Libraries. Methods Mol. Biol. 1263, 225–242. 10.1007/978-1-4939-2269-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.