Abstract
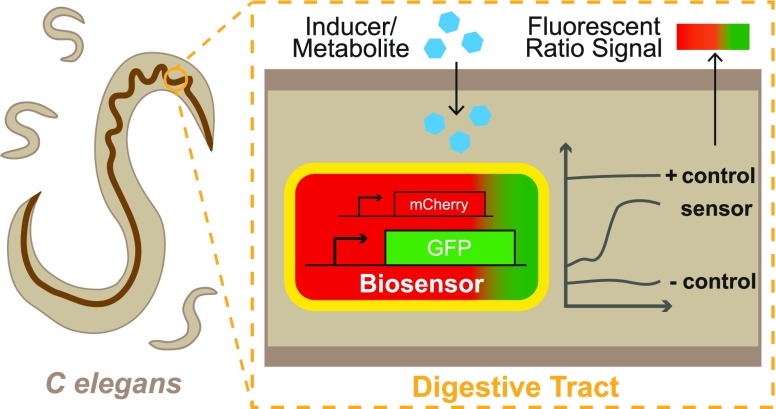
Caenorhabditis elegans has become a key model organism within biology. In particular, the transparent gut, rapid growing time, and ability to create a defined gut microbiota make it an ideal candidate organism for understanding and engineering the host microbiota. Here we present the development of an experimental model that can be used to characterize whole-cell bacterial biosensors in vivo. A dual-plasmid sensor system responding to isopropyl β-d-1-thiogalactopyranoside was developed and fully characterized in vitro. Subsequently, we show that the sensor was capable of detecting and reporting on changes in the intestinal environment of C. elegans after introducing an exogenous inducer into the environment. The protocols presented here may be used to aid the rational design of engineered bacterial circuits, primarily for diagnostic applications. In addition, the model system may serve to reduce the use of current animal models and aid in the exploration of complex questions within general nematode and host–microbe biology.
Keywords: synthetic biology, biosensors, microbiome engineering, E. coli Nissle 1917, Caenorhabditis elegans
As synthetic biology is increasingly applied to develop microbiome engineering tools such as biosensors1 and live biotherapeutics,2,3 animal models will be vital for building systems that function robustly in complex in vivo environments. As such, there is a demand for cheap, robust, tractable model systems that can serve both as a development platform and to probe host–microbe interactions.
The mouse is a model frequently used to study the intestinal microbiome.4 The similar taxonomic levels of the microbiota to humans, an extensive knowledge of genetic backgrounds, the custom genotypes and phenotypes, plus the use of humanized gnotobiotic systems all lead to a mimicry of the human gut microbiota phylogenic composition and allow researchers to investigate perturbations in a human-like system.5−8 To date, the majority of synthetic biology approaches to engineer or monitor the intestinal microbiota in vivo have been demonstrated within a mouse model.9−15 However, there is widespread concern regarding the reproducibility of findings due to genetic variability, handling techniques, mouse vendors, and diet.16,17 Furthermore, the majority of the cross-talk between the gut microbiota and the host has been shown to be host-specific; therefore, findings in mice may not always translate to humans.18
The invertebrate Caenorhabditis elegans is a transparent nematode worm that is 1 mm in length, lives in temperate soil environments, and feeds on soil bacteria. It was the first multicellular organism to have its whole genome sequenced,19 and a combination of a short 2 to 3 week lifespan, a transparent cell wall, and genetic tractability have enabled it to become an extremely versatile model system used to study energy metabolism, immunity, and aging.20 Whereas the C. elegans worm demonstrates a diverse microbiota in the wild,21,22 it is typically monoxenically grown with one species in the lab; this enables researchers to easily create and maintain a defined intestinal microbiota. The intestines are one of their major organs and constitute roughly a third of their somatic mass.23 The transparent cell wall and aerobic lumen also enable the simple visualization of fluorescent proteins and markers. The worm possesses an innate immune system that is used to regulate the intestinal bacterial load as it ages.24 The peak transition of bacteria through the intestines can be as short as 2 min during young adulthood, although the bacteria eventually colonize the lumen in a number of days as the worm ages.25 The emerging need for both convenient and robust tools to investigate host–microbiota interactions has resulted in a growing interest in the use of C. elegans as a live animal model for both host–microbiota interactions and synthetic biology. Examples include high-throughput screens to elucidate the complexity underlying host–microbe–drug interactions,26 understanding how bacterial-produced metabolites affect the worm gene expression and its lifespan,27 and the role of stochasticity in the colonization of the gut by microbiota.28 In synthetic biology, C. elegans has been used as a target for engineered nematicidal bacteria29 and to characterize a sense-and-kill synthetic circuit in E. coli Nissle 1917 (EcN) that could colonize the worm to prevent a Pseudomonas aeruginosa gut infection.30
Here we describe the development of protocols for the characterization of bacterial whole-cell biosensors that function within the C. elegans gastrointestinal tract. We first constructed a dual-plasmid-based biosensor system in EcN that responds to isopropyl β-d-1-thiogalactopyranoside (IPTG). This biosensor was then characterized in vitro at 37 and 20 °C. We then describe experimental protocols and an automated imaging pipeline that allowed us to examine quantitatively how this biosensor strain reports on changes in environmental levels of IPTG. Taken together, these results outline how C. elegans can be used as a novel model organism for further characterizing biosensors and host–microbe interactions within the digestive tract.
Results and Discussion
Ratiometric, Dual-Plasmid IPTG Biosensor System
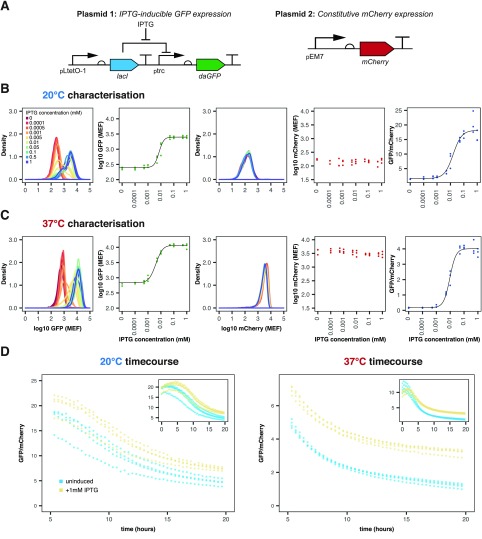
A dual reporter system using mCherry and GFP was created in EcN (Figure 1A) with mCherry constitutively expressed and GFP expression under the control of the inducible pLac promoter, allowing for the calculation of a ratiometric fold increase in GFP induction. (Plasmid maps are given in Figure S1.) This approach is considered to be more effective in reducing the background noise and accounting for fluctuations in diverse conditions in vivo compared with a single, absolute measurement.31,32 The pLac-inducible system showed a robust increase in the GFP/mCherry ratio upon the induction of the dual reporter system (Figure 1B,C). In comparison, the GFP/mCherry ratios in the negative promoterless EcN_OG241_ mCherry and positive constitutive EcN_OXB19_GFP_mCherry controls were seen to remain relatively constant (Figure S2).
Figure 1.
EcN_pLac_GFP_mCherry in vitro characterization within LB media at 20 and 37 °C. (A) Plasmids in the biosensor system based on constitutive mCherry and inducible GFP expression. (B,C) Characterization of the strain at 20 and 37 °C, respectively. From left to right: density plot of GFP induction, median GFP fluorescence, density plot of mCherry fluorescence, median mCherry fluorescence, and GFP/mCherry ratios over all IPTG concentrations. Flow cytometry data with 10 000 events (n = 3). (D) Plate reader time series showing the ratio for both uninduced (blue) and induced (yellow) cells (n = 4, circles indicate individual data points).
To try and mimic conditions of the C. elegans digestive tract in vitro, characterization was additionally performed at 20 °C (room temperature). At 37 °C, the fitted median GFP fluorescence increased from 685 ± 254 MEF (molecules of equivalent fluorophore, fitted value ± standard error, for 30 000 events from three biological replicates) when uninduced to 11 720 ± 270 MEF after the induction with 1 mM IPTG. A change in GFP expression was also detected at 20 °C, increasing from 250 ± 61 MEF to 2510 ± 65 MEF with 1 mM IPTG induction. The fitted median GFP/mCherry ratio induction with 1 mM IPTG was much greater at 20 °C than at 37 °C: 18.2 ± 0.84 and 4.01 ± 0.08, respectively.
The threshold for detection, Kd, and the dynamic range (illustrated in Figure S3) of the EcN_pLac_GFP_mCherry strain were adversely affected at 20 °C (Table 1). This was particularly prominent for the dynamic range of the GFP/mCherry ratios, which was approximately halved at 20 °C when compared with 37 °C (10.4 and 21.1, respectively). This may be because of slower growth and expression rates at the lower temperature. However, the linear range of the GFP/mCherry ratio was much larger at 20 °C than at 37 °C: At 20 °C, the linear range spanned 72.9 μM, from 0 to 72.9 μM; at 37 °C the linear range covered only 35.6 μM, from 0.2 to 35.8 μM. This suggests that the ratiometric output can be used to distinguish between a larger range of inducer concentrations at 20 °C than at 37 °C. Furthermore, EcN_OG241_mCherry produced lower ratios at both 20 and 37 °C compared with the uninduced EcN_pLac_GFP_mCherry strain (see Figure 1B,C and Figure S2), showing that the EcN_pLac_GFP_mCherry biosensor is leaky, with low basal levels of GFP expression, fmin, even in the uninduced state. Figure 1D displays plate reader data for the GFP/mCherry ratio over time for both induced and uninduced biosensor bacteria. These data illustrate that there is a maturation period before a stable ratio is approached, which is not clear from the flow cytometry data alone.
Table 1. Hill Parameter Fitting to GFP Induction and GFP/mCherry Ratio Curves for Both 20 and 37 °C.
| GFP induction
curves |
GFP/mCherry ratios |
|||
|---|---|---|---|---|
| parameter | 20 °C | 37 °C | 20 °C | 37 °C |
| fmin (MEF) | 250 ± 61 | 685 ± 254 | 1.60 ± 0.60 | 0.18 ± 0.07 |
| fmax (MEF) | 2510 ± 65 | 11720 ± 270 | 18.2 ± 0.84 | 4.01 ± 0.08 |
| Kd (μM) | 11.2 ± 1.17 | 8.68 ± 0.76 | 15.9 ± 3.47 | 9.39 ± 0.70 |
| n | 2.61 ± 0.83 | 1.88 ± 0.38 | 1.35 ± 0.30 | 1.98 ± 0.35 |
| dynamic range | 9.0 | 16.1 | 10.4 | 21.1 |
| linear range (μM) | 34.7 | 34.2 | 72.9 | 35.6 |
Fitted values ± standard error where appropriate.
In summary, whereas there appear to be differences seen in biosensor performance between the two temperatures, the characterization at 20 °C showed that the EcN_pLac_GFP_mCherry biosensor was capable of detecting changes in IPTG concentration under conditions more commensurate with the temperature within the C. elegans digestive tract. Therefore, the biosensor was compatible with the handling and culture of the C. elegans nematode.
Imaging of the Colonized C. elegans Intestinal Environment
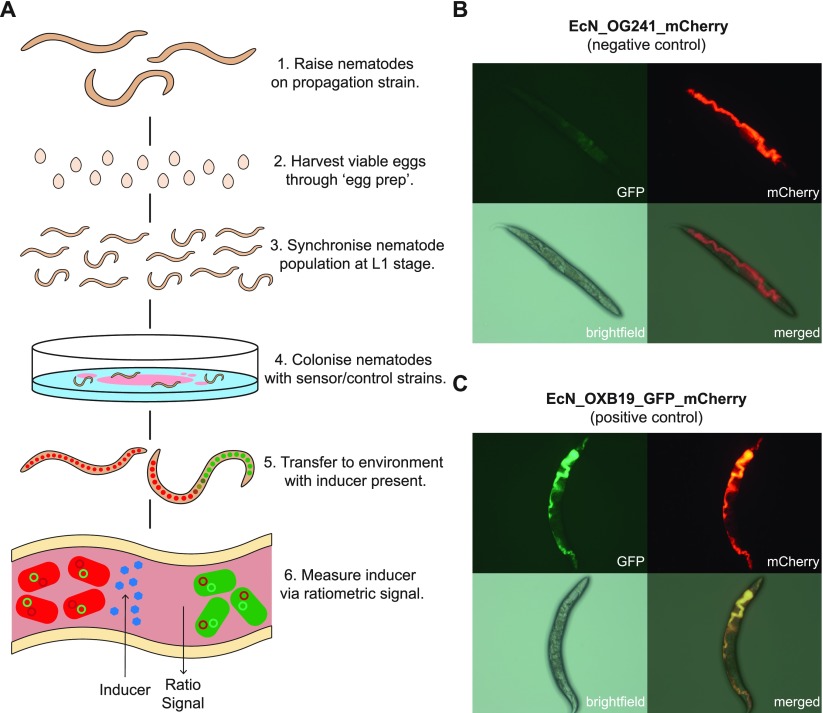
To test the hypothesis that C. elegans can be used to characterize bacterial biosensors, we developed the experimental protocol depicted in Figure 2A. Preliminary experiments confirmed that the wild-type lab N2 C. elegans strain could indeed grow and successfully develop on both the EcN_OG241_mCherry and EcN_OXB19_GFP_mCherry control strains constitutively expressing mCherry or GFP and mCherry, respectively. In addition, the GFP and mCherry signals could be used to image the C. elegans intestine. This is supported by the images within Figure S4 and the Supplementary Video, which show that the mCherry signal of the biosensor bacteria is contained only within the intestinal lumen. The negative GFP control EcN_OG241_mCherry showed only high levels of mCherry (Figure 2B). EcN_OXB19_GFP_mCherry showed direct colocalization with the strong expression of both GFP and mCherry throughout the digestive tract (Figure 2C).
Figure 2.
(A) Graphical representation of the C. elegans induction assay protocol with EcN–NGM plates and sensor induction quantification using the GFP/mCherry ratio. Representative images of nematodes colonized with (B) EcN_OG241_mCherry (negative control strain) expressing only mCherry and (C) EcN_OXB19_GFP_mCherry (positive control strain) constitutively expressing both mCherry and GFP. Panel labels refer to the imaging method.
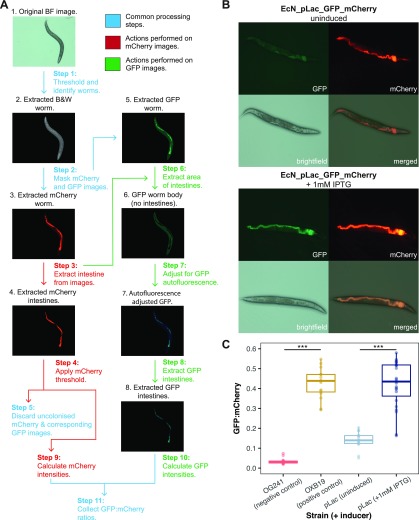
Once it was clear that the nematodes could survive on the engineered EcN strains and that the reporter proteins could be detected, an image processing pipeline was developed that could be used to quantify the GFP/mCherry ratios (Figure 3A). The pipeline, developed in MATLAB, was able to successfully identify worm bodies from the images (ignoring noise from the plate background) and extract the colonized intestines (Figure 3B). Steps were added that allowed for the rejection of uncolonized worm images (using a threshold on minimum mCherry fluorescence) and the removal of GFP autofluorescence (the subtraction of mean nematode body GFP intensity). It should be noted that for images that contained more than one nematode, the GFP/mCherry ratio was calculated over the entirety of the image. In addition, it was seen that not all nematodes became colonized; therefore, some images were discarded during the automated analysis. (A representative image of an uncolonized nematode can be seen in Figure S5.) This pipeline was initially trialed on nematodes that were grown on plates of EcN_pLac_GFP_mCherry with and without 1 mM IPTG for 7 days before imaging. (Representative images can be seen in Figure 3B.) The mean GFP/mCherry ratio of nematodes exposed to IPTG and unexposed was 0.42 ± 0.12 and 0.14 ± 0.04, respectively (Figure 3C). In addition, the uninduced EcN_pLac_GFP_mCherry demonstrated relatively high basal levels of GFP expression in accordance with the in vitro characterization.
Figure 3.
(A) Image analysis pipeline developed to automate the quantification of biosensor induction. (The brightness of the images has been adjusted.) (B) Representative images of nematodes colonized with the pLac biosensor strain. Top: uninduced, bottom: induced with 1 mM IPTG. (C) Preliminary characterization of the pLac biosensor. The first two columns refer to the negative and positive controls, respectively. These are then followed by EcN_pLac_GFP_mCherry colonized worms, both uninduced and induced (n ≥ 15 images, p values: ***<0.05, two-sided Mann–Whitney).
Characterization of the pLac (IPTG-Inducible) Biosensor in vivo
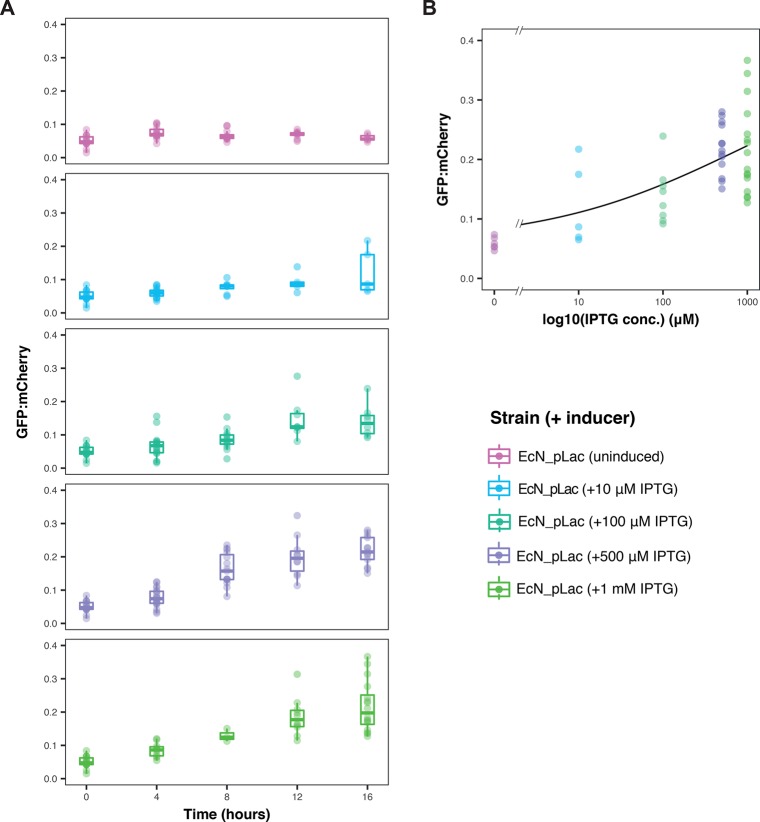
Our next goal was to demonstrate that the EcN_pLac_GFP_mCherry strain could detect and report on an environmental signal from within the C. elegans intestines in a quantitative manner. To ensure the separation of the biosensor from the external environment, an induction assay was carried out whereby worms were grown for 7 days on nematode growth medium (NGM) agar plates spread with the pLac strain. These were then transferred to plain NGM agar plates supplemented with varying concentrations of IPTG. Overall, ∼17% of the worm images collected were deemed uncolonized based on our mCherry threshold. The GFP/mCherry ratios collected in vivo were also lower than those from in vitro characterization, which was observed in the uninduced controls. The results are provided in Figure 4A. From these results, it can be seen that the GFP/mCherry ratios increased in both a time- and dose-dependent manner. This was not seen in the EcN_OG241_mCherry or EcN_OXB19_GFP_mCherry control strains. (See Figure S6.) Substantial increases in the GFP/mCherry ratios could be seen in as little as 4 h for as low as 10 μM IPTG. Figure 4B shows the induction at the 16 h time point as a function of the log of IPTG concentration; superimposed is a Hill function fit showing the dose-dependent response with increasing levels of IPTG.
Figure 4.
Full characterization of the pLac biosensor in vivo. (A) Timecourse of GFP/mCherry ratios in individual 7 day old C. elegans worms grown on the EcN_pLac_GFP_mCherry sensor strain and transferred to inducer plates supplemented with varying IPTG concentrations (n ≥ 4 images). At 16 h, all induced EcN_plac_GFP_mCherry ratios were found to be significantly higher than at 0 h (p value <0.05, two-sided Mann–Whitney). (B) GFP/mCherry ratios of the pLac biosensor, with various IPTG concentrations at the 16 h time point, fit with a Hill function (as in Figure 1).
Conclusions
Collectively, the results reported here indicate that the live C. elegans model can be used to characterize engineered biological systems and to design precise microbiota investigations. One limitation to our approach, highlighted in Figure 1D, is that there is a relatively long transient period in the GFP/mCherry ratios over time. After a period of hours, the measured ratio begins to stabilize. However, because the C. elegans are colonized for 7 days before they are induced in vivo, it is safe to assume that the bacteria have passed through this maturation period and the GFP/mCherry ratios have stabilized. Another potential limitation is that although these data were gathered under well-controlled conditions (same media, age-matched worms), the biosensor might not be useful in less controlled conditions (for example, when there are changes in other parameters not related to IPTG), which may also change the fluorescence ratio. Further work could explore this effect in detail and identify biosensors that are robust to these effects.
The use of this model can also provide valuable insights into general nematode biology. Currently, there is some debate within the nematode field as to whether E. coli become dormant within the digestive tracts of nematodes after ingestion. As with the report by Hwang et al.,30 the behavior of the EcN dual-plasmid strains provides evidence that this is not the case. Data presented here suggest that EcN is able to remain active even within the intestines of aged nematodes, with the EcN_pLac_GFP_mCherry strain capable of detecting and responding to environmental cues after a colonization period of 7 days. Our results also suggest that because GFP and mCherry require the presence of oxygen to fluoresce, the C. elegans intestinal environment is capable of transporting oxygen to some extent, even in older, fully colonized nematodes. Although this could hinder the ability of the model to predict the sensor behavior within the mouse or human intestines, the model can still be used to understand the performance of systems under normoxic conditions and could be further developed using nematodes that can survive under anoxic conditions.33 Another finding is that some nematodes remained uncolonized (in terms of fluorescence output) using the protocol in Figure 2A. A possible explanation could be the aerobic state of the digestive tract, as densely colonized nematodes may not be capable of providing sufficient oxygen transport through their digestive tracts. These effects may also give rise to the lower GFP/mCherry ratio observed in vivo, and may be explored further by investigating a range of colonization periods. Future work will explore the live C. elegans intestinal environment using engineered bacterial sensors.
The protocols described here have been used to characterize an IPTG-inducible biosensor. However, the system is readily adaptable to other possible metabolites or biomarkers of interest. In principle, this model could be used to detect inducers of interest introduced exogenously or derived from the host itself. IPTG is a molecule that is widely used as an inducer in synthetic genetic circuits. As the results of Figure 4 show, the pLac promoter can be induced while in the digestive tract of C. elegans. Therefore, this promoter may be used in future studies to produce targeted expression within the digestive tract of the C. elegans nematode, in an analogous manner to how an aTc-inducible system in Bacteroides was used in mice.15
With minimal regulatory and time constraints, our approach allows for experiments to be carried out on a powerful yet simple and defined host–microbiota model system. In comparison with mouse models, the C. elegans system both is more cost-effective and requires far less specialized facilities to maintain. Furthermore, C. elegans lends itself to high-throughput experimental techniques. Taken together, these factors illustrate that this model has the potential to complement the underlying approach of the “design, build, test, learn” cycle that is fundamental to synthetic biology and provides a useful alternative that may be considered alongside current approaches. More generally, the C. elegans model will allow the exploration of host–microbe interactions and bacterial competition within an in vivo environment. We believe that the methodology reported here will help expand our knowledge of the microbiome and allow for the reduction and replacement of current animal models used for testing in vivo synthetic biology approaches.
Methods
Flow Cytometry Analysis and Plotting
Flow cytometry was performed on an Attune NxT acoustic focusing cytometer with an Attune NxT autosampler (Thermo Fisher Scientific, U.K.). One μL of the appropriate strain culture was transferred into 200 μL of sterile phosphate-buffered saline (PBS) in a shallow polystyrene 96-well plate. The Attune NxT autosampler was used to record 10 000 events (for each individual sample) with four washes between samples. 10 000 events were recorded for all samples, across all experiments, to keep the total number of events constant. GFP was excited using the blue laser (488 nm) and detected using a 530/30 nm bandpass filter. mCherry was excited with the yellow laser (561 nm) and detected using a 620/15 nm bandpass filter. Additionally, a sample of a 1:300 dilution of rainbow calibration particles in PBS (Spherotec, U.K.) was recorded, allowing for the conversion of arbitrary units to MEF using Python scripts based on the FlowCal software.34
Collected flow cytometry standard (FCS) data were analyzed and plotted using custom Python and R scripts. Visualization and curve fitting were performed in R using the “ggplot2” package and the “nls” fitting function. GFP induction and ratiometric increase data were fit using Hill functions
where f is the observed value (either fluorescence or ratio), fmin is the minimum fitted value, fmax is the maximum fitted value, [x] is the inducer concentration, Kd is the threshold sensitivity, and n is the cooperativity. (These parameters are illustrated in Figure S3.) The errors reported are the standard errors from the least-squares fit, provided through the “nls” function in R.
The dynamic range was calculated using the expression
The linear range was calculated by taking the derivative of the Hill function over the length of the Hill fit
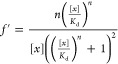 |
and then defined as the range of concentrations over which the derivative of the Hill function was >5% of the max value. Hill functions were fit to median values of replicates for the characterization data presented within Table 1 and Figures 1B,C and 4B.
C. elegans Strains and Handling
All C. elegans experiments were carried out in the wild-type lab strain N2 (provided by the Caenorhabditis Genetics Center, USA). Unless stated otherwise, worms were maintained and raised at 20 °C on nematode growth medium (NGM) seeded with E. coli OP50, an auxotroph lab strain whose growth is limited on NGM.
Adult worms were maintained and passaged according to normal protocols. In brief, this involved seeding NGM agar plates with 150 μL of overnight E. coli OP50 culture and incubating for 48 h at 20 °C to create a bacterial lawn for food. Five to six L4-stage worms were picked and transferred to seeded NGM plates and incubated for 24 h, allowing them to reach the adult stage and begin laying eggs. After 24 h, the adult worms were removed from each plate. The plates were then incubated for a further 48 h to enable a large number of eggs to hatch, feed on the bacterial lawn, and become fertilized with eggs.
After 48 h, an “egg prep” was carried out to isolate the eggs from the worms. This involved washing NGM plates with M9 media to collect all worms. Once settled, the M9 media were aspirated, and the remaining worms were washed with 400 μL of bleach/NaOH at a ratio of 7:8. This mixture was vortexed for 3 to 4 min (allowing the release of the fertile eggs) and then neutralized with 13 mL of M9. The tube was then centrifuged until a firm pellet had formed, and once again the M9 media were aspirated without disturbing the egg pellet. This wash was repeated twice more before the egg pellet was resuspended with 10 mL of M9 media and transferred to a sterile, empty Petri dish for 24 h of incubation. This incubation allowed the eggs to hatch and arrested the worms at the first larval stage (L1). This provided an entirely synchronized nematode population. After 24 h of incubation, 400 L1s were seeded on the appropriate bacterial NGM plate for experiments or repassaging. Before imaging, worms were passaged as sterile and nonreproductive adults with the drug fluorodeoxyuridine (FUdR). This involved picking worms at the L4 stage and transferring them to seeded NGM plates, supplemented with FUdR at a concentration of 20 μM. This enabled the maintenance of a synchronously aging population of worms.
Individual EcN–NGM plates were prepared in the same manner as with E. coli OP50 but with an overnight antibiotic culture of the respective EcN sensor strain or control instead. After preparing the worms as previously described, 400 synchronized L1 stage worms were transferred to NGM plates seeded with the respective EcN strain. These included EcN_OXB19_GFP_mCherry and EcN_OG241_mCherry as positive and negative controls, respectively. The inducible EcN_pLac_GFP_mCherry was used to investigate the sensor assay. After 48 h of growth on the respective EcN–NGM plates, 45–50 L4 worms were picked and transferred to FUdR-coated NGM plates seeded with the same EcN strain as before.
EcN Biosensor Induction Assays in C. elegans
As worms age, the peristaltic movements in their intestines decrease, and they eventually become constipated while the bacteria proliferates in the gut.20 Induction assays were carried out on 7 day old adult worms as they became colonized by the fast-growing EcN. Initially, plain NGM plates were prepared and seeded with the overnight culture of the relevant strain. These were then incubated at 37 °C overnight. Approximately 50 sterile nematode eggs (gained from the egg prep mentioned above) were added to these seeded plates and incubated at 20 °C for 2 days. Around 30 nematodes were then collected at random and transferred to EcN–NGM plates supplemented with the same strain, supplemented with FUdR to prevent egg maturation (as detailed above). These were then incubated for a further 5 days (to a total of 7 days) to allow for full colonization of the majority of nematodes. It should be noted that antibiotics were not used in the EcN–NGM plates; instead, it was assumed that the majority of the EcN bacteria would retain the dual plasmid system over this 7 day period. Approximately 25 worms were then picked from the respective EcN–NGM plates and transferred to either an unseeded plain NGM agar plate or an unseeded NGM agar plate containing the relevant IPTG concentration. Plates were then stored at 20 °C for the duration of the assay. After induction, worms from either the control NGM plate or the assay plate were anaesthetized for imaging with 0.2% levamisole.
C. elegans Imaging
Anaesthetized worms were imaged with a Zeiss Axio Scope using GFP (excitation: 470 nm; emission: 525 nm) and mCherry (excitation: 560 nm; emission: 630 nm) filters. All worms on the inducer plates were imaged after being transferred at random from the original colonization plates. Exposure times were set at 500 ms for each, and laser intensities were kept constant. Images were acquired using the Zen software and analyzed using the developed MATLAB pipeline. Further details of the materials and methods can be found in the Supporting Information.
Acknowledgments
J.W.R. and T.O. were funded through the BBSRC LIDo Doctoral Training Partnership. E.R.G. and L.M.Q. were funded through the MRC. L.R. was funded jointly through a BBSRC studentship and a Microsoft Research Scholarship. G.M.T. was supported by funding from the BBSRC and EPSRC. F.C. was supported through the Wellcome Trust/Royal Society (grant no. 102531/Z/13/A) and the MRC (grant no. MC-A654-5QC80). C.P.B. was supported through the Wellcome Trust (grant no. 097319/Z/11/Z) and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant no. 770835).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.9b00166.
Author Contributions
⊥ J.W.R. and T.O. contributed equally. F.C. and C.P.B. conceived this study. J.W.R, T.O., E.R.G., L.M.Q., and L.R. conducted the experiments. J.W.R. and T.O. performed the data analysis. J.W.R., F.C., and C.P.B. wrote the manuscript. F.C., G.M.T., and C.P.B. supervised the projects. All authors edited the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Riglar D. T.; Silver P. A. (2018) Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 16, 214–225. 10.1038/nrmicro.2017.172. [DOI] [PubMed] [Google Scholar]
- Chang H.-J.; Voyvodic P. L.; Zúñiga A.; Bonnet J. (2017) Microbially derived biosensors for diagnosis, monitoring and epidemiology. Microb. Biotechnol. 10, 1031–1035. 10.1111/1751-7915.12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir T.; Fedorec A. J.; Danino T.; Barnes C. P. (2018) Synthetic Biology and Engineered Live Biotherapeutics: Toward Increasing System Complexity. Cell Syst 7, 5–16. 10.1016/j.cels.2018.06.008. [DOI] [PubMed] [Google Scholar]
- Hugenholtz F.; de Vos W. M. (2018) Mouse models for human intestinal microbiota research: a critical evaluation. Cell. Mol. Life Sci. 75, 149–160. 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J.; Ridaura V. K.; Faith J. J.; Rey F. E.; Knight R.; Gordon J. I. (2009) The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci. Transl. Med. 1, 6ra14–6ra14a. 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J. J.; McNulty N. P.; Rey F. E.; Gordon J. I. (2011) Predicting a Human Gut Microbiota’s Response to Diet in Gnotobiotic Mice. Science 333, 101–104. 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A. L.; Kallstrom G.; Faith J. J.; Reyes A.; Moore A.; Dantas G.; Gordon J. I. (2011) Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. U. S. A. 108, 6252–7. 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. L. A.; Vieira-Silva S.; Liston A.; Raes J. (2015) How informative is the mouse for human gut microbiota research?. Dis. Models & Mech. 8, 1–16. 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F.; March J. C. (2010) Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc. Natl. Acad. Sci. U. S. A. 107, 11260–4. 10.1073/pnas.1001294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Guo L.; Zhang Y.; Walzem R. L.; Pendergast J. S.; Printz R. L.; Morris L. C.; Matafonova E.; Stien X.; Kang L.; Coulon D.; McGuinness O. P.; Niswender K. D.; Davies S. S. (2014) Incorporation of therapeutically modified bacteria into Gut microbiota inhibits obesity. J. Clin. Invest. 124, 3391–3406. 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotula J. W.; Kerns S. J.; Shaket L. A.; Siraj L.; Collins J. J.; Way J. C.; Silver P. A. (2014) Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. U. S. A. 111, 4838–43. 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino T.; Prindle A.; Kwong G. A.; Skalak M.; Li H.; Allen K.; Hasty J.; Bhatia S. N. (2015) Programmable probiotics for detection of cancer in urine. Sci. Transl. Med. 7, 289ra84. 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhrvold C.; Kotula J. W.; Hicks W. M.; Conway N. J.; Silver P. A. (2015) A distributed cell division counter reveals growth dynamics in the gut microbiota. Nat. Commun. 6, 10039. 10.1038/ncomms10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daeffler K. N.-M.; Galley J. D.; Sheth R. U.; Ortiz-Velez L. C.; Bibb C. O.; Shroyer N. F.; Britton R. A.; Tabor J. J. (2017) Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol. 13, 923. 10.15252/msb.20167416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B.; Zimmermann M.; Barry N. A.; Goodman A. L. (2017) Engineered Regulatory Systems Modulate Gene Expression of Human Commensals in the Gut. Cell 169, 547–558. 10.1016/j.cell.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukens D.; Brinkman B. M.; Raes J.; De Vos M.; Vandenabeele P. (2016) Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol. Rev. 40, 117–32. 10.1093/femsre/fuv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson A. J.; McCoy K. D. (2015) Standardised animal models of host microbial mutualism. Mucosal Immunol. 8, 476–86. 10.1038/mi.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls J. F.; Mahowald M. A.; Ley R. E.; Gordon J. I. (2006) Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127, 423–33. 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1998) Genome Sequence of the Nematode C. elegans: A Platform for Investigating Biology. Science 282, 2012–2018. 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Cabreiro F.; Gems D. (2013) Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol. Med. 5, 1300–10. 10.1002/emmm.201100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen P.; Marsh S. A.; Braker I.; Heitland N.; Wagner S.; Nakad R.; Mader S.; Petersen C.; Kowallik V.; Rosenstiel P.; Félix M.-A.; Schulenburg H. (2016) The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 14, 38. 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel B. S.; Rowedder H.; Braendle C.; Félix M.-A.; Ruvkun G. (2016) Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci. U. S. A. 113, E3941–E3949. 10.1073/pnas.1607183113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L.; Shtonda B. B. (2003) Food transport in the C. elegans pharynx. J. Exp. Biol. 206, 2441–57. 10.1242/jeb.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal-Celhay C.; Bradley E. R.; Blaser M. J. (2012) Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 12, 49. 10.1186/1471-2180-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafouri S.; McGhee J. (2007) Bacterial residence time in the intestine of Caenorhabditis elegans. Nematology 9, 87–91. 10.1163/156854107779969718. [DOI] [Google Scholar]
- Scott T. A.; et al. (2017) Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell 169, 442–456. 10.1016/j.cell.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E.; MacNeil L. T.; Ritter A. D.; Yilmaz L. S.; Rosebrock A. P.; Caudy A. A.; Walhout A. J. M. (2014) Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156, 759–70. 10.1016/j.cell.2014.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega N. M.; Gore J. (2017) Stochastic assembly produces heterogeneous communities in the Caenorhabditis elegans intestine. PLoS Biol. 15, e2000633 10.1371/journal.pbio.2000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracho O. R.; Manchery C.; Haskell E. C.; Blanar C. A.; Smith R. P. (2016) Circumvention of Learning Increases Intoxication Efficacy of Nematicidal Engineered Bacteria. ACS Synth. Biol. 5, 241–249. 10.1021/acssynbio.5b00192. [DOI] [PubMed] [Google Scholar]
- Hwang I. Y.; Koh E.; Wong A.; March J. C.; Bentley W. E.; Lee Y. S.; Chang M. W. (2017) Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 8, 15028. 10.1038/ncomms15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Su S.; Wei J.; Bahgi R.; Hope-Weeks L.; Qiu J.; Wang S. (2015) Ratio-metric sensor to detect riboflavin via fluorescence resonance energy transfer with ultrahigh sensitivity. Phys. E (Amsterdam, Neth.) 72, 17–24. 10.1016/j.physe.2015.04.006. [DOI] [Google Scholar]
- Shynkar V. V.; Klymchenko A. S.; Kunzelmann C.; Duportail G.; Muller C. D.; Demchenko A. P.; Freyssinet J.-M.; Mely Y. (2007) Fluorescent Biomembrane Probe for Ratiometric Detection of Apoptosis. J. Am. Chem. Soc. 129, 2187–2193. 10.1021/ja068008h. [DOI] [PubMed] [Google Scholar]
- Kitazume H.; Dayi M.; Tanaka R.; Kikuchi T. (2018) Assessment of the behaviour and survival of nematodes under low oxygen concentrations. PLoS One 13, e0197122 10.1371/journal.pone.0197122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Hair S. M.; Sexton J. T.; Landry B. P.; Olson E. J.; Igoshin O. A.; Tabor J. J. (2016) FlowCal: A User-Friendly, Open Source Software Tool for Automatically Converting Flow Cytometry Data from Arbitrary to Calibrated Units. ACS Synth. Biol. 5, 774–780. 10.1021/acssynbio.5b00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.