Abstract
Transforming growth factor-beta (TGF-β) signaling is one of the important cellular pathways that play key roles for tissue maintenance. In particular, it is important in the context of inflammation and tumorigenesis by modulating cell growth, differentiation, apoptosis, and homeostasis. TGF-β receptor type 2 (TGFBR2) mutations affected by a mismatch repair deficiency causes colorectal cancers (CRCs) with microsatellite instability, which is, however, associated with relatively better survival rates. On the other hand, loss of SMAD4, a transcription factor in the TGF-β superfamily signaling, promotes tumor progression. Loss of heterozygosity on chromosome 18 can case SMAD4-deficient CRC, which results in poorer patients’ survival. Such bidirectional phenomenon driven by TGF-β signaling insufficiency reflects the complexity of this signaling pathway in CRC. Moreover, recent understanding of CRC at the molecular level (consensus molecular subtype classification) provides deep insight into the important roles of TGF-β signaling in the tumor microenvironment. Here we focus on the TGF-β signaling in CRC and its interaction with the tumor microenvironment. We summarize the molecular mechanisms of CRC tumorigenesis and progression caused by disruption of TGF-β signaling by cancer epithelial cells and host stromal cells.
Keywords: TGF-β signaling, colorectal cancer, SMAD4, tumor microenvironment
1. Introduction
Transforming growth factor-beta (TGF-β) signaling pathway plays critical roles in controlling tissue development, proliferation, differentiation, apoptosis, and homeostasis [1]. As such, disruption of this signaling pathway leads to several diseases including cancers. TGF-β signaling regulates many target genes—either positively or negatively—in a context-dependent manner [2]. Although TGF-β signaling inhibits epithelial growth in normal tissues, it promotes tumor cell progression in tissues with advanced cancer [3]. This phenomenon is known as TGFβ paradox. These context-dependent and paradoxical dynamics of the signaling complicate the understanding of the role of TGF-β signaling in cancer biology. As the effect on the colonic epithelial cells, TGF-β signaling exhibits reduction in cell proliferation, along with promotion of differentiation and apoptosis [4]. In addition to its effect on epithelial cells, TGF-β plays protective roles against luminal bacterial antigens by suppressing intestinal immune cells in the stroma and inducing immune tolerance [5,6]. Therefore, disruption of TGF-β signaling in the colon prompts tumor progression not only via epithelial cells transformation but also via tumor-stromal interactions [7,8,9,10,11].
Recent advances in DNA sequencing technology, such as next-generation sequencing and digital polymerase chain reaction (PCR), re-focuses the importance of DNA alteration in cancer cells. Recently, a novel classification for colorectal cancer (CRC) was advocated as consensus molecular subtype (CMS) based on the following molecular features: CMS1 (microsatellite instability (MSI)-immune) as hypermutated, microsatellite unstable, and strong immune activation; CMS2 (canonical) as epithelial, marked WNT and MYC signaling activation; CMS3 (metabolic) as epithelial and evident metabolic dysregulation; and CMS4 (mesenchymal) as prominent TGF-β activation, stromal invasion, and angiogenesis [12]. Most CRC cells with high levels of MSI (MSI-H) accumulate mutations in TGF-β receptor type 2 (TGFBR2) as it carries microsatellite sequences [13,14]. As these findings indicated, disruption of TGF-β signaling plays a pivotal role in CRC pathogenesis in several molecular types of CRC. Moreover, clinical evidence has revealed its strong involvement in patients’ prognosis after curative resection [15]. In this review, we summarize the proposed mechanisms of TGF-β signaling disruption involved in CRC development, progression, and invasion/metastasis.
2. TGF-β Signaling Pathway
2.1. TGF-β Signaling in Cell Biology
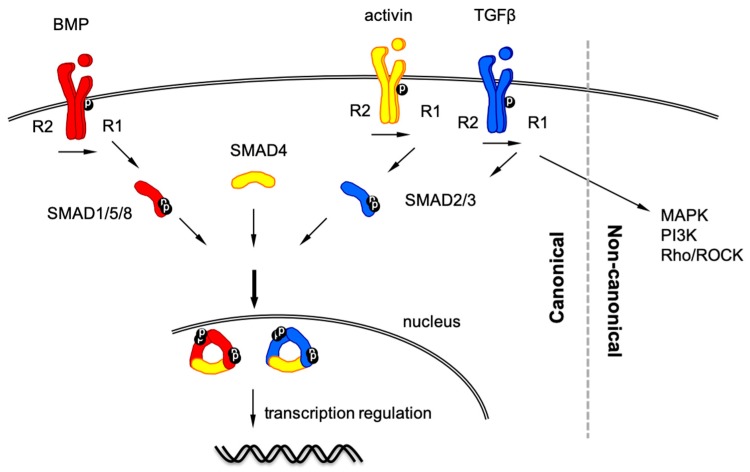
TGF-β superfamily signaling involves > 30 components, mainly divided into two subfamilies: the TGF-β-activin-nodal subfamily and the bone morphogenetic protein (BMP) subfamily [1,2,16] (Figure 1). TGF-β ligands assemble their corresponding receptors: two type 1 components and two type 2 components. Type 2 receptors serve as activators to phosphorylate type I receptors, whereas type 1 receptors function as propagators to transduce the signal downstream to cytoplasmic proteins. Components of both receptors are serine/threonine kinases. After ligand binding, BMP type 1 receptors phosphorylate SMAD1/5/8 (the abbreviation of SMAD refers to the homologies of SMA (small worm phenotype of Caenorhabditis elegans) and Drosophila MAD (Mothers Against Decapentaplegic), whereas TGF-β type I receptors and activin type 1 receptors phosphorylate SMAD2/3. These sets of SMAD proteins are known as receptor-regulated SMAD (R-SMAD). Phosphorylation of two C-terminal serine residues on R-SMAD facilitates trimerization with two R-SMAD molecules and one SMAD4 (also known as common-mediator SMAD, Co-SMAD). This SMAD trimer plays a central role in the TGF-β superfamily signaling to translocate to the nucleus and bind DNA via their DNA binding site. The CAGAC motif or its complementary sequence TCTG motif is referred to as the SMAD binding element (SBE) where SMAD2/3/4 mainly bind. In addition, SMAD1/5/8 and SMAD2/3/4 can also bind the GC-rich element when activated [4,17]. R-SMAD molecules in the SMAD4-R-SMAD complex can also bind other transcription factors as partners to regulate their transcription. In addition to the canonical SMAD-dependent pathway, TGF-β superfamily ligands transduce non-canonical, SMAD-independent pathways such as various mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)/Akt and Rho/Rho-associated protein kinase (ROCK) pathways [18].
Figure 1.
Schematic representation of Transforming growth factor-beta (TGF-β) superfamily signaling pathway.
Similar to most signaling pathways, TGF-β signaling is regulated from the ligand level to the effector level. Most of the TGF-β ligands act as paracrine fashion, and their access to the cognate receptors is regulated by ligand-binding proteins such as soluble proteins and extracellular matrix [19,20]. In addition to the signal-transducing SMAD members (R-SMAD and Co-SMAD), another type of SMAD inhibits TGF-β signaling pathway (inhibitory SMAD, I-SMAD). SMAD6/7 (I-SMAD) inhibits signal transduction by interfering with the phosphorylation of R-SMAD from type 1 receptors. Ubiquitination of R-SMAD/Co-SMAD is also one mechanism for signal degradation [21].
2.2. TGF-β Signaling in Clinical Situation of CRC Patients
Molecular classification of CRC can provide biological interpretability of CRC. In CMS1 CRC, MSI-H provides accumulation of many somatic gene mutations including TGFBR2, which results in the release of tumor neoantigens and activation of tumor immunity. It was reported that MSI-H was an independent favorable prognostic factor for Stage II/III (no distant metastasis) CRC patients after curative resection [15]. In fact, CMS1 CRC exhibits a low metastatic rate compared with other subtypes [22]. Although TGFBR2 mutation itself is not a favorable prognostic factor within MSI-H CRC population, MSI-H CRC patients exhibit better prognosis compared to microsatellite-stable ones [15,23]. However, once metastasized, CMS1 CRC exhibits poorer survival. One of the reasons of poor prognosis after metastases would be that MSI-H confers resistance to 5-fluorouracil (5-FU)-based chemotherapy [24].
MSI-H derived from deficient mismatch repair (dMMR) accounts for only 15% of CRCs. Alternatively, approximately 85% of invasive CRCs exhibit chromosomal instability (CIN) and loss of heterozygosity (LOH) in some chromatin areas [25]. CIN CRCs accumulate driver mutations such as adenomatous polyposis coli (APC), TP53, SMAD4, KRAS, and PI3K catalytic subunit-α (PIK3CA), which Vogelstein and colleagues advocated as the adenoma-carcinoma sequence [26]. In this sequence, TGF-β signaling pathway also plays a pivotal role in CRC progression. In fact, absent expression of SMAD4 (a key transcription factor for TGF-β signaling) is an independent poor prognostic factor after curative surgery for Stage II/III CRC and CRC liver metastases [15,27,28,29]. Moreover, CMS4 carrying the mesenchymal phenotype with TGF-β-activated stroma showed the worst prognosis with low benefit from chemotherapy among all molecular classes [12,30,31,32]. In this scenario, TGF-β signaling causes epithelial-to-mesenchymal transition (EMT) in cancer cells, resulting in an aggressive phenotype [18]. To this end, TGF-β signaling directs serrated adenomas to the mesenchymal CRC subtype, while TGFBR2 mutation impairs EMT [33,34]. Collectively, these basic and clinical data indicate that disruption of TGF-β signaling, especially in advanced CRC, results in an aggressive phenotype of CRC and, consequently, poor prognosis.
3. TGF-β Signaling in Cancer Cells
3.1. TGFBR2 Mutation in Cancer Cells
For appropriate function of TGF-β signaling, active TGF-β receptors (both type 1 and 2 receptors) are mandatory [19]. TGFBR2 mutations are frequently found in MSI-H CRC [35]. MSI-H CRC cells carrying dMMR harbor silent expression of mismatch repair genes through either germline mutations of MMR genes such as MutL homolog 1 (MLH1), MutS homolog 2 (MSH2), MSH6, and Postmeiotic segregation increased 2 (PMS2), or MLH1 promoter hypermethylation [36,37]. Lynch syndrome is an autosomal dominant hereditary cancer syndrome that carries germline mutations of one of these 4 MMR genes, resulting in the development of many types of cancers including CRC, endometrial, ovarian, gastric, small intestine, pancreatic, and urothelial tract cancers [38]. Lynch syndrome accounts for approximately 3% of CRCs, whereas approximately 12–15% of CRCs is sporadic MSI-H CRC, resulting from hypermethylation of the MLH1 promoter [39]. Because TGFBR2, carrying a 10-adenine repeat, is only one of the genes affected by dMMR, it is possible that TGFBR2 mutation is merely a bystander event [13,40]. However, according to several mouse studies, TGFBR2 mutation itself does have the potential to transform normal colonic epithelial cells to malignant cells [41,42]. In addition to TGFBR2 mutation, BMP receptor type 2 (BMPR2) can be also affected by dMMR as it also carries a 7-adenine repeat that is also affected by dMMR [43].
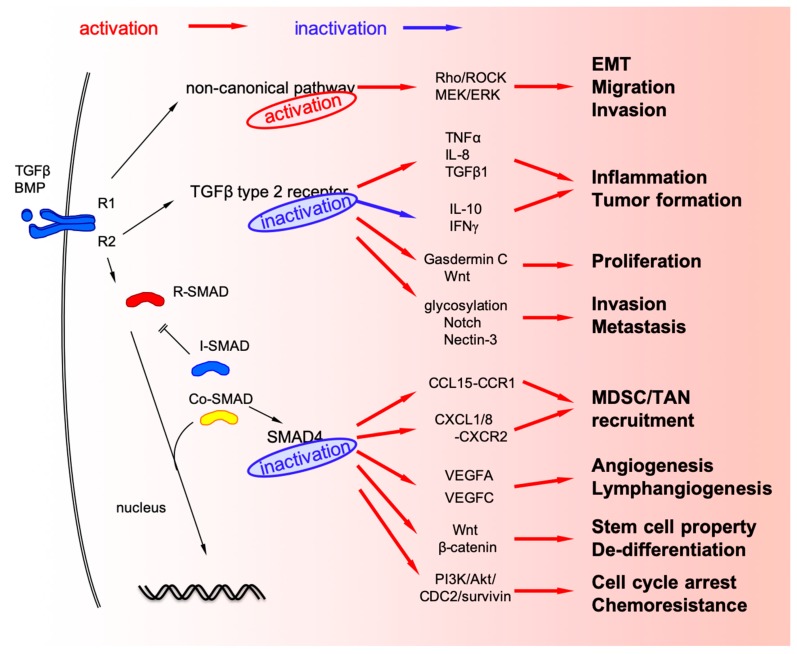
MSI-H CRC exhibits high immune response that can be targeted for immune checkpoint inhibitors [44]. Although high immune response in MSI-H CRC is the result of tumor neoantigen load caused by hypermutation, TGFBR2 impairment can also directly promote inflammation in the tumor microenvironment of CRC. TGFBR2 deficiency in an APC-deleted mouse model of intestinal adenoma increased inflammatory burden and promoted tumor progression via producing tumor necrosis factor-α (TNF-α), interleukin (IL)-8, and TGF-β1 as well as suppressing of anti-inflammatory cytokines such as IL-10 and interferon (IFN)-γ, which resulted in increased infiltration of CD11b+Gr1+ granulocyte population into the tumor microenvironment (Figure 2) [45]. Moreover, TGFBR2 disruption in combination with inflammation in the colon causes invasive CRC via tumor-associated macrophage (TAM) infiltration [46].
Figure 2.
TGF-β alteration in colorectal cancer (CRC) cells.
TGFBR2 inactivation in CRC cells contributes to the malignant phenotype via multiple pathways such as Wnt-β-catenin, Hippo, and MAPK [47]. In a mouse model designed to conditionally knockdown TGFBR2 in the proximal colon, TGFBR2 impairment in combination with Wnt-β-catenin pathway activation promoted the upregulation of Gasdermin C, which stimulated the proliferation of CRC cells [48].
Glycosylation is an important post-translational protein modification and also affects malignant phenotype of cancer cells [49]. TGF-β signaling can also modulate the protein glycosylation pattern in MSI-H CRC. TGFBR2 impairment in MSI-H CRC cell line, HCT116, can upregulate some of glycosylation-related genes and affect important cell signaling pathways such as Notch [50]. In this cell line model, Nectin-3 (a cell surface glycoprotein that modulates cancer cell invasion and metastasis) was also upregulated when TGFBR2 was impaired, and reconstitution of TGFBR2 upregulated growth differentiation factor-15 (GDF15; one of the ligands of TGF-β superfamily signaling) in a cell line model [51]. TGFBR2 mutation in CRC can also cause changes in the components secreted by cancer cells. For example, in vitro experiments with MSI-H TGFBR2-deficient HCT116 cells revealed that extracellular matrix and nucleosome-related proteins were upregulated, while proteasome-associated proteins in the extracellular vesicles were downregulated [52,53]. Clinical implications of these cell line-based experiments are, however, not fully understood.
3.2. Mutations and Deletions of SMAD4 (Co-SMAD) in CRC Cells
LOH is one of the common features of carcinogenesis that causes cancer cells to lose tumor suppressor genes and acquire malignant phenotype [54]. Chromosome 18q21 is frequently affected by LOH in microsatellite-stable CRC. There are many genes in this chromosomal region including SMAD2, SMAD4, and deletion of colon cancer (DCC) that may contribute to form malignant phenotype of CRC. Among the genes on chromosome 18q21, SMAD4 is the established tumor suppressor gene, and loss of SMAD4 disrupts canonical TGF-β signaling because it is a transcription factor for the signaling [54]. Moreover, The Cancer Genome Atlas (TCGA) database revealed that SMAD4 is one of the most frequently mutated genes in CRC [55].
SMAD4 is one of the key driver genes that contribute to CRC progression and metastasis [56,57]. Consequently, SMAD4 loss onto APC mutation in intestinal epithelial cells causes malignant invasive phenotype in mouse models [58,59]. Loss of SMAD4 protein expression is found approximately 20–40% of human CRCs [8,9,10,11,60,61]. Although LOH can be the main cause of SMAD4 loss in CRC, there are other proposed mechanisms that contribute to SMAD4 defect in post-transcriptional and post-translational regulation: ubiquitylation, sumoylation, and mircoRNA interference [62,63].
SMAD4 acts as a transcription factor by forming trimers with R-SMAD components and directly regulates target genes. R-SMAD-SMAD4 complexes can also associate with DNA-binding partners to act as a transcription co-factor [4]. Therefore, there are many target genes regulated by SMAD4, indicating that several changes in cancer cells happen when R-SMAD-SMAD4 complexes are disrupted [64,65,66]. RNA sequencing comparing SMAD4-proficient and SMAD4-deficient colonic epithelial cells in mice revealed upregulation of many inflammation-related genes [66]. Among them, Ccl9, one of the C-C motif chemokine ligands, is upregulated in a SMAD4-deficient intestinal tumor mouse model [59]. In this mouse model, chemokine expression recruits myeloid cells with corresponding receptors, Ccr1, to promote tumor invasion and metastases [59,67,68]. Similar interactions between SMAD4-deficient CRC cells and surrounding myeloid-derived cells via chemokine signaling was observed in human CRC samples [8,9,10,11]. Namely, loss of SMAD4 from CRC cells promotes upregulation of C-C motif chemokine CCL15 (human orthologue of mouse Ccl9) to recruit CCR1+ myeloid-derived suppressor cells (MDSC) [8,9,10,69]. SMAD4-deficient CRC cells also produce C-X-C motif chemokine ligand CXCL1/8 to recruit its corresponding receptor CXCR2+ tumor-associated neutrophils (TAN) [11]. MDSC accumulation is characteristic of CMS4 immune contexture [70]. These experimental and observational results suggest that SMAD4-deletion in CRC causes both cancer cell phenotype and tumor microenvironment to switch to the more aggressive cancer phenotype.
TGF-β signaling plays a crucial role in angiogenesis in the tumor microenvironment [71,72]. SMAD4 can also regulate the expression of vascular endothelial growth factor (VEGF)-A and VEGF-C, the main angiogenic factors for tumor angiogenesis and lymphangiogenesis. Disruption of SMAD4 promotes upregulation of these angiogenic factors, resulting in promoting angiogenesis and lymphangiogenesis in CRC [73,74]. TGF-β signaling also plays a critical role in the differentiation of epithelial cells. Loss of SMAD4 promotes β-catenin expression, and simultaneous SMAD4 loss and Wnt activation in the intestinal epithelium trigger the acquisition of stem cell properties and lead to de-differentiation and rapid adenoma formation in the differentiated intestinal epithelium of the Cre-driven conditional mouse model [75,76].
Chemoresistance is also an important property of malignant phenotype of cancer cells, and loss of SMAD4 is a predictive biomarker for 5-FU-based chemotherapy [77,78]. SMAD4 deficiency activates PI3K/Akt/cell-division cycle 2 (CDC2)/survivin pathway to attenuate G1/2 cell cycle arrest, providing resistance to 5-FU-based chemotherapy because 5-FU basically acts as a thymidylate synthase inhibitor to block DNA replication [79,80].
SMAD4 loss-induced changes in vivo and in vitro were summarized in Table 1 and Table 2.
Table 1.
SMAD4 loss-induced changes in vivo (animal models).
| Models | Factors Changed | Function Observed |
|---|---|---|
| GEMM 1 [59,67] | Ccl9 upregulation | CCR1+ myeloid cell recruitment |
| Xenograft [8,9,10] | CCL15 upregulation | CCR1+ TAN/MDSC recruitment |
| GEMM [75] | Wnt activation | Dedifferentiation Stem cell characteristics |
| Allograft [80] | PI3K/Akt/CDC2/survivin activation | 5-FU resistance |
1 GEMM, genetically-engineered mouse model.
Table 2.
SMAD4 loss-induced changes in vitro.
3.3. Non-Canonical TGF-β Signaling Pathways in CRC
Non-canonical TGF-β signaling pathways also modulate important cellular physiology. Disruption of non-canonical TGF-β pathways as well as canonical ones is also frequently found in CRC. Although loss of SMAD4 may lead to the blockade of canonical TGF-β signaling, it alters BMP signaling via non-canonical pathway to promote CRC metastasis through activation of Rho/ROCK pathway, leading to EMT, migration, and invasion [81]. Loss of SMAD4 also activates alternative MEK/ERK pathways to promote cell mortality, migration, and invasion [82,83].
4. TGF-β Signaling in Stromal Cells in the Tumor Microenvironment
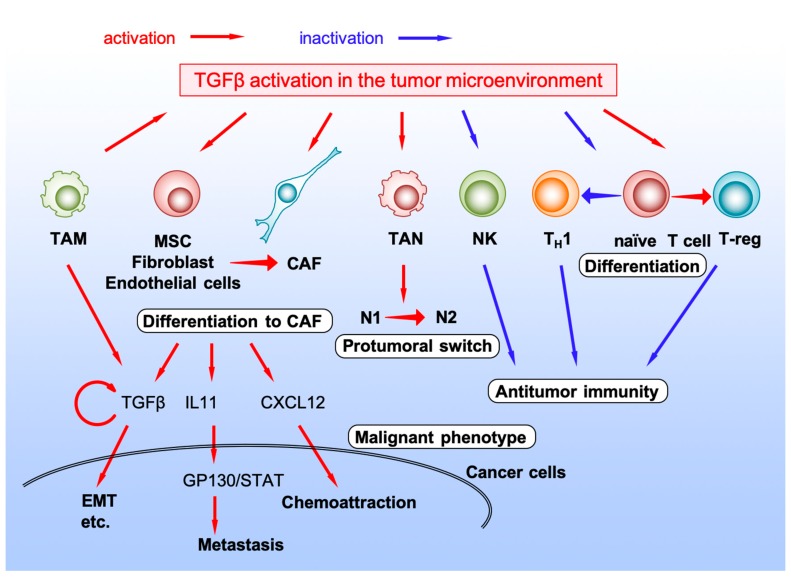
Cancer tissues contain abundant stromal cells in addition to cancer epithelial cells. Interaction between cancer cells and surrounding stromal cells can either promote or inhibit cancer progression. In the tumor microenvironment, various cytokine/chemokine networks play pivotal roles in controlling the interaction between cancer cells and stromal cells [84,85]. As described above, CMS4 mesenchymal phenotype with poor prognosis characterizes prominent TGF-β activation [12]. These observations suggest that TGF-β activation in the tumor microenvironment can promote the tumor-stromal interaction to induce a malignant CRC phenotype and poorer prognosis (Figure 3).
Figure 3.
TGF-β activation in the tumor microenvironment.
4.1. Cancer-Associated Fibroblast
Cancer-associated fibroblast (CAF) is one of the major components of the tumor microenvironment and act as an important factor in tumor progression and metastasis [86,87,88]. TGF-β activation in the tumor microenvironment promotes differentiation of mesenchymal stem cells (MSCs) to CAFs, activating phosphorylation of signal transducer and activator of transcription 3 (STAT3) and nuclear localization of p-STAT3 via Janus kinase (JAK)/STAT pathway [86]. Normal fibroblasts or endothelial cells can also be converted to CAFs by stimulation of TGF-β superfamily ligands, such as nodal or TGF-β2, to support tumor growth [89,90]. TGF-β ligands also directly activate CAFs; activated CAFs produce chemokine CXCL12 and interact CXCR4+ CRC cells, or IL-11 to activate CRC cells through GP130/STAT signaling to metastasize to distant organs [91,92,93]. CAFs secrete TGF-β ligand from themselves to accelerate malignant phenotype of CRC in the hypoxic condition that normally occurs in vivo within the tumor microenvironment [94,95].
4.2. Natural Killer Cell
Natural killer (NK) cells can be expanded ex vivo, activated and transferred to cancer patients to effectively kill cancer cells. However, highly immunosuppressive tumor microenvironment caused, in part, by excessive TGF-β signaling, may limit the activity of NK cells. Exposure to TGF-β ligand decreases the ability of activated NK cells to kill cancer cells ex vivo, while inhibition of TGF-β signaling in the tumor microenvironment preserves the function of highly activated, in vitro expanded NK cells in CRC in vivo [96].
4.3. TANs
Peripheral neutrophil-to-lymphocyte ratio (NLR) in patients with CRC predicts the prognosis after treatment [97,98,99,100]. Although higher neutrophil level could be just bystander in patients with poor prognosis, neutrophils may contribute directly to the tumor progression. In the tumor microenvironment of SMAD4-deficient CRC, TANs are recruited via the chemokine CXCL1/8-CXCR2 axis [11]. Tumor microenvironment contains at least two types of neutrophils: N1 with an anti-tumoral phenotype and N2 with a protumoral phenotype [101,102,103]. TGF-β signaling in the tumor microenvironment regulates phenotypical changes in neutrophils to N2 protumoral ones, while TGF-β blockade with a small molecule inhibitor results in the recruitment and activation of N1 antitumoral TANs [101].
4.4. TAMs
TAMs in the tumor microenvironment were known to release pro-inflammatory cytokines there to form inflamed environment, which engages in bidirectional interaction between cancer cells and host immune cells [104]. SMAD4 deletion in CRC cells decreased the number of S100A8+ monocytes or CD68+ TAMs in the tumor microenvironment; this is associated with unfavorable prognoses in CRC patients [105,106]. Recruited TAM in the CRC microenvironment produces TGF-β ligand to promote proliferation and invasion via EMT or VEGF [107,108]. In compound mutant mice that have mutations in Apc and Tgfbr2 in the intestinal epithelial cells, TAM infiltrates into the invasive tumors express membrane-type1-matrix metalloproteinase (MMP), causing MMP2 activation [46]. Although TAMs can be one of the sources of TGF-β ligand expression, the effects of TGF-β signaling on TAMs in CRC is not fully understood [108].
4.5. T Lymphocyte
As NLR can predict CRC patients’ prognosis, low levels of T cell infiltration or low activity of type 1 T-helper cells (TH1) can also predict poor outcome of CRC patients [109]. Infiltrating T cells are activated through both signals from major histocompatibility complex (MHC)-presented immunogenic peptide antigens to the T cell receptor (TCR) and signals from CD80/86 on the antigen-presenting cells (APC) to T cell surface receptor CD28. However, once activated, T cell expressed co-inhibitory receptors, such as cytotoxic T lymphocyte antigen 4 (CTLA4) and programmed cell death 1 (PD1), and become exhausted T cell [110]. Abundant TGF-β in the tumor microenvironment promotes T cell exclusion and prevents them to acquire TH1-effector phenotype [95]. Inhibition of TGF-β using a small molecule inhibitor, galunisertib, unleashed a potent and enduring cytotoxic T cell response against CRC cells and rendered tumors susceptible to anti-PD-L1-PD-1 therapy.
Regulatory T cells (T-regs) are a subpopulation of T cells that modulate immune system and self-antigen tolerance. T-regs express biomarkers such as CD4, forkhead box P3 (FOXP3), and CD25, and differentiates from naïve CD4+ T cells by TGF-β stimulation [111]. In the tumor microenvironment, T-regs play a critical role in suppressing tumor immunity and promoting cancer progression [112]. Therefore, enriched TGF-β in the tumor microenvironment of CRC promotes phenotypical changes of T cells to T-regs and tumor progression [113]. A meta-analysis comparing T-reg infiltration and patients’ prognosis reported that high FOXP3+ T-reg density was associated with poor overall and disease-free survival in patients with all types of cancer [114]. However, the prognostic role of T-reg infiltration is tumor-dependent, and, among CRC patients, a higher number of tumor-infiltrating FOXP3+ T-regs predicts favorable outcome of CRC patients [114,115].
5. Discussion
CRC is the second to third most common cause of cancer-related deaths worldwide [116,117]. Recent developments in surgical technology (e.g., laparoscopic or robotic surgery) and novel therapeutic compounds (small molecule inhibitors and molecular targeted antibodies) are promising, and the mortality rate of CRC is gradually decreasing in some western countries [116,117]. However, once metastasized, treatment strategies can be limited, and, if unresectable, outcomes in CRC patients are unfavorable. TGF-β signaling may contribute to this unfavorable outcome in CMS4 and metastasized CMS1 subpopulations. Therefore, comprehensive understanding of TGF-β signaling in both tumor cells and the tumor microenvironment is mandatory for the construction of novel therapeutic strategies.
As discussed above, the disruption of TGF-β signaling in CRC cells generally promotes tumor formation in the early stage, while its activation may promote cancer invasion and metastasis. Moreover, its activation in the tumor microenvironment generally suppresses tumor immunity and supports cancer cell survival. The bidirectional function of TGF-β signaling within cancer cells and multi-directional functions between cancer cells and their microenvironment make effective drug discovery for CRC treatment difficult. This is because simple blockade of TGF-β signaling can activate tumor immunity in the tumor microenvironment but may alter cancer cell phenotypes to more aggressive ones.
Acknowledgments
The authors thank Susumu Inamoto, Rei Mizuno, Takamasa Yamamoto, Gen Nishikawa. Ryotaro Ogawa and Yoshiyuki Kiyasu for data acquisition.
Abbreviations
| 5-FU | 5-fluorouracil |
| APC | Adenomatous Polyposis Coli |
| APC | antigen-presenting cell |
| BMP | bone morphogenetic protein |
| BMPR2 | BMP receptor type 2 |
| CAF | cancer-associated fibroblast |
| CCL | C-C motif chemokine ligand |
| CDC | cell-division cycle |
| CMS | consensus molecular subtype |
| Co-SMAD | common-mediator SMAD |
| CRC | colorectal cancer |
| CTLA4 | cytotoxic T lymphocyte antigen 4 |
| CXCL | C-X-C motif chemokine ligand |
| dMMR | mismatch repair deficiency |
| EMT | epithelial to mesenchymal transition |
| FOXP3 | forkhead box P3 |
| GDF15 | growth differentiation factor-15 |
| GEMM | genetically-engineered mouse model |
| IL | interleukin |
| INFγ | interferon-γ |
| I-SMAD | inhibitory SMAD |
| JAK | Janus kinase |
| LOH | loss of heterozygosity |
| MAPK | mitogen-activated protein kinase |
| MDSC | myeloid-derived suppressor cells |
| MHC | major histocompatibility complex |
| MLH | MutL homolog |
| MMP | matrix metalloproteinase |
| MSC | mesenchymal stem cell |
| MSH | MutS homolog |
| MSI | microsatellite instability |
| MSI-H | high level of MSI |
| PCR | polymerase chain reaction |
| PD1 | programmed cell death 1 |
| PI3K | phosphoinositide 3-kinase |
| PIK3CA | PI3K catalytic subunit-α |
| PMS | Postmeiotic segregation increased |
| R-SMAD | receptor-regulated SMAD |
| ROCK | Rho-associated protein kinase |
| SMAD | Caenorhabditis elegans SMA and Drosophila MAD |
| STAT | signal transducer and activator of transcription |
| TAM | tumor-associated macrophages |
| TAN | tumor-associated neutrophils |
| TCR | T-cell receptor |
| TGFβ | Transforming growth factor-beta |
| TGFBR2 | TGFβ receptor type 2 |
| TH1 | type 1 T-helper cell |
| TNFα | tumor necrosis factor-α |
| T-reg | regulatory T-cell |
| VEGF | vascular endothelial growth factor |
Author Contributions
K.K. and Y.S. conceptualize this study. Y.I. and K.K. wrote the manuscript. All authors reviewed the manuscript for important intellectual content.
Funding
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.K. and Y.I.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Jung B., Staudacher J.J., Beauchamp D. Transforming Growth Factor beta Superfamily Signaling in Development of Colorectal Cancer. Gastroenterology. 2017;152:36–52. doi: 10.1053/j.gastro.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Principe D.R., Doll J.A., Bauer J., Jung B., Munshi H.G., Bartholin L., Pasche B., Lee C., Grippo P.J. TGF-β: Duality of Function Between Tumor Prevention and Carcinogenesis. J. Natl. Cancer Inst. 2014;106 doi: 10.1093/jnci/djt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Massague J., Seoane J., Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 5.Ihara S., Hirata Y., Koike K. TGF-β in inflammatory bowel disease: A key regulator of immune cells, epithelium, and the intestinal microbiota. J. Gastroenterol. 2017;52:777–787. doi: 10.1007/s00535-017-1350-1. [DOI] [PubMed] [Google Scholar]
- 6.Batlle R., Andrés E., Gonzalez L., Llonch E., Igea A., Gutierrez-Prat N., Berenguer-Llergo A., Nebreda A.R. Regulation of tumor angiogenesis and mesenchymal–endothelial transition by p38α through TGF-β and JNK signaling. Nat. Commun. 2019;10:3071. doi: 10.1038/s41467-019-10946-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muñoz N.M., Upton M., Rojas A., Washington M.K., Lin L., Chytil A., Sozmen E.G., Madison B.B., Pozzi A., Moon R.T., et al. Transforming Growth Factor β Receptor Type II Inactivation Induces the Malignant Transformation of Intestinal Neoplasms Initiated by Apc Mutation. Cancer Res. 2006;66:9837. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- 8.Itatani Y., Kawada K., Fujishita T., Kakizaki F., Hirai H., Matsumoto T., Iwamoto M., Inamoto S., Hatano E., Hasegawa S., et al. Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1+ myeloid cells and facilitate liver metastasis. Gastroenterology. 2013;145:1064–1075. doi: 10.1053/j.gastro.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 9.Inamoto S., Itatani Y., Yamamoto T., Minamiguchi S., Hirai H., Iwamoto M., Hasegawa S., Taketo M.M., Sakai Y., Kawada K. Loss of SMAD4 Promotes Colorectal Cancer Progression by Accumulation of Myeloid-Derived Suppressor Cells through the CCL15-CCR1 Chemokine Axis. Clin. Cancer Res. 2016;22:492–501. doi: 10.1158/1078-0432.CCR-15-0726. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto T., Kawada K., Itatani Y., Inamoto S., Okamura R., Iwamoto M., Miyamoto E., Chen-Yoshikawa T.F., Hirai H., Hasegawa S., et al. Loss of SMAD4 Promotes Lung Metastasis of Colorectal Cancer by Accumulation of CCR1+ Tumor-Associated Neutrophils through CCL15-CCR1 Axis. Clin. Cancer Res. 2017;23:833–844. doi: 10.1158/1078-0432.CCR-16-0520. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa R., Yamamoto T., Hirai H., Hanada K., Kiyasu Y., Nishikawa G., Mizuno R., Inamoto S., Itatani Y., Sakai Y., et al. Loss of SMAD4 Promotes Colorectal Cancer Progression by Recruiting Tumor-Associated Neutrophils via the CXCL1/8-CXCR2 Axis. Clin. Cancer Res. 2019;25:2887–2899. doi: 10.1158/1078-0432.CCR-18-3684. [DOI] [PubMed] [Google Scholar]
- 12.Guinney J., Dienstmann R., Wang X., de Reynies A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Miranda N.F.C.C., van Dinther M., van den Akker B.E.W.M., van Wezel T., ten Dijke P., Morreau H. Transforming Growth Factor β Signaling in Colorectal Cancer Cells With Microsatellite Instability Despite Biallelic Mutations in TGFBR2. Gastroenterology. 2015;148:1427–1437. doi: 10.1053/j.gastro.2015.02.052. [DOI] [PubMed] [Google Scholar]
- 14.Grady W.M. Polymerase Slippage Restoration of Frameshifted TGFBR2 in Colorectal Cancer: A Novel Paradigm. Gastroenterology. 2015;148:1276–1279. doi: 10.1053/j.gastro.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 15.Roth A.D., Delorenzi M., Tejpar S., Yan P., Klingbiel D., Fiocca R., d’Ario G., Cisar L., Labianca R., Cunningham D., et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J. Natl. Cancer Inst. 2012;104:1635–1646. doi: 10.1093/jnci/djs427. [DOI] [PubMed] [Google Scholar]
- 16.Weiss A., Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev. Dev. Biol. 2013;2:47–63. doi: 10.1002/wdev.86. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Malpartida P., Batet M., Kaczmarska Z., Freier R., Gomes T., Aragón E., Zou Y., Wang Q., Xi Q., Ruiz L., et al. Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nat. Commun. 2017;8:2070. doi: 10.1038/s41467-017-02054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao Y., Baker D., Ten Dijke P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 2019;20:2767. doi: 10.3390/ijms20112767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi Y., Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 20.Chang C. Agonists and Antagonists of TGF-beta Family Ligands. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 22.Simoneaux R. The Four Colorectal Cancer Consensus Molecular Subtypes. Oncol. Times. 2018;40:10–11. doi: 10.1097/01.COT.0000531932.39051.dd. [DOI] [Google Scholar]
- 23.Shima K., Morikawa T., Yamauchi M., Kuchiba A., Imamura Y., Liao X., Meyerhardt J.A., Fuchs C.S., Ogino S. TGFBR2 and BAX mononucleotide tract mutations, microsatellite instability, and prognosis in 1072 colorectal cancers. PLoS ONE. 2011;6:e25062. doi: 10.1371/journal.pone.0025062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sargent D.J., Marsoni S., Monges G., Thibodeau S.N., Labianca R., Hamilton S.R., French A.J., Kabat B., Foster N.R., Torri V., et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dienstmann R., Vermeulen L., Guinney J., Kopetz S., Tejpar S., Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer. 2017;17:79–92. doi: 10.1038/nrc.2016.126. [DOI] [PubMed] [Google Scholar]
- 26.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voorneveld P.W., Jacobs R.J., Kodach L.L., Hardwick J.C. A Meta-Analysis of SMAD4 Immunohistochemistry as a Prognostic Marker in Colorectal Cancer. Transl. Oncol. 2015;8:18–24. doi: 10.1016/j.tranon.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan P., Klingbiel D., Saridaki Z., Ceppa P., Curto M., McKee T.A., Roth A., Tejpar S., Delorenzi M., Bosman F.T., et al. Reduced Expression of SMAD4 Is Associated with Poor Survival in Colon Cancer. Clin. Cancer Res. 2016;22:3037–3047. doi: 10.1158/1078-0432.CCR-15-0939. [DOI] [PubMed] [Google Scholar]
- 29.Mizuno T., Cloyd J.M., Vicente D., Omichi K., Chun Y.S., Kopetz S.E., Maru D., Conrad C., Tzeng C.D., Wei S.H., et al. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur. J. Surg. Oncol. 2018;44:684–692. doi: 10.1016/j.ejso.2018.02.247. [DOI] [PubMed] [Google Scholar]
- 30.Okita A., Takahashi S., Ouchi K., Inoue M., Watanabe M., Endo M., Honda H., Yamada Y., Ishioka C. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9:18698–18711. doi: 10.18632/oncotarget.24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mooi J.K., Wirapati P., Asher R., Lee C.K., Savas P., Price T.J., Townsend A., Hardingham J., Buchanan D., Williams D., et al. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: Molecular analysis of the AGITG MAX clinical trial. Ann. Oncol. 2018;29:2240–2246. doi: 10.1093/annonc/mdy410. [DOI] [PubMed] [Google Scholar]
- 32.Sveen A., Bruun J., Eide P.W., Eilertsen I.A., Ramirez L., Murumägi A., Arjama M., Danielsen S.A., Kryeziu K., Elez E., et al. Colorectal Cancer Consensus Molecular Subtypes Translated to Preclinical Models Uncover Potentially Targetable Cancer Cell Dependencies. Clin. Cancer Res. 2018;24:794. doi: 10.1158/1078-0432.CCR-17-1234. [DOI] [PubMed] [Google Scholar]
- 33.Pino M.S., Kikuchi H., Zeng M., Herraiz M.T., Sperduti I., Berger D., Park D.Y., Iafrate A.J., Zukerberg L.R., Chung D.C. Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology. 2010;138:1406–1417. doi: 10.1053/j.gastro.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fessler E., Drost J., van Hooff S.R., Linnekamp J.F., Wang X., Jansen M., De Sousa E Melo F., Prasetyanti P.R., Ijspeert J.E., Franitza M., et al. TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol. Med. 2016;8:745–760. doi: 10.15252/emmm.201606184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markowitz S., Wang J., Myeroff L., Parsons R., Sun L., Lutterbaugh J., Fan R.S., Zborowska E., Kinzler K.W., Vogelstein B., et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 36.Rubenstein J.H., Enns R., Heidelbaugh J., Barkun A. American Gastroenterological Association Institute Guideline on the Diagnosis and Management of Lynch Syndrome. Gastroenterology. 2015;149:777–782. doi: 10.1053/j.gastro.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 37.Vasen H.F., Blanco I., Aktan-Collan K., Gopie J.P., Alonso A., Aretz S., Bernstein I., Bertario L., Burn J., Capella G., et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): Recommendations by a group of European experts. Gut. 2013;62:812–823. doi: 10.1136/gutjnl-2012-304356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K., Kaneda M., Futagawa M., Takeshita M., Kim S., Nakama M., Kawashita N., Tatsumi-Miyajima J. Genetic and genomic basis of the mismatch repair system involved in Lynch syndrome. Int. J. Clin. Oncol. 2019;24:999–1011. doi: 10.1007/s10147-019-01494-y. [DOI] [PubMed] [Google Scholar]
- 39.Haraldsdottir S., Hampel H., Wu C., Weng D.Y., Shields P.G., Frankel W.L., Pan X., de la Chapelle A., Goldberg R.M., Bekaii-Saab T. Patients with colorectal cancer associated with Lynch syndrome and MLH1 promoter hypermethylation have similar prognoses. Genet. Med. 2016;18:863–868. doi: 10.1038/gim.2015.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y., Pasche B. TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum. Mol. Genet. 2007;16 Spec No 1:R14–R20. doi: 10.1093/hmg/ddl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai E., Nakayama M., Oshima H., Kouyama Y., Niida A., Fujii S., Ochiai A., Nakayama K.I., Mimori K., Suzuki Y., et al. Combined Mutation of Apc, Kras, and Tgfbr2 Effectively Drives Metastasis of Intestinal Cancer. Cancer Res. 2018;78:1334–1346. doi: 10.1158/0008-5472.CAN-17-3303. [DOI] [PubMed] [Google Scholar]
- 42.Takeda H., Kataoka S., Nakayama M., Ali M.A.E., Oshima H., Yamamoto D., Park J.-W., Takegami Y., An T., Jenkins N.A., et al. CRISPR-Cas9–mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proc. Natl. Acad. Sci. USA. 2019;116:15635. doi: 10.1073/pnas.1904714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinheiro M., Pinto C., Peixoto A., Veiga I., Lopes P., Henrique R., Baldaia H., Carneiro F., Seruca R., Tomlinson I., et al. Target gene mutational pattern in Lynch syndrome colorectal carcinomas according to tumour location and germline mutation. Br. J. Cancer. 2015;113:686–692. doi: 10.1038/bjc.2015.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogino S., Giannakis M. Immunoscore for (colorectal) cancer precision medicine. Lancet. 2018;391:2084–2086. doi: 10.1016/S0140-6736(18)30953-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Principe D.R., DeCant B., Staudacher J., Vitello D., Mangan R.J., Wayne E.A., Mascariñas E., Diaz A.M., Bauer J., McKinney R.D., et al. Loss of TGFβ signaling promotes colon cancer progression and tumor-associated inflammation. Oncotarget. 2017;8:3826. doi: 10.18632/oncotarget.9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oshima H., Nakayama M., Han T.S., Naoi K., Ju X., Maeda Y., Robine S., Tsuchiya K., Sato T., Sato H., et al. Suppressing TGFbeta signaling in regenerating epithelia in an inflammatory microenvironment is sufficient to cause invasive intestinal cancer. Cancer Res. 2015;75:766–776. doi: 10.1158/0008-5472.CAN-14-2036. [DOI] [PubMed] [Google Scholar]
- 47.Morris S.M., Davison J., Carter K.T., O’Leary R.M., Trobridge P., Knoblaugh S.E., Myeroff L.L., Markowitz S.D., Brett B.T., Scheetz T.E., et al. Transposon mutagenesis identifies candidate genes that cooperate with loss of transforming growth factor-beta signaling in mouse intestinal neoplasms. Int. J. Cancer. 2017;140:853–863. doi: 10.1002/ijc.30491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miguchi M., Hinoi T., Shimomura M., Adachi T., Saito Y., Niitsu H., Kochi M., Sada H., Sotomaru Y., Ikenoue T., et al. Gasdermin C Is Upregulated by Inactivation of Transforming Growth Factor β Receptor Type II in the Presence of Mutated Apc, Promoting Colorectal Cancer Proliferation. PLoS ONE. 2016;11:e0166422. doi: 10.1371/journal.pone.0166422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferreira I.G., Pucci M., Venturi G., Malagolini N., Chiricolo M., Dall’Olio F. Glycosylation as a Main Regulator of Growth and Death Factor Receptors Signaling. Int. J. Mol. Sci. 2018;19:580. doi: 10.3390/ijms19020580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J., Katzenmaier E.M., Kopitz J., Gebert J. Reconstitution of TGFBR2 in HCT116 colorectal cancer cells causes increased LFNG expression and enhanced N-acetyl-d-glucosamine incorporation into Notch1. Cell. Signal. 2016;28:1105–1113. doi: 10.1016/j.cellsig.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 51.Lee J., Fricke F., Warnken U., Schnölzer M., Kopitz J., Gebert J. Reconstitution of TGFBR2-Mediated Signaling Causes Upregulation of GDF-15 in HCT116 Colorectal Cancer Cells. PLoS ONE. 2015;10:e0131506. doi: 10.1371/journal.pone.0131506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fricke F., Michalak M., Warnken U., Hausser I., Schnolzer M., Kopitz J., Gebert J. SILAC-Based Quantification of TGFBR2-Regulated Protein Expression in Extracellular Vesicles of Microsatellite Unstable Colorectal Cancers. Int. J. Mol. Sci. 2019;20:4162. doi: 10.3390/ijms20174162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fricke F., Mussack V., Buschmann D., Hausser I., Pfaffl M.W., Kopitz J., Gebert J. TGFBR2-dependent alterations of microRNA profiles in extracellular vesicles and parental colorectal cancer cells. Int. J. Oncol. 2019;55:925–937. doi: 10.3892/ijo.2019.4859. [DOI] [PubMed] [Google Scholar]
- 54.Weinberg R.A. Tumor suppressor genes. Biol. Cancer. 2013;2:231–274. [Google Scholar]
- 55.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 57.Huang D., Sun W., Zhou Y., Li P., Chen F., Chen H., Xia D., Xu E., Lai M., Wu Y., et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018;37:173–187. doi: 10.1007/s10555-017-9726-5. [DOI] [PubMed] [Google Scholar]
- 58.Takaku K., Oshima M., Miyoshi H., Matsui M., Seldin M.F., Taketo M.M. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/S0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 59.Kitamura T., Kometani K., Hashida H., Matsunaga A., Miyoshi H., Hosogi H., Aoki M., Oshima M., Hattori M., Takabayashi A., et al. SMAD4-deficient intestinal tumors recruit CCR1+ myeloid cells that promote invasion. Nat. Genet. 2007;39:467–475. doi: 10.1038/ng1997. [DOI] [PubMed] [Google Scholar]
- 60.Riggins G.J., Kinzler K.W., Vogelstein B., Thiagalingam S. Frequency of Smad gene mutations in human cancers. Cancer Res. 1997;57:2578–2580. [PubMed] [Google Scholar]
- 61.Salovaara R., Roth S., Loukola A., Launonen V., Sistonen P., Avizienyte E., Kristo P., Jarvinen H., Souchelnytskyi S., Sarlomo-Rikala M., et al. Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut. 2002;51:56–59. doi: 10.1136/gut.51.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng D., Zhao S., Tang H., Zhang D., Sun H., Yu F., Jiang W., Yue B., Wang J., Zhang M., et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget. 2016;7:45199–45213. doi: 10.18632/oncotarget.9900. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Xu P., Lin X., Feng X.H. Posttranslational Regulation of Smads. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fei T., Xia K., Li Z., Zhou B., Zhu S., Chen H., Zhang J., Chen Z., Xiao H., Han J.-D.J., et al. Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination. Genome Res. 2010;20:36–44. doi: 10.1101/gr.092114.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kennedy B.A., Deatherage D.E., Gu F., Tang B., Chan M.W., Nephew K.P., Huang T.H., Jin V.X. ChIP-seq defined genome-wide map of TGFbeta/SMAD4 targets: Implications with clinical outcome of ovarian cancer. PLoS ONE. 2011;6:e22606. doi: 10.1371/journal.pone.0022606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Means A.L., Freeman T.J., Zhu J., Woodbury L.G., Marincola-Smith P., Wu C., Meyer A.R., Weaver C.J., Padmanabhan C., An H., et al. Epithelial Smad4 Deletion Up-Regulates Inflammation and Promotes Inflammation-Associated Cancer. Cell. Mol. Gastroenterol. Hepatol. 2018;6:257–276. doi: 10.1016/j.jcmgh.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitamura T., Fujishita T., Loetscher P., Revesz L., Hashida H., Kizaka-Kondoh S., Aoki M., Taketo M.M. Inactivation of chemokine (C-C motif) receptor 1 (CCR1) suppresses colon cancer liver metastasis by blocking accumulation of immature myeloid cells in a mouse model. Proc. Natl. Acad. Sci. USA. 2010;107:13063–13068. doi: 10.1073/pnas.1002372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirai H., Fujishita T., Kurimoto K., Miyachi H., Kitano S., Inamoto S., Itatani Y., Saitou M., Maekawa T., Taketo M.M. CCR1-mediated accumulation of myeloid cells in the liver microenvironment promoting mouse colon cancer metastasis. Clin. Exp. Metastasis. 2014;31:977–989. doi: 10.1007/s10585-014-9684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao M., Mishra L., Deng C.-X. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018;14:111–123. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roelands J., Kuppen P.J.K., Vermeulen L., Maccalli C., Decock J., Wang E., Marincola F.M., Bedognetti D., Hendrickx W. Immunogenomic Classification of Colorectal Cancer and Therapeutic Implications. Int. J. Mol. Sci. 2017;18:2229. doi: 10.3390/ijms18102229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zonneville J., Safina A., Truskinovsky A.M., Arteaga C.L., Bakin A.V. TGF-β signaling promotes tumor vasculature by enhancing the pericyte-endothelium association. BMC Cancer. 2018;18:670. doi: 10.1186/s12885-018-4587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Itatani Y., Kawada K., Yamamoto T., Sakai Y. Resistance to Anti-Angiogenic Therapy in Cancer-Alterations to Anti-VEGF Pathway. Int. J. Mol. Sci. 2018;19:1232. doi: 10.3390/ijms19041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y., Sheng J., Dai D., Liu T., Qi F. Smad4 acts as tumor suppressor by antagonizing lymphangiogenesis in colorectal cancer. Pathol. Res. Pract. 2015;211:286–292. doi: 10.1016/j.prp.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 74.Li X., Li X., Lv X., Xiao J., Liu B., Zhang Y. Smad4 Inhibits VEGF-A and VEGF-C Expressions via Enhancing Smad3 Phosphorylation in Colon Cancer. Anat. Rec. 2017;300:1560–1569. doi: 10.1002/ar.23610. [DOI] [PubMed] [Google Scholar]
- 75.Perekatt A.O., Shah P.P., Cheung S., Jariwala N., Wu A., Gandhi V., Kumar N., Feng Q., Patel N., Chen L., et al. SMAD4 Suppresses WNT-Driven Dedifferentiation and Oncogenesis in the Differentiated Gut Epithelium. Cancer Res. 2018;78:4878–4890. doi: 10.1158/0008-5472.CAN-18-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Freeman T.J., Smith J.J., Chen X., Washington M.K., Roland J.T., Means A.L., Eschrich S.A., Yeatman T.J., Deane N.G., Beauchamp R.D. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of beta-catenin. Gastroenterology. 2012;142:562–571. doi: 10.1053/j.gastro.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang B., Zhang B., Chen X., Bae S., Singh K., Washington M.K., Datta P.K. Loss of Smad4 in colorectal cancer induces resistance to 5-fluorouracil through activating Akt pathway. Br. J. Cancer. 2014;110:946–957. doi: 10.1038/bjc.2013.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papageorgis P., Cheng K., Ozturk S., Gong Y., Lambert A.W., Abdolmaleky H.M., Zhou J.R., Thiagalingam S. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71:998–1008. doi: 10.1158/0008-5472.CAN-09-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Alexander P.B., Wang X.F. TGF-beta Family Signaling in the Control of Cell Proliferation and Survival. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang B., Leng C., Wu C., Zhang Z., Dou L., Luo X., Zhang B., Chen X. Smad4 sensitizes colorectal cancer to 5-fluorouracil through cell cycle arrest by inhibiting the PI3K/Akt/CDC2/survivin cascade. Oncol. Rep. 2016;35:1807–1815. doi: 10.3892/or.2015.4479. [DOI] [PubMed] [Google Scholar]
- 81.Voorneveld P.W., Kodach L.L., Jacobs R.J., Liv N., Zonnevylle A.C., Hoogenboom J.P., Biemond I., Verspaget H.W., Hommes D.W., de Rooij K., et al. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology. 2014;147:196–208. doi: 10.1053/j.gastro.2014.03.052. [DOI] [PubMed] [Google Scholar]
- 82.Ai X., Wu Y., Zhang W., Zhang Z., Jin G., Zhao J., Yu J., Lin Y., Zhang W., Liang H., et al. Targeting the ERK pathway reduces liver metastasis of Smad4-inactivated colorectal cancer. Cancer Biol. Ther. 2013;14:1059–1067. doi: 10.4161/cbt.26427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang B., Halder S.K., Kashikar N.D., Cho Y.J., Datta A., Gorden D.L., Datta P.K. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138:969–980. doi: 10.1053/j.gastro.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hinshaw D.C., Shevde L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nagarsheth N., Wicha M.S., Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017;17:559–572. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tan H.X., Cao Z.B., He T.T., Huang T., Xiang C.L., Liu Y. TGFbeta1 is essential for MSCs-CAFs differentiation and promotes HCT116 cells migration and invasion via JAK/STAT3 signaling. Onco Targets Ther. 2019;12:5323–5334. doi: 10.2147/OTT.S178618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koliaraki V., Pallangyo C.K., Greten F.R., Kollias G. Mesenchymal Cells in Colon Cancer. Gastroenterology. 2017;152:964–979. doi: 10.1053/j.gastro.2016.11.049. [DOI] [PubMed] [Google Scholar]
- 88.Lamprecht S., Sigal-Batikoff I., Shany S., Abu-Freha N., Ling E., Delinasios G.J., Moyal-Atias K., Delinasios J.G., Fich A. Teaming Up for Trouble: Cancer Cells, Transforming Growth Factor-beta1 Signaling and the Epigenetic Corruption of Stromal Naive Fibroblasts. Cancers. 2018;10:61. doi: 10.3390/cancers10030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Z., Zhang J., Zhou J., Lu L., Wang H., Zhang G., Wan G., Cai S., Du J. Nodal Facilitates Differentiation of Fibroblasts to Cancer-Associated Fibroblasts that Support Tumor Growth in Melanoma and Colorectal Cancer. Cells. 2019;8:538. doi: 10.3390/cells8060538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ciszewski W.M., Sobierajska K., Wawro M.E., Klopocka W., Chefczynska N., Muzyczuk A., Siekacz K., Wujkowska A., Niewiarowska J. The ILK-MMP9-MRTF axis is crucial for EndMT differentiation of endothelial cells in a tumor microenvironment. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:2283–2296. doi: 10.1016/j.bbamcr.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 91.Calon A., Lonardo E., Berenguer-Llergo A., Espinet E., Hernando-Momblona X., Iglesias M., Sevillano M., Palomo-Ponce S., Tauriello D.V., Byrom D., et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015;47:320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 92.Hawinkels L.J., Paauwe M., Verspaget H.W., Wiercinska E., van der Zon J.M., van der Ploeg K., Koelink P.J., Lindeman J.H., Mesker W., ten Dijke P., et al. Interaction with colon cancer cells hyperactivates TGF-beta signaling in cancer-associated fibroblasts. Oncogene. 2014;33:97–107. doi: 10.1038/onc.2012.536. [DOI] [PubMed] [Google Scholar]
- 93.Calon A., Espinet E., Palomo-Ponce S., Tauriello D.V., Iglesias M., Cespedes M.V., Sevillano M., Nadal C., Jung P., Zhang X.H., et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tang Y.A., Chen Y.F., Bao Y., Mahara S., Yatim S., Oguz G., Lee P.L., Feng M., Cai Y., Tan E.Y., et al. Hypoxic tumor microenvironment activates GLI2 via HIF-1alpha and TGF-beta2 to promote chemoresistance in colorectal cancer. Proc. Natl. Acad. Sci. USA. 2018;115:E5990–E5999. doi: 10.1073/pnas.1801348115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tauriello D.V.F., Palomo-Ponce S., Stork D., Berenguer-Llergo A., Badia-Ramentol J., Iglesias M., Sevillano M., Ibiza S., Canellas A., Hernando-Momblona X., et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554:538–543. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 96.Otegbeye F., Ojo E., Moreton S., Mackowski N., Lee D.A., de Lima M., Wald D.N. Inhibiting TGF-beta signaling preserves the function of highly activated, in vitro expanded natural killer cells in AML and colon cancer models. PLoS ONE. 2018;13:e0191358. doi: 10.1371/journal.pone.0191358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dell’Aquila E., Cremolini C., Zeppola T., Lonardi S., Bergamo F., Masi G., Stellato M., Marmorino F., Schirripa M., Urbano F., et al. Prognostic and predictive role of neutrophil/lymphocytes ratio in metastatic colorectal cancer: A retrospective analysis of the TRIBE study by GONO. Ann. Oncol. 2018;29:924–930. doi: 10.1093/annonc/mdy004. [DOI] [PubMed] [Google Scholar]
- 98.Haram A., Boland M.R., Kelly M.E., Bolger J.C., Waldron R.M., Kerin M.J. The prognostic value of neutrophil-to-lymphocyte ratio in colorectal cancer: A systematic review. J. Surg. Oncol. 2017;115:470–479. doi: 10.1002/jso.24523. [DOI] [PubMed] [Google Scholar]
- 99.Li H., Zhao Y., Zheng F. Prognostic significance of elevated preoperative neutrophil-to-lymphocyte ratio for patients with colorectal cancer undergoing curative surgery: A meta-analysis. Medicine. 2019;98:e14126. doi: 10.1097/MD.0000000000014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Inamoto S., Kawada K., Okamura R., Hida K., Sakai Y. Prognostic impact of the combination of neutrophil-to-lymphocyte ratio and Glasgow prognostic score in colorectal cancer: A retrospective cohort study. Int. J. Colorectal Dis. 2019;34:1303–1315. doi: 10.1007/s00384-019-03316-z. [DOI] [PubMed] [Google Scholar]
- 101.Fridlender Z.G., Sun J., Kim S., Kapoor V., Cheng G., Ling L., Worthen G.S., Albelda S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mizuno R., Kawada K., Itatani Y., Ogawa R., Kiyasu Y., Sakai Y. The Role of Tumor-Associated Neutrophils in Colorectal Cancer. Int. J. Mol. Sci. 2019;20:529. doi: 10.3390/ijms20030529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Giese M.A., Hind L.E., Huttenlocher A. Neutrophil plasticity in the tumor microenvironment. Blood. 2019;133:2159–2167. doi: 10.1182/blood-2018-11-844548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mantovani A., Allavena P. The interaction of anticancer therapies with tumor-associated macrophages. J. Exp. Med. 2015;212:435–445. doi: 10.1084/jem.20150295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ang C.W., Nedjadi T., Sheikh A.A., Tweedle E.M., Tonack S., Honap S., Jenkins R.E., Park B.K., Schwarte-Waldhoff I., Khattak I., et al. Smad4 loss is associated with fewer S100A8-positive monocytes in colorectal tumors and attenuated response to S100A8 in colorectal and pancreatic cancer cells. Carcinogenesis. 2010;31:1541–1551. doi: 10.1093/carcin/bgq137. [DOI] [PubMed] [Google Scholar]
- 106.Gulubova M., Ananiev J., Yovchev Y., Julianov A., Karashmalakov A., Vlaykova T. The density of macrophages in colorectal cancer is inversely correlated to TGF-beta1 expression and patients’ survival. J. Mol. Histol. 2013;44:679–692. doi: 10.1007/s10735-013-9520-9. [DOI] [PubMed] [Google Scholar]
- 107.Zhang D., Qiu X., Li J., Zheng S., Li L., Zhao H. TGF-beta secreted by tumor-associated macrophages promotes proliferation and invasion of colorectal cancer via miR-34a-VEGF axis. Cell Cycle. 2018;17:2766–2778. doi: 10.1080/15384101.2018.1556064. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 108.Cai J., Xia L., Li J., Ni S., Song H., Wu X. Tumor-Associated Macrophages Derived TGF-betaInduced Epithelial to Mesenchymal Transition in Colorectal Cancer Cells through Smad2,3-4/Snail Signaling Pathway. Cancer Res. Treat. 2019;51:252–266. doi: 10.4143/crt.2017.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ling A., Lundberg I.V., Eklöf V., Wikberg M.L., Öberg Å., Edin S., Palmqvist R. The infiltration, and prognostic importance, of Th1 lymphocytes vary in molecular subgroups of colorectal cancer. J. Pathol. Clin. Res. 2015;2:21–31. doi: 10.1002/cjp2.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Havel J.J., Chowell D., Chan T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer. 2019;19:133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sun X., Cui Y., Feng H., Liu H., Liu X. TGF-beta signaling controls Foxp3 methylation and T reg cell differentiation by modulating Uhrf1 activity. J. Exp. Med. 2019 doi: 10.1084/jem.20190550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shitara K., Nishikawa H. Regulatory T cells: A potential target in cancer immunotherapy. Ann. N. Y. Acad. Sci. 2018;1417:104–115. doi: 10.1111/nyas.13625. [DOI] [PubMed] [Google Scholar]
- 113.Yamada N., Kuranaga Y., Kumazaki M., Shinohara H., Taniguchi K., Akao Y. Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-beta1-mediated suppression. Oncotarget. 2016;7:27033–27043. doi: 10.18632/oncotarget.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shang B., Liu Y., Jiang S.-J., Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: A systematic review and meta-analysis. Sci. Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hu G., Li Z.A., Wang S. Tumor-infiltrating FoxP3(+) Tregs predict favorable outcome in colorectal cancer patients: A meta-analysis. Oncotarget. 2017;8:75361–75371. doi: 10.18632/oncotarget.17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cronin K.A., Lake A.J., Scott S., Sherman R.L., Noone A.-M., Howlader N., Henley S.J., Anderson R.N., Firth A.U., Ma J., et al. Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics. Cancer. 2018;124:2785–2800. doi: 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Malvezzi M., Carioli G., Bertuccio P., Boffetta P., Levi F., La Vecchia C., Negri E. European cancer mortality predictions for the year 2019 with focus on breast cancer. Ann. Oncol. 2019;30:781–787. doi: 10.1093/annonc/mdz051. [DOI] [PubMed] [Google Scholar]