Abstract
Oligosaccharyltransferase (OST) is a multi-span membrane protein complex that catalyzes the addition of glycans to selected Asn residues within nascent polypeptides in the lumen of the endoplasmic reticulum. This process, termed N-glycosylation, is a fundamental post-translational protein modification that is involved in the quality control, trafficking of proteins, signal transduction, and cell-to-cell communication. Given these crucial roles, N-glycosylation is essential for homeostasis at the systemic and cellular levels, and a deficiency in genes that encode for OST subunits often results in the development of complex genetic disorders. A growing body of evidence has also demonstrated that the expression of OST subunits is cell context-dependent and is frequently altered in malignant cells, thus contributing to tumor cell survival and proliferation. Importantly, a recently developed inhibitor of OST has revealed this enzyme as a potential target for the treatment of incurable drug-resistant tumors. This review summarizes our current knowledge regarding the functions of OST in the light of health and tumor progression, and discusses perspectives on the clinical relevance of inhibiting OST as a tumor treatment.
Keywords: endoplasmic reticulum, N-glycosylation, oligosaccharyltransfease, tumors
1. Introduction
Most, if not all, of the secretory and membrane proteins synthesized in the endoplasmic reticulum (ER) of eukaryotes are modified with N-glycans (Figure 1) [1]. This post-translational protein modification is highly conserved throughout evolution and is catalyzed by the oligosaccharyltransferase (OST) complex [2]. The biological relevance of N-glycosylation has been elucidated in a number of genetic and biochemical studies that demonstrate that N-glycosylation plays an indispensable role in protein folding, degradation, trafficking, cell signaling, and intercellular communication [3,4,5].
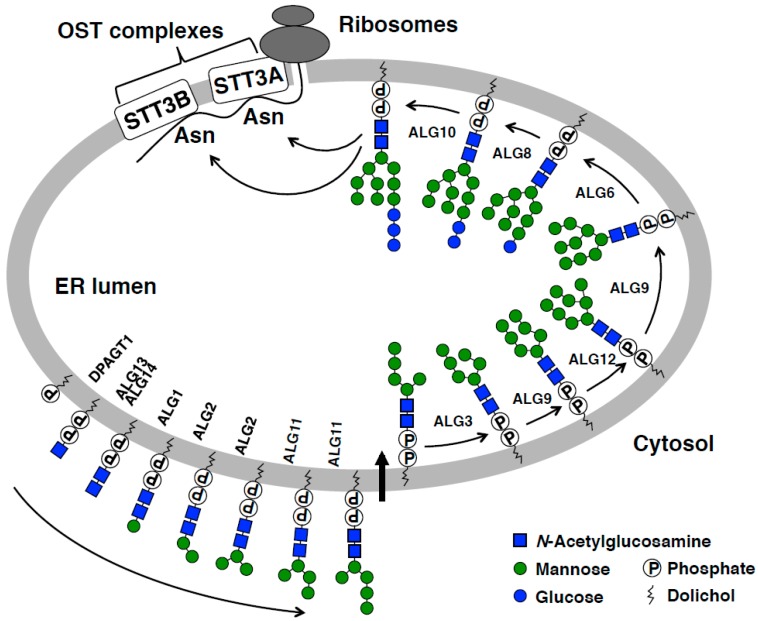
Figure 1.
A model of N-glycosylation in the endoplasmic reticulum (ER). The biosynthesis of dolichol-linked oligosaccharides (DLOs) is initiated on the cytosolic side of the ER membrane by a series of membrane-anchored glycosyltransferases. A DLO intermediate (Man5GlcNAc2-PP-dolichol) is then transported to the ER lumen and further modified with mannose (Man) and glucose (Glc) units, resulting in the synthesis of the fully assembled DLO (Glc3Man9GlcNAc2-PP-dolichol). OST transfers the glycan moiety of DLO en bloc to amide group of the side chain of Asn residue within consensus sequences (Asn-Xaa-Ser/Thr, Xaa ≠ Pro), thus forming an N-glycosidic linkage. Mammalian cells that express STT3A and STT3B as catalytic subunits of OST and incorporated into distinct OST complexes (STT3A-OST and STT3B-OST). N-glycosylation reaction catalyzed by STT3A-OST is coupled with translation and translocation of nascent polypeptides (co-translational N-glycosylation), whereas STT3B-OST mediates the N-glycosylation of sites that are skipped by STT3A-OST (post-translational N-glycosylation) [17].
When glycoproteins exit the ER and traffic through the Golgi apparatus for maturation, the N-glycans are subjected to dramatic structural remodeling by the concerted action of various glycosyltransferases and glycosidases [5]. The expression and function of these enzymes are tightly regulated by genetic and epigenetic factors, as well as by the supply of acceptor and donor substrates, which are often dysregulated in tumor cells [6,7]. As a consequence, tumor cells express aberrant N-glycans, which have been shown to be associated with malignancy and poor prognosis. The correlation between tumor progression and the terminal modification of N-glycans has long been an extensive focus of basic and clinical research studies in glycobiology and have been sophisticatedly reviewed elsewhere [6,7,8,9,10,11,12,13,14,15,16]. In contrast, the association of OST with tumor progression has only recently been explored. In this review, we summarize the functions of OST in health and diseases with a focus on genetic disorders and tumors.
2. Overview of N-Glycosylation in the ER
N-glycosylation in the ER is divided into glycan biosynthesis and transfer phases (Figure 1) [3,18,19]. Dolichol-linked oligosaccharides (DLOs) are glycolipids that function as glycan donor substrates for N-glycosylation. The biosynthesis of DLOs begins on the cytosolic side of the ER membrane and ends in the luminal side. In this process, monosaccharides are assembled onto dolichyl phosphate one at a time directly or indirectly from nucleotide sugars. The fully assembled DLO consists of three glucose (Glc), nine mannose (Man), and two N-acetylglucosamine (GlcNAc) residues that are covalently linked to dolichyl pyrophosphate. Once assembled, the tetradecaoligosaccharide (Glc3Man9GlcNAc2) of DLO is transferred by OST en bloc to Asn residues within the consensus sequences (Asn-Xaa-Ser/Thr, Xaa ≠ Pro) of nascent polypeptides. Genes that are involved in N-glycan biosynthesis in the ER are highly conserved in eukaryotes, whereas eubacteria and archaebacteria use different sets of genes to synthesize N-glycans [20,21,22]. It is important to note that the catalytic subunit of OST, that is, STT3, is evolutionarily conserved among the three domains of life [2]. In mammals, the STT3 gene is duplicated and the gene products (STT3A and STT3B) are expressed to mediate N-glycosylation in a mutually complementary manner (see below) (Figure 1) [23].
3. OST and Its Action
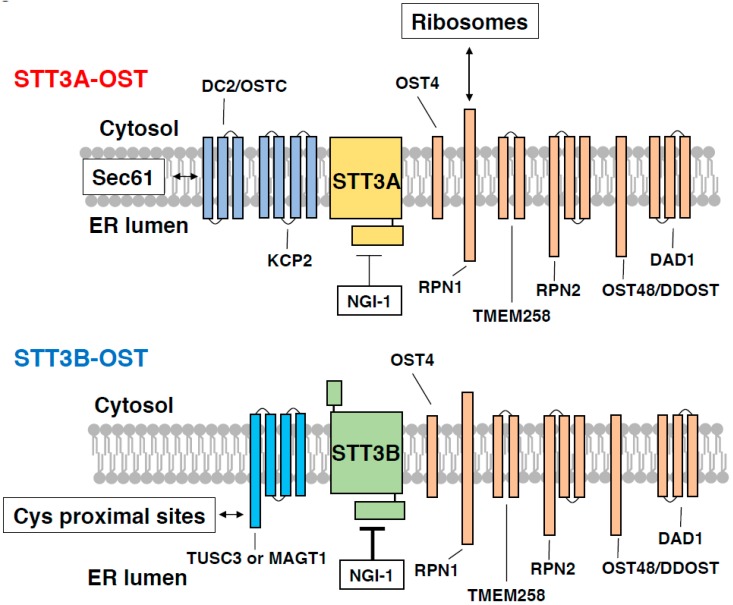
Mammalian cells express two distinct OST complexes that contain STT3A or STT3B as the catalytic subunits and several accessory proteins (Figure 2 and Table 1; STT3A-OST and STT3B-OST) [24,25,26,27]. These accessory proteins include six common subunits (RPN1, RPN2, OST48/DDOST, OST4, TMEM258 and DAD1), STT3A-OST-specific subunits (DC2/OSTC and KCP2) [28] and STT3B-OST-specific subunits (TUSC3 and MAGT1) [17,29]. The two OST complexes are known to have distinct, but partially overlapping specificity to DLO glycans and acceptor sites [23,24,30,31,32,33]. Regarding DLO glycans, it has been reported that in vitro, STT3A-OST shows a strict specificity to the fully assembled DLO, whereas STT3B-OST can also accept DLOs that are completely devoid of glucose residues [24]. The glucose residues of DLO are required for the efficient binding of STT3A-OST, but not STT3B-OST, to acceptor peptides, indicating that the glycan moiety of the fully assembled DLO promotes N-glycosylation by STT3A-OST [24]. STT3 orthologs contain an evolutionarily conserved external loop 5 (EL5), which binds to both donor and acceptor substrates via its N-terminal and C-terminal regions, respectively [34]. It has been proposed that the EL5 loop of Campylobacter lari PglB, a bacterial ortholog of STT3, controls the accessibility of the glycan moiety of lipid-linked oligosaccharides to the active site of PglB. Although the precise role of the EL5 loop of mammalian STT3 proteins in catalysis remains unknown, it is attractive to speculate that the microenvironment surrounding the EL5 loop is distinctly different between STT3A-OST and STT3B-OST, which could limit the full activation of STT3A-OST by incompletely assembled DLOs. In support of this hypothesis, DC2, a STT3A-OST-specific subunit, is in contact with the second transmembrane domain of STT3A, which is located close to the EL5 loop [35].
Figure 2.
Subunit composition of STT3A-OST and STT3B-OST. STT3A-OST (upper side) and STT3B-OST (lower side) contain six shared subunits (RPN1, RPN2, DAD1, OST48, OST4, and TMEM258; shown in orange) and specific subunits (DC2/OSTC and KCP2 for STT3A-OST; shown in dark blue, and TUSC3 and MAGT1 for STT3B-OST; shown in cyan). The cytosolic domain of RPN1 in complex with STT3A-OST makes contact with the 60S subunit of membrane-bound ribosomes [35]. In contrast, DC2/OSTC mediates interaction between STT3A-OST and Sec61 protein-conducting channel [35], allowing co-translational N-glycosylation. STT3B-OST contains either one of TUSC3 or MAGT1, which has an oxidoreductase activity and facilitates N-glycosylation of Cys proximal sites [33]. N-glycosylation inhibitor 1 (NGI-1) inhibits STT3B-OST more efficiently than STT3A-OST (represented by thick and thin T bars) [43].
Table 1.
Subunit compositions and functions of oligosaccharyltransferase (OST).
| OST Subunits | Type of OST | Functions in OST Complexes | Phenotypes Caused by Mutations | Phenotypes Caused by Downregulation | References |
|---|---|---|---|---|---|
| STT3A | STT3A-OST | Catalytic subunit, co-translational glycosylation | STT3A-CDG | Impaired co-translational glycosylation | [42] |
| STT3B | STT3B-OST | Catalytic subunit, post-translational glycosylation | STT3B-CDG | Impaired post-translational glycosylation | [42] |
| RPN1 | Shared | Binding to ribosome | Not known | Reduced expression of STT3A and STT3B | [23,35] |
| RPN2 | Shared | Binding to ribosome? | Not known | Hypoglycosylation of P-glycoprotein and CD63 | [44,45,46] |
| TMEM258 | Shared | Association with RPN1 | Not known | Increased intestinal inflammation in dextran sodium sulfate-treated haploinsufficient mice | [26] |
| DAD1 | Shared | Stabilization of RPN1, RPN2 and OST48 | Increased susceptibility to apoptotic cell death at non-permissive temperature | Reduced expression of STT3A, STT3B, OST48/DDOST and KCP2 | [47,48,49] |
| OST48/DDOST | Shared | Stabilization of STT3A-OST and STT3B-OST | DDOST-CDG | Reduced expression of STT3A, STT3B, DAD1 and KCP2 | [49,50] |
| OST4 | Shared | Stabilization of STT3A-OST, but not STT3B-OST | Not known | Hypoglycosylation of prosaposin and reduced expression of STT3A and KCP2 | [27] |
| DC2/OSTC and KCP2 | STT3A-OST | Association with the Sec61 protein-conducting channel, co-translational glycosylation | Not known | Impaired co-translational glycosylation | [28,35] |
| TUSC3 | STT3B-OST | Thioredoxin, glycosylation at Cys-proximal sites | Autosomal recessive mental retardation | Impaired Mg2+ uptake and tumor progression | [33,51,52,53,54,55,56,57,58,59,60,61,62,63,64] |
| MAGT1 | STT3B-OST | Thioredoxin, glycosylation at Cys-proximal sites | X-linked immunodeficiency | Impaired Mg2+ uptake | [33,52,65,66] |
OST catalyzes the transfer of glycans to acceptor sites only when the sites are located in the extended region of a polypeptide [23] and are located a distance of 12–14 amino-acid units from the luminal surface of the ER membrane [36]. Because nascent polypeptides begin the folding process as soon as they emerge into the ER lumen, N-glycosylation needs to occur before the acceptor sites are embedded in the folded region of the proteins. In mammalian cells, STT3A-OST is associated with membrane-bound ribosomes and the protein-conducting channel [23,25,35]. This supra-molecular complex formation enables STT3A-OST to catalyze N-glycosylation coupled with translation and translocation, that is, co-translational N-glycosylation. However, STT3A-OST skips some of the acceptor sites when they are located too close to each other [31], near signal peptides [23], and in the extreme C-terminus of the polypeptide [32]. These skipped sites may be modified by STT3B-OST in a post-translational manner [23], thereby maximizing the efficiency of N-glycosylation. It should be noted that STT3B-OST can also modify some of the STT3A-sites in the absence of STT3A-OST [29], raising the possibility that these sites are located in the polypeptide region that remains as an extended structure. It is also possible that STT3B-OST becomes associated with the translocon to mediate co-translational N-glycosylation in the absence of STT3A-OST. The association of STT3B with the ribosome-associated membrane protein fraction is much weaker than that for STT3A [23], and no clear density of STT3B-OST has been found in the cryo-electron tomography map of the translocon in micorosomes from STT3A-knocked out cells [35]. However, the yeast OST complex, which is classified as a STT3B type [2], has the ability to directly bind to ribosomes in vitro [37]. Given the above findings, we speculate that, in the absence of STT3A-OST, STT3B-OST may be transiently associated with the translocon and mediate co-translational N-glycosylation with a much lower efficiency than STT3A-OST.
Transthyretin (TTR), a soluble secretory protein that is involved in familial amyloidosis, is not modified with N-glycans under normal conditions. Interestingly, however, a disease-related mutation (D18G) of TTR destabilizes the polypeptides and exposes a cryptic N-glycosylation site (N98-D-S), where STT3B-OST adds N-glycans post-translationally. This unusual glycan modification facilitates the clearance of mutant TTR by bringing the protein to the ER-associated degradation (ERAD) machinery [38]. The issue of whether cryptic STT3B-dependent N-glycosylation sites are present in other proteins to promote their ERAD is unclear. Interestingly, however, hemophilia-related missense mutations around the unused N-glycosylation site N582 of coagulation factor VIII (FVIII) have been identified as promoting the N-glycosylation of the site by STT3B, thus impairing the secretion of the aberrantly glycosylated FVIII via, at least in part, the binding of the mutant protein to calreticulin [39,40]. Although the mechanisms by which mutant proteins are trapped in the ER are different between TTR and FVIII, it is reasonable to speculate that STT3B is responsible for the N-glycosylation of cryptic sites that are exposed upon protein-destabilizing, disease-causing mutations. It has also been reported that single-nucleotide variants that occur in somatic cells, for example, tumor cells, can create new N-glycosylation sites in several proteins [41]. It would be interesting to investigate whether these N-glycosylation sites have an impact on tumor progression and which OST complex is responsible for their modification.
A deficiency of either the STT3A or STT3B gene causes type I congenital disorders of glycosylation (CDGs) with similar symptoms [42], highlighting the need of both N-glycosylation activities for health. The N-glycosylation status of serum transferrin has been used to identify type I CDGs. Transferrin contains two N-glycosylation sites, which are modified by STT3A [32], and is therefore heavily hypoglycosylated in STT3A-CDG [42]. Consistent with this substrate specificity of OST, the N-glycosylation of transferrin is affected only moderately in STT3B-CDG [42]. The identification of other serum glycoproteins that have STT3B-dependent sites will be required for the routine identification of patients with STT3B-CDG.
4. Roles of Accessory OST Subunits in N-Glycosylation and Health
Although accessory subunits of OST are required for structural integrity and the maximal activity of OST, their biological roles are not fully understood. Here we summarize key proposed functions of the accessory subunits of OST in N-glycosylation and complex formation (Table 1). Genetic disorders caused by OST deficiency are also discussed.
4.1. Shared Subunits
RPN1 and RPN2 (Ost1 and Swp1 in yeast, respectively) are OST subunits shared by both STT3A-OST and STT3B-OST. These proteins were originally identified as potential receptors of membrane-bound ribosomes [44], referred to as “ribophorins”. A cryo-electron microscopy (EM) model of STT3A-OST complexed with the membrane-bound ribosomes and Sec61 protein-conducting channel revealed the presence of a clear contact between the 60S ribosome subunit and the cytosolic domain of RPN1 [35]. In contrast, the density of the cytosolic domain of RPN2 was not visible in the cryo-EM map and the role of this subunit in the binding to ribosomes remains unknown. It should also be noted that RPN1 and RPN2 both contain large luminal domains with no known functions. Interestingly, STT3B-OST contains RPN1, but this OST complex is not associated with the membrane-bound ribosomes [23]. STT3B has a longer cytosolic domain than STT3A, which may interfere with the binding of RPN1 to ribosomes [35].
TMEM258 is a shared subunit of STT3A-OST and STT3B-OST. The TMEM258 gene was originally identified as a gene associated with Crohn’s disease, rheumatoid arthritis, and elevated arachidonic acid levels in blood [26]. In mice, a haploinsufficiency of TMEM258 worsens dextran sodium sulfate-induced colitis. TMEM258 is associated with RPN1 and is required for the maximal N-glycosylation of α1 anti-trypsin [26].
DAD1 was initially identified as a negative regulator of apoptosis in temperature-sensitive baby hamster kidney 21 (BHK21) mutant cells [47] and later as an essential OST subunit [48,67]. DAD1 forms heterotetrameric OST subcomplexes with RPN1, RPN2, and OST48/DDOST [67]. Moreover, in BHK21 mutant cells, DAD1 is rapidly degraded at non-permissive temperatures, which decreases the protein stability of RPN1, RPN2, and OST48/DDOST [48]. These findings indicate that DAD1 has a role in maintaining the structural integrity of the OST complex.
OST48/DDOST was first identified as a mammalian ortholog of yeast Wbp1, an essential OST subunit in this organism, and was later found to be associated with the mammalian OST complex [24]. A short cytosolic tail of OST48/DDOST has a functional ER-retention di-lysine motif, serving as a mechanism for retaining OST in the ER [68]. As with the case of DAD1, OST48/DDOST is required for the stability of both STT3A-OST and STT3B-OST [49]. Patients with DDOST-CDG have been identified from untyped CDGs by a combination of whole genome sequencing and biochemical studies [50].
OST4 is a very small, 37 amino acid protein in humans and was identified by its sequence similarity to the yeast ortholog [2]. Although OST4 is associated with both STT3A-OST and STT3B-OST, the knockdown of OST4 destabilizes only STT3A-OST [27]. This is functionally relevant, as the knockdown of OST4 or STT3A, but not STT3B, shows a similar hypoglycosylation phenotype to prosaposin [27], a typical STT3A-dependent substrate [23].
4.2. Subunits Specific for STT3A-OST and STT3B-OST
DC2/OSTC and KCP2 are STT3A-OST-specific subunits and are found only in vertebrates [35]. These subunits are not required for the enzymatic activity of STT3A-OST, but are required for the association with the Sec61 channel and, therefore, co-translational N-glycosylation [28,35]. The cryo-EM map of the mammalian ribosome translocon complex located DC2 at the interface between STT3A and Sec61, physically bridging the protein-conducting channel and glycosylation machinery [35]. The mechanism by which STT3B-OST excludes these adaptor proteins is currently unknown, largely due to unavailability of the structural information of STT3B-OST.
STT3B-OST contains either one TUSC3 or one MAGT1 (formerly N33 and IAP [24]) that have a large luminal thioredoxin domain required for N-glycosylation at sites near cysteine residues [33]. Patients with a deficiency in the MAGT1 and TUSC3 genes have been reported to show distinct symptoms. Mutations in the MAGT1 gene are associated with an X-linked immunodeficiency [65] and causes apparent N-glycosylation defects in immunity-related proteins [66]. In contrast, a TUSC3 deficiency has been associated with autosomal recessive mental retardation with no obvious hypoglycosylation of serum transferrin [51]. Apart from their functions in N-glycosylation, the TUSC3 and MAGT1 genes were also identified in a screening of genes that can complement growth defects of yeast mutant cells lacking the plasma membrane Mg2+ transporter Alr1 [52]. The TUSC3 and MAGT1 genes are reportedly involved in the uptake of Mg2+ in mammalian cells, although their Mg2+ transport activity has not been demonstrated. The MAGT1 gene is ubiquitously expressed in various tissues and upregulated when extracellular Mg2+ levels are low [52]. On the other hand, the expression of TUSC3 gene is insensitive to extracellular Mg2+ levels and is limited to the ovary, placenta, prostate, testis, adipose tissue, and the lung [52]. The distinct expression patterns of the MAGT1 and TUSC3 genes may explain the differences in symptoms of the patients with these genes.
5. Roles of OST in Tumor Progression
The N-glycosylation reaction catalyzed by OST has been implicated in the mechanism of immune evasion where tumor cells survive in the tumor microenvironment [9]. The programmed death-ligand 1 (PD-L1) is an immune checkpoint molecule expressed on the surface of various tumor cells and binds to the programmed death-1 (PD-1) receptor on T cells to inactivate them [69]. PD-L1 is a single-pass transmembrane protein with four potential N-glycosylation sites—one (N35) being located at a distance of 17 amino acids from the predicted signal sequence cleavage site, while the other three (N192, N200 and N219) are located adjacent to the transmembrane domain. The addition of glycans to the latter three sites in the ER prevents PD-L1 phosphorylation by glycogen synthase kinase 3β, followed by ERAD, thereby ensuring the cell surface expression of PD-L1 [70].
Epithelial-to-mesenchymal transition (EMT) is a process in which tumor cells lose their contact and acquire migratory and invasive properties by altering protein expression profiles [71,72,73]. Accompanied by the large increase in glycoprotein production in the ER, EMT also programs cells to increase capacity of protein folding and ERAD, and upregulate the metabolic flux of the hexosamine biosynthetic pathway to produce UDP-GlcNAc as substrate for N-glycan biosynthesis [73]. Upon the induction of EMT by transforming growth factor β in breast cancer stem cells, the expression of PD-L1 is upregulated along with STT3A and STT3B through β-catenin signaling [74]. This cell context-dependent regulation of STT3 expression fulfills the N-glycosylation demands of increased PD-L1, thus ensuring the cell surface expression of PD-L1 [74]. As discussed above, however, STT3 proteins require other subunits to be functional, strongly suggesting the presence of mechanisms that control the expression levels of accessory subunits of OST on demand as well. EMT can also influence the expression of glycosyltransferases that are involved in the terminal modification of N-glycans [75], which include the formation of poly-N-acetyllactosamine (poly-LacNAc) repeats [76]. Interestingly, β1,3-GlcNAc transferase 3, an enzyme that initiates the formation of poly-LacNAc repeats on N-glycans of PD-L1, is essential for the functional binding of PD-L1 to PD-1 [77]. These findings suggest that EMT reprograms the N-glycosylation machinery of tumor cells toward the activation of PD-L1 to establish PD-L1-mediated immune escape.
6. Association of RPN2 and TUSC3 with Tumor Progression
RPN2 was identified as a gene that confers docetaxel resistance to breast cancers by regulating, as a part of OST, the N-glycosylation of the P-glycoprotein and CD63 [45,46]. The expression of RPN2 is clinically relevant, showing a positive correlation with the degree of aggressiveness of breast cancers in patients [78]. An aberrantly high expression of RPN2 is also associated with the progression of non-small cell lung [79], colorectal [80], gastric [81], and esophageal cancers [82]. RPN2 is also required for the N-glycosylation of the epidermal growth factor receptor (EGFR), aiding the cell surface expression of EGFR and downstream signaling [83]. Although the molecular mechanisms underlying the abnormal expression of RPN2 in various cancer cells are unknown, RPN2 has been shown to be a target gene of microRNA-128 in colorectal cancer cells [84]. It has been reported that the expression of this small non-coding RNA changes during postnatal development [85,86], raising the possibility that RPN2 expression is cell-context-dependent and regulated dynamically during development and oncogenesis.
TUSC3 has been identified as a candidate tumor suppressor [53]. The TUSC3 gene is located in a region of chromosome 8p22, which is frequently deleted in various epithelial cancers, including prostate, colon, lung, and liver cancers [54,55]. Moreover, the expression of the TUSC3 gene can also be downregulated by promoter methylation in the colon, lung, and ovarian cancers [56,57,58]. Suppressing TUSC3 expression has a large impact on the morphology and homeostasis of the ER [59,60] and promotes EMT and tumor growth [61,62,63]. However, target glycoproteins that are affected by the loss of TUSC3 and are responsible for tumor progression have not been identified. It should be noted that the overexpression of TUSC3 has also been observed in clinical samples of non-small cell lung cancers (NSCLC) compared to benign controls [61,64]. The contradicting expression profiles of TUSC3 in tumors may reflect the complexity of its gene regulation mechanisms. These collective findings suggest that altered TUSC3 expression has an impact on the glycosylation of a subset of STT3B-dependent substrates, leading to the acquisition of aggressive phenotypes within the established tumors.
7. OST as a Potential Druggable Target for Cancer Treatment
Although N-glycosylation represents an attractive target for cancer therapy, drugs that are capable of targeting this pathway have not yet been identified. However, the recent development of a novel class of N-glycosylation inhibitors has provided insights into the clinical relevance of OST in cancer treatment. One inhibitor in this class, termed the N-glycosylation inhibitor 1 (NGI-1), was found in the cell-based high-throughput screening of small compounds that can deactivate an ER-targeted, inactivated form of a luciferase mutant (ERLucT) by inhibiting its N-glycosylation [43]. NGI-1 has the potency to inhibit both OST complexes with a preference for STT3B-OST over STT3A-OST [43,87]. Interestingly, the chemical derivatization of NGI-1 increases the selectivity of the analogs to STT3B-OST [87], raising the possibility of structure-based drug design with the objective of separately modulating the functions of individual OST complexes.
It has been shown that NGI-1 inhibits the proliferation of a subset of non-small cell lung cancer (NSCLC) cells that depend on receptor tyrosine kinases (RTKs) for proliferation, such as EGFR and fibroblast growth factor receptor [87]. The N-glycosylation of human EGFR at Asn420 is known to be a negative regulator of ligand-independent oligomerization and the tyrosine phosphorylation of the receptor [88]. However, blocking N-glycosylation with NGI-1 results in the internalization of EGFR and the disruption of EGFR signaling, ultimately inducing cellular senescence, but not apoptosis [43].
EGFR tyrosine kinase inhibitors (TKIs) have been clinically used to treat NSCLC cells harboring EGFR-activating kinase domain mutations [89]. However, drug resistance develops by acquiring secondary and tertiary mutations in the kinase domain [90,91,92] or the amplification of the hepatocyte growth factor receptor MET [93,94,95], leading to tumor progression. N-glycosylation blockade with NGI-1 is able not only to re-sensitize TKI-resistant NSCLC cells to TKIs, but also to result in the dissociation of EGFR from amplified MET signaling by inducing the internalization of EGFR [96]. Thus, the unique action of NGI-1 may offer therapeutic advantages in the treatment of drug-resistant tumors.
8. Concluding Remarks
Genetic and biochemical evidence has demonstrated that N-glycosylation in the ER is essential for homeostasis at the systemic and cellular levels, leading to the assumption that the attachment of N-glycans to proteins takes place constitutively and optimally without strict regulation. However, it has become evident that the expression of OST subunits is cell context-dependent and the regulation involves genetic and epigenetic mechanisms. The on-demand-type regulation of OST expression significantly contributes to tumor progression and possibly to physiological processes where cells need to dramatically change their characteristics.
A novel OST inhibitor has opened up an avenue for fighting incurable drug-resistant tumors. Importantly, the action of NGI-1 is distinct from any other existing anti-cancer drug. However, the efficacy and target molecules in each tumor type remain to be defined if undesired side effects are to be prevented and the effects of OST blockade on possible tumor treatment is to be maximized. In this regard, the toxicity associated with the inhibition of OST should be considered carefully, as OST inhibition affects global N-glycosylation, which may generate undesired proteins, result in detrimental effects to healthy cells, and complicate cancers [97]. Thus, efforts should be made to understand the inhibitory mechanisms of NGI-1, as well as the precise functions of accessory subunits of OST in N-glycosylation and tumor progression, which will accelerate structure-based drug design and discovery.
In summary, OST is now considered to be a gatekeeper of health and tumor progression. Further studies will be necessary to explore molecular links that connect OST to physiology and pathology to expand our knowledge on the functions of this mysterious enzyme.
Acknowledgments
We wish to thank our laboratory members for fruitful discussions.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Apweiler R., Hermjakob H., Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the swiss-prot database. Biochim. Biophys. Acta. 1999;1473:4–8. doi: 10.1016/S0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 2.Kelleher D.J., Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- 3.Harada Y., Hirayama H., Suzuki T. Generation and degradation of free asparagine-linked glycans. Cell Mol. Life Sci. 2015;72:2509–2533. doi: 10.1007/s00018-015-1881-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley P., Taniguchi N., Aebi M. N-glycans. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., et al., editors. Essentials of Glycobiology. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2015. pp. 99–111. [Google Scholar]
- 5.Ohtsubo K., Marth J.D. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Pinho S.S., Reis C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi N., Kizuka Y. Glycans and cancer: Role of n-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv. Cancer Res. 2015;126:11–51. doi: 10.1016/bs.acr.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Chiaradonna F., Ricciardiello F., Palorini R. The nutrient-sensing hexosamine biosynthetic pathway as the hub of cancer metabolic rewiring. Cells. 2018;7:53. doi: 10.3390/cells7060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu J.M., Li C.W., Lai Y.J., Hung M.C. Posttranslational modifications of pd-l1 and their applications in cancer therapy. Cancer Res. 2018;78:6349–6353. doi: 10.1158/0008-5472.CAN-18-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsico G., Russo L., Quondamatteo F., Pandit A. Glycosylation and integrin regulation in cancer. Trends Cancer. 2018;4:537–552. doi: 10.1016/j.trecan.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Pang X., Li H., Guan F., Li X. Multiple roles of glycans in hematological malignancies. Front. Oncol. 2018;8:364. doi: 10.3389/fonc.2018.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.RodrIguez E., Schetters S.T.T., van Kooyk Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 2018;18:204–211. doi: 10.1038/nri.2018.3. [DOI] [PubMed] [Google Scholar]
- 13.Chandler K.B., Costello C.E., Rahimi N. Glycosylation in the tumor microenvironment: Implications for tumor angiogenesis and metastasis. Cells. 2019;8:544. doi: 10.3390/cells8060544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mereiter S., Balmana M., Campos D., Gomes J., Reis C.A. Glycosylation in the era of cancer-targeted therapy: Where are we heading? Cancer Cell. 2019;36:6–16. doi: 10.1016/j.ccell.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Peixoto A., Relvas-Santos M., Azevedo R., Santos L.L., Ferreira J.A. Protein glycosylation and tumor microenvironment alterations driving cancer hallmarks. Front. Oncol. 2019;9:380. doi: 10.3389/fonc.2019.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reily C., Stewart T.J., Renfrow M.B., Novak J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019;15:346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrimal S., Cherepanova N.A., Gilmore R. Cotranslational and posttranslocational n-glycosylation of proteins in the endoplasmic reticulum. Semin. Cell Dev. Biol. 2015;41:71–78. doi: 10.1016/j.semcdb.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aebi M. N-linked protein glycosylation in the er. Biochim. Biophys. Acta. 2013;1833:2430–2437. doi: 10.1016/j.bbamcr.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Harada Y. Biosynthesis and degradation of dolichol-linked oligosaccharides. Trends Glycosci. Glycotechnol. 2016;28:E91–E96. doi: 10.4052/tigg.1512.1E. [DOI] [Google Scholar]
- 20.Dell A., Galadari A., Sastre F., Hitchen P. Similarities and differences in the glycosylation mechanisms in prokaryotes and eukaryotes. Int. J. Microbiol. 2010;2010:148178. doi: 10.1155/2010/148178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larkin A., Imperiali B. The expanding horizons of asparagine-linked glycosylation. Biochemistry. 2011;50:4411–4426. doi: 10.1021/bi200346n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magidovich H., Eichler J. Glycosyltransferases and oligosaccharyltransferases in archaea: Putative components of the n-glycosylation pathway in the third domain of life. FEMS Microbiol. Lett. 2009;300:122–130. doi: 10.1111/j.1574-6968.2009.01775.x. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz-Canada C., Kelleher D.J., Gilmore R. Cotranslational and posttranslational n-glycosylation of polypeptides by distinct mammalian ost isoforms. Cell. 2009;136:272–283. doi: 10.1016/j.cell.2008.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelleher D.J., Karaoglu D., Mandon E.C., Gilmore R. Oligosaccharyltransferase isoforms that contain different catalytic stt3 subunits have distinct enzymatic properties. Mol. Cell. 2003;12:101–111. doi: 10.1016/S1097-2765(03)00243-0. [DOI] [PubMed] [Google Scholar]
- 25.Shibatani T., David L.L., McCormack A.L., Frueh K., Skach W.R. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain sec61, trap, and two potential new subunits. Biochemistry. 2005;44:5982–5992. doi: 10.1021/bi047328f. [DOI] [PubMed] [Google Scholar]
- 26.Graham D.B., Lefkovith A., Deelen P., de Klein N., Varma M., Boroughs A., Desch A.N., Ng A.C.Y., Guzman G., Schenone M., et al. Tmem258 is a component of the oligosaccharyltransferase complex controlling er stress and intestinal inflammation. Cell Rep. 2016;17:2955–2965. doi: 10.1016/j.celrep.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumax-Vorzet A., Roboti P., High S. Ost4 is a subunit of the mammalian oligosaccharyltransferase required for efficient n-glycosylation. J. Cell Sci. 2013;126:2595–2606. doi: 10.1242/jcs.115410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shrimal S., Cherepanova N.A., Gilmore R. Dc2 and kcp2 mediate the interaction between the oligosaccharyltransferase and the er translocon. J. Cell Biol. 2017;216:3625–3638. doi: 10.1083/jcb.201702159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherepanova N.A., Gilmore R. Mammalian cells lacking either the cotranslational or posttranslocational oligosaccharyltransferase complex display substrate-dependent defects in asparagine linked glycosylation. Sci. Rep. 2016;6:20946. doi: 10.1038/srep20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bas T., Gao G.Y., Lvov A., Chandrasekhar K.D., Gilmore R., Kobertz W.R. Post-translational n-glycosylation of type i transmembrane kcne1 peptides: Implications for membrane protein biogenesis and disease. J. Biol. Chem. 2011;286:28150–28159. doi: 10.1074/jbc.M111.235168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrimal S., Gilmore R. Glycosylation of closely spaced acceptor sites in human glycoproteins. J. Cell Sci. 2013;126:5513–5523. doi: 10.1242/jcs.139584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrimal S., Trueman S.F., Gilmore R. Extreme c-terminal sites are posttranslocationally glycosylated by the stt3b isoform of the ost. J. Cell Biol. 2013;201:81–95. doi: 10.1083/jcb.201301031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherepanova N.A., Shrimal S., Gilmore R. Oxidoreductase activity is necessary for n-glycosylation of cysteine-proximal acceptor sites in glycoproteins. J. Cell Biol. 2014;206:525–539. doi: 10.1083/jcb.201404083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Napiorkowska M., Boilevin J., Sovdat T., Darbre T., Reymond J.L., Aebi M., Locher K.P. Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase. Nat. Struct. Mol. Biol. 2017;24:1100–1106. doi: 10.1038/nsmb.3491. [DOI] [PubMed] [Google Scholar]
- 35.Braunger K., Pfeffer S., Shrimal S., Gilmore R., Berninghausen O., Mandon E.C., Becker T., Forster F., Beckmann R. Structural basis for coupling protein transport and n-glycosylation at the mammalian endoplasmic reticulum. Science. 2018;360:215–219. doi: 10.1126/science.aar7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilsson I.M., von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- 37.Harada Y., Li H., Li H., Lennarz W.J. Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. Proc. Natl. Acad. Sci. USA. 2009;106:6945–6949. doi: 10.1073/pnas.0812489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato T., Sako Y., Sho M., Momohara M., Suico M.A., Shuto T., Nishitoh H., Okiyoneda T., Kokame K., Kaneko M., et al. Stt3b-dependent posttranslational n-glycosylation as a surveillance system for secretory protein. Mol. Cell. 2012;47:99–110. doi: 10.1016/j.molcel.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 39.Wei W., Zheng C., Zhu M., Zhu X., Yang R., Misra S., Zhang B. Missense mutations near the n-glycosylation site of the a2 domain lead to various intracellular trafficking defects in coagulation factor viii. Sci. Rep. 2017;7:45033. doi: 10.1038/srep45033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei W., Misra S., Cannon M.V., Yang R., Zhu X., Gilmore R., Zhu M., Zhang B. Molecular mechanisms of missense mutations that generate ectopic n-glycosylation sites in coagulation factor viii. Biochem. J. 2018;475:873–886. doi: 10.1042/BCJ20170884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan Y., Hu Y., Yan C., Goldman R., Pan Y., Mazumder R., Dingerdissen H.M. Loss and gain of n-linked glycosylation sequons due to single-nucleotide variation in cancer. Sci. Rep. 2018;8:4322. doi: 10.1038/s41598-018-22345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shrimal S., Ng B.G., Losfeld M.E., Gilmore R., Freeze H.H. Mutations in stt3a and stt3b cause two congenital disorders of glycosylation. Hum. Mol. Genet. 2013;22:4638–4645. doi: 10.1093/hmg/ddt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez-Sambrooks C., Shrimal S., Khodier C., Flaherty D.P., Rinis N., Charest J.C., Gao N., Zhao P., Wells L., Lewis T.A., et al. Oligosaccharyltransferase inhibition induces senescence in rtk-driven tumor cells. Nat. Chem. Biol. 2016;12:1023–1030. doi: 10.1038/nchembio.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreibich G., Czako-Graham M., Grebenau R., Mok W., Rodriguez-Boulan E., Sabatini D.D. Characterization of the ribosomal binding site in rat liver rough microsomes: Ribophorins i and ii, two integral membrane proteins related to ribosome binding. J. Supramol. Struct. 1978;8:279–302. doi: 10.1002/jss.400080307. [DOI] [PubMed] [Google Scholar]
- 45.Honma K., Iwao-Koizumi K., Takeshita F., Yamamoto Y., Yoshida T., Nishio K., Nagahara S., Kato K., Ochiya T. Rpn2 gene confers docetaxel resistance in breast cancer. Nat. Med. 2008;14:939–948. doi: 10.1038/nm.1858. [DOI] [PubMed] [Google Scholar]
- 46.Tominaga N., Hagiwara K., Kosaka N., Honma K., Nakagama H., Ochiya T. Rpn2-mediated glycosylation of tetraspanin cd63 regulates breast cancer cell malignancy. Mol. Cancer. 2014;13:134. doi: 10.1186/1476-4598-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakashima T., Sekiguchi T., Kuraoka A., Fukushima K., Shibata Y., Komiyama S., Nishimoto T. Molecular cloning of a human cdna encoding a novel protein, dad1, whose defect causes apoptotic cell death in hamster bhk21 cells. Mol. Cell Biol. 1993;13:6367–6374. doi: 10.1128/MCB.13.10.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanjay A., Fu J., Kreibich G. Dad1 is required for the function and the structural integrity of the oligosaccharyltransferase complex. J. Biol. Chem. 1998;273:26094–26099. doi: 10.1074/jbc.273.40.26094. [DOI] [PubMed] [Google Scholar]
- 49.Roboti P., High S. The oligosaccharyltransferase subunits ost48, dad1 and kcp2 function as ubiquitous and selective modulators of mammalian n-glycosylation. J. Cell Sci. 2012;125:3474–3484. doi: 10.1242/jcs.103952. [DOI] [PubMed] [Google Scholar]
- 50.Jones M.A., Ng B.G., Bhide S., Chin E., Rhodenizer D., He P., Losfeld M.E., He M., Raymond K., Berry G., et al. Ddost mutations identified by whole-exome sequencing are implicated in congenital disorders of glycosylation. Am. J. Hum. Genet. 2012;90:363–368. doi: 10.1016/j.ajhg.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garshasbi M., Hadavi V., Habibi H., Kahrizi K., Kariminejad R., Behjati F., Tzschach A., Najmabadi H., Ropers H.H., Kuss A.W. A defect in the tusc3 gene is associated with autosomal recessive mental retardation. Am. J. Hum. Genet. 2008;82:1158–1164. doi: 10.1016/j.ajhg.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H., Clapham D.E. Mammalian magt1 and tusc3 are required for cellular magnesium uptake and vertebrate embryonic development. Proc. Natl. Acad. Sci. USA. 2009;106:15750–15755. doi: 10.1073/pnas.0908332106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasickova K., Horak P., Vanhara P. Tusc3: Functional duality of a cancer gene. Cell Mol. Life Sci. 2018;75:849–857. doi: 10.1007/s00018-017-2660-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacGrogan D., Levy A., Bova G.S., Isaacs W.B., Bookstein R. Structure and methylation-associated silencing of a gene within a homozygously deleted region of human chromosome band 8p22. Genomics. 1996;35:55–65. doi: 10.1006/geno.1996.0322. [DOI] [PubMed] [Google Scholar]
- 55.Bova G.S., MacGrogan D., Levy A., Pin S.S., Bookstein R., Isaacs W.B. Physical mapping of chromosome 8p22 markers and their homozygous deletion in a metastatic prostate cancer. Genomics. 1996;35:46–54. doi: 10.1006/geno.1996.0321. [DOI] [PubMed] [Google Scholar]
- 56.Pils D., Horak P., Vanhara P., Anees M., Petz M., Alfanz A., Gugerell A., Wittinger M., Gleiss A., Auner V., et al. Methylation status of tusc3 is a prognostic factor in ovarian cancer. Cancer. 2013;119:946–954. doi: 10.1002/cncr.27850. [DOI] [PubMed] [Google Scholar]
- 57.Duppel U., Woenckhaus M., Schulz C., Merk J., Dietmaier W. Quantitative detection of tusc3 promoter methylation—A potential biomarker for prognosis in lung cancer. Oncol. Lett. 2016;12:3004–3012. doi: 10.3892/ol.2016.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taniue K., Hayashi T., Kamoshida Y., Kurimoto A., Takeda Y., Negishi L., Iwasaki K., Kawamura Y., Goshima N., Akiyama T. Uhrf1-kat7-mediated regulation of tusc3 expression via histone methylation/acetylation is critical for the proliferation of colon cancer cells. Oncogene. 2019 doi: 10.1038/s41388-019-1032-y. [DOI] [PubMed] [Google Scholar]
- 59.Horak P., Tomasich E., Vanhara P., Kratochvilova K., Anees M., Marhold M., Lemberger C.E., Gerschpacher M., Horvat R., Sibilia M., et al. Tusc3 loss alters the er stress response and accelerates prostate cancer growth in vivo. Sci. Rep. 2014;4:3739. doi: 10.1038/srep03739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeon Y.J., Kim T., Park D., Nuovo G.J., Rhee S., Joshi P., Lee B.K., Jeong J., Suh S.S., Grotzke J.E., et al. Mirna-mediated tusc3 deficiency enhances upr and erad to promote metastatic potential of nsclc. Nat. Commun. 2018;9:5110. doi: 10.1038/s41467-018-07561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng S., Zhai J., Lu D., Lin J., Dong X., Liu X., Wu H., Roden A.C., Brandi G., Tavolari S., et al. Tusc3 accelerates cancer growth and induces epithelial-mesenchymal transition by upregulating claudin-1 in non-small-cell lung cancer cells. Exp. Cell Res. 2018;373:44–56. doi: 10.1016/j.yexcr.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 62.Kratochvilova K., Horak P., Esner M., Soucek K., Pils D., Anees M., Tomasich E., Drafi F., Jurtikova V., Hampl A., et al. Tumor suppressor candidate 3 (tusc3) prevents the epithelial-to-mesenchymal transition and inhibits tumor growth by modulating the endoplasmic reticulum stress response in ovarian cancer cells. Int. J. Cancer. 2015;137:1330–1340. doi: 10.1002/ijc.29502. [DOI] [PubMed] [Google Scholar]
- 63.Gu Y., Wang Q., Guo K., Qin W., Liao W., Wang S., Ding Y., Lin J. Tusc3 promotes colorectal cancer progression and epithelial-mesenchymal transition (emt) through wnt/beta-catenin and mapk signalling. J. Pathol. 2016;239:60–71. doi: 10.1002/path.4697. [DOI] [PubMed] [Google Scholar]
- 64.Gu Y., Pei X., Ren Y., Cai K., Guo K., Chen J., Qin W., Lin M., Wang Q., Tang N., et al. Oncogenic function of tusc3 in non-small cell lung cancer is associated with hedgehog signalling pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:1749–1760. doi: 10.1016/j.bbadis.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 65.Li F.Y., Chaigne-Delalande B., Kanellopoulou C., Davis J.C., Matthews H.F., Douek D.C., Cohen J.I., Uzel G., Su H.C., Lenardo M.J. Second messenger role for Mg2+ revealed by human t-cell immunodeficiency. Nature. 2011;475:471–476. doi: 10.1038/nature10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuda-Lennikov M., Biancalana M., Zou J., Ravell J.C., Zheng L., Kanellopoulou C., Jiang P., Notarangelo G., Jing H., Masutani E., et al. Magnesium transporter 1 (magt1) deficiency causes selective defects in n-linked glycosylation and expression of immune-response genes. J. Biol. Chem. 2019;294:13638–13656. doi: 10.1074/jbc.RA119.008903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kelleher D.J., Gilmore R. Dad1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase. Proc. Natl. Acad. Sci. USA. 1997;94:4994–4999. doi: 10.1073/pnas.94.10.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu J., Kreibich G. Retention of subunits of the oligosaccharyltransferase complex in the endoplasmic reticulum. J. Biol. Chem. 2000;275:3984–3990. doi: 10.1074/jbc.275.6.3984. [DOI] [PubMed] [Google Scholar]
- 69.Mahoney K.M., Freeman G.J., McDermott D.F. The next immune-checkpoint inhibitors: Pd-1/pd-l1 blockade in melanoma. Clin. Ther. 2015;37:764–782. doi: 10.1016/j.clinthera.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li C.W., Lim S.O., Xia W., Lee H.H., Chan L.C., Kuo C.W., Khoo K.H., Chang S.S., Cha J.H., Kim T., et al. Glycosylation and stabilization of programmed death ligand-1 suppresses t-cell activity. Nat. Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 72.Pang X.X., Bai Q., Wu F., Chen G.J., Zhang A.H., Tang C.S. Urotensin ii induces er stress and emt and increase extracellular matrix production in renal tubular epithelial cell in early diabetic mice. Kidney Blood Press Res. 2016;41:434–449. doi: 10.1159/000443445. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J., Jamaluddin M., Zhang Y., Widen S.G., Sun H., Brasier A.R., Zhao Y. Type ii epithelial-mesenchymal transition upregulates protein n-glycosylation to maintain proteostasis and extracellular matrix production. J. Proteome Res. 2019;18:3447–3460. doi: 10.1021/acs.jproteome.9b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsu J.M., Xia W., Hsu Y.H., Chan L.C., Yu W.H., Cha J.H., Chen C.T., Liao H.W., Kuo C.W., Khoo K.H., et al. Stt3-dependent pd-l1 accumulation on cancer stem cells promotes immune evasion. Nat. Commun. 2018;9:1908. doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X., Wang X., Tan Z., Chen S., Guan F. Role of glycans in cancer cells undergoing epithelial-mesenchymal transition. Front. Oncol. 2016;6:33. doi: 10.3389/fonc.2016.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lucena M.C., Carvalho-Cruz P., Donadio J.L., Oliveira I.A., de Queiroz R.M., Marinho-Carvalho M.M., Sola-Penna M., de Paula I.F., Gondim K.C., McComb M.E., et al. Epithelial mesenchymal transition induces aberrant glycosylation through hexosamine biosynthetic pathway activation. J. Biol. Chem. 2016;291:12917–12929. doi: 10.1074/jbc.M116.729236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li C.W., Lim S.O., Chung E.M., Kim Y.S., Park A.H., Yao J., Cha J.H., Xia W., Chan L.C., Kim T., et al. Eradication of triple-negative breast cancer cells by targeting glycosylated pd-l1. Cancer Cell. 2018;33:187–201.e110. doi: 10.1016/j.ccell.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ono M., Tsuda H., Kobayashi T., Takeshita F., Takahashi R.U., Tamura K., Akashi-Tanaka S., Moriya T., Yamasaki T., Kinoshita T., et al. The expression and clinical significance of ribophorin ii (rpn2) in human breast cancer. Pathol. Int. 2015;65:301–308. doi: 10.1111/pin.12297. [DOI] [PubMed] [Google Scholar]
- 79.Fujita Y., Yagishita S., Takeshita F., Yamamoto Y., Kuwano K., Ochiya T. Prognostic and therapeutic impact of rpn2-mediated tumor malignancy in non-small-cell lung cancer. Oncotarget. 2015;6:3335–3345. doi: 10.18632/oncotarget.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bi C., Jiang B. Downregulation of rpn2 induces apoptosis and inhibits migration and invasion in colon carcinoma. Oncol. Rep. 2018;40:283–293. doi: 10.3892/or.2018.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fujimoto D., Goi T., Koneri K., Hirono Y. Rpn2 is effective biomarker to predict the outcome of combined chemotherapy docetaxel and cisplatin for advanced gastric cancer. Oncotarget. 2018;9:15208–15218. doi: 10.18632/oncotarget.24622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y., Huang C., Bai Q., Yu J. Ribophorin ii promotes cell proliferation, migration, and invasion in esophageal cancer cells in vitro and in vivo. Biosci. Rep. 2019;39:BSR20182448. doi: 10.1042/BSR20182448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H., Al-Japairai K., Tao Y., Xiang Z. Rpn2 promotes colorectal cancer cell proliferation through modulating the glycosylation status of egfr. Oncotarget. 2017;8:72633–72651. doi: 10.18632/oncotarget.20005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou T., Wu L., Wang Q., Jiang Z., Li Y., Ma N., Chen W., Hou Z., Gan W., Chen S. Microrna-128 targeting rpn2 inhibits cell proliferation and migration through the akt-p53-cyclin pathway in colorectal cancer cells. Oncol. Lett. 2018;16:6940–6949. doi: 10.3892/ol.2018.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan C.L., Plotkin J.L., Veno M.T., von Schimmelmann M., Feinberg P., Mann S., Handler A., Kjems J., Surmeier D.J., O’Carroll D., et al. Microrna-128 governs neuronal excitability and motor behavior in mice. Science. 2013;342:1254–1258. doi: 10.1126/science.1244193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang W., Feng Y., Liang J., Yu H., Wang C., Wang B., Wang M., Jiang L., Meng W., Cai W., et al. Loss of microrna-128 promotes cardiomyocyte proliferation and heart regeneration. Nat. Commun. 2018;9:700. doi: 10.1038/s41467-018-03019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rinis N., Golden J.E., Marceau C.D., Carette J.E., Van Zandt M.C., Gilmore R., Contessa J.N. Editing n-glycan site occupancy with small-molecule oligosaccharyltransferase inhibitors. Cell Chem. Biol. 2018;25:1231–1241. doi: 10.1016/j.chembiol.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsuda T., Ikeda Y., Taniguchi N. The asn-420-linked sugar chain in human epidermal growth factor receptor suppresses ligand-independent spontaneous oligomerization. Possible role of a specific sugar chain in controllable receptor activation. J. Biol. Chem. 2000;275:21988–21994. doi: 10.1074/jbc.M003400200. [DOI] [PubMed] [Google Scholar]
- 89.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 90.Kwak E.L., Sordella R., Bell D.W., Godin-Heymann N., Okimoto R.A., Brannigan B.W., Harris P.L., Driscoll D.R., Fidias P., Lynch T.J., et al. Irreversible inhibitors of the egf receptor may circumvent acquired resistance to gefitinib. Proc. Natl. Acad. Sci. USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pao W., Miller V.A., Politi K.A., Riely G.J., Somwar R., Zakowski M.F., Kris M.G., Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the egfr kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thress K.S., Paweletz C.P., Felip E., Cho B.C., Stetson D., Dougherty B., Lai Z., Markovets A., Vivancos A., Kuang Y., et al. Acquired egfr c797s mutation mediates resistance to azd9291 in non-small cell lung cancer harboring egfr t790m. Nat. Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Engelman J.A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J.O., Lindeman N., Gale C.M., Zhao X., Christensen J., et al. Met amplification leads to gefitinib resistance in lung cancer by activating erbb3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 94.Turke A.B., Zejnullahu K., Wu Y.L., Song Y., Dias-Santagata D., Lifshits E., Toschi L., Rogers A., Mok T., Sequist L., et al. Preexistence and clonal selection of met amplification in egfr mutant nsclc. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bean J., Brennan C., Shih J.Y., Riely G., Viale A., Wang L., Chitale D., Motoi N., Szoke J., Broderick S., et al. Met amplification occurs with or without t790m mutations in egfr mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl. Acad. Sci. USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lopez Sambrooks C., Baro M., Quijano A., Narayan A., Cui W., Greninger P., Egan R., Patel A., Benes C.H., Saltzman W.M., et al. Oligosaccharyltransferase inhibition overcomes therapeutic resistance to egfr tyrosine kinase inhibitors. Cancer Res. 2018;78:5094–5106. doi: 10.1158/0008-5472.CAN-18-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tax G., Lia A., Santino A., Roversi P. Modulation of erqc and erad: A broad-spectrum spanner in the works of cancer cells? J. Oncol. 2019;2019:8384913. doi: 10.1155/2019/8384913. [DOI] [PMC free article] [PubMed] [Google Scholar]