Abstract
The plant-specific transcription factor gene family, YABBY, belongs to the subfamily of zinc finger protein superfamily and plays an essential regulatory role in lateral organ development. In this study, nine YABBY genes were identified in the pineapple genome. Seven of them were located on seven different chromosomes and the remaining two were located on scaffold 1235. Through protein structure prediction and protein multiple sequence alignment, we found that AcYABBY3, AcYABBY5 and AcYABBY7 lack a C2 structure in their N-terminal C2C2 zinc finger protein structure. Analysis of the cis-acting element indicated that all the seven pineapple YABBY genes contain multiple MYB and MYC elements. Further, the expression patterns analysis using the RNA-seq data of different pineapple tissues indicated that different AcYABBYs are preferentially expressed in various tissues. RT-qPCR showed that the expression of AcYABBY2, AcYABBY3, AcYABBY6 and AcYABBY7 were highly sensitive to abiotic stresses. Subcellular localization in pineapple protoplasts, tobacco leaves and Arabidopsis roots showed that all the seven pineapple YABBY proteins were nucleus localized. Overexpression of AcYABBY4 in Arabidopsis resulted in short root under NaCl treatment, indicating a negative regulatory role of AcYABBY4 in plant resistance to salt stress. This study provides valuable information for the classification of pineapple AcYABBY genes and established a basis for further research on the functions of AcYABBY proteins in plant development and environmental stress response.
Keywords: YABBY, pineapple, expression pattern, subcellular localization, abiotic stress
1. Introduction
Plants are often exposed to extreme environments during their development and growth. Abiotic stresses such as salt, drought, high temperature and cold lead to adverse effect on growth and development of plants, resulting in yield loss. In plants, a series of defense systems play a vital role in survival under extreme external environmental changes. Transcription factors play a crucial role in plant defense system regulating gene expression, some of which are associated with the abiotic stress response [1].
A number of transcription factors are plant-specific. Structurally, transcription factors are divided into four functional regions, namely DNA binding domain, oligomeric site, transcriptional regulatory domain and nuclear localization signal region. These functional domains determine the characteristics, function, regulation and nuclear localization of transcription factors. The YABBY transcription factor family is widely present in plants and is a subfamily of the zinc finger protein superfamily. YABBY transcription factor possess two conserved domains, the N-terminal zinc finger domain and the C-terminal YABBY domain. The amino acid residues in these two domains are highly conserved and these domains are involved in the specific binding of DNA [2].
The evolutionary history of YABBY gene family is consistent with the origin of the leaves of seed plants. These transcription factors are specific to seed plants [3] and play important regulatory roles in the development of plants lateral organ. There are five YABBY subfamilies in angiosperms, namely INO, CRC, YABBY2, FIL/YABBY3 and YABBY5 [4]. The YABBY genes are well studied in Arabidopsis thaliana, where the six members of YABBY genes revealed overlapping functions [2,5]. All AtYABBYs promote the differentiation of the abaxial surface cells of lateral organs and participate in the establishment of dorsal-ventral polarity, leaf expansion and flower organ development [6,7]. Although they display similar functions, yet different AtYABBY genes have diversified expression pattern and function. For example, AtYABBY2, AtYABBY3 and AtFIL genes are specifically expressed in the distal region of the aerial part of plants and Arabidopsis overexpression of AtYABBY3 and AtFIL exhibited leaf-rolling [8,9]. AtYABBY2, AtYABBY3, AtFIL and AtYABBY5 show functional redundancy during leaf development [10]. While AtCRC regulates the development of carpel and nectary [3,10]. AtINO mainly regulates the development of ovules [11]. In rice, DL and OsYABBY1, the homologs of AtYAB2 and AtCRC are not expressed in a polar manner in the lateral organs and their functions are not associated with polarity regulation of lateral organ development [12,13]. OsDL mainly regulates the carpel identity and the formation of main veins of the leaves by promoting cell proliferation in the central region of rice leaves [14,15] whereas, OsYABBY1 was found to be involved in the feedback regulation of gibberellin (GA) biosynthesis and metabolism resulting semi-dwarf phenotype in overexpression lines. [16,17]. Although OsYABBY3 is involved in leaf development, unlike its function in Arabidopsis, it does not affect the establishment of leaf polarity [18]. Other than above developmental processes, the YABBY family Shattering1 gene is responsible for seed shattering in cereals including sorghum, rice and maize [19].
Pineapple (Ananas comosus L.), a tropical edible fruit, is a perennial monocotyledon belonging to the Bromeliaceae. The completed assembly of the whole genome of pineapple [20] provides an opportunity for the systematic study of the pineapple YABBY family. Here, we identified 9 YABBY genes in pineapple and analyzed the gene structure, motif pattern of AcYABBYs, phylogenetic relationship of YABBYs between pineapple, Arabidopsis and rice. We found 7 AcYABBY genes located on seven different chromosomes. Through protein structure prediction and protein multiple sequence alignment analysis, we discovered that AcYABBY3, AcYABBY5 and AcYABBY7 lack a C2 at the N-terminus and thus could not constitute a C2C2 zinc finger domain. We further performed subcellular localization analysis for the seven YABBY proteins in pineapple protoplasts, tobacco leaves and Arabidopsis roots and found that the AcYABBY proteins were mainly localized in the nucleus. Moreover, we also analyzed the promoter cis-acting element and found that all the seven AcYABBYs have MYB and MYC domains, those are involved in drought, low temperature, salt and ABA stress responses. We further performed RT-qPCR analysis and showed that the expression levels of the seven AcYABBYs were changed under different abiotic stresses, including salt, drought, cold, hot, ABA and ethephon stresses at different time points. Finally, we found that over-expression of AcYABBY4 in Arabidopsis resulted in increased susceptibility to salt stress. This study provides comprehensive information about pineapple AcYABBY proteins and a basis for studying the function of AcYABBY family members in plant development and response to environmental stresses.
2. Results
2.1. Identification and Characterization of the Pineapple YABBY Transcription Factors
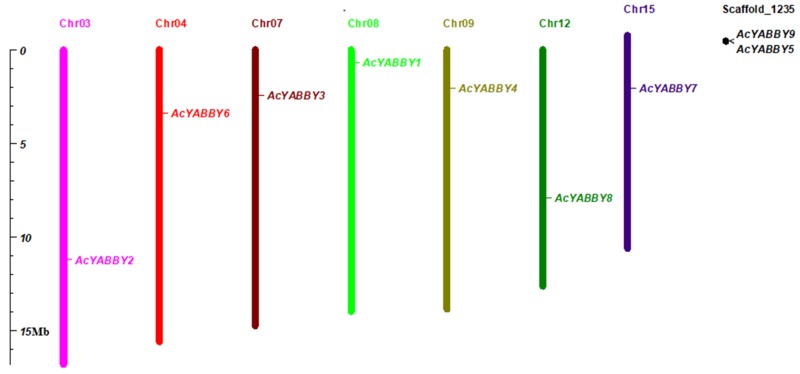
To identify AcYABBY genes, BLAST and Hidden Markov Model searches were used to search the pineapple genome with YABBY sequences from Arabidopsis as query. A total of 9 AcYABBY genes were identified from pineapple genome and named AcYABBY1 to AcYABBY9. The protein lengths of these genes ranged from 49 aa (AcYABBY8) to 226 aa (AcYABBY2) with the corresponding molecular weight ranging from 5383.98 to 24706.13 Da. The additional information about AcYABBYs transcript ID, gene name, proteins size, protein isoelectric point and exons are listed in Table 1. There are two genes, AcYABBY8 and AcYABBY9, with incomplete N-terminal sequence. We found that the CDSs of AcYABBY8 and AcYABBY9 lack ATG, which codes the initiation codon, suggesting we did not obtain the full length genes. Unfortunately, the conserved YABBY domain was located at the N terminal of YABBY proteins according to the reported studies and our conversation assay. To obtain more exact results, we considered the remaining seven genes for further studies. According to the mapping results, seven AcYABBY genes were localized on seven different chromosomes and two genes were located on scaffold1235 (Figure 1).
Table 1.
Protein information of pineapple YABBYs, including sequenced ID, chromosome locations, isoelectric point (pI), molecular weight (MW), protein length, CDS length and exon number.
| Name | Gene Locus | Chromosome Location |
PI | MW (Da) |
Protein Length(aa) |
CDS Length(bp) |
Exon |
|---|---|---|---|---|---|---|---|
| AcYABBY1 | Aco007606 | 8 | 9.45 | 20,042.95 | 182 | 549 | 5 |
| AcYABBY2 | Aco016279 | 3 | 6.88 | 24,706.13 | 226 | 681 | 7 |
| AcYABBY3 | Aco005138 | 7 | 9.25 | 19,962.60 | 178 | 537 | 6 |
| AcYABBY4 | Aco008751 | 9 | 8.64 | 20,161.95 | 178 | 537 | 6 |
| AcYABBY5 | Aco028479 | scaffold1235 | 9.05 | 15,073.06 | 136 | 411 | 5 |
| AcYABBY6 | Aco002202 | 4 | 7.71 | 20,872.43 | 188 | 567 | 6 |
| AcYABBY7 | Aco003917 | 15 | 9.05 | 21,453.75 | 190 | 573 | 7 |
| AcYABBY8 | Aco026269 | 12 | 7.01 | 5383.98 | 49 | 150 | 2 |
| AcYABBY9 | Aco028478 | scaffold1235 | 5.82 | 5988.82 | 53 | 162 | 2 |
Figure 1.
Distribution of YABBY genes in pineapple genome. Pineapple chromosomes and scaffolds having YABBY genes are shown here. The length of bar represents the size of the chromosome.
2.2. Phylogenetic Analysis of YABBY Family Genes
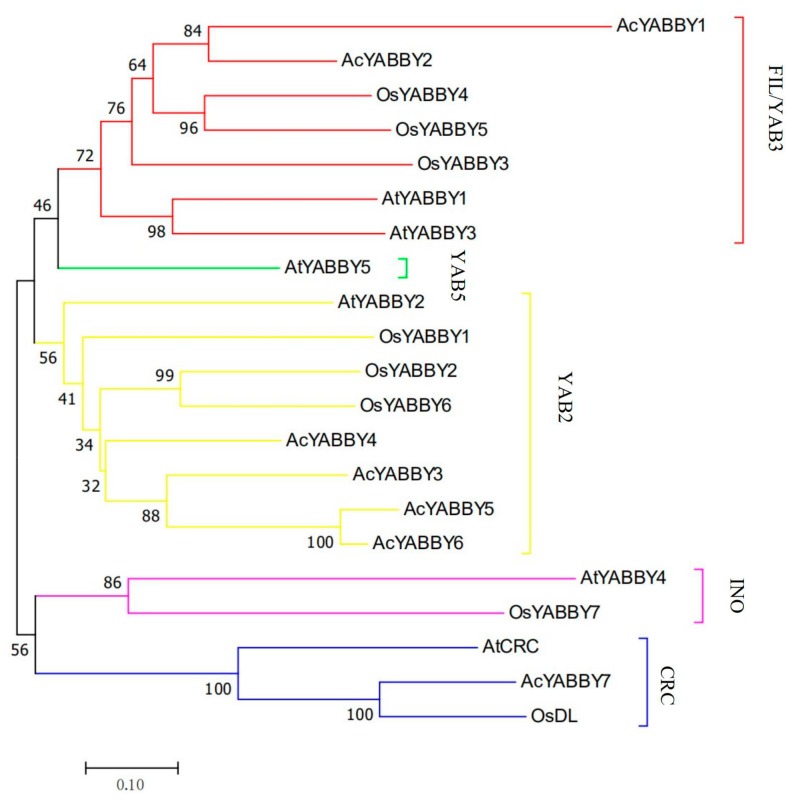
A phylogenetic tree was constructed to determine the phylogenetic relationships of YABBY genes in pineapple, Arabidopsis and rice. Phylogenetic analysis of 7 pineapple YABBY genes, 6 Arabidopsis YABBY genes and 8 rice YABBY genes was performed by generating a neighbor-joining phylogenetic tree (Figure 2). The result showed that the YABBY genes of these three species could be divided into five subfamilies—YAB2, CRC, YAB5, FIL/YAB3 and INO. However, the AcYABBYs were divided to three subfamilies—FIL/YAB3, CRC and YAB2 (Figure 3a). Among these subfamilies, YAB2 had the largest members with four pineapple genes, one Arabidopsis gene and three rice genes. FIL/YAB3 was the second largest subgroup, containing two pineapple genes, two Arabidopsis genes and three rice genes. CRC contained three genes, including one pineapple gene (AcYABBY7), one Arabidopsis thaliana gene (AtCRC) and one rice gene (OsDL). However, INO contained one Arabidopsis gene (AtYABBY4) and one rice gene (OsYABBY7), yet no pineapple gene was categorized into this group. The smallest subgroup, YAB5, only had one Arabidopsis gene (AtYABBY5), suggesting that YAB5 may have a particular function in the Arabidopsis thaliana.
Figure 2.
Phylogenetic relationship of the YABBYs proteins of Ac (Pineapple), At (Arabidopsis thaliana), Os (Oryza sativa). The phylogenetic tree was made using neighbor-joining with a bootstrap values of 1000. Phylogenetic tree divides YABBYs into five subfamilies (YAB2, CRC, YAB5, FIL/YAB3 and INO). Each subfamily is represented with different color.
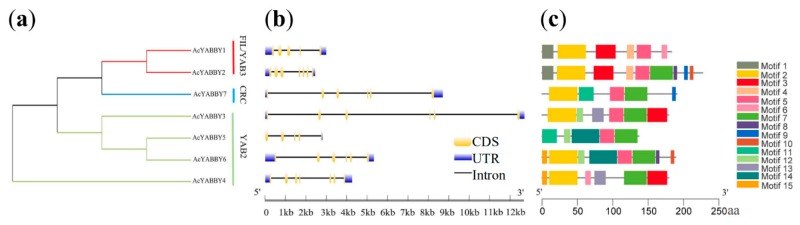
Figure 3.
Phylogenetic relationships, gene structure and architecture of conserved protein motifs in pineapple YABBY genes. (a) The phylogenetic tree was constructed based on the full-length sequences of pineapple YABBY proteins using MEGA 7 software. Details of clusters are shown in different colors. (b) Exon-intron structure of pineapple YABBY genes. Blue boxes represent untranslated 5′- and 3′-regions; yellow boxes represent exons; black lines represent introns. (c) The motif composition of pineapple YABBY proteins. The motifs, numbers 1–15, are displayed with different colored boxes.
2.3. Gene Structure Analysis and Identification of Conserved Motifs
To investigate the structure diversity, we used Gene Structure Display Server [21]. The results showed that the exon number of the AcYABBY genes ranged from a minimum of 2 and a maximum of 7. AcYABBY2 and AcYABBY7 had the maximum exons numbers (Figure 3b). The schematic diagram representing the structure of AcYABBY proteins was constructed from the results of the MEME motif analysis (Figure 3c). The number of motifs ranged from 1 to 15 [22]. AcYABBY5 and AcYABBY7 contained five motifs. AcYABBY1, AcYABBY3 and AcYABBY4 had six motifs, while AcYABBY6 had eight motifs. AcYABBY2 had the largest number of motifs, containing nine motifs. The similar motif arrangements among AcYABBY proteins indicate that the protein architecture is conserved within a particular subfamily. The functions of these conserved motifs remain to be elucidated.
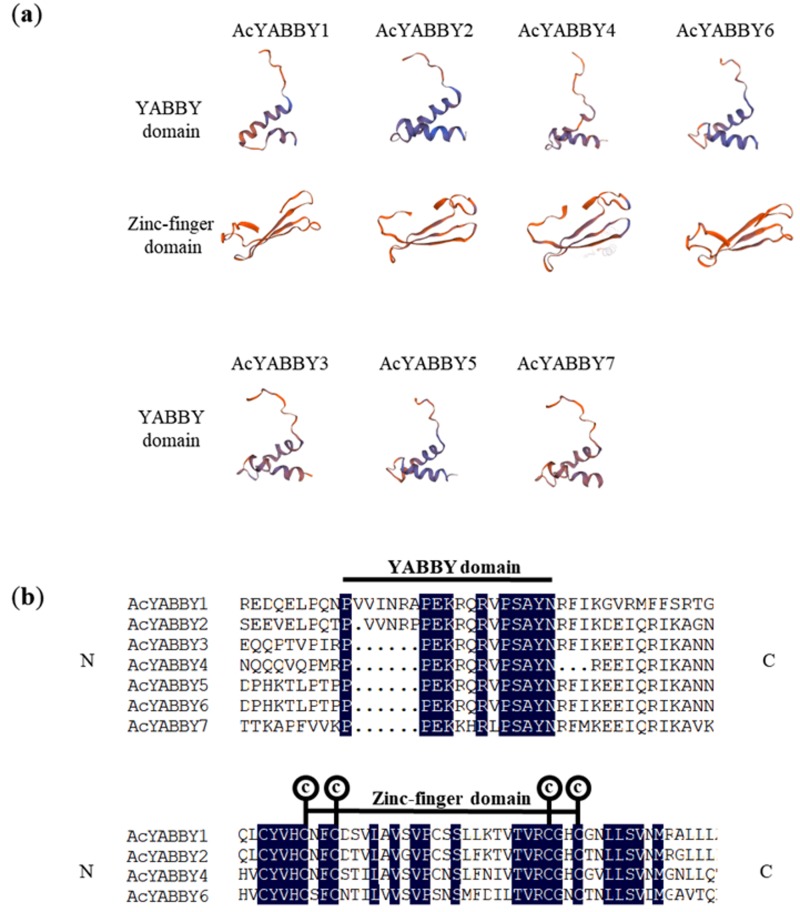
2.4. AcYABBY Protein Homology Modeling and Sequence Alignment
The YABBY family possesses a C2C2 zinc finger domain at the N-terminus and a YABBY domain at the C-terminus. To study AcYABBY protein conformation, seven pineapple YABBY protein were analyzed by SWISS-MODEL to identify the best template with known structures and similar sequence. All the seven AcYABBY proteins were predicted with a YABBY domain. Whereas, only four AcYABBY proteins contained a zinc-finger domain (Figure 4a). The zinc-finger domain was not detected in the three AcYABBY genes (AcYABBY3, AcYABBY5, AcYABBY7). To analyze further, we used DNAMAN for domain analysis of protein sequence alignment (Figure 4b). We found that one C2 structure was missing from the C2C2 structure in the protein sequences of the these three AcYABBY genes. Therefore, we concluded that zinc finger structure was incomplete and unpredictable in AcYABBY3, AcYABBY5, AcYABBY7. The specific cause of this phenomenon is still unclear and may be related to the evolution of the pineapple YABBY genes.
Figure 4.
Predicated structures and multiple sequence alignment of pineapple YABBY proteins. (a) Predicted protein structures of AcYABBY proteins. (b) Multiple sequence alignment of AcYABBYs, zinc-finger domain and YABBY domain amino. ′N′ and ′C′ indicate the N-terminal and C-terminal. The four cysteine residues putatively responsible of the zinc-finger structure are also indicated. Identical amino acids are highlighted in blue.
2.5. Expression Profiling and Subcellular Localization of AcYABBYs
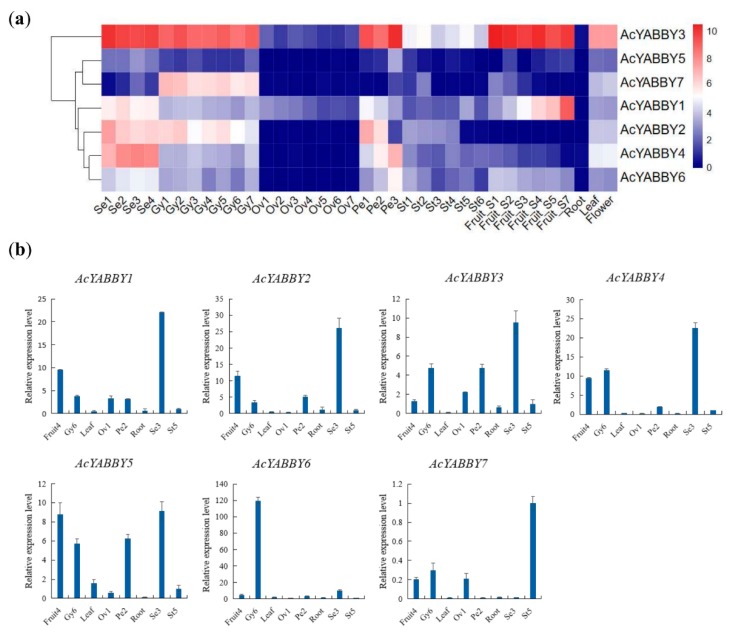
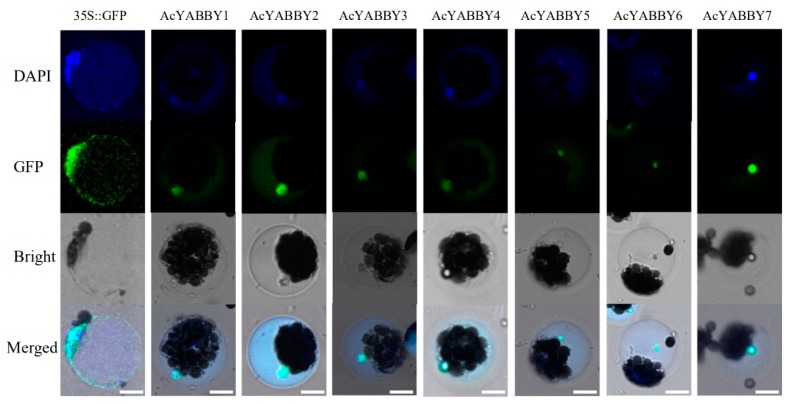
Transcriptome data from different developmental stages of pineapple tissue were used to study the expression patterns of seven AcYABBY genes to understand the transcriptional diversity of the AcYABBY genes. The expression levels of AcYABBY genes were analyzed by FPKM values from RNA sequence data. Hierarchical clusters and expression patterns of each gene were generated based on the average log values of each gene in each tissue. According to Heat-map analysis, the AcYABBY3 expressed ubiquitously in most organs. The AcYABBY3 was highly and specifically expressed at all stages of development of the organs (sepal Se1-4, gynoecium Gy1-7, petal Pe1-3, fruit S1-7, flower and leaf). Conversely, the AcYABBY5 and AcYABBY6 showed relatively low expression levels in almost all organs. Besides, some genes were highly expressed in specific organs. For example, AcYABBY1 was highly expressed in sepal (Se2 stage) and fruit (S4-7 stages). AcYABBY2 was highly expressed in sepal (Se1-4 stages), pistil (Gy1/2/4/5 stages) and petal (Pe1-2 stages). AcYABBY4 was highly expressed in sepal (Se1-4 stages) and petal (Pe3 stage). AcYABBY7 was highly expressed in gynoecium (Gy1-7 stages) (Figure 5a). The reliability of transcriptome data was further verified by RT-qPCR experiment, which was carried out on eight representative samples for seven YABBY genes. The results revealed that expression patterns of the YABBY genes detected by RT-qPCR were partially consistent with the results of RNA-seq analysis (Figure 5b). The differences between RT-qPCR and RNA-seq may be caused by sampling and it is difficult to keep the samples’ stage exactly at the same as RNA-seq sequencing samples. To understand molecular characteristics of AcYABBYs, seven pineapple YABBY genes (YABBY1-7) were selected for subcellular localization. In pineapple protoplasts, AcYABBYs-GFP were co-localized with a DAPI signal, suggesting that the AcYABBY proteins were mainly localized in the nucleus (Figure 6). Also, the AcYABBYs-GFP fusion proteins were also localized in the nucleus of tobacco leaves (Figure S1) and Arabidopsis roots (Figure S2). These localization results were consistent with each other, suggesting that AcYABBY proteins were localized to the nucleus.
Figure 5.
Expression profiles of the pineapple YABBY genes. (a) Heat-map of AcYABBY genes expression profiles in different tissue generated from RNA-seq data. The AcYABBYs were clustered according to their expression patterns. Red color indicates high levels of transcript abundance and blue indicates low transcript abundance. The color scale is shown on the right. Samples are mentioned at the bottom of each lane: Se (sepal) Se1-Se4, Gy (gynoecium) Gy1-7 Ov (ovule) Ov1-Ov7, Pe (petal) Pe1-Pe3, St (stamen) St1-St5, Fruit S1-S7, Root, Leaf, Flower. (b) Expression analysis of 7 pineapple YABBY genes in eight representative samples by RT-qPCR. RT-qPCR data were normalized using pineapple PP2A gene and vertical bars indicate standard deviation.
Figure 6.
Subcellular localization of pineapple YABBY proteins in pineapple protoplasts. The 35S::AcYABBYs-GFP and 35S::GFP control vectors were transiently expressed in pineapple protoplasts. Results were visualized by a confocal microscope after 16 h transformation. Bar = 10 μm.
2.6. Cis-Acting Elements and RT-qPCR Analysis of AcYABBY Ggenes
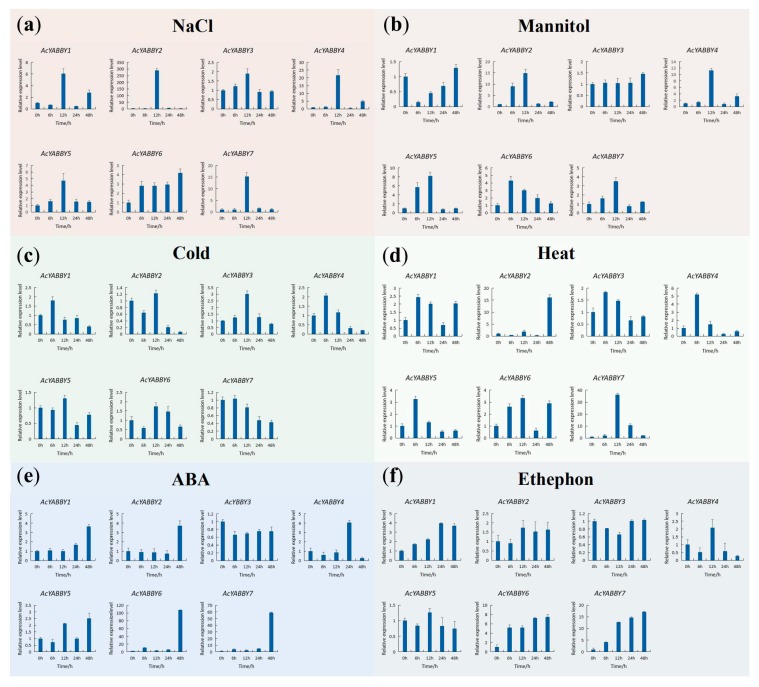
The study of the cis-element indicated that each of the pineapple genes comprised the MYB and MYC elements in their promoter regions. MYB elements have been reported to be associated with drought, low temperature, salt and ABA stress responses and MYC elements are associated with drought and salt stress response [1]. Besides, the majority of the pineapple YABBYs except AcYABBT3, AcYABBT4 and AcYABBT7 also contained at least an ERE element, which responds to ethylene. The identified Motif of the cis-acting elements and the sequences are listed in Table 2. To investigate the expression pattern of AcYABBY genes in response to different abiotic stresses, RT-qPCR was carried out using pineapple leaves exposed to various abiotic stresses for different time intervals such as 0, 6, 12, 24 and 48 h (h). The AcYABBY genes expressed diversely under the six types of abiotic stresses. Under salt stress (150 mM NaCl), the transcription level of AcYABBYs increased gradually. Among them, AcYABBY1, AcYABBY2, AcYABBY4, AcYABBY5 and AcYABBY7 transcript levels were highly up-regulated with the highest expression at 12 h. Notably, AcYABBY2 increased to a peak (~250-fold) at 12 h and then rapidly declined to a level similar to the control (Figure 7a). Under drought stress, AcYABBY2 increased to a peak of ~15-fold at 12 h. AcYABBY4 increased to a maximum of ~10-fold at 12 h. AcYABBY5 increased to a maximum of ~8-fold at 12 h. After 12 h, the expression level of AcYABBY2, AcYABBY4 and AcYABBY5 rapidly declined to a level similar to the control when plants were subjected to drought stress (Figure 7b). Expression level of AcYABBY3 increased slowly to a peak of ~3-fold at 12 h and then declined to a level similar to the control under cold stress (Figure 7c). Under heat stress, AcYABBY7 increased to a peak of ~30-fold at 12 h and then rapidly declined to a level similar to the control. AcYABBY2 rose to a maximum of ~15-fold at 48 h (Figure 7d). Under ABA stress, AcYABBY6 and AcYABBY7 transcript levels were highly up-regulated at 48h. AcYABBY6 increased to a peak of ~100-fold and AcYABBY7 increased to a maximum of ~60-fold at 48h (Figure 7e). Under ethephon stress, AcYABBY6 increased to a maximum of ~8-fold at 48h and AcYABBY7 increased to a maximum of ~17-fold at 48h (Figure 7f). These results together suggested that AcYABBYs are involved in plant response to abiotic stresses.
Table 2.
Distribution of MYB, MYC and ERE cis-acting element in pineapple YABBY promoters.
| Gene | Motif Sequence | AcYABBY1 | AcYABBY2 | AcYABBY3 | AcYABBY4 | AcYABBY5 | AcYABBY6 | AcYABBY7 |
|---|---|---|---|---|---|---|---|---|
| MYB | TAACCA | 6 | 5 | 6 | 9 | 2 | 4 | 2 |
| CAACAG | ||||||||
| CAACCA | ||||||||
| MYC | CATTTG | 4 | 3 | 4 | 7 | 8 | 5 | 5 |
| CATGTG | ||||||||
| CAATTG | ||||||||
| ERE | ATTTCATA | 3 | 2 | 0 | 0 | 1 | 3 | 0 |
| ATTTTAAA |
Figure 7.
Expression profiles of 7 selected AcYABBY genes in response to various abiotic stress treatments. (a) NaCl treatment. (b) Mannitol treatment. (c) Cold treatment. (d) Heat treatment. (e) ABA treatment. (f) Ethephon treatment. RT-qPCR data were normalized using pineapple PP2A gene as reference gene. Error bars indicate Standard Deviation.
2.7. AcYABBY4 Negatively Regulates the High Salinity Tolerance in Arabidopsis
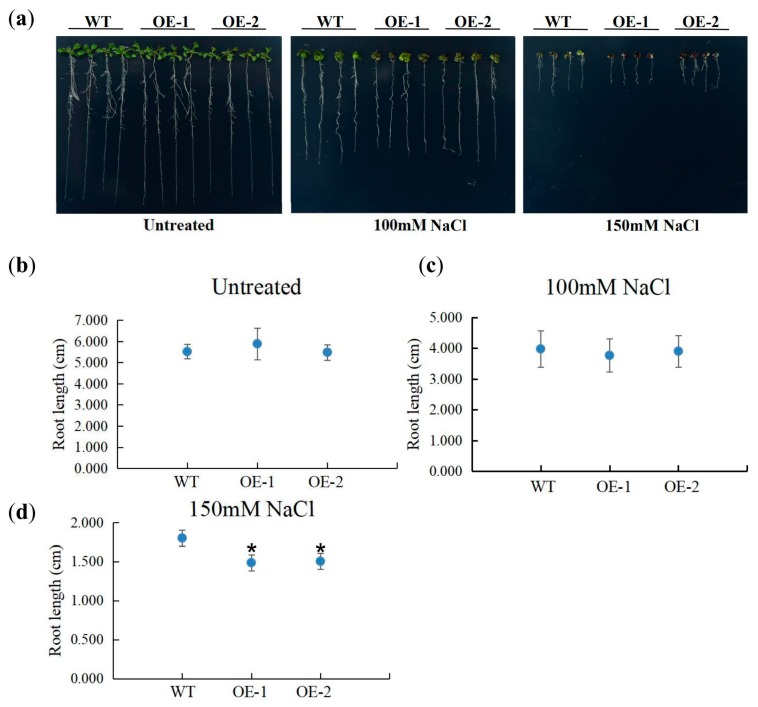
To further investigate the function of AcYABBYs upon abiotic stress, we overexpressed AcYABBY4 driven by 35S promoter in Arabidopsis plants and compared the growth phenotype of AcYABBY4-overexpression (AcYABBY4-OE) lines with the wild-type plants under optimum and salinity condition. For the phenotype comparison under salt stress, seedlings were cultured vertically on ½ MS medium for three days and then transferred to ½ MS medium supplemented with or without NaCl and allowed to grow for additional seven days. The root length of all lines showed a similar phenotype when grown on ½ MS medium without NaCl (Figure 8a,b). Under the 100 mM NaCl treatment, the root length of AcYABBY4-OE seedlings were also comparable to that of wild-type (Figure 8c). Whereas, under 150 mM NaCl treatment, the root length of AcYABBY4-OE seedlings were significantly reduced compared to wild-type seedlings (Figure 8d). Collectively, the results indicate that AcYABBY4-OE plants are susceptible to high salinity stress, suggesting that AcYABBY4 may be a negative regulator in plant response to high salinity stress.
Figure 8.
Phenotypic comparison of wild-type (WT) and AcYABBY-over-exression (AcYABBY-OE) Arabidopsis seedling under salt treatment. (a) Seedlings grown on ½ MS medium for three days were transferred to ½ MS medium supplemented with NaCl or without NaCl and allowed to grow for additional seven days. (b) Comparative root lengths of WT and transgenic lines in control conditions. (c) Comparative root length between WT and transgenic lines under 100mM NaCl treatment. (d) Comparative root lengths of WT and transgenic lines under 150 mM NaCl treatment. * denotes the significant difference (* p < 0.05) of t-tests between the transgenic lines and wild type.
3. Discussion
3.1. Diversity of YABBY Transcription Factors in Plants
The plant-specific YABBY transcription factor family is involved in early embryonic development and lateral organ development. In particular, it plays an important role in the establishment of the near-distal axis polarity of leaves and is also involved in biological processes, such as plant development and stress response [23]. The YABBY genes are well studied in dicotyledonous plants such as Arabidopsis. However, relatively less information is available about monocot YABBY genes though it is reported in rice. In Arabidopsis, there are six members in YABBY family, namely AtCRC, AtYABBY1, AtFIL, AtYABBY3, AtINO/AtYABBY4, AtABBY5 [2,5], with their unique and overlapping functions [17,24]. Various evidences suggest that they promote the differentiation of the distal axis of lateral organ cells [6,7] and have important effect on leaf expansion and floral organ development. In rice, there are eight YABBY members, OsDL, OsYABBY1, OsYABBY2, OsYABBY3, OsYABBY4, OsYABBY5, OsYABBY6 and OsYABBY7 [10]. They play important roles in regulating the development of lateral organs such as leaves and flowers [25]. Pineapple (Ananas comosus L.) is the third most economically important tropical fruit crop in the world after bananas [26]. Scientists are trying to improve its quality by improving resistance to environmental changes, increase the yield and improve its taste [27]. Therefore, the functional study of YABBY genes in regulation of plant development and stress response is important for breeding programs and agricultural production [25]. However, there are no reports on the characterization of pineapple YABBY proteins until now.
Here, we identified nine YABBY genes in pineapple genome and named them from AcYABBY1 to AcYABBY9. According to phylogenetic analysis, pineapple YABBY genes were more closely related to rice YABBY genes, because pineapple as a perennial monocot is evolutionarily more related to grasses, including corn, rice and wheat. The YABBY transcription factor family is a subfamily of the zinc finger protein superfamily, with a zinc finger domain at the N-terminus and a “helix-loop-helix” YABBY domain at the C-terminus similar to HMG-box, which have been confirmed to be associated with specific binding of DNA [2]. Some of the pineapple YABBY proteins are highly conserved with Arabidopsis and rice YABBY proteins. We found that some pineapple YABBY genes, including AcYABBY3, AcYABBY5 and AcYABBY7, lack a N-terminal C2C2 zinc finger domain. It could be due to a technical issue in the process of genome assembly. To have more exact results, we only selected seven YABBY genes for further studies with YABBY gene features.
3.2. AcYABBY Gene Expression Profiles Analysis
The completion of the pineapple genome sequence has provided us an opportunity to explore the specific genes responsible for specific traits [20]. As shown in Figure 5, hierarchical clusters and expression patterns shows that the AcYABBY genes have distinct expression pattern. For example, AcYABBY7 is highly expressed in pistil. It was reported that OsDL is involved in the regulation of floral meristem [13]. Phylogenetic analysis indicate that AcYABBY7 and OsDL are in the same subcluster, implying that AcYABBY7 may also be involved in the tissue development of pineapple pistil. Interestingly, some AcYABBYs are preferentially expressed in floral organs. For example, AcYABBY3 and AcYABBY2 are enriched in sepals, pistils and petals. The specific enrichments of AcYABBY4 in sepals and petals suggests its probable role in floral development of pineapple. OsYABBY1 was found to be involved in the feedback regulation of gibberellin (GA) metabolism in rice [16]. Phylogenetic analysis shows that AcYABBY3 and AcYABBY4 and OsYABBY1 are clustered together. It will be interesting to test whether AcYABBY3 and AcYABBY4 are also involved in the feedback regulation of gibberellin metabolism. Besides, we found that AcYABBY3 and AcYABBY1 are highly expressed in the fruit. It will be worth to investigate their potential role in the pineapple fruit development. The expression pattern of AcYABBYs and its corresponding function of rice homologous genes provide us a clue for understanding the function of AcYABBYs in pineapples.
3.3. AcYABBY4 Inhibits Root Growth of Seedlings under Salt Stress
Pineapple plant growth is affected by many factors during its growth and development, such as the uneven distribution of hormones, drought, salt and other adverse environmental conditions. In soybean, YABBY genes are involved in abiotic stress response and GmYABBY10 act as a negative regulator of salt and drought stress in Arabidopsis [1]. However, the function of AcYABBY in plant response to abiotic stresses remains unknown. We analyzed the cis-acting elements of the pineapple YABBY family and found that the AcYABBY genes also comprise cis-acting elements such as MYB, MYC and ERE in their promoters. These cis-acting elements are involved in abiotic stress response in plants such as MYB elements are involved in salt, drought, low temperature and response to ABA [28,29]. MYC elements are involved in salt, drought and ABA stress response and ERE is related to ethylene response. Plant abiotic stress-responsive transcription factors could bind to these cis-acting elements [1]. The presence of these cis-elements in the promoter of AcYABBYs indicate that AcYABBYs are associated with stress response and may also be involved in plant adaptation to the environmental changes. Studying the function of the YABBY genes in plant response to abiotic stresses may help us to improve the agriculture production of pineapple. Using RT-qPCR we analyzed the response of the AcYABBY gene family under abiotic stress conditions such as NaCl, drought, cold, heat, ABA and ethephon treatments. The results showed that the expression patterns of pineapple YABBY genes were different under six stress conditions. For example, the expression of AcYABBY2, AcYABBY3, AcYABBY4, AcYABBY6 and AcYABBY7 was induced by NaCl, cold, drought, ABA and heat stress, respectively (Figure 7). These results suggest that AcYABBYs may play an important role in response to abiotic stress. The response of plants to abiotic stress is a complex process that is regulated by different molecular and cellular pathways. Here, the response of the YABBY genes to six different stresses laid the foundation for further functional study of the pineapple YABBY genes. To investigate the AcYABBYs’ functions, we constructed all the YABBYs’ over expression vectors and transformed it into Arabidopsis. We noted that the AcYABBY4 over expression lines showed obvious phenotype with vegetative growth and development. Considering that the AcYABBY4 basically had an early response than AcYABBY2 during different stress conditions and the AcYABBY4 indeed have a significant response to NaCl stress, so we chose AcYABBY4 as our target gene for the functional analysis. However, Arabidopsis overexpressing AcYABBY4 showed delayed growth and small seedlings size than wild-type (WT). We also found that the root growth of AcYABBY4 overexpression lines were like WT and shows a normal phenotype in early stages of growth. Therefore, to avoid the effect of developmental changes on interpretation of our results of salt stress, we selected root growth as the parameter to study the salt stress. We found that the Arabidopsis overexpressing AcYABBY4 is more sensitive to salt stress, indicating that AcYABBY4 facilitates plants to perceive changes in the external salt environment more quickly and then inhibits plant growth through other complex regulatory mechanisms. Therefore, AcYABBY4 may be a negative regulator of salt tolerance in transgenic Arabidopsis (Figure 8), providing a reliable basis for understanding YABBY participation in the biological process of plant stress response.
There is a close relationship between hormonal signal transduction and stress response. The response elements related to ABA and ethylene have important functions in plant abiotic stress and disease resistance. These observations also suggests that the YABBY genes may play an important role in response to ABA and ethylene accumulation of pineapple plants. The regulation mechanism of AcYABBY proteins under biological stress and in hormone signal transduction still remains elusive and could be a key part of further study of AcYABBYs.
4. Materials and Methods
4.1. Identification of YABBY Transcription Factors in Pineapple
The pineapple YABBY protein and genome sequences were downloaded from Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html) and PlantTFDB (http://planttfdb.cbi.pku.edu.cn/) (Table S4). To identify the pineapple YABBY genes, HMMER3.02 (http://hmmer.wustl.edu/) with default parameters settings were used to search for the PFAM YABBY domain (PF04690) (http://pfam.sanger.ac.uk/). Also, BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgiwas) was used to search for potential pineapple genes containing the YABBY transcription factor. To achieve the accuracy of the analysis, we further analyzed the conserved sequence using NCBI-CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) with an E-value threshold of 0.013 and abandoned any sequences lacking the YABBY annotation [30]. The isoelectric point (pI) and molecular weight (MW) of the AcYABBY proteins were predicted using ExPASy-Compute pI/MW (https://web.expasy.org/compute_pi/).
4.2. Chromosome Locations of Pineapple YABBY Genes
The information about the chromosome location of the AcYABBY genes were obtained from Phytozome. Based on the starting position of the gene and the length of the relevant chromosome, Mapchart [31] was used to visualize the localization of the pineapple YABBY gene on chromosomes and scaffolds.
4.3. Construction of Phylogenetic Tree
To generate phylogenetic trees, amino acid sequences of YABBYs in Arabidopsis, rice and pineapple were compared and analyzed by MEGA7 [32]. The protein sequences were aligned by MUSCLE and phylogenetic tree was constructed using Neighbor-Joining method with a bootstrap value of 1000.
4.4. Gene Structure Analysis and Identification of Conserved Motifs
The exon-intron substructure map of the pineapple YABBY genes were analyzed using online tool GSDS (http://gsds.cbi.pku.edu.cn/). The motifs of the AcYABBY proteins were determined using the MEME Suite 5.0.5 with classic mode, zoops for selecting the site distribution and 15 motifs. (http://meme-suite.org/).
4.5. Modeling of Protein 3D Structures and Sequence Alignment of AcYABBY Proteins
SWISS-MODEL (https://www.swissmodel.expasy.org) was used to predict the structures of seven YABBY proteins. The templates used in the study are listed in Table S1. The alignment of the AcYABBY protein sequences were carried out using DNAMAN [33].
4.6. RNA-Seq and RT-qPCR Data Analysis
To understand the expression pattern of YABBY genes in different development stages of pineapple, sepal, pistil, ovule, petal, stamen, fruit, root, leaf and flower RNA-Seq data (SRA315090) was downloaded from the NCBI database [20]. The trimmed pair-end reads of all tissues were aligned with the pineapple genome using TopHat v2.1.1 [34] with default parameter settings. The FPKM values were derived from Cuffdiff v2.2.1 and then a heatmap of YABBY genes expression was generated by the Rmap software package. To determine the relative transcript level of the selected AcYABBY genes, total RNA was extracted from different tissues (sepal, pistil, ovule, petal, stamen, fruit, root, leaf) of pineapple according to the procedure in the description of the RNA Plant Extraction Kit (OMEGA, Guangzhou, China). RNA quality was tested using gel electrophoresis and NanoDrop2000c (Thermo Fisher Scientific, Fujian, China). One μg of purified total RNA was reverse transcribed into cDNA in a 20 μL reaction volume using AMV reverse transcriptase (Takara, Beijing, China) according to the supplier′s instructions. Quantitative primers were designed by IDT (http://www.idtdna.com/pages/products/gene-expression/primetime-qpcr-assays-and-primers), with G+C content between 45%–55%, primer length between 17–25 bases, Tm value between 58–62 and quantitative product size between 80–150 bp. The reaction volume of RT-qPCR was 20 μL (10 μL 2×TransStart Top Green qPCR SuperMix, 8 μL nuclease-free water, 0.5 μL forward primer, 0.5 μL reverse primer and 1ul cDNA) according to the supplier′s instructions (TransGen Biotech, Beijing, China). The RT-qPCR parameters were as follows: 95 °C for 30 s; 95 °C for 5 s and 60 °C for 40 s for 40 cycles; 95 °C for 15 s. In each case, three technical replicates and at least three independent biological replicates were used. The primers used in this study are listed in Table S2. For RT-qPCR data analysis, the quantification cycle (Cq)value was automatically calculated by the Bio-Rad CFX Manager 3.1 system software and the delta-delta Cq method was used to calculate the relative expression levels.
4.7. Vector Construction and Subcellular Localization
The full-length of coding sequences of the AcYABBY genes were amplified using the primers listed in Table S3. The PCR fragments were cloned into the pENTR/D-TOPO vectors (Invitrogen) and sequenced separately. The positive clones were recombined into the destination vector pGWB505 using LR reaction. The vectors harboring the AcYABBY genes were transformed into Agrobacterium tumefaciens (GV3101) and infiltrated to tobacco leaves. Subcellular localizations in tobacco leaves were observed with confocal microscope using GFP and DAPI stain. The A. tumefaciens (GV3101) with AcYABBY genes were also used to transform wild-type Arabidopsis by a floral dip procedure [35]. Transgenic Arabidopsis lines were selected on ½ MS medium supplemented with 50 mg/L hygromycin. All the experiments were carried out in the T2 generation. The roots of the positive transgenic plants were stained by PI solution and subcellular localization was observed with confocal microscope. All AcYABBY genes were also used to study the protein localization in the protoplast of pineapple [36].
4.8. Cis-Acting Elements in the Pineapple YABBY Genes Promoter region
For each pineapple YABBY gene, 2000 bp upstream of the translational start codon was selected from Phytozome 12 as promoter region and analyzed for presence of cis-acting elements using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The main cis-acting elements are listed in Table 2.
4.9. Stress Treatment
One month old tissue culture raised seedlings of pineapple (MD2 variety) were maintained under 3000 Lux light intensity and a day/night cycle of 16/8 h at 25 ± 2 °C in a controlled environment [36]. For different treatments, the seedlings were transferred to solutions containing 150 mM NaCl for salt stress, 350 mM Mannitol for osmotic stress, 100 μM ABA, 100 μM Ethephon. Cold and heat stress were performed by placing seedlings in the 4 °C and 45 °C chambers. Samples were collected at 0, 6, 12, 24 and 48h [37] after each treatment and immediately stored in liquid nitrogen.
4.10. Tolerance Assays under Stress Conditions
To assess phenotype under salt stress, the seeds of wild type and transgenic Arabidopsis lines were surface sterilized and seeded on ½ MS plates, then kept at 4 °C for 48 h in the dark before germination. About 50 seeds of each transgenic line were seeded on ½ MS medium for three days were transferred to ½ MS medium supplemented with NaCl or without NaCl and kept at 22 ± 2 °C under 16 h light/8 h dark for seven days and root length was measured.
5. Conclusions
In this study, nine pineapple YABBY genes were identified those are preferentially expressed in different tissues. Transient expression analysis in pineapple protoplasts, tobacco leaves and Arabidopsis roots showed that pineapple YABBY protein were localized in the nucleus. RT-qPCR results demonstrate that AcYABBY2, AcYABBY4 and AcYABBY7 are regulated by drought, NaCl, cold and heat stress. Functional analysis of AcYABBY4 suggests that AcYABBY4 is a negative regulator of salt stress.
Acknowledgments
We would like to thank the reviewers for their helpful comments on the original manuscript.
Abbreviations
| ABA | Abscisic acid; |
| GSDS | Gene Structure Display Server |
| HMM | Hidden Markov Models |
| MEGE | Molecular Evolutionary Genetics Analysis |
| MEME | Multiple Em for Motif Elicitation |
| RT-qPCR | Real-time Quantitative PCR |
| RNA-Seq | RNA sequencing |
| SMART | Simple Modular Architecture Research Tool |
| MS | Murashige and Skoog |
| PEG | Polyethylene glycol |
| WT | Wild type |
| GFP | Green fluorescent protein |
| AtYABBY | Arabidopsis thaliana YABBY |
| OsYABBY | rice (Oryza sativa) YABBY |
| AcYABBY | pineapple (Ananas comosus) YABBY |
| CDS | Coding sequence |
| ATG | Starting codon |
| FPKM | Fragments Per Kilobase Million |
| DAPI | 4′,6-Diamidino-2-Phenylindole |
| MS | Murashige and Skoog |
| HMG | High mobility group |
| MYB | V-myb Myeloblastosis Viral Oncogene Homolog |
| MYC | Myelocytomatosis oncogenes |
| ERE | Ethylene Responsive Element |
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/20/23/5863/s1.
Author Contributions
Z.L. and G.L. designed the experiment and wrote the manuscript. M.C. helped in photography and figure arrangement. S.V.G.N.P. provided the pineapple materials and helped in tissue culture. M.A. helped with article modification. Q.Z. helped in protoplast isolation. X.H. helped in plant culturing. X.W. and Y.L. provided reagents. Y.Q. conceived the study and revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by NSFC (U1605212, 31761130074, 31970333 to Y.Q.), a Guangxi Distinguished Experts Fellowship and a Newton Advanced Fellowship (NA160391) to Y.Q., Science and Technology Major Project of Guangxi--Research and application of ecological and high efficient cultivation techniques for dominant and characteristic fruits (AA17204097-6) and Project of Guangxi featured fruit innovation team on pineapple breeding and cultivation post under national modern agricultural industry technology system (nycytxgxcxtd-17-05).
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Zhao S.P., Lu D., Yu T.F., Ji Y.J., Zheng W.J., Zhang S.X., Chai S.C., Chen Z.Y., Cui X.Y. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol. Biochem. 2017;119:132–146. doi: 10.1016/j.plaphy.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 2.Sawa S., Watanabe K., Goto K., Liu Y.G., Shibata D., Kanaya E., Morita E.H., Okada K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999;13:1079–1088. doi: 10.1101/gad.13.9.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarojam R., Sappl P.G., Goldshmidt A., Efroni I., Floyd S.K., Eshed Y., Bowman J.L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell. 2010;22:2113–2130. doi: 10.1105/tpc.110.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada T., Ito M., Kato M. Expression pattern of INNER NO OUTER homologue in Nymphaea (water lily family, Nymphaeaceae) Dev. Genes Evol. 2003;213:510–513. doi: 10.1007/s00427-003-0350-8. [DOI] [PubMed] [Google Scholar]
- 5.Siegfried K.R., Eshed Y., Baum S.F., Otsuga D., Drews G.N., Bowman J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- 6.Balasubramanian S., Schneitz K. NOZZLE links proximal-distal and adaxial-abaxial pattern formation during ovule development in Arabidopsis thaliana. Development. 2002;129:4291–4300. doi: 10.1242/dev.129.18.4291. [DOI] [PubMed] [Google Scholar]
- 7.Eshed Y., Baum S.F., Bowman J.L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99:199–209. doi: 10.1016/S0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- 8.Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- 9.Yamada T., Yokota S., Hirayama Y., Imaichi R., Kato M., Gasser C.S. Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. Plant J. 2011;67:26–36. doi: 10.1111/j.1365-313X.2011.04570.x. [DOI] [PubMed] [Google Scholar]
- 10.Eckardt N.A. YABBY genes and the development and origin of seed plant leaves. Plant Cell. 2010;22:2103. doi: 10.1105/tpc.110.220710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stahle M.I., Kuehlich J., Staron L., von Arnim A.G., Golz J.F. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell. 2009;21:3105–3118. doi: 10.1105/tpc.109.070458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang S., Hur J., Kim S.J., Han M.J., Kim S.R., An G. Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Mol. Biol. 2004;56:133–143. doi: 10.1007/s11103-004-2648-y. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H.Y. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell. 2004;16:500–509. doi: 10.1105/tpc.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha C.M., Jun J.H., Fletcher J.C. Control of Arabidopsis leaf morphogenesis through regulation of the YABBY and KNOX families of transcription factors. Genetics. 2010;186:197–206. doi: 10.1534/genetics.110.118703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X.L., Yang Z.P., Zhang J., Zhang L.G. Ectopic expression of BraYAB1–702, a member of YABBY gene family in Chinese cabbage, causes leaf curling, inhibition of development of shoot apical meristem and flowering stage delaying in Arabidopsis thaliana. Int. J. Mol. Sci. 2013;14:14872–14891. doi: 10.3390/ijms140714872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toriba T., Harada K., Takamura A., Nakamura H., Ichikawa H., Suzaki T., Hirano H.Y. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet Genom. 2007;277:457–468. doi: 10.1007/s00438-006-0202-0. [DOI] [PubMed] [Google Scholar]
- 17.Villanueva J.M., Broadhvest J., Hauser B.A., Meister R.J., Schneitz K., Gasser C.S. INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev. 1999;13:3160–3169. doi: 10.1101/gad.13.23.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohmori Y., Toriba T., Nakamura H., Ichikawa H., Hirano H.Y. Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice. Plant J. 2011;65:77–86. doi: 10.1111/j.1365-313X.2010.04404.x. [DOI] [PubMed] [Google Scholar]
- 19.Lin Z., Li X., Shannon L.M., Yeh C.T., Wang M.L., Bai G., Peng Z., Li J., Trick H.N., Clemente T.E., et al. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 2012;44:720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ming R., VanBuren R., Wai C.M., Tang H., Schatz M.C., Bowers J.E., Lyons E., Wang M.L., Chen J., Biggers E., et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015;47:1435–1442. doi: 10.1038/ng.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo A.Y., Zhu Q.H., Chen X., Luo J.C. GSDS: A gene structure display server. Yi Chuan. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- 22.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 23.Bowman J.L. The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol. 2000;3:17–22. doi: 10.1016/S1369-5266(99)00035-7. [DOI] [PubMed] [Google Scholar]
- 24.Finet C., Floyd S.K., Conway S.J., Zhong B., Scutt C.P., Bowman J.L. Evolution of the YABBY gene family in seed plants. Evol. Dev. 2016;18:116–126. doi: 10.1111/ede.12173. [DOI] [PubMed] [Google Scholar]
- 25.Dai M., Hu Y., Zhao Y., Liu H., Zhou D.X. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007;144:380–390. doi: 10.1104/pp.107.095737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., Liu J., Ming R. Genomic analyses of the CAM plant pineapple. J. Exp. Bot. 2014;65:3395–3404. doi: 10.1093/jxb/eru101. [DOI] [PubMed] [Google Scholar]
- 27.Su Z.X., Wang L.L., Li W.M., Zhao L.H., Huang X.Y., Azam S.M., Qin Y. Genome-Wide Identification of Auxin Response Factor (ARF) Genes Family and its Tissue-Specific Prominent Expression in Pineapple (Ananas comosus) Trop Plant Biol. 2017;10:86–96. doi: 10.1007/s12042-017-9187-6. [DOI] [Google Scholar]
- 28.He Y.A., Li W., Lv J., Jia Y.B., Wang M.C., Xia G.M. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2012;63:1511–1522. doi: 10.1093/jxb/err389. [DOI] [PubMed] [Google Scholar]
- 29.Yang A., Dai X.Y., Zhang W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold and dehydration tolerance in rice. J. Exp. Bot. 2012;63:2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C., Wang D., Yang C., Kong N., Shi Z., Zhao P., Nan Y., Nie T., Wang R., Ma H., et al. Genome-wide identification of the potato WRKY transcription factor family. PLoS ONE. 2017;12:e0181573. doi: 10.1371/journal.pone.0181573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W., Yan M., Hu B., Priyadarshani S.V.G.N., Hou Z., Ojolo S.P., Xiong J., Zhao H., Qin Y. Characterization and the Expression Analysis of Nitrate Transporter (NRT) Gene Family in Pineapple. Trop. Plant Biol. 2018;11:177–191. doi: 10.1007/s12042-018-9209-z. [DOI] [Google Scholar]
- 32.Zhang M., Liu Y., Shi H., Guo M., Chai M., He Q., Yan M., Cao D., Zhao L., Cai H., et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genomics. 2018;19:159. doi: 10.1186/s12864-018-4511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Hua X., Xu J., Chen Z., Fan T., Zeng Z., Wang H., Hour A.L., Yu Q., Ming R., et al. Comparative genomics revealed the gene evolution and functional divergence of magnesium transporter families in Saccharum. BMC Genomics. 2019;20:1471–2164. doi: 10.1186/s12864-019-5437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 36.Priyadarshani S., Hu B., Li W., Ali H., Jia H., Zhao L., Ojolo S.P., Azam S.M., Xiong J., Yan M., et al. Simple protoplast isolation system for gene expression and protein interaction studies in pineapple (Ananas comosus L.) Plant Methods. 2018;14:95. doi: 10.1186/s13007-018-0365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie T., Chen C., Li C., Liu J., Liu C., He Y. Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics. 2018;19:490. doi: 10.1186/s12864-018-4880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.