Abstract
Background
Euphorbiaceae is one of the largest families of flowering plants. Due to its exceptional growth form diversity and near-cosmopolitan distribution, it has attracted much interest since ancient times. SBP-box (SBP) genes encode plant-specific transcription factors that play critical roles in numerous biological processes, especially flower development. We performed genome-wide identification and characterization of SBP genes from four economically important Euphorbiaceae species.
Results
In total, 77 SBP genes were identified in four Euphorbiaceae genomes. The SBP proteins were divided into three length ranges and 10 groups. Group-6 was absent in Arabidopsis thaliana but conserved in Euphorbiaceae. Segmental duplication played the most important role in the expansion processes of Euphorbiaceae SBP genes, and all the duplicated genes were subjected to purify selection. In addition, about two-thirds of the Euphorbiaceae SBP genes are potential targets of miR156, and some miR-regulated SBP genes exhibited high intensity expression and differential expression in different tissues. The expression profiles related to different stress treatments demonstrated broad involvement of Euphorbiaceae SBP genes in response to various abiotic factors and hormonal treatments.
Conclusions
In this study, 77 SBP genes were identified in four Euphorbiaceae species, and their phylogenetic relationships, protein physicochemical characteristics, duplication, tissue and stress response expression, and potential roles in Euphorbiaceae development were studied. This study lays a foundation for further studies of Euphorbiaceae SBP genes, providing valuable information for future functional exploration of Euphorbiaceae SBP genes.
Keywords: Euphorbiaceae, SBP-box, miR156, Tissue expression, Stress response, Gene duplication
Background
Transcription factors (TFs) are DNA-binding proteins that play essential roles in the regulatory networks of critical developmental processes [1]. According to the specific protein structure, TFs can be divided into distinct families. SQUAMOSA promoter-binding protein (SBP)-box (briefly: SBP) or SBP-like (SPL) genes encode a type of TF family that is uniquely conserved in plants. SBP genes were first identified in Antirrhinum majus, and they were found to regulate the expression of MADS-box genes, which are critical in floral development [2]. Since then, studies on SBP genes have continually been carried out. As a result, SBP genes have continually been identified in plants ranging from monocyte algae to flowering plants [3, 4]. It has been reported that SBP genes play critical roles in regulating flowering, fruit ripening, phase transition, and other physiological processes. In Arabidopsis thaliana, AtSPL3, AtSPL4, and AtSPL5 are direct upstream activators of LEAFY, FRUITFULL, and APETALA1, and they redundantly promote flowering [5]. They also integrate developmental aging and photoperiodic signals in a process that involves the flowering locus T (FT)-flowering locus D (FD) module in A. thaliana [6]. In addition, AtSPL9 and AtSPL15 as well as AtSPL2, AtSPL10, and AtSPL11 are regarded as regulators of plastochron and branching [7, 8]. AtSPL1 and AtSPL12 have been reported to play roles in plant thermotolerance during the reproductive stage [9]. AtSPL7 is a regulator of copper homeostasis and responses to light and copper [10]. There are also reports on SBP genes of other species: an SBP gene in Solanum lycopersicum (tomato) is critical for normal ripening [11]; OsSPL16 of Oryza sativa (rice) is a regulator of grain size, shape, and quality [12]; and OsSPL14 plays a role in controlling tiller growth in rice [13].
SBP genes encode a class of proteins that have a conserved DNA-binding domain (SBP-specific domain) that contains about 75 amino acid residues (aa). The SBP-specific domain is sufficient to bind to the GTAC core motif [2, 14–16]. There are three common structures in all SBP-specific domains: two zinc fingers and a nuclear localization signal (NLS). The NLS and the second zinc finger partly overlap [16]. Additionally, some SBP genes can be regulated by miRNAs (about 22–24 nt), which reduce protein levels at the transcriptional or translational stage by complementarily binding to their target mRNAs [17–19]. MiR156 plays the most important regulatory roles out of almost all the miRNAs that regulate SBP genes (with target sites located either in the coding region [CDS] or 3′ untranslated region [UTR]) [20, 21]. It has been predicted that 10 of the 16 AtSPL genes are potential targets of miR156/157 (collectively known as miR156). Due to regulation by miRNAs, some SBP genes are involved in complex regulatory processes. For example, miR156 improves the drought tolerance of Medicago sativa by silencing SPL13 [22] and it regulates the juvenile-to-adult phase transition by regulating downstream target SBP genes [5, 6, 23]. Additionally, via miR156 regulation, AtSPL3 temporally regulates shoot development in A. thaliana [24].
Euphorbiaceae is a large and widespread plant family that consists of more than 8000 species, including herbs, perennial shrubs, and trees. They are evolutionarily diverse, and have various traits that allow them to adapt to dynamic environmental conditions. With the increasing demand for food, industrial raw materials, ornamental plants, and herbal medicines, Euphorbiaceae plants have become increasingly attractive. There are many agri-economically important Euphorbiaceae species that have been widely cultivated, such as Ricinus communis (castor bean), Manihot esculenta (cassava), Jatropha curcas (physic nut), and Hevea brasiliensis (rubber tree). Castor bean can be cultivated at a large range of latitudes, and its oil is an important industrial raw material for producing lubricants and paints [25, 26]. Cassava has a starch-enriched root, and it has been a crucial food crop and is also ideal for bioethanol production [27, 28]. Physic nut has seeds with a high oil content that can be processed into biodiesel [29, 30]. The rubber tree is the most important source of natural rubber production, which is indispensable in daily life [31]. However, there are few studies on these non-model plants. More in-depth research, such as understanding the structure, evolution, and function of key gene families, is required to improve crop productivity and commercialization.
The SBP-box gene family has been identified and characterized in different plant species, such as A. thaliana [14], Malus domesrica (apple) [32], Physcomitrella patens (a moss species) [4], and Zea mays (maize) [33]. However, the SBP genes in Euphorbiaceae, and their evolutionary and functional characteristics, are rarely studied. Fortunately, the continuous publication of genome sequencing data [34–37] allows more in-depth research to be conducted on the Euphorbiaceae SBP-box gene family. Herein, we performed a genome-wide investigation of the SBP-box gene family in four Euphorbiaceae species. 77 SBP genes were identified using both local protein–protein Basic Local Alignment Search Tool (BLASTP) and hidden Markov model (HMM) searches. These genes were divided into three length ranges, and into 10 well-defined groups based on total sequence similarity and structural conservation. Duplication events and synteny blocks also supported our grouping scheme and revealed the details of the expansion process of Euphorbiaceae SBP genes. Additionally, a large amount of Euphorbiaceae SBP genes can be regulated by miR156. According to the expression profiles associated with different tissues and stress treatments, a large amount of miR-regulated SBP genes are highly differentially expressed in different tissues and the stress responses are ubiquitous among either miR-regulated or non-regulated SBP genes. Thus, we conducted a comprehensive analysis of Euphorbiaceae SBP genes, and provided valuable evolutionary information for further research.
Results
Identification and characterization
Previous studies on the SBP-box gene family have mainly focused on the model plant A. thaliana. There are few studies on non-model plants such as Euphorbiaceae plants. Zhang and Ling reported on the identification and structural analysis of castor bean SBP genes, but they provided little function prediction information [38]. Here, we performed a comparative analysis of SBP genes from four representative Euphorbiaceae species: cassava, rubber tree, physic nut, and castor bean (Table 1). We systematically identified and characterized the SBP genes of Euphorbiaceae, and predicted their potential functions.
Table 1.
SBP gene members and data sources
To comprehensively identify the SBP genes of each Euphorbiaceae species, we performed a whole-genome scan to identify protein-coding genes containing the SBP-specific domain by using both BLASTP and HMM search, and we then removed the proteins with incomplete SBP-specific domains. A total of 77 SBP genes containing 145 transcripts were identified (Additional file 1: Table S1). For each Euphorbiaceae species, the number of SBP genes varied from 15 to 26, comprising 15 in physic nut, 15 in castor bean, 21 in cassava, and 26 in rubber tree. The number of SBP genes was closely associated with genome size. For example, rubber tree and cassava had a relatively large number of SBP genes and they both experienced a recent genome duplication event [34, 39].
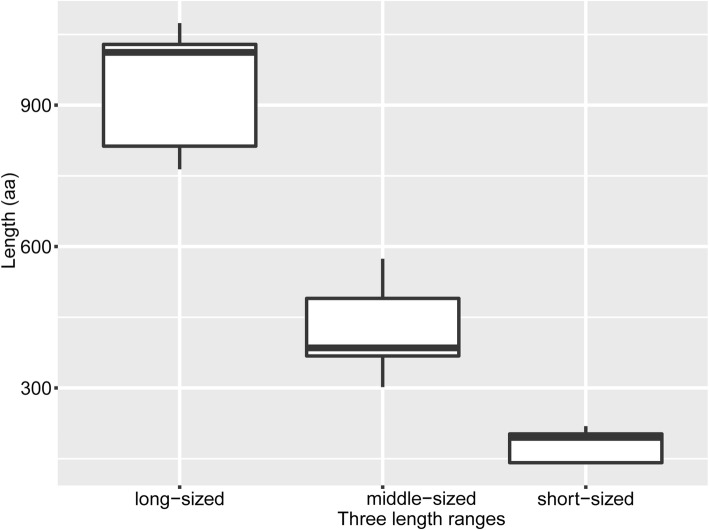
To further characterize the SBP proteins, the basic properties including protein length, isoelectric point value, and molecular weight were analyzed (Additional file 1: Table S2). The Euphorbiaceae SBPs covered a large range of lengths (140–1074 aa). Notably, the lengths exhibited a trimodal distribution (Fig. 1, Additional file 1: Table S2). The short-sized SBPs contained 140–219 aa with an average length of 182 aa; the middle-sized SBPs contained 302–557 aa with an average length of 418 aa; and the long-sized SBPs contained > 780 aa with an average length of 956 aa. The number of SBP genes in the short-, middle-, and long-sized length categories were: 15, 41, and 21, respectively. The corresponding molecular masses were 15.69–24.4, 33.94–63.49, and 85.6–119.32 kDa, respectively.
Fig. 1.
The distribution of three length ranges of SBPs. Y-axis represents protein length (aa); X-axis lists three length ranges
Phylogenetic analysis and classification
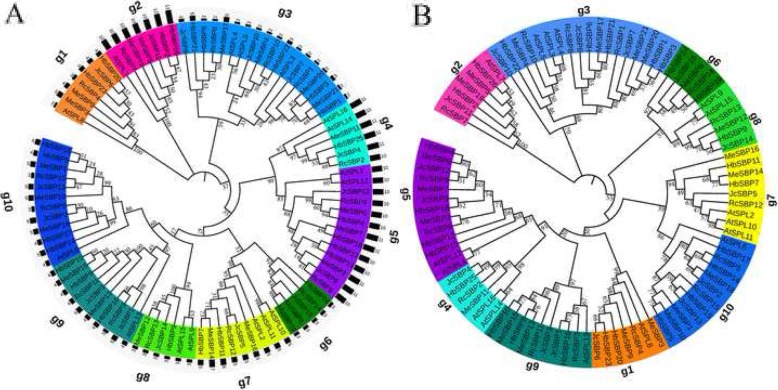
To better understand the functions and evolutionary trajectory of the Euphorbiaceae SBP genes, a phylogenetic analysis of the 77 Euphorbiaceae SBPs plus 16 A. thaliana SPLs was implemented (Fig. 2). We first constructed a neighbor-joining phylogenetic tree involving the 93 SBPs. (Fig. 2a). The SBPs were divided into 10 distinct groups according to the phylogenetic analysis, namely, g1, g2, g3, g4, g5, g6, g7, g8, g9, and g10. This phylogenetic relationship was further confirmed by the maximum likelihood analysis showing that each group was supported by a bootstrap value > 60% (Fig. 2b). Nine groups (all except g6) contained A. thaliana SPLs, which is consistent with previous results [14, 40]. In addition, for the groups containing AtSPL genes, the Euphorbiaceae SBP genes were often close together, while the A. thaliana SBP genes were also close together. The protein characteristics of each group are summarized in Table 2. The exon number in each group exhibited a uniform tendency that was consistent with protein length (Fig. 2a).
Fig. 2.
The phylogenetic tree. The neighbor-joining tree (a) was created using the MEGA7.0 program (bootstrap value set at 1000). The maximum likelihood tree (b) was constructed by PAUP* program. All these SBP proteins were divided into 10 groups, respectively are: g1, g2, g3, g4, g5, g6, g7, g8, g9, g10. The SBP genes in a specific group were marked with a specific color. The bootstrap values were marked by percentage, ‘%’ was omited. The intron number for each SBP gene was displayed in a black bar outmost (a)
Table 2.
The physicochemcial properties of 10 Euphorbiaceae SBP groups
| Groups | Mean Length (aa) |
Mean Mw | Mean Pi | Target site |
|---|---|---|---|---|
| g1 | 304.7 | 34,075.1 | 8.95 | None |
| g2 | 782.7 | 87,961.2 | 6.52 | None |
| g3 | 181.1 | 20,208.9 | 8.55 | 3’UTR |
| g4 | 1072 | 118,801.4 | 8.82 | None |
| g5 | 1009.2 | 111,898.3 | 6.86 | None |
| g6 | 403 | 44,980.9 | 7.97 | CDS |
| g7 | 483.3 | 52,934.7 | 9.24 | CDS |
| g8 | 374.3 | 39,878.7 | 9.24 | CDS |
| g9 | 376.2 | 41,260.6 | 8.66 | CDS |
| g10 | 512.5 | 56,049.3 | 7.55 | CDS |
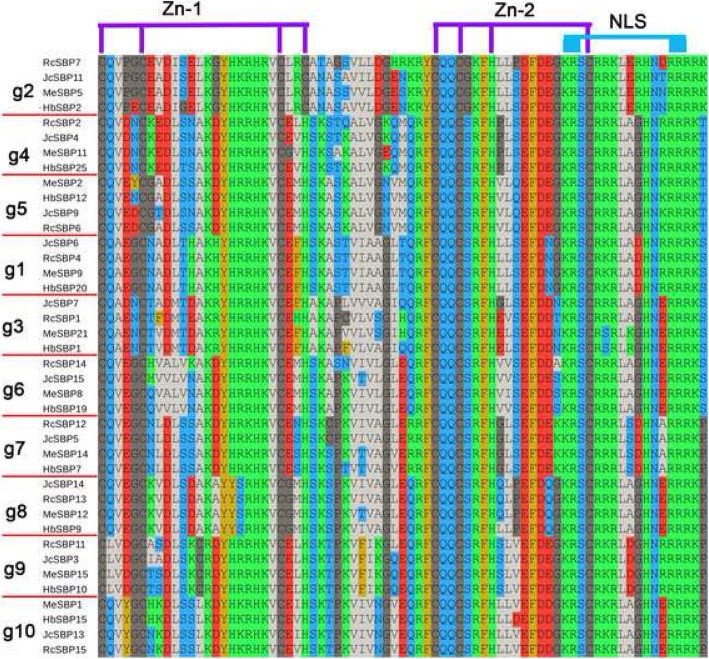
We also conducted multiple sequence alignment for the conserved SBP-specific domain, which contained approximately 75 aa. Due to high structural similarity, we selected only one SBP gene per species per group for better visualization. All SBP-specific domains contained two zinc finger motifs and one nuclear localization signal (NLS) motif (Fig. 3). Nevertheless, the first zinc finger motif for g2 (Cys-Cys-Cys-Cys) was different from that in the other groups (Cys-Cys-Cys-His). For all the members of the 10 groups, compared with the first zinc finger, there was no structural difference in the second zinc finger (which was typically Cys-Cys-His-Cys). Moreover, each group had its own sequence features. For example, the second amino acid residue in g9 was L, while the fifth amino acid residue was K in g4 and G in its sister group g5.
Fig. 3.
The multiple alignment of SBP-specific domain. One gene in each group for per species was chosen. Zn-1, Zn-2 and one NLS are highlighted on the top
Gene structure and conserved motif analysis
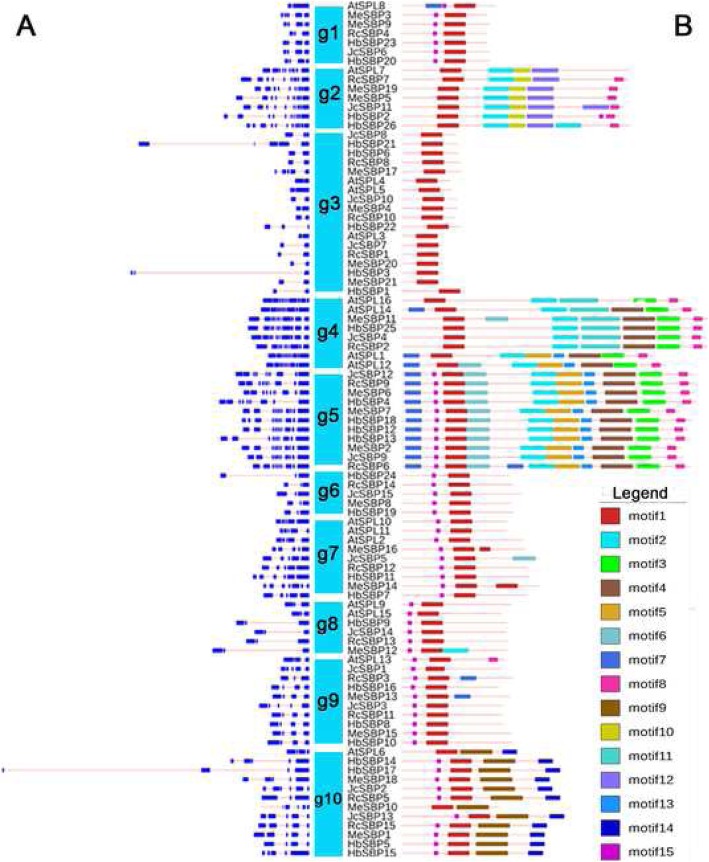
We further examined the structures of all SBP genes, comprising 77 in Euphorbiaceae and 16 in A. thaliana (Fig. 4a). The structural patterns were similar within each group but distinct between any two groups. In addition, the intron lengths of AtSPL genes were shorter than those in Euphorbiaceae genes. To identify the structural similarities and differences in SBPs between groups, a conserved motif analysis was performed. A total of 15 conserved motifs, including the SBP-specific domain (motif1), were found (Fig. 4b, Additional file 2: Fig. S1). The motif number was consistent with the protein length (Fig. 4b); the proteins in g2/4/5 were rich in motifs, sharply contrasting with the proteins in g3, which had only one motif. Some motifs were conserved across groups of different length ranges. For example, motif15 was shared for each middle-sized group and long-sized g5. Some motifs were group-specific: motif9 and motif14 were unique to g10, which was different from other middle-sized groups that contained only 2–3 motifs. Moreover, g4 and g5 shared many motifs, while motif5/13/4 were g5-specific and motif6 was g4-specific. Among the long-sized groups, g2 exhibited many differences in motifs compared to g4 and g5. In addition, g5 always contained both Ankyrin (ANK) and transmembrane regions, and the g5 proteins may be involved in protein–protein interactions.
Fig. 4.
SBP gene structures and motifs. Exons are indicated by blue box; introns are indicated by pink lines; UTR sequences are indicated by black boxes. The motifs are highlighted in different colored boxes with numbers 1 to 15. The phylogenetic groups of g1 to g10 are indicated in the middle. a Schematic representation of intron-exon composition of Euphorbiaceae SBP genes. b Schematic representation of conserved motifs of Euphorbiaceae SBP transcription factors
Chromosomal locations and gene duplication events
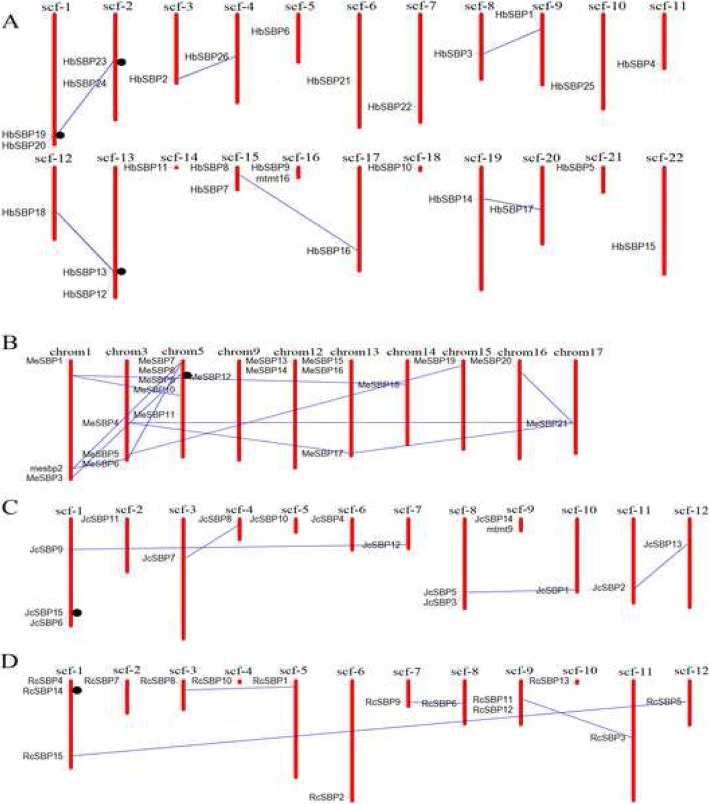
The chromosomal distribution of the Euphorbiaceae SBP genes throughout the four Euphorbiaceae genomes was plotted using MapInspect software. Because of the lack of chromosome-level assembly data for physic nut, castor bean, and rubber tree, we plotted their SBP gene distribution at the scaffold level instead of the chromosome level (Fig. 5, Additional file 1: Table S3). Gene duplication events among the Euphorbiaceae SBP genes were also examined (Fig. 5, Additional file 1: Table S4.1). MCScan searching combined with micro-fragment comparison was used to find accurate duplicate gene pairs. Based on these two methods, 26 segment duplications were found: 12 in cassava, 6 in rubber tree, 4 in physic nut, and 4 in castor bean (Additional files 1: Table S4.1). The rubber tree contained the largest number of SBP genes but a relatively low number of duplications. Imperfect sequencing data partly led to the incomplete linear relationship between the number of duplicate gene pairs and the genome size. Segment duplications made a greater contribution to the Euphorbiaceae SBP gene expansions than tandem duplications (Additional file 1: Table S4.2). Six tandem duplication gene pairs were identified (Fig. 5). Interestingly, each SBP gene in g6 had one tandem duplication gene in g1 (HbSBP19-HbSBP20, HbSBP24-HbSBP23, JcSBP15-JcSBP6, RcSBP14-RcSBP4, and MeSBP8-MeSBP9), which suggests that these tandem duplication SBP genes may result in functional differentiation.
Fig. 5.
Chromosomal locations and gene duplication events of Euphorbiaceae SBP genes. For cassava, the sequence number represents the chromosome number. For physic, rubber tree and castor bean, the scaffold numbers are indicated on the top and their detail scaffold IDs are recorded in Additional file 1: Table S3. SBP gene pairs from segmental duplications are linked by blue lines; tandem duplications are marked by black circle. Each species are plotted in a unique part of (a) rubber tree, (b) cassava, (c) physic nut, (d) castor bean
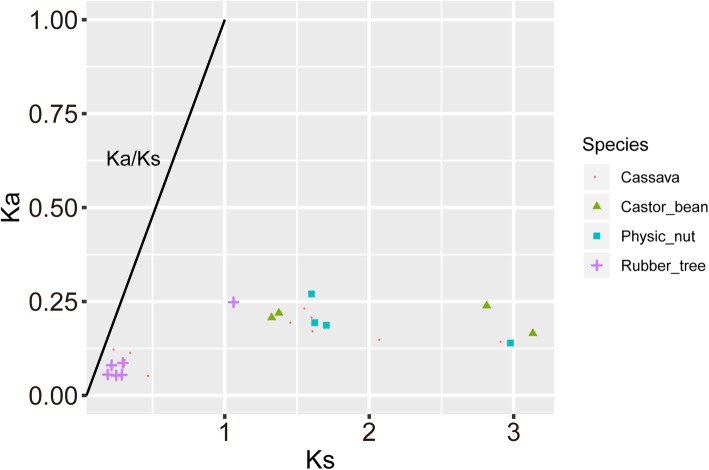
All the predicted segment duplications were found within group, and they support our grouping scheme well. To further understand the evolutionary constraints on the Euphorbiaceae SBP genes, synonymous (Ks) and nonsynonymous (Ka) substitutions per site and their ratio (Ka/Ks) were calculated for the segment duplication gene pairs to explore their roles in the expansionary processes of SBP genes. The time to a certain duplication event can be calculated using the Ks value, as synonymous mutations accumulate at a relatively constant rate over time. Some Ks values were < 1 (marked –S) while others were 1–3 (marked –L) (Fig. 6). The bimodal distribution of the Ks values indicates that there were two large-scale duplication events. Ks-S duplications only existed in cassava and rubber tree, whereas Ks-L duplications were shared by all four Euphorbiaceae species (Additional file 1: Table S4.1). Given the Ks-L values in rubber tree, the –L duplications are likely to be associated with the triplication event related to all core eudicots [41]. The –L duplications generated branches consisting of conserved Euphorbiaceae genes. All the Ka-L values were greater than the Ka-S values (Fig. 6). However, the Ka-L/Ks-L values were lower than the Ka-S/Ks-S ones, which mean that selection pressure on Ka was higher than Ks for SBP genes (Fig. 6). All Ka/Ks values were < 0.5 (Fig. 6), suggesting that the Euphorbiaceae SBP-box gene family underwent strong purifying selection to reduce detrimental mutations after duplication.
Fig. 6.
Ka, Ks and their ratio. Gene pairs from different species are indicated by different scatter. The x and y axes denote Ks and Ka for each gene pair and the black line represents Ka/Ks ratio = 1. The –S range are the gene pairs whose Ks value less than 1, and the –L range are the gene pairs whose Ks value more than 1. Detailed values of Ka, Ks and Ka/Ks listed in Additional file 1: Table S4
Synteny analysis
To explore the evolutionary process of the Euphorbiaceae SBP-box gene family, we conducted a comparative analysis of synteny blocks of genomes among the four Euphorbiaceae species and A. thaliana (Additional file 3: Fig. S2). Here, 141 syntenic blocks between Euphorbiaceae species were discovered (Additional file 3: Fig. S2). A high level of synteny relationships were found at both the species level (21/21 SBP genes in cassava, 15/15 in physic nut, 13/15 in castor bean, and 17/26 in rubber tree) and group level (all 10 groups were covered). Moreover, no intergroup synteny blocks were found (Additional file 1: Table S5), which is in accordance with the segment duplication results and validated our grouping scheme.
Prediction of microRNA target sites
We found the target sites of miR156 either in the CDS or 3’UTR (Table 3). For both A. thaliana and Euphorbiaceae, there was a similar ratio (2/1) of with- to without-target SBP genes. Long-sized SBP genes had no target sites, while both the middle- and short-sized SBP genes had target sites located either in CDS or 3’UTR (Table 2). However, one exception was that g1, a middle-sized group, contained no miR156 target (neither in A. thaliana nor in the Euphorbiaceae species).
Table 3.
The miR156 target information of Euphorbiaceae SBP genes
| Location | ID | CDS/3’UTR length | Target site | miR site |
|---|---|---|---|---|
| CDS | JcSBP1 | 1014 | 818 GUGCUCUCUCUCUUCUGUCA 837 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | JcSBP2 | 1590 | 1148 GUGCUCUCUCUCUUCUGUCA 1167 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | JcSBP3 | 954 | 683 GUGCUCUCUCUCUUCUGUCA 702 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | JcSBP5 | 1443 | 1154 GUGCUCUCUCUCUUCUGUCA 1173 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | JcSBP13 | 1725 | 1289 GUGCUCUCUCUCUUCUGUCA 130 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | JcSBP14 | 1119 | 230 AAGGGUGUAAAGUGGAUCUGA 250 | 21 UACCCAUAAUUCAUCUAGACU 1 |
| CDS | JcSBP15 | 1260 | 830 GUGCUCUCUCUCUUCUGUCA 849 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | JcSBP7 | 237 | 150 CUGCUCUCUCUCUUCUGUCA 169 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | JcSBP8 | 530 | 4 UGCUCCCUCUCUUCUGUCAU 23 | 20 ACGAGAGAGAGAAGACAGUU 1 |
| 3’UTR | JcSBP10 | 214 | 25 UGCUCCCUCUCUUCUGUCAU 44 | 20 ACGAGAGAGAGAAGACAGUU 1 |
| CDS | RcSBP3 | 1149 | 968 GUGCUCUCUCUCUUCUGUCA 987 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | RcSBP5 | 1674 | 1229 CUGCUCUCUCUCUUCUGUCA 1248 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | RcSBP11 | 1155 | 884 GUGCUCUCUCUCUUCUGUCA 903 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | RcSBP12 | 1452 | 1163 GUGCUCUCUCUCUUCUGUCA 1182 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | RcSBP13 | 1134 | 782 GUGCUCUCUCUCUUCUGUCA 801 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | RcSBP14 | 1167 | 809 GUGCUCUCUCUCUUCUGUCA 828 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | RcSBP15 | 1542 | 1094 GUGCUCUCUCUCUUCUGUCA 1113 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | RcSBP1 | 214 | 122 AUGCUCUCUCUCUUCUGUCA 141 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | RcSBP8 | 235 | 6 UUGCUCUCUCUCUUCUGUCA 25 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | RcSBP10 | 325 | 32 AUGCUCCCUCUCUUCUGUCA 51 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP5 | 1518 | 1073 GUGCUCUCUCUCUUCUGUCA 1092 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP7 | 1446 | 1160 GUGCUCUCUCUCUUCUGUCA 1179 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP8 | 1152 | 881 GUGCUCUCUCUCUUCUGUCA 900 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP9 | 1125 | 773 GUGCUCUCUCUCUUCUGUCA 792 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP10 | 1149 | 878 GUGCUCUCUCUCUUCUGUCA 897 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP11 | 1473 | 1181 GUGCUCUCUCUCUUCUGUCA 1200 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP14 | 1596 | 1151 GUGCUCUCUCUCUUCUGUCA 1170 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP15 | 1500 | 1073 GUGCUCUCUCUCUUCUGUCA 1092 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP16 | 1107 | 917 GUGCUCUCUCUCUUCUGUCA 936 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP17 | 1674 | 1229 GUGCUCUCUCUCUUCUGUCA 1248 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP19 | 1224 | 818 GUGCUCUCUCUCUUCUGUCA 837 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | HbSBP24 | 1197 | 791 GUGCUCUCUCUCUUCUGUCA 810 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | HbSBP1 | 263 | 156 AUGCUCUCUCUCUUCUGUCA 175 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | HbSBP3 | 266 | 114 AUGCUCUCUCUCUUCUGUCA 133 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | HbSBP6 | 389 | 18 UUGCUCUCUAUCUUCUGUCA 37 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | HbSBP21 | 2797 | 18 UUGCUCCCUCUCUUCUGUCA 37 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | HbSBP22 | 318 | 19 ACGCUCCCUCUCUUCUGUCA 38 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | MeSBP1 | 1518 | 1073 GUGCUCUCUCUCUUCUGUCA 1092 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | MeSBP8 | 1212 | 818 GUGCUCUCUCUCUUCUGUCA 837 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | MeSBP10 | 1050 | 869 GUGCUCUCUCUCUUCUGUCA 888 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | MeSBP12 | 1125 | 773 GUGCUCUCUCUCUUCUGUCA 792 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | MeSBP13 | 1146 | 875 GUGCUCUCUCUCUUCUGUCA 894 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | MeSBP14 | 1467 | 1178 GUGCUCUCUCUCUUCUGUCA 1197 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | MeSBP15 | 1158 | 881 GUGCUCUCUCUCUUCUGUCA 900 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | MeSBP16 | 1437 | 1151 GUGCUCUCUCUCUUCUGUCA 1170 | 20 CACGAGAGAGAGAAGACAGU 1 |
| CDS | MeSBP18 | 1563 | 1118 GUGCUCUCUCUCUUCUGUCAU 1138 | 21 CACGAGAGAGAGAAGACAGUU 1 |
| 3’UTR | MeSBP4 | 211 | 16 AUGCUCCCUCUCUUCUGUCA 35 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | MeSBP17 | 996 | 18 UUGCUCCCUCUCUUCUGUCA 37 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | MeSBP20 | 218 | 171 GUGCUCUCUCUCGUAUGUCA 190 | 20 CACGAGAGAGAGAAGACAGU 1 |
| 3’UTR | MeSBP21 | 384 | 122 AUGCUCUCUAUCUUCUGUCA 141 | 20 CACGAGAGAGAGAAGACAGU 1 |
Tissue expression profiles of JcSBP genes
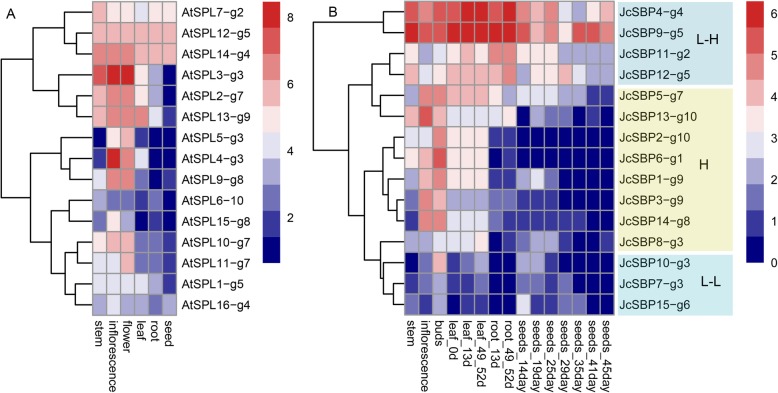
To further illustrate the potential functions of each SBP gene, we conducted a comparative analysis of the expression data (from stem, inflorescence, buds, leaf, root, and seed) of physic nut and A. thaliana (Fig. 7). Because of the high similarity of SBP genes among the four Euphorbiaceae species, the analysis of the SBP genes of physic nut is very representative. Hierarchical clustering was used to visualize the global expression profile of the JcSBP genes (Fig. 7b). The expression patterns of the JcSBP genes could be divided into low differential expression between tissues (JcSBP4, JcSBP9, JcSBP11, JcSBP12, JcSBP10, JcSBP7, and JcSBP15) and high differential expression between tissues (JcSBP5, JcSBP13, JcSBP2, JcSBP6, JcSBP1, JcSBP3, JcSBP14, and JcSBP8). The former could be further divided into low expression genes (JcSBP10, JcSBP7, and JcSBP15) and high expression genes (JcSBP4, JcSBP9, JcSBP11, and JcSBP12).
Fig. 7.
The tissue expression profiles. The tissue expression profiles of A. thaliana (a). Expression profiles of physic nut SBP genes among different tissues and development stages (b). The low expression differential groups were highlighted in blue (marked with L), and the high expression differential groups were highlighted in orange (marked with H). The blue groups can be further divided into high expressional and low expressional groups that marked with L-H and L-L respectively
There were significant differences in the expression profiles of JcSBP genes between the with- and without-target genes (Fig. 7b). The JcSBP genes of g2/4/5 (long-sized groups) contained no target sites, and they were highly expressed without differential expression between tissues. In contrast, the with-target JcSBP genes in the middle-sized groups were highly differentially expressed in different tissues (with high expression in the buds and inflorescences, though several genes also played roles in the stem, leaf, or root). However, the tissue expression differences of the other with-target JcSBP genes (in the short-sized groups) were not as significant as the with-target JcSBP genes in the middle-sized groups.
The expression patterns of AtSPL genes in g3 and g10 were significantly different from those in physic nut (Fig. 7). Regarding g3, the relative expression intensity of AtSPL genes was higher than those in physic nut, and they were highly expressed in more tissues. In contrast, regarding g10, the relative expression intensity of JcSBP genes was higher than AtSPL genes. The expression signal of AtSPL6 was barely observable. However, JcSBP2 and 13 were redundantly expressed in the stem, inflorescence, and root.
Stress response expression profiles of JcSBP genes
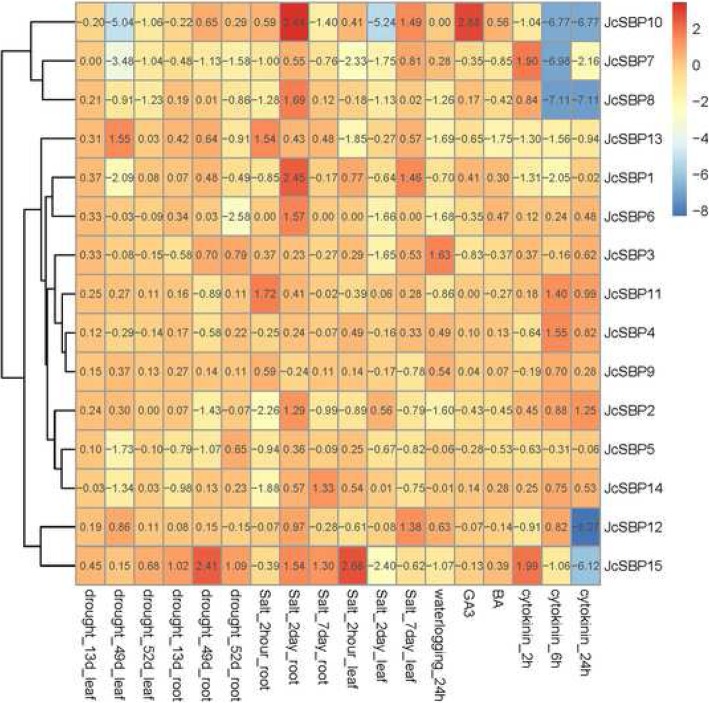
To further explore the possible physiological processes in which Euphorbiaceae SBP genes participate, the expression levels in physic nut in response to various abiotic stresses (salt, drought, and waterlogging) and hormonal treatments (gibberellin 3 [GA3], 6-benzylaminopurine [BA], and cytokinin) were obtained. Log2 transformations of the ratio of the treatment group data to their corresponding control group data are displayed in Fig. 8; log2 transformed values > 1 or < − 1 were viewed as representing differential expression.
Fig. 8.
Expression profiles of physic nut SBP genes in response to abiotic stress and hormone stress treatments. The numerical values in different color scales represent log2 transform of the ratio of the experimental group and control group in a specific treatment condition
First, in response to drought (Fig. 8), JcSBP7 and JcSBP10 showed > 4-fold decreased expression in the leaves. In the roots, JcSBP7, JcSBP6, JcSBP2, and JcSBP5 were down-regulated, while JcSBP15 was up-regulated under all drought treatments. Second, in response to salt (Fig. 8), eight JcSBP genes (JcSBP1, JcSBP2, JcSBP6, JcSBP8, JcSBP10, JcSBP11, JcSBP13, and JcSBP15) were up-regulated in the roots. Six JcSBP genes (JcSBP2, JcSBP6, JcSBP8, JcSBP7, JcSBP10, and JcSBP14) showed > 2-fold decreased expression in the roots. In the leaves, there were six down-regulated JcSBP genes (JcSBP10, JcSBP7, JcSBP13, JcSBP6, JcSBP3, and JcSBP15) and four up-regulated JcSBP genes (JcSBP10, JcSBP1, JcSBP12, and JcSBP15), while JcSBP10 and JcSBP15 showed both up- and down-regulated patterns. Third, in response to waterlogging treatment, several JcSBP genes were down-regulated (JcSBP8, JcSBP13, JcSBP6, JcSBP2, and JcSBP15) or up-regulated (JcSBP3).
We further assessed the expression level of JcSBP genes in response to GA3, BA, and cytokinin treatments (Fig. 8). Compared to the control groups, JcSBP10 was increased almost 8-fold in response to GA3. JcSBP13 decreased by > 2-fold in response to BA. Compared with the response to GA3 and BA, more JcSBP genes were up-regulated in response to cytokinin. Five JcSBP genes (JcSBP10, JcSBP8, JcSBP13, JcSBP1, and JcSBP12) decreased in response to cytokinin and three increased (JcSBP11, JcSBP4, and JcSBP2). Additionally, two JcSBP genes (JcSBP7 and JcSBP15) displayed both up- and down-regulated expression.
Discussion
In view of their excellent agricultural traits, several Euphorbiaceae species have become important food sources or industrial raw materials. Cassava [27, 28], physic nut [29, 30], castor bean [25, 42], and rubber tree [31] have been widely domesticated and cultivated. The continuously increasing quantity of genome sequencing data, genetic linkage maps, and abundance of high-throughput transcriptome sequencing data make further exploration of gene functions in non-model plants like Euphorbiaceae species possible. Previous studies on SBP genes have revealed their crucial roles in plant development, especially in flower development, signal transduction, and defense processes [5–10]. However, the functions of Euphorbiaceae SBPs are still unknown. In this study, genome-wide analyses (including the analyses of the evolutionary trajectory, miR156 regulation, and expression profiles) of the Euphorbiaceae SBP-box gene family were conducted to shed new light on Euphorbiaceae SBP genes.
The phylogenetic relationships, synteny analysis, and tissue expression profiles showed that the SBP genes of Euphorbiaceae and A. thaliana are similar in structure, evolutionary trajectory, and functions. In light of the high similarity between SBP genes of Euphorbiaceae and A. thaliana, we can predict the functions of some of the SBP genes of Euphorbiaceae based on the well-studied AtSPL genes. Regarding the long-sized groups, AtSPL7 (in g2) has been reported to be related to Cu homeostasis in A. thaliana, and it regulates the expression of Cu-responsive genes and is considered to be a central regulator of copper homeostasis [43, 44]. The gene that is homologous to AtSPL7 was conserved in Euphorbiaceae and, similar to A. thaliana, it exhibited significantly high expression in the roots. Mutations of AtSPL14 (in g4) result in resistance to the fungal toxin fumonisin B1 [45]. AtSPL1 and AtSPL12 (in g5) play redundant roles in thermotolerance at the reproductive stage [9].
Regarding the middle-sized groups (g1/6/7/8/9/10), one of their remarkable characteristics is that they can be regulated by miR156 (all except g1). Due to regulation by miR156, these SBP genes play critical roles in plant development. AtSPL13 (in g9) has been implicated in delaying leaf outgrowth during germination [46]. AtSPL2, AtSPL10, and AtSPL11 (in g7) affect the morphological features associated with phase change [7]. AtSPL9 and AtSPL15 (in g8) play redundant roles in reproductive transition and vegetative phase change [8, 47]. AtSPL8 (in g1) is related to seed formation, root development, and petal trichome [48, 49]. As in A. thaliana, all the middle-sized JcSBP genes were differentially expressed between different tissues and exhibited high intensity expression, which suggests that they may be involved in different physiological processes and play critical roles in plant development and reproduction.
As we know, A. thaliana is monoecious, while physic nut is diecious; A. thaliana is a kind of biennial herb, while physic nut is a kind of perennial woody plant. It is worth exploring the functions of Euphorbiaceae SBP genes regarding the flowering process, phase transformation, seed development, etc. We found that the expression patterns of the SBP genes in g3 were significantly different between A. thaliana and physic nut, and there may be functional differences between them. In addition, regarding g10, the tissue expression profiles of A. thaliana were significantly different from those of physic nut in both relative expression intensity and the differential expression between different tissues. Moreover, g6 was absent from A. thaliana but conserved in Euphorbiaceae, and it was highly expressed in seeds and exhibited a relatively high response to salt, drought, and cytokinin. These results suggest that there may be some new functions or regulatory forms of SBP genes in Euphorbiaceae, and understanding these genes is helpful to further reveal the physiological regulation processes in Euphorbiaceae.
Sometimes plants are cultivated for their roots, sometimes for their seeds, and sometimes for their fruits. The formation of different tissues and organs may be related to different regulatory processes. Our study suggests that some SBP genes are differentially expressed in different tissues and organs, and may be associated with specific physiological processes. For example, physic nut and castor bean are cultivated for their seeds, so flower development and seed formation are important for a higher crop production. Both middle- and small-sized SBP genes are related to inflorescence or bud development according to their tissue expression profiles (Fig. 7b). In addition, several SBP genes were found to be related to seed development, such as JcSBP5/13/1/8, which express relative high in seeds (Fig. 7b). On the other hand, unlike physic nut and castor bean, cassava is cultivated for its roots, and JcSBP5/13 are highly expressed in the roots (Fig. 7b). Therefore, increasing the study of these SBP genes may contribute to the deeper understanding of specific physiological processes and subsequent agricultural genetics studies.
Conclusions
SBP-box genes encode a series of plant-specific TFs, which have been identified and characterized in several species. Significant progress has been achieved regarding the identification of the functions of some SBP genes in several species, but little attention has been paid to non-model plants. In the present study, we identified 77 putative SBP genes in the genomes of four Euphorbiaceae species. From the results of the phylogeny analysis, we divided the Euphorbiaceae SBP genes into 10 independent groups, and the subsequent results regarding the structural analysis and the distribution of duplication gene pairs supported our grouping scheme. The genome comparison indicated that segment duplication played crucial roles in Euphorbiaceae SBP gene expansion, and all the duplication gene pairs were subjected to purify selection. In addition, two-thirds of Euphorbiaceae SBP genes may be regulated by miR156, and these miR-regulated genes all belonged to the middle- or short-sized groups. Comparative synteny analysis between the genomes of five species (including A. thaliana) showed that a large number of SBP genes were located in syntenic regions, implying that these SBP genes probably come from common ancestors. Furthermore, to illustrate the probable functions of these SBP genes, we conducted a comparative analysis of the expression profiles of JcSBP and AtSPL genes in various tissues/organs. Most miR-regulated JcSBP genes were more differentially expressed than miR-nonregulated JcSBP genes. G6 is conserved in Euphorbiaceae but not in A. thaliana, and we assume that it is functionally active as it was highly expressed in the buds and stems. However, the short-sized JcSBP genes were not as active as their homologous AtSPL genes, indicating there may be some functional differences between A. thaliana and Euphorbiaceae. Lastly, many JcSBP genes were up- or down-regulated in response to certain abiotic or phytohormone stresses, implying that they may be involved in the responses to various stresses or in physic nut development. Our data provide valuable information for further functional studies of Euphorbiaceae SBP genes. The flowering mechanism between A. thaliana and Euphorbiaceae and the high demand for increases in crop yield make the exploration of Euphorbiaceae SBP genes highly valuable.
Methods
Data sources
Genomic and proteomic sequences were obtained from the Phytozome portal for cassava (manihot_esculenta_v6, JGI; https://phytozome.jgi.doe.gov/pz/portal.html), National Center for Biotechnology Information (NCBI) for castor bean (JCVI_RCG_v1.1; https://www.ncbi.nlm.nih.gov/), NCBI for rubber tree (ASM165405v1; https://www.ncbi.nlm.nih.gov/), and NCBI for physic nut (JatCur_1.0; https://www.ncbi.nlm.nih.gov/). The A. thaliana genomic and proteomic sequences were obtained from TAIR (TAIR10 release; https://www.arabidopsis.org/). Gene expression data for physic nut were obtained from the NCBI (https://www.ncbi.nlm.nih.gov/).
Identification, characterization, and phylogenetic analysis
Both HMM [50] and BLASTP [51] searches were performed to accurately identify the SBP TFs in the Euphorbiaceae species. The well-characterized A. thaliana SBP protein sequences were used as queries for BLASTP searches (e-value ≤1e-10). The SBP-specific HMM profile (PF03110) was used for queries, and the HMMER toolkit was used in the HMM searches. The conserved SBP-specific domain was confirmed using Simple Modular Architecture Research Tool (SMART) [52] (http://smart.embl-heidelberg.de/), and the incomplete SBP-specific domains were discarded. In the cases involving multiple transcripts of the same gene, a dot followed by a serial number was added at the end of each name. The physicochemical properties, including protein length, molecular weight (MW), and isoelectric point (Pi), for the identified SBP proteins were predicted using the ExPASy Proteomics Server (https://prosite.expasy.org/) [53]. Multiple sequence alignment of SBP protein sequences was performed by Multiple Sequence Comparison by Log-Expectation (MUSCLE) in MEGA v7.0 [54]. A neighbor-joining tree was constructed using MEGA v7.0. The maximum likelihood tree was generated using the PAUP* program, employing the JTT substitution model and 100 bootstrap replicates [55].
Conserved motifs and gene structure analysis
The online Multiple Expectation Maximization for Motif Elucidation (MEME) toolkit was used to identify additional motifs (http://meme-suite.org/) [56], which were conserved and located outside the SBP-specific domain region. All SBP protein sequences were used for the queries. The parameters were set as follows: minimum width was 6, maximum width was 150, motif number was 15, and minimum number of sites was 2. Both SBP gene sequences and the corresponding coding sequences were uploaded to the online Gene Structure Display Server (GSDS v2.0; http://gsds.cbi.pku.edu.cn/) to obtain intron/exon structure information [57].
Chromosomal localization
A gene location map for each Euphorbiaceae species based on the chromosomal position of each SBP gene was generated by MapInspect (https://mapinspect.software.informer.com/). SBP gene locations of cassava were mapped into chromosomes, and SBP gene locations of the other three species were mapped into scaffolds due to their incomplete genome assembly information.
Detection of gene duplication events and synteny relationships
Duplicated gene pairs derived from tandem or segmental duplication were identified according to the method described in the Plant Genome Duplication Database [58]. An all-against-all BLASTP comparison (e-value ≤1e-10) provided gene pairs for syntenic clustering using MCScan v1.1 (e-value ≤1e-10) [59]. Segment duplication was also predicted by the micro-fragment comparison method. The SBP duplicate gene pairs from the above analysis were again examined by BLASTP (e-value ≤1e-10), and all the SBP genes obtained from the above analysis were used as anchors of micro-fragments generated by the collection of 20 upstream and 20 downstream coding genes. Tandem duplications were identified if two SBP genes were next to each other or they had one unrelated gene between them [60].
Estimation of synonymous (Ks) and nonsynonymous (Ka) substitutions per site and their ratio (Ka/Ks)
SBP gene pairs caused by segmental duplication were used to estimate Ka, Ks, and their ratio. Coding sequences from segmentally duplicated SBP gene pairs were aligned using webPRANK (https://www.ebi.ac.uk/goldman-srv/webprank/) [61]. KaKs_Calculator v2.0 [62] was used to compute Ka, Ks, and Ka/Ks. All the counting processes followed the YN model [63] (a simple model of voting). The Ka/Ks value can reveal the selective pressure of duplicated genes [64], and the Ks value can reflect the divergence time for duplication events. All-against-all BLASTP searches (e-value ≤1e-10) were conducted to investigate the synteny relationships of the proteomes of the four Euphorbiaceae species and A. thaliana. The synteny blocks were then calculated using MCScan v1.1 [59], and the synteny relationships were visualized using Circos v0.69–5 [65].
MicroRNA target prediction
MiR156 and miR157 were combined into the miR156 family in miRBase (https://www.mirbase.org/) [66], due to their highly similar structures. The well-characterized miR156 mature sequences from miRBase were set as the background data to search against the mRNA sequences of Euphorbiaceae SBP genes using psRNATarget program (http://plantgrn.noble.org/v1_psRNATarget/) [67] with default parameters. The detailed positions of miRNA (located in the CDS or 3’UTR region) were further determined on the basis of the locations of target sites and the CDS length.
Expression analysis
SBP gene expression data in six tissues (stem, inflorescence, bud, root, and seed) and under various treatments (gibberellin [GA3], 6-benzylaminopurine [BA], cytokinin, high salt concentration, drought, and waterlogging) of the four Euphorbiaceae species were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/). A. thaliana expression data were obtained from TAIR (TAIR10 release; https://www.arabidopsis.org/). All data were analyzed using the Tuxedo suite (TopHat and Cufflinks; http://post.queensu.ca/~rc91/NGS/TuxedoTutorial.html) [68], and they were then upper quartile normalized and log2 transformed. The gene expression profiles were displayed in heatmaps using the R package pheatmap [69].
Supplementary information
Additional file 1. This file contains the additional tables (Table S1-S5) associated with the manuscript. Table numbers and titles were listed as follows: Table S1: The information of Euphorbiaceae SBP genes. Table S2: The protein physicochemical properties of Euphorbiaceae SBP proteins. Table S3: The parallel table of scaffold IDs and serial number. Table S4: The information of duplications. Table S5: The identified synteny relationships between Euphorbiaceae species.
Additional file 2: Fig. S1: The sequence logos of 15 motifs.
Additional file 3: Fig. S2: The synteny relationships among Euphorbiaceae and A. thaliana.
Acknowledgments
We thank all colleagues in our laboratory for helpful discussions and technical assistance.
About this supplement
This article has been published as part of BMC Genomics, Volume 20 Supplement 9, 2019: 18th International Conference on Bioinformatics. The full contents of the supplement are available at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-20-supplement-9
Abbreviations
- ANK
Ankyrin
- BA
6-Benzylaminopurine
- CDS
Coding DNA sequence
- GA
Gibberellins
- HMM
Hidden Markov Model
- Ka
Nonsynonymous substitutions per non-synonymous site
- Ks
Synonymous substitutions per synonymous site
- MEME
Multiple Expectation Maximization for Motif Elucidation
- ML
Maximum likelihood
- MW
Molecular weight
- NJ
Neighbor-joining
- NLS
Nuclear localization signal
- Pi
Isoelectric point
- UTR
Untranslated regions
Authors’ contributions
CL conceived, designed, and supervised this study. JL performed the experiments, analyzed the data and wrote the paper, JL, XG, SS, CL revised the paper. All authors reviewed and approved the manuscript.
Funding
The publication cost of this article was funded by the National Natural Science Foundation of China (No. 31970609), Start-up Fund from Xishuangbanna Tropical Botanical Garden, ‘Top Talents Program in Science and Technology’ from Yunnan Province.
Availability of data and materials
The datasets supporting the conclusions of this article are included in the article and in its additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jing Li, Email: lijing2@xtbg.ac.cn.
Xiaoyang Gao, Email: gaoxiaoyang@xtbg.ac.cn.
Shiye Sang, Email: sangshiye@xtbg.ac.cn.
Changning Liu, Email: liuchangning@xtbg.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6319-4.
References
- 1.Gong W, Shen YP, Ma LG, Pan Y, Du YL, Wang DH, et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 2004;135(2):773–782. doi: 10.1104/pp.104.042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein J, Saedler H, Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet. 1996;250(1):7–16. doi: 10.1007/BF02191820. [DOI] [PubMed] [Google Scholar]
- 3.Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418(1–2):1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Riese M, Hohmann S, Saedler H, Munster T, Huijser P. Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene. 2007;401(1–2):28–37. doi: 10.1016/j.gene.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell. 2009;17(2):268–278. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung JH, Lee HJ, Ryu JY, Park CM. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol Plant. 2016;9(12):1647–1659. doi: 10.1016/j.molp.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009;50(12):2133–2145. doi: 10.1093/pcp/pcp148. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67(1–2):183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao LM, Liu YQ, Chen DY, Xue XY, Mao YB, Chen XY. Arabidopsis transcription factors SPL1 and SPL12 confer plant Thermotolerance at reproductive stage. Mol Plant. 2017;10(5):735–748. doi: 10.1016/j.molp.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Zhao X, Li J, Cai H, Deng XW, Li L. MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell. 2014;26(12):4933–4953. doi: 10.1105/tpc.114.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38(8):948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44(8):950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 13.Luo L, Li W, Miura K, Ashikari M, Kyozuka J. Control of tiller growth of rice by OsSPL14 and Strigolactones, which work in two independent pathways. Plant Cell Physiol. 2012;53(10):1793–1801. doi: 10.1093/pcp/pcs122. [DOI] [PubMed] [Google Scholar]
- 14.Cardon G, Höhmann S, Klein J, Nettesheim K, Saedler H, Huijser P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene. 1999;237(1):91–104. doi: 10.1016/S0378-1119(99)00308-X. [DOI] [PubMed] [Google Scholar]
- 15.Cardon GH, Höhmann S, Nettesheim K, Saedler H, Huijser P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 1997;12(2):367–377. doi: 10.1046/j.1365-313X.1997.12020367.x. [DOI] [PubMed] [Google Scholar]
- 16.Birkenbihl RP, Jach G, Saedler H, Huijser P. Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol. 2005;352(3):585–596. doi: 10.1016/j.jmb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 18.Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25(7):2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18(10):758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant MicroRNA targets. Cell. 2002;110(4):513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 21.Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13(7):343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Arshad M, Feyissa BA, Amyot L, Aung B, Hannoufa A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017;258:122–136. doi: 10.1016/j.plantsci.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133(18):3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogunniyi DS. Castor oil: a vital industrial raw material. Bioresour Technol. 2006;97(9):1086–1091. doi: 10.1016/j.biortech.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Mubofu EB. Castor oil as a potential renewable resource for the production of functional materials. Sustain Chem Process. 2016;4(1):11. doi: 10.1186/s40508-016-0055-8. [DOI] [Google Scholar]
- 27.Balat M, Balat H. Recent trends in global production and utilization of bio-ethanol fuel. Appl Energy. 2009;86(11):2273–2282. doi: 10.1016/j.apenergy.2009.03.015. [DOI] [Google Scholar]
- 28.Schmitz PM, Kavallari A. Crop plants versus energy plants--on the international food crisis. Bioorg Med Chem. 2009;17(12):4020–4021. doi: 10.1016/j.bmc.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 29.King AJ, He W, Cuevas JA, Freudenberger M, Ramiaramanana D, Graham IA. Potential of Jatropha curcas as a source of renewable oil and animal feed. J Exp Bot. 2009;60(10):2897–2905. doi: 10.1093/jxb/erp025. [DOI] [PubMed] [Google Scholar]
- 30.Achten WMJ, Mathijs E, Verchot L, Singh VP, Aerts R, Muys B. Jatropha biodiesel fueling sustainability? Biofuels Bioprod Biorefin. 2007;1(4):283–291. doi: 10.1002/bbb.39. [DOI] [Google Scholar]
- 31.Lau NS, Makita Y, Kawashima M, Taylor TD, Kondo S, Othman AS, et al. The rubber tree genome shows expansion of gene family associated with rubber biosynthesis. Sci Rep. 2016;6:28594. doi: 10.1038/srep28594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Hou H, Li X, Xiang J, Yin X, Gao H, et al. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus x domestica Borkh.) Plant Physiol Biochem. 2013;70:100–114. doi: 10.1016/j.plaphy.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 33.Mao H-D, Yu L-J, Li Z-J, Yan Y, Han R, Liu H, et al. Genome-wide analysis of the SPL family transcription factors and their responses to abiotic stresses in maize. Plant Gene. 2016;6:1–12. doi: 10.1016/j.plgene.2016.03.003. [DOI] [Google Scholar]
- 34.Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, et al. Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol. 2010;28(9):951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang C, Yang M, Fang Y, Luo Y, Gao S, Xiao X, et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat Plants. 2016;2(6):16073. doi: 10.1038/nplants.2016.73. [DOI] [PubMed] [Google Scholar]
- 36.Wu P, Zhou C, Cheng S, Wu Z, Lu W, Han J, et al. Integrated genome sequence and linkage map of physic nut (Jatropha curcas L.), a biodiesel plant. Plant J. 2015;81(5):810–821. doi: 10.1111/tpj.12761. [DOI] [PubMed] [Google Scholar]
- 37.Prochnik S, Marri PR, Desany B, Rabinowicz PD, Kodira C, Mohiuddin M, et al. The cassava genome: current progress, future directions. Trop Plant Biol. 2012;5(1):88–94. doi: 10.1007/s12042-011-9088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang SD, Ling LZ. Genome-wide identification and evolutionary analysis of the SBP-box gene family in castor bean. PLoS One. 2014;9(1):e86688. doi: 10.1371/journal.pone.0086688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren R, Wang H, Guo C, Zhang N, Zeng L, Chen Y, et al. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol Plant. 2018;11(3):414–428. doi: 10.1016/j.molp.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z, Wang X, Gu S, Hu Z, Xu H, Xu C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene. 2008;407(1–2):1–11. doi: 10.1016/j.gene.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 41.Jiao Y, Leebens-Mack J, Ayyampalayam S, Bowers JE, McKain MR, McNeal J, et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13(1):R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fidias P, Grossbard M, Lynch TJ. A phase II study of the Immunotoxin N901-blocked ricin in small-cell lung cancer. Clin Lung Cancer. 2002;3(3):219–222. doi: 10.3816/CLC.2002.n.006. [DOI] [PubMed] [Google Scholar]
- 43.Lännenpää M, Jänönen I, Hölttä-Vuori M, Gardemeister M, Porali I, Sopanen T. A new SBP-box gene BpSPL1 in silver birch (Betula pendula) Physiol Plant. 2004;120(3):491–500. doi: 10.1111/j.0031-9317.2004.00254.x. [DOI] [PubMed] [Google Scholar]
- 44.Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. SQUAMOSA promoter binding protein–Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell. 2009;21(1):347. doi: 10.1105/tpc.108.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone JM, Liang X, Nekl ER, Stiers JJ. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005;41(5):744–754. doi: 10.1111/j.1365-313X.2005.02334.x. [DOI] [PubMed] [Google Scholar]
- 46.Martin RC, Asahina M, Liu P-P, Kristof JR, Coppersmith JL, Pluskota WE, et al. The microRNA156 and microRNA172 gene regulation cascades at post-germinative stages in Arabidopsis. Seed Sci Res. 2010;20(2):79–87. doi: 10.1017/S0960258510000085. [DOI] [Google Scholar]
- 47.Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev Cell. 2016;37(3):254–266. doi: 10.1016/j.devcel.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Unte US, Sorensen A-M, Pesaresi P, Gandikota M, Leister D, Saedler H, et al. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell. 2003;15(4):1009. doi: 10.1105/tpc.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Schwarz S, Saedler H, Huijser P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol Biol. 2007;63(3):429–439. doi: 10.1007/s11103-006-9099-6. [DOI] [PubMed] [Google Scholar]
- 50.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 51.Mount DW. Using the Basic Local Alignment Search Tool (BLAST) Cold Spring Harb Protoc. 2007;2007(7):pdb.top17. doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- 52.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. PNAS. 1998;95(11):5857. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(Web Server issue):W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). 2003.
- 56.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41(Database issue):D1152–D1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320(5875):486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 60.The Arabidopsis Genome I Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 61.Löytynoja A, Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinf. 2010;11(1):579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinf. 2010;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17(1):32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 64.Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18(9):486–487. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 65.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39(Web Server issue):W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Protocols. 2012;7:562. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng Z, et al. RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014;14(1):169. doi: 10.1186/1471-2229-14-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. This file contains the additional tables (Table S1-S5) associated with the manuscript. Table numbers and titles were listed as follows: Table S1: The information of Euphorbiaceae SBP genes. Table S2: The protein physicochemical properties of Euphorbiaceae SBP proteins. Table S3: The parallel table of scaffold IDs and serial number. Table S4: The information of duplications. Table S5: The identified synteny relationships between Euphorbiaceae species.
Additional file 2: Fig. S1: The sequence logos of 15 motifs.
Additional file 3: Fig. S2: The synteny relationships among Euphorbiaceae and A. thaliana.
Data Availability Statement
The datasets supporting the conclusions of this article are included in the article and in its additional files.