Abstract
Background
Most HIV infections originate from individuals who are undiagnosed and unaware of their infection. Estimation of this quantity from surveillance data is hard because there is incomplete knowledge about (i) the time between infection and diagnosis (TI) for the general population, and (ii) the time between immigration and diagnosis for foreign-born persons.
Methods
We developed a new statistical method for estimating the incidence of HIV-1 and the number of undiagnosed people living with HIV (PLHIV), based on dynamic modelling of heterogeneous HIV-1 surveillance data. The methods consist of a Bayesian non-linear mixed effects model using multiple biomarkers to estimate TI of HIV-1-positive individuals, and a novel incidence estimator which distinguishes between endogenous and exogenous infections by modelling explicitly the probability that a foreign-born person was infected either before or after immigration. The incidence estimator allows for direct calculation of the number of undiagnosed persons. The new methodology is illustrated combining heterogeneous surveillance data from Sweden between 2003 and 2015.
Results
A leave-one-out cross-validation study showed that the multiple-biomarker model was more accurate than single biomarkers (mean absolute error 1.01 vs ≥1.95). We estimate that 816 [95% credible interval (CI) 775-865] PLHIV were undiagnosed in 2015, representing a proportion of 10.8% (95% CI 10.3-11.4%) of all PLHIV.
Conclusions
The proposed methodology will enhance the utility of standard surveillance data streams and will be useful to monitor progress towards and compliance with the 90–90-90 UNAIDS target.
Keywords: HIV-1, incidence estimation, undiagnosed HIV-1 infections, BED assay, pol sequences
Key Messages
Combined heterogeneous HIV-1 surveillance data and biomarker data can be used to estimate both local incidence and the number of undiagnosed persons living with HIV.
Explicit modelling of the dynamics, heterogeneity and correlation of multiple biomarkers over time improved estimation of time between infection and diagnosis.
Explicit modelling of the probability that foreign-born persons were infected before or after immigration improves accuracy of estimates of endogenous incidence and undiagnosed persons living with HIV.
The endogenous incidence of HIV-1 in Sweden is declining, despite continued immigration of HIV-1 infected persons.
The proportion of undiagnosed PLHIV decreased over 2010–15 and was estimated to be 10.8% (95% CI, 10.3-11.4%) in 2015.
Introduction
The majority of new HIV-1 infections originate from individuals who are undiagnosed and unaware of their infection, especially in countries with good access and adherence to antiretroviral therapy (ART), which greatly reduces infectiousness.1–4 Thus, knowledge about the size of the undiagnosed HIV-1 population is highly relevant for public health and HIV prevention. In 2014, the Joint United Nations Programme on HIV and AIDS (UNAIDS) launched the 90–90-90 target, which states that by 2020: (i) 90% of all persons living with HIV (PLHIV) should know their status; (ii) 90% of all diagnosed HIV cases should receive ART; and (iii) 90% of all people receiving ART should have achieved viral suppression.5 The European Center for Disease Prevention and Control (ECDC) estimates that one in seven (14%) PLHIV in Europe are unaware of their infection.6
Estimation of the size of the undiagnosed population is challenging, because the time of infection usually is unknown and the time until diagnosis (TI) is highly variable due to differences in testing behaviour, risk awareness and rate of disease progression. Until now, most estimates of HIV-1 incidence and undiagnosed population have been based on methods that classify patients as recently or long-term infected rather than estimate TI directly. Several such methods based on CD4+ T lymphocyte (CD4) counts, HIV-1 antibody tests and viral sequence diversity have been described. CD4 counts are commonly used,7 but their rate of decline is variable8–10 limiting their utility as a single biomarker. HIV-1 antibody concentrations increase over time from infection approaching an asymptote in long-term infections.11 The BED IgG-capture enzyme immunoassay (BED assay) has been used to estimate incidence12,13 by classifying recent and long-term infections. Sequence-based methods exploit the increase in intra-patient sequence diversity following infection, which can be approximated by the fraction of polymorphic nucleotides in the partial HIV-1 pol gene sequences that are used for detecting drug resistance mutations (referred to as pol polymorphisms).14,15
Fewer studies have attempted to estimate TI continuously, rather than distinguishing between recent and long-term infections, and to account for individual variations. Sommen et al.16 used antibody levels to two HIV-1 antigens (IDE and V3) to calculate the posterior distribution of HIV-1 infection times and incidence in France. Romero-Severson et al.11 used BED assay results to combine a model of within-host time-continuous IgG dynamics17 with a Bayesian estimator for the incidence of HIV-1 in Sweden in 2002–09. However, these studies neither explicitly accounted for immigration of infected persons, nor attempted to estimate the size of the undiagnosed HIV-1-infected population.
Here we present a statistical method to estimate HIV-1 incidence and the number of undiagnosed PLHIV. The method is based on: (i) a model of the joint dynamics of multiple-biomarker levels from the time of infection that allow a more accurate estimation of individual TIs; and (ii) the estimation of the time from immigration to diagnosis for exogenous infections using unlinked surveillance data. The TI model uses BED, CD4 and pol polymorphisms, but can be easily generalized to include other or additional biomarkers. The estimation of the time from immigration to diagnosis allows the assessment of how endogenous and exogenous infections contribute to the undiagnosed fraction.
In the application of the model to surveillance data from Sweden, we estimated that endogenous HIV-1 infections have decreased over 2010–15 and that 10.8% (95% CI, 10.3-11.4%) of all infected persons in Sweden were undiagnosed in 2015. This estimate is in line with previously published results,18 and adds uncertainty quantification.
Methods
The methodology to estimate HIV incidence and number of undiagnosed PLHIV was developed for countries like Sweden, i.e. countries with reliable HIV surveillance systems, availability of biomarker data on (a subset of) newly diagnosed patients, and non-negligible immigration.
Data
Multiple surveillance data sources in Sweden were collated for this work. We used published data on 1357 HIV-1 infected patients diagnosed in Sweden between 2003 and 201011,17,19,20 with complete biomarker data on CD4 counts, BED levels and pol polymorphism counts at diagnosis. These patients represented 39% of all patients diagnosed in Sweden during this period. These data included likely country of infection, transmission route, last negative and first positive HIV-1 test, laboratory evidence of primary HIV-1 infection and plasma HIV-1 RNA levels, but not data on time of arrival in Sweden for foreign-born persons. A subset of 31 treatment-naïve patients. having longitudinal biomarker data and known infection times, was used to build and validate the multiple biomarker model.
Data on the yearly number of diagnosed HIV-1 cases in Sweden between 2003 and 2015 were collected as part of mandatory national case reporting, and was stratified by reported transmission route, place of infection (Sweden or abroad) and AIDS at HIV diagnosis. In total 5777 patients were diagnosed in Sweden during 2003–15. Data on linkage to care (diagnosed HIV-1 patients regularly attending scheduled visits) were obtained from the Swedish National Quality Assurance Registry InfCareHIV, as previously described.18 For 2466 of 2978 (83%) of the foreign-born patients, data on the time between first arrival and diagnosis in Sweden were available; however, these data were anonymized and could not be linked to the other data. For details on the patients and laboratory methods, see Supplementary Table S1 and Section 1, available as Supplementary data at IJE online.
Modelling approach
Our modelling approach to estimate the incidence of new HIV-1 infections and the size of the undiagnosed population consists of the following four main steps: (i) estimate parameters of our novel multiple-biomarker model using longitudinal biomarker data on 31 individuals with known infection dates; (ii) apply the multiple-biomarker model to estimate TI of the HIV-1 infected population, stratified by transmission group and country of origin using data from case reports, and use bootstrap for patients with missing biomarkers; (iii) estimate HIV incidence trends over the period 2000 to 2015, using the time distribution from TI to diagnosis obtained in the previous step and including the probabilistic allocation of foreign-born infected individuals to endogenous or exogenous infections; and iv) calculation of the undiagnosed fraction with its uncertainty using bootstrap.
Multiple-biomarker model
The biomarkers , are modelled jointly as a function of TI as follows:
where denotes the individual with biomarkers measured at calendar time after infection date . The dynamics are described by biomarker specific curves , fixed and random effects and as i.i.d. error terms such that . The model is formulated in a Bayesian framework and fitted using Markov chain Monte Carlo (MCMC) to longitudinal data with known TI. Full specification on the prior distributions assigned to the unknown parameters can be found in Supplementary Section 2.1, available as Supplementary data at IJE online. The biomarker model used data from three biomarkers and was trained on a subset of 31 treatment-naïve patients having longitudinal biomarker data and known infection times. For comparison, the three biomarkers were also modelled separately. To assess the representativeness of the dataset, we compared the estimated parameter values with values reported in the literature.
Estimation of TI and handling of foreign-born cases
The Bayesian formulation allows us to use our model to estimate directly the unknown time of infection of new individuals, by treating their TI as a ‘latent’ variable, described by a prior distribution. This approach was used in a leave-one-out cross-validation analysis using the 31-patient longitudinal dataset (where TI was represented by any of the ), as well as in the prediction of TI for newly infected individuals (1357 patients between 2003 and 2010), with biomarkers measured only at time of diagnosis, i.e. .
To estimate HIV-1 incidence and undiagnosed PLHIV, foreign-born persons infected before first arrival should only be counted after arrival to the country of investigation, e.g. Sweden. As immigration dates were not available for the above-mentioned 1357 patients, we first estimated a typical distribution for the time interval between immigration and HIV diagnosis using independent and unlinked surveillance data, and then applied this distribution to foreign-born patients among the 1357 patients.
Incidence estimation
We extended the model proposed by Sommen et al.16 to estimate HIV-1 incidence, defined as the number of new infections. Our modifications allow us: i) to consider all reported cases within a moving window of size (i.e. [) in the incidence estimation at time , denoted as (rather than only cases reported at time ); and (ii) to account for imported infections. Typically, the time period of interestis 1 year, and we denote the incidence as where . The incidence is expressed as a weighted sum of posterior densities of infection (or entry) times:
| (1) |
where and denote the number of endogenous and exogenous infections diagnosed in , respectively, and and are equal to the inverse of the probability of being diagnosed in for individuals infected at or immigrated at , respectively. Here, represents the posterior distribution of the infection time for an individual (diagnosed at ) obtained using observed biomarkers, and is the ‘backward’ distribution used to generate an entry time for a foreign-born case given diagnosis at in the country (by year of diagnosis). The time of the latest negative test that individual may have is denoted by . The second term in (1) is evaluated only when the generated immigration time is more recent than the infection time as estimated by the biomarkers. This comparison allows for the definition of the sets and . For unobserved years (i.e. upper end of the moving window falls after the present time) the number of diagnoses is assumed to follow a Poisson distribution with mean equal to the reported cases in the last observed year.
Estimation of undiagnosed fraction
The calculation of the HIV-infected undiagnosed individuals in a specific year follows directly from the incidence estimation (both endogenous and exogenous). Here, we define more generally the incidence of infection at time as and the number of undiagnosed individuals at time can be written as:
where are the diagnosis times for individuals estimated to have been infected (or immigrated) at time . That is, the number of undiagnosed patients in year is the cumulative sum of the product of the number of endogenous/exogenous infections that occurred/were imported years ago and the probability of being not yet diagnosed after years. This calculation is stratified by transmission route.
Credible intervals are obtained by generating 100 bootstrap samples of size 1000 from the distributions of infection times. All statistical analyses were carried out using R version 3.3.21 and JAGS.22 For further details on the model and parameters, see the Supplementary Section 2 and Section 3, available as Supplementary data at IJE online.
Results
The multiple-biomarker model improved estimation of TI
The multiple-biomarker model for estimating TI based on BED levels, CD4 counts and pol polymorphisms, was trained on longitudinal data from 31 HIV-1 patients with known infection times. The parameter values describing the growth or decline of the biomarkers for the training data (Supplementary Figures S1–S3 and Table S2, available as Supplementary data at IJE online) agreed with published values,7,14,23–25 which justified the use of the model on biomarker data from other HIV-1 patients. A leave-one-out cross-validation analysis showed that the multiple biomarker model gave more accurate estimates of TI than each of the single biomarkers according to four different measures of precision (Table 1). TI estimates were more precise if biomarkers were measured sooner after infection (MAE = 0.91 at <1 year; 1.20 at 1–2 years; 1.40 at >2 years, see Supplementary Table S3, available as Supplementary data at IJE online). Biomarker measurements at two or three time points, rather than a single time point, only slightly improved TI estimation (Supplementary Table S4, available as Supplementary data at IJE online). This motivated the use of single time point biomarker measurements in the application to newly diagnosed patients in Sweden.
Table 1.
Mean predictive performance of single and multiple biomarker models assessed with a leave-one-out cross-validation analysis evaluated by four measures of precision
| Model | Bias | MAE | RMSE | Coverage |
|---|---|---|---|---|
| CD4 | −2.62 | 3.12 | 3.77 | 0.81 |
| BED | −1.80 | 1.95 | 2.21 | 0.90 |
| POL | −2.16 | 2.22 | 3.20 | 0.81 |
| MBM | −0.68 | 1.01 | 1.38 | 0.93 |
CD4, CD4+ T lymphocyte count; BED, antibody levels measured using the BED IgG-capture enzyme immunoassay; POL, fraction of polymorphisms in HIV-1 pol gene sequences; MBM, multiple biomarker model based on CD4, BED and pol; MAE, mean absolute error; RMSE, root mean square error. All values are in years.
Time between infection and diagnosis varied by transmission route
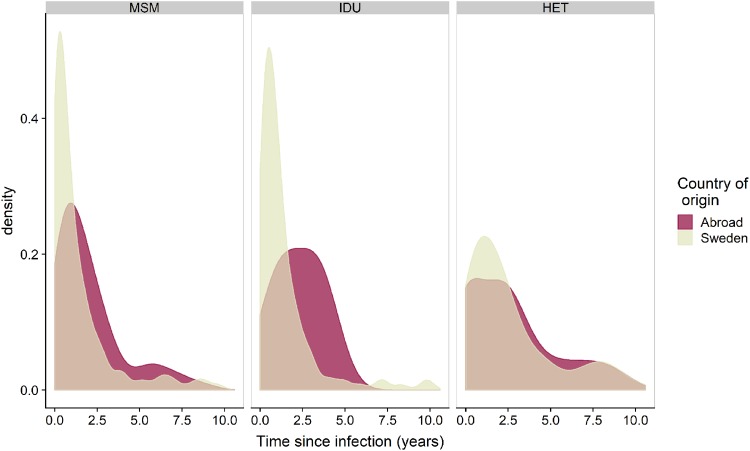
Figure 1 shows the distribution of the estimated time from infection until diagnosis in the three main transmission groups [men who have sex with men (MSM), intravenous drug users (IDU) and heterosexual transmission route (HET)] broken down by place of origin (Sweden or abroad). For MSM and IDU, the estimated time between infection and diagnosis showed similar distributions, with around 60% of individuals being diagnosed within 1 year after infection and 8% being diagnosed >5 years after infection. Heterosexually infected persons had longer time until diagnosis, with around 31% and 19% being diagnosed within 1 year and after >5 years after estimated TI, respectively. As expected, persons reported to have been born abroad also had longer time between estimated TI and diagnosis in each transmission group, as some of them are likely to be exogenous infections, i.e. infected before first arrival to Sweden (overall median 2.0 years vs 0.89 years, Mann-Whitney test, P <0.05).
Figure 1.
Distribution of estimated time from infection (TI) to diagnosis by transmission route and country of origin, modelled using CD4, BED and pol polymorphism data obtained at or close to diagnosis from 1357 patients diagnosed in Sweden 2003-10. MSM, men who have sex with men; IDU, intravenous drug users; HET, heterosexual transmission route.
Decreasing HIV-1 incidence in Sweden
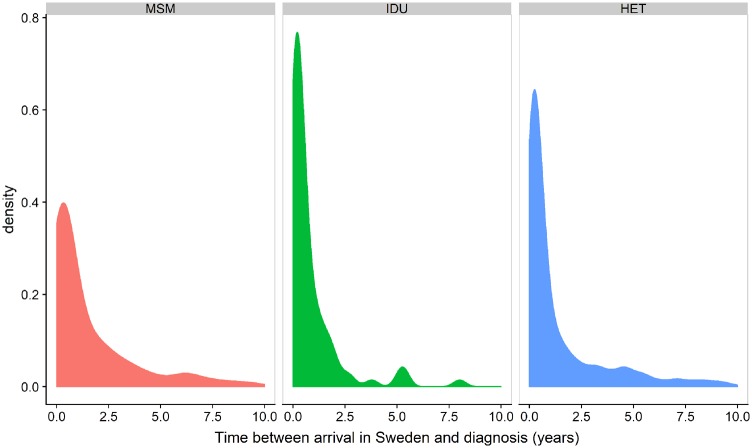
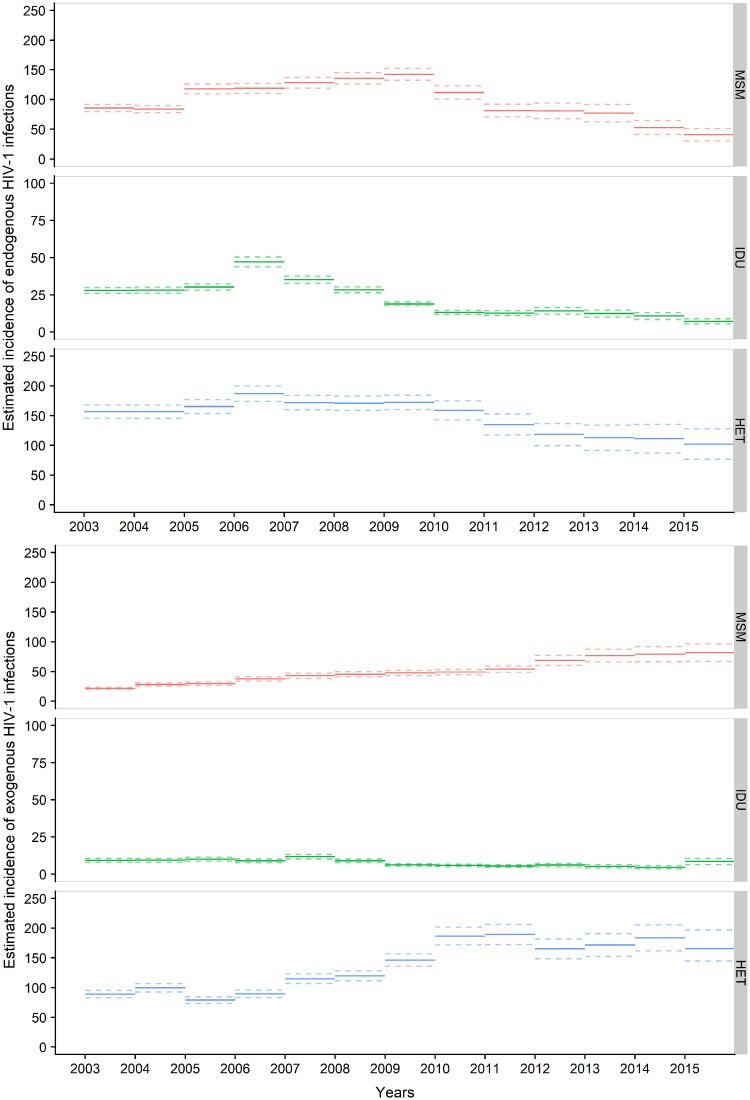
To estimate HIV-1 incidence in Sweden, we explicitly modelled the impact of migration so that patients estimated to have been infected before immigrating to Sweden contributed to incidence only from the estimated date of first entry into the country (Figure 2). Figure 3 shows that the incidence of endogenous infections decreased in all three transmission groups, especially among MSM and IDU. We estimate that 150 (95% CI, 112-187) infections occurred in Sweden in 2015 (41 MSM, 7 IDU and 102 HET), compared with 284 (95% CI, 255-312) in 2010 (112 MSM, 13 IDU and 159 HET), P <0.01. In contrast, the incidence of persons entering Sweden already infected (i.e. exogenous infections) was estimated to have been stable or slightly increasing, with 257 (95% CI, 218-305) persons in this category in 2015 (82 MSM, nine IDU and 166 HET), compared with 242 (95% CI, 221-262) in 2010 (P = 0.15).
Figure 2.
Distribution of estimated time from arrival in Sweden to diagnosis by transmission route, modelled using anonymized data on time between first entry into Sweden and diagnosis for 2466 patients, obtained from the Public Health Agency of Sweden. MSM, men who have sex with men; IDU, intravenous drug users; HET, heterosexual transmission route.
Figure 3.
Estimated HIV-1 incidence in Sweden 2003-15, per year and transmission route. The model explicitly accounts for exogenous infections. Thus, persons estimated to have been infected before first entry in Sweden only contribute to incidence in Sweden from the estimated date of entry. The upper panels show estimated incidence of HIV-1-infection among persons residing in Sweden. The lower panels show the incidence of HIV-1-infected persons entering Sweden for the first time. MSM, men who have sex with men; IDU, intravenous drug users; HET, heterosexual transmission route. Note the scale on the y-axis is different for IDU.
Proportion of undiagnosed HIV-1-infected persons close to 10% UNAIDS target
Our model also provided estimates on the number of undiagnosed PLHIV in Sweden at the end of the years 2010 to 2015. Table 2 shows that number of undiagnosed PLHIV in Sweden was estimated to have decreased from 907 (95% CI, 880-934) persons in 2010 to 816 (95% CI, 775-865) in 2015. During the same period, the number of diagnosed patients linked to care increased from 5281 to 6747, which means that the proportion of undiagnosed PLHIV decreased from 14.7% (95% CI, 14.3-15.0%) in 2010 to 10.8% (95% CI, 10.3-11.4%) in 2015. Of the undiagnosed PLHIV in 2015, 341 persons were estimated to have been infected while living in Sweden (78 MSM, 8 IDU and 255 HET) and 475 persons were estimated to have been infected before first entering Sweden (165 MSM, 11 IDU and 299 HET).
Table 2.
Estimated number and proportion of undiagnosed HIV-1-infected persons in Sweden 2010–15. The number undiagnosed in 2015 was calculated using the estimated incidence divided by main transmission route. The number undiagnosed in previous years (2014–10) was calculated assuming the incidence in the years before 2003 constant and equal to that of 2003
| Year | No. of PLHIV linked to care | No. of undiagnosed PLHIV (95% CI) | Proportion undiagnosed PLHIV (95% CI) |
|---|---|---|---|
| 2010 | 5281 | 907 (880-935) | 14.7% (14.3-15.0%) |
| 2011 | 5616 | 894 (863-925) | 13.8% (13.3-14.1%) |
| 2012 | 5918 | 877 (837-916) | 12.9% (12.3-13.4%) |
| 2013 | 6205 | 865 (826-912) | 12.2% (11.7-12.8%) |
| 2014 | 6469 | 845 (802-896) | 11.5% (11.0-12.1%) |
| 2015 | 6747 | 816 (775-865) | 10.8% (10.3-11.4%) |
Discussion
In this manuscript we present a new method for estimating HIV incidence and size of undiagnosed population which differentiates between exogenous and endogenous infection of foreign-born cases. Our method builds on (i) a Bayesian model to estimate time of infection (TI) using multiple biomarkers, and (ii) an extension of the incidence estimator proposed by Sommen et al.16 to explicitly model the probability that an HIV-1 infection in an immigrating person occurred before or after immigration. Our methodology was designed for use on heterogeneous and unlinked surveillance data.
We applied the method to data from Sweden and estimated TI using three biomarkers (CD4 counts, BED assay results and proportion of polymorphic sites in HIV-1 pol sequences) as well as data on main transmission route, presence of a last negative test, AIDS at diagnosis and diagnosed primary HIV-1 infection (PHI). We treated TI as a continuous random variable rather than as a simple binary state of being recently or long-term infected, which increases power and reduces bias. We found that a combination of the three biomarkers gave more precise TI estimates than one or two of these biomarkers. Each of the three biomarkers have certain advantages and limitations and show considerable inter-individual variability. A well-known disadvantage of the BED assay is that it can give false-positive recent results for patients with low CD4 counts.26,27 This problem is reduced by our model because the inclusion of CD4 counts and pol polymorphisms partially corrects false-recent BED results, and because persons with AIDS at diagnosis where assigned a prior distribution with an average time from infection to diagnosis of 8 years28 (Supplementary Figure S4, available as Supplementary data at IJE online). Furthermore, low BED results in AIDS patients were rare in our dataset (Supplementary Figure S7 and Table S5, available as Supplementary data at IJE online).
We observed a decrease in the estimated incidence of HIV-1 infections and number of undiagnosed PLHIV in Sweden. The proportion of undiagnosed PLHIV was estimated to be 10.8% in 2015, which is close to the 10% UNAIDS 90–90-90 target. We estimated that the incidence of HIV-1-infections among persons residing in Sweden decreased by almost two-thirds from 2010 to 2015. The decrease was more pronounced in MSM and IDU than among heterosexually infected persons. This agrees with the fact that 87% of persons with diagnosed HIV-1-infection were on effective ART in 2015,18 which means that they were effectively non-infectious,29 and that the time between infection and diagnosis was shorter among MSM and IDUs than among heterosexuals (Figure 2). Data from InfCareHIV shows that the coverage of effective ART is very high (87% in 201518) and similar across transmission groups, and thus differences in ART coverage cannot explain the difference in incidence between transmission groups. The important distinction between endogenous and exogenous infections would not be noted if we had not accounted for immigration, as the overall incidence was almost constant over the observation period (Supplementary Figure S9, available as Supplementary data at IJE online).
Despite the training dataset being relatively small (31 patients), the modelled biomarkers trajectories agreed with previously published estimates obtained using larger datasets,7,14,23–25 which gives us confidence that the model design and parameter values are valid for other patients. For the purpose of illustrating the methodology, we assumed a constant incidence of infection before 2003, as official data from the Public Health Institute of Sweden show that the number of new diagnoses has been fairly stable between 1990 and 2003.30 Violations of this assumption would have minimal impact on our estimates of incidence and fraction undiagnosed during 2010–15, which is the focus of the paper. However, our methodology can easily be extended to include a wider time window estimating incidence in past years (e.g. before 2003).
The strengths of our approach are that it relies on routine or easy-to-collect data and provides estimates of the size of undiagnosed population, stratified by HIV transmission group, explicitly accounting for exogenous infections. In addition, the method considers the several sources of uncertainty involved, such as differences in HIV-1 testing behaviour, differences in disease progression and biomarker measurement error. Because the method estimates TI for each investigated person, it allows for detailed HIV-1 epidemiological investigations, such as the causes and consequences of late presentation of HIV infection31 and the effectiveness of HIV-1 prevention (e.g. pre-exposure prophylaxis).32,33 The method can easily be adapted to other biomarkers like the LAg avidity assay and viral diversity.34–36
Our study suggests a way forward for generating up-to-date reports on the key parameters (incidence and number of undiagnosed persons) needed to understand the effectiveness of HIV control methods, the application of public health triage and the progress towards the UNAIDS 90–90-90 targets. The global movement of people in response to a changing world means that health systems will be challenged with increasing numbers of foreign-born persons; surveillance methods will need to correctly account for infections in those populations which occur both before and after migration. Integrating computational modules into public health surveillance streams that can account for complex, heterogeneous data with missing values will greatly increase the utility of surveillance, not only as a passive monitoring tool but as an active intervention aid.
Funding
This work was supported by the Swedish Research Council (grant numbers 340–2013-5003 and K2014-57X-09935) and the National Institutes of Health (NIH) (grant number R01AI087520).
Conflict of interest: None declared.
Supplementary Material
References
- 1. Marks G, Crepaz N, Janssen RS.. Estimating sexual transmission of HIV from persons aware and unaware that they are infected with the virus in the USA. AIDS 2006;20:1447–50. [DOI] [PubMed] [Google Scholar]
- 2. Hall HI, Holtgrave DR, Maulsby C.. HIV transmission rates from persons living with HIV who are aware and unaware of their infection. AIDS 2012;26:893–96. [DOI] [PubMed] [Google Scholar]
- 3. Pharris A, Quinten C, Noori T. et al. Estimating HIV incidence and number of undiagnosed individuals living with HIV in the European Union/European Economic Area, 2015. Euro Surveill 2016, Dec 1. doi: 10.2807/1560-7917.ES.2016.21.48.30417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen MS, Chen YQ, McCauley M. et al. Antiretroviral therapy for the prevention of HIV-1 transmission. N Engl J Med 2016;375:830–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UNAIDS. 90-90-90: An Ambitious Treatment Target to Help End the AIDS Epidemic. Geneva: UNAIDS, 2014. .
- 6.ECDC/WHO. Surveillance Report: HIV/AIDS Surveillance in Europe 2015 Stockholm: ECDC/WHO, 2016.
- 7. Cori A, Pickles M, van SA. et al. CD4+ cell dynamics in untreated HIV-1 infection: overall rates, and effects of age, viral load, sex and calendar time. AIDS 2015;29:2435.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goujard C, Bonarek M, Meyer L. et al. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin Infect Dis 2006;42:709–15. [DOI] [PubMed] [Google Scholar]
- 9. Lodi S, Guiguet M, Costagliola D, Fisher M, De Luca A, Porter K.. Kaposi sarcoma incidence and survival among HIV-infected homosexual men after HIV seroconversion. J Natl Cancer Inst 2010;102:784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Minga A, Lewden C, Gabillard D. et al. CD4 eligibility thresholds: an analysis of the time to antiretroviral treatment in West African HIV-1 seroconverters. AIDS 2011;25:819.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Romero-Severson EO, Petrie CL, Ionides E, Albert J, Leitner T.. Trends of HIV-1 incidence with credible intervals in Sweden 2002–09 reconstructed using a dynamic model of within-patient IgG growth. Int J Epidemiol 2015;44:998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hall HI, Song R, Rhodes P. et al. Estimation of HIV incidence in the United States. JAMA 2008;300:520–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bätzing-Feigenbaum J, Loschen S, Gohlke-Micknis S. et al. Country-wide HIV incidence study complementing HIV surveillance in Germany. Eurosurveillance 2008;13:18971. [PubMed] [Google Scholar]
- 14. Kouyos RD, von WV, Yerly S. et al. Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin Infect Dis 2011;52:532–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giorgi EE, Funkhouser B, Athreya G, Perelson AS, Korber BT, Bhattacharya T.. Estimating time since infection in early homogeneous HIV-1 samples using a Poisson model. BMC Bioinformatics 2010;11:532.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sommen C, Commenges D, Vu SL, Meyer L, Alioum A.. Estimation of the distribution of infection times using longitudinal serological markers of HIV: implications for the estimation of HIV incidence. Biometrics 2011;67:467–75. [DOI] [PubMed] [Google Scholar]
- 17. Skar H, Albert J, Leitner T.. Towards estimation of HIV-1 date of infection: a time-continuous IgG-model shows that seroconversion does not occur at the midpoint between negative and positive tests. PLoS One 2013;8:e60906.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gisslén M, Svedhem V, Lindborg L. et al. Sweden, the first country to achieve the Joint United Nations Programme on HIV/AIDS. UNAIDS/World Health Organization (WHO) 90-90-90 continuum of HIV care targets. HIV Med 2017;18:305–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karlsson A, Björkman P, Bratt G. et al. Low prevalence of transmitted drug resistance in patients newly diagnosed with HIV-1 infection in Sweden 2003–2010. PLoS One 2012;7:e33484.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Widgren K, Skar H, Berglund T, Kling A-M, Tegnell A, Albert J.. Delayed HIV diagnosis common in Sweden, 2003–2010. Scand J Infect Dis 2014;46:862–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Core Development Team. R: A Language and Environment for Statistical Computing Vienna: R Foundation for Statistical Computing, 2016. https://www.R-project.org/.
- 22. Plummer M. JAGS: a program for analysis of Bayesian graphical models using Gibbs sampling. Proceedings of the 3rd International Workshop on Distributed Statistical Computing; 200320-22 March Vienna, 2003; 125.
- 23. Lange N, Carlin BP, Gelfand AE.. Hierarchical Bayes models for the progression of HIV infection using longitudinal CD4 T-cell numbers. J Am Stat Assoc 1992;87:615–26. [Google Scholar]
- 24. Lynch ML, DeGruttola V.. Predicting time to threshold for initiating antiretroviral treatment to evaluate cost of treatment as prevention of human immunodeficiency virus. J R Stat Soc C 2015;64:359–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parekh BS, Hanson DL, Hargrove J. et al. Determination of mean recency period for estimation of HIV type 1 Incidence with the BED-capture EIA in persons infected with diverse subtypes. AIDS Res Hum Retroviruses 2011;27:265–73. [DOI] [PubMed] [Google Scholar]
- 26. Busch MP, Pilcher CD, Mastro TD. et al. Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. AIDS 2010;24:2763–71. [DOI] [PubMed] [Google Scholar]
- 27. Guy R, Gold J, Calleja JMG. et al. Accuracy of serological assays for detection of recent infection with HIV and estimation of population incidence: a systematic review. Lancet Infect Dis 2009;9:747–59. [DOI] [PubMed] [Google Scholar]
- 28. Longini IM Jr, Clark WS, Gardner LI, Brundage JF.. The dynamics of CD4+ T-lymphocyte decline in HIV-infected individuals: a Markov modeling approach. J Acquir Immune Defic Syndr 1991;4:1141–47. [PubMed] [Google Scholar]
- 29. Cohen MS, Chen YQ, McCauley M. et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med 2011;365:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Public Health Agency of Sweden. HIVinfektion (HIV infection), 2018. https://www.folkhalsomyndigheten.se/folkhalsorapportering-statistik/statistikdatabaser-och-visualisering/sjukdomsstatistik/hivinfektion/? p=6087.
- 31. Antinori A, Coenen T, Costagiola D. et al. Late presentation of HIV infection: a consensus definition. HIV Med 2011;12:61–64. [DOI] [PubMed] [Google Scholar]
- 32. Molina J-M, Capitant C, Spire B. et al. On-demand preexposure prophylaxis in men at high risk for HIV-1 infection. N Engl J Med 2015;373:2237–46. [DOI] [PubMed] [Google Scholar]
- 33. Marrazzo JM, Ramjee G, Richardson BA. et al. Tenofovir-based preexposure prophylaxis for HIV infection among African women. N Engl J Med 2015;372:509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wei X, Liu X, Dobbs T. et al. Development of two avidity-based assays to detect recent HIV type 1 seroconversion using a multisubtype gp41 recombinant protein. AIDS Res Hum Retroviruses 2010;26:61–71. [DOI] [PubMed] [Google Scholar]
- 35. Duong YT, Qiu M, De AK. et al. Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 2012;7:e33328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Puller V, Neher R, Albert J.. Estimating time of HIV-1 infection from next-generation sequence diversity. PLoS Comput Biol 2017;13:e1005775.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.