Summary
We previously identified an evolutionarily conserved protein named transmembrane protein 119 (TMEM119) as the most reliable maker for human microglia. Recent studies showed that under homeostatic conditions, microglia intensely express TMEM119, whereas the expression levels are greatly reduced in disease-associated microglia (DAM) activated at the site of neurodegeneration. Nasu-Hakola disease (NHD) is a rare autosomal recessive disorder, pathologically characterized by leukoencephalopathy, astrogliosis, axonal spheroids, and accumulation of microglia. However, it remains unknown whether microglia are homeostatic or activated in NHD brains. In the present study, we identified TMEM119 on microglia in NHD brains by immunohistochemistry. TMEM119 was expressed on microglia in NHD brains as well as in the brains of non-neurological controls (NC) and Alzheimer's disease (AD) patients, although TMEM119-immunolabeled areas exhibited great variability from case to case without significant differences among the study population. These results suggest that TMEM119 expression on microglia might play a key role in steady-state brain maintenance in NHD, AD and controls.
Keywords: Alzheimer's disease, disease-associated microglia (DAM), microglia, Nasu-Hakola disease, TMEM119
1. Introduction
Microglia are resident myeloid cells of the central nervous system (CNS) that play a principal role in the maintenance of normal tissue homeostasis and plasticity (1). Microglia are ontogenetically and functionally distinct from monocyte-derived macrophages that infiltrate the CNS under pathological conditions (2-4). Microglia are originated from primitive hematopoietic progenitor cells present in the yolk sac during embryonic development (5). The establishment of specific markers that distinguish resident microglia from circulating blood-derived macrophages in human brains is essential for exact evaluation of microglial contributions to the human brain pathology.
To identify microglia-specific markers, we performed a comparative analysis of five comprehensive datasets of mouse microglia transcriptome characterized by microarray and RNA-Seq technologies (6). Then, we identified transmembrane protein 119 (TMEM119) as the most reliable marker for human microglia. The resting microglia presenting with a ramified morphology have a capacity to constantly migrate and scavenge invading pathogens, apoptotic debris, unwanted synapses, and pathologically accumulated protein aggregates by sensing them with a panel of pattern recognition receptors (PRRs) (7-9). The homeostatic mouse microglia express high levels of Cx3cr1, P2ry12, and Tmem119 (3,9). At sites of neurodegeneration, they are transformed into disease-associated microglia (DAM), also called as the microglial neurodegenerative phenotype (MGnD), which upregulate the expression of Tyrobp, Apoe, and Trem2 and reduce the expression of Cx3cr1, P2ry12, and Tmem119 (9-11). These cells are named as the stage 1 (Trem2-independent) DAM (9,10). Trem2 signaling further induces the expression of Lpl, Cst7, Axl, and Itgax. They are termed as the stage 2 (Trem2-dependent) DAM (9,10).
Nasu-Hakola disease (NHD), also designated polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL), is a rare autosomal recessive disorder, characterized by progressive presenile dementia and formation of multifocal bone cysts, caused by loss-of-function mutations of either TYROBP (DAP12) or TREM2 (12). TREM2 and DAP12 constitute a receptor/adaptor signaling complex expressed exclusively on osteoclasts, dendritic cells, macrophages, and microglia. Although NHD patients are clustered in Japan and Finland, approximately 200 NHD cases are presently reported worldwide. Clinically, NHD patients show recurrent bone fractures during the third decade of life, a frontal lobe syndrome during the fourth decade of life, and progressive dementia and death until the fifth decade of life (13). Pathologically, the brains of NHD patients exhibit extensive demyelination designated leukoencephalopathy, astrogliosis, axonal spheroids, and accumulation of microglia predominantly in the frontal and temporal lobes and the basal ganglia (14). At present, molecular mechanisms responsible for development of leukoencephaolpathy in NHD brains remain unknown. Because NHD is a pathological entity of microgliopathy where microglia act as a key regulator of leukoencephalopathy, we propose the hypothesis that TMEM119 expression on microglia might play a central role in maintenance of homeostasis in NHD brains. In the present study, we have attempted to clarify the expression pattern of TMEM119 on microglia in NHD brains, compared with AD brains.
2. Materials and Methods
2.1. Human brain tissues
The brain autopsies were performed at the National Center Hospital, National Center of Neurology and Psychiatry (NCNP), Japan, Kohnodai Hospital, National Center for Global Health and Medicine (NCGM), Japan, and affiliated hospitals of Research Resource Network (RRN), Japan. The comprehensive examination by established neuropathologists (YS and TI) validated the pathological diagnosis. The Ethics Committee of NCNP for the Human Brain Research, the Ethics Committee of NCGM on the Research Use of Human Samples, and the Human Research Ethics Committee of Meiji Pharmaceutical University approved the present study. Written informed consent was obtained in all cases at autopsy, following the regulation of the institutional ethics committees.
For immunohistochemical studies, serial sections of the frontal lobe were prepared from four subjects who died of non-neurological causes (NC), ten Alzheimer's disease (AD) patients, and five NHD patients, as listed in Table 1. The homozygous mutation of a single base deletion of 141G (c.141delG) in exon 3 of DAP12 was identified in NHD1, NHD2, and NHD5, while the genetic analysis was not performed in NHD3 or NHD4. All AD cases were satisfied with the Consortium to Establish a Registry for Alzheimer's Disease (CERAD) criteria for diagnosis of definite AD, categorized into the stage C of amyloid deposition (15). They were classified into the stage VI of neurofibrillary degeneration, following the Braak's staging (16).
Table 1. Study Population.
| Case No. | Age (year-old) | Sex (male, female) | Pathological Diagnosis |
|---|---|---|---|
| NC1 | 63 | M | prostate cancer and acute myocardial infarction |
| NC2 | 67 | M | dissecting aortic aneurysm |
| NC3 | 57 | M | alcoholic liver cirrhosis |
| NC4 | 61 | M | rheumatoid arthritis and interstitial pneumonia |
| NHD1 | 42 | M | Nasu-Hakola disease (DAP12 c.141delG) |
| NHD2 | 48 | F | Nasu-Hakola disease (DAP12 c.141delG) |
| NHD3 | 44 | M | Nasu-Hakola disease (mutation not analyzed) |
| NHD4 | 32 | F | Nasu-Hakola disease (mutation not analyzed) |
| NHD5 | 38 | M | Nasu-Hakola disease (DAP12 c.141delG) |
| AD1 | 68 | F | Alzheimer's disease (C/VI) |
| AD2 | 70 | F | Alzheimer's disease (C/VI) |
| AD3 | 68 | F | Alzheimer's disease (C/VI) |
| AD4 | 56 | M | Alzheimer's disease (C/VI) |
| AD5 | 59 | M | Alzheimer's disease (C/VI) |
| AD6 | 81 | M | Alzheimer's disease (C/VI) |
| AD7 | 68 | F | Alzheimer's disease (C/VI) |
| AD8 | 80 | M | Alzheimer's disease (C/VI) |
| AD9 | 72 | M | Alzheimer's disease (C/VI) |
| AD11 | 77 | F | Alzheimer's disease (C/VI) |
The study population is composed of four subjects who died of non-neurological causes (NC), five Nasu-Hakola disease (NHD) patients, and ten Alzheimer's disease (AD) patients. The homozygous mutation of c.141delG in exon 3 of DAP12 was identified in NHD1, NHD2, and NHD5. All AD cases were satisfied with the stage C of amyloid deposition in the CERAD criteria and the stage VI of neurofibrillary degeneration in the Braak's staging.
2.2. Immunohistochemistry
After deparaffination, tissue sections were heated in 10 mM sodium citrate buffer, pH 6.0 by autoclave at 110℃ for 15 min in a temperature-controlled pressure chamber (Biocare Medical, Pacheco, CA, USA). They were treated at room temperature (RT) for 15 min with 3% hydrogen peroxide-containing methanol to block the endogenous peroxidase activity. They were then incubated with phosphate-buffered saline (PBS) containing 10% normal goat serum at RT for 15 min to block non-specific staining, followed by incubation in a moist chamber at 4℃ overnight with a rabbit polyclonal anti-human TMEM119 antibody numbered HPA051870 (Sigma, St. Louis, MO, USA) (6) or a rabbit polyclonal anti-Iba1 antibody (FUJIFILM Wako Pure Chemical, Osaka, Japan).
After washing with PBS, tissue sections were incubated at RT for 30 min with a horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (Nichirei, Tokyo, Japan), followed by incubation with diaminobenzidine tetrahydrochloride (DAB) substrate (Vector, Burlingame, CA, USA). They were processed for a counterstain with hematoxylin. Negative controls underwent all the steps except for exposure to primary antibody. In limited experiments, double immunolabeling was performed using HPA051870 followed by incubation with a HRP-conjugated anti-rabbit secondary antibody and exposure to DAB, and in combination with a mouse monoclonal antibody against amyloid-β peptide (12B2; Immunobiological Laboratories, Gunma, Japan), followed by incubation with an alkaline phosphatase-conjugated anti-mouse secondary antibody (Nichirei) and exposure to Warp Red chromogen (Biocare Medical).
2.3. Quantification of TMEM119 immunoreactivity
To quantify immunolabeled areas, the images derived from three fields each of the frontal cortex were captured at a 200 X magnification on the Olympus BX51 universal microscope. They were then processed for quantification by using ImageJ software (National Institute of Health, Bethesda, MD, USA). The differences in the TMEM119-positive areas among NC, NHD, and AD subjects were evaluated statistically by one-way analysis of variance (ANOVA) followed by post-hoc Tukey's test.
3. Results and Discussion
By immunohistochemistry, we found that TMEM119 was intensely expressed predominantly on ramified microglia located in the frontal cortex and white matter of NHD (Figure 1, panels A, C, E), AD and NC brains (Figure 2, panels A, C, F). Iba1 immunolabeling was always more intense when compared with that of TMEM119 in these brains (Figure 1, panels B, D, F; Figure 2, panels B, D). Multipolar fine process-bearing ramified microglia were well stained by TMEM119 antibody (Figure 2, panel F), while amoeboid microglia exhibited a less intense immunoreactivity for TMEM119. Clusters of TMEM119-expressing microglia were often incorporated into the amyloid-beta (Aβ) deposition in AD brains (Figure 2, panel E). Overall, TMEM119- immunolabeled areas or Iba1-positive areas were not significantly different among NHD, AD and NC brains (Figure 3, panels A, B). Approximately 32.5% of Iba1+ microglia expressed TMEM119 in NHD brains, 35.0% in AD brains, and 32.8% in NC brains.
Figure 1.
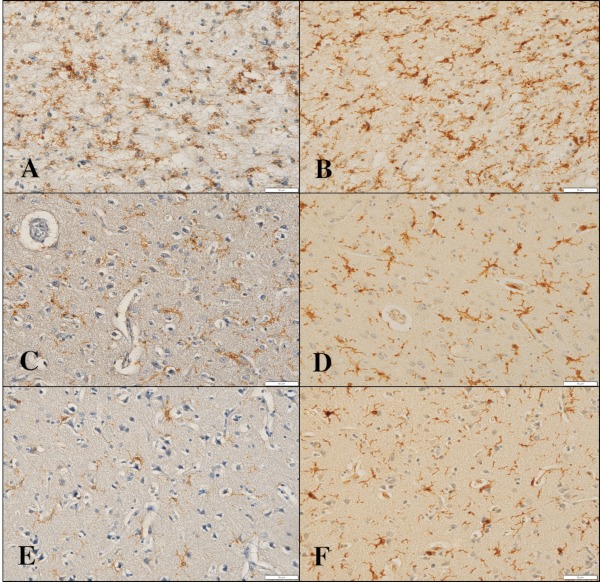
TMEM119 expression in the frontal cortex of NHD brains. (A) TMEM119, the frontal cortex, NHD5, (B) Iba1, the same field of (A), (C) TMEM119, the frontal cortex, NHD2, (D) Iba1, the same field of (C), (E) TMEM119, the frontal cortex, NHD4, and (F) Iba1, the same field of (E). Scale bars indicate (A-F) 50 μm.
Figure 2.
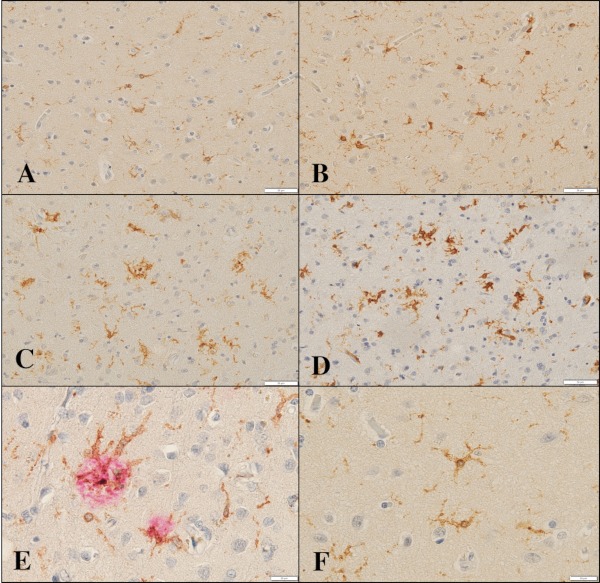
TMEM119 expression in the frontal cortex of NC and AD brains. (A) TMEM119, the frontal cortex, NC1, (B) Iba1, the same field of (A), (C) TMEM119, the frontal cortex, AD8, (D) Iba1, the same field of (C), (E) TMEM119 (brown) and Aβ (red), the frontal cortex, AD8, and (F) TMEM119, the frontal cortex, NC1. Scale bars indicate (A-D) 50 μm and (E, F) 20 μm.
Figure 3.
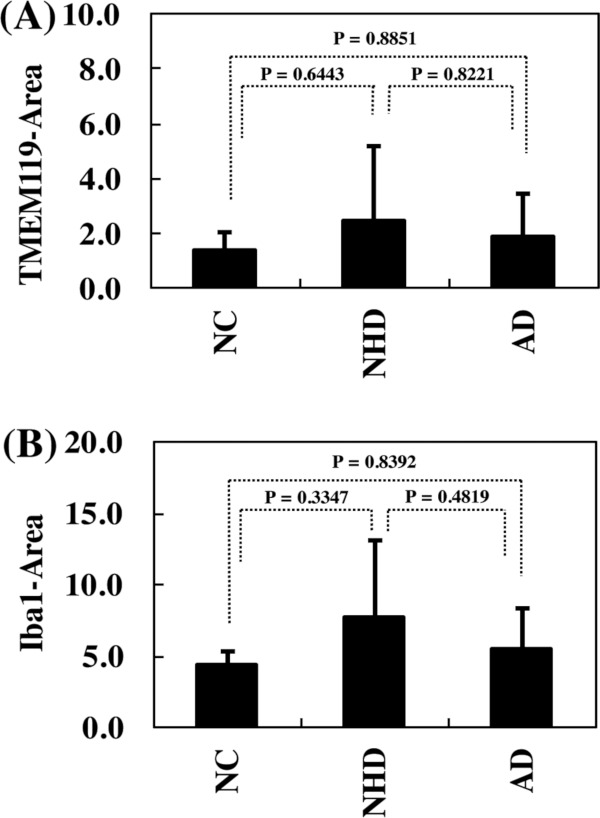
TMEM119-immunolabeled areas of the frontal cortex. The differences in the TMEM119-immunolabeled or Iba1-immunolabeled areas (pixel2) of the frontal cortex among NC, NHD and AD subjects were evaluated statistically by one-way ANOVA followed by post-hoc Tukey's test. (A) TMEM119 and (B) Iba1.
By the data-mining approach, we previously identified an evolutionarily conserved protein TMEM119 as the most promising marker specific for human microglia (6). TMEM119 discriminates resident microglia from blood-derived macrophages in the human brain (6,17). Several recent studies revealed that TMEM119 is highly expressed on microglia under homeostatic conditions, while its immunoreactivity is greatly reduced in disease-associated microglia named DAM, accumulating at sites of neurodegeneration (9-11). DAM cells are activated sequentially by TREM2-independent and TREM2-dependent pathways (9,10). DAM cells sense neurodegeneration-associated molecular patterns (NAMPs), which serve as danger signals expressed on apoptotic bodies of dying neural cells, myelin debris, lipid degradation products, and extracellular protein aggregates (9). On the other hand, NAMPs trigger resting microglia into a DAM phenotype with decreased expression of TMEM119 (9). We found that TMEM119 is expressed on a subset of microglia profoundly in NHD brains as well as in NC and AD brains, and TMEM119- immunolabeled areas exhibited great variability from case to case without significant differences among the study population. Furthermore, Iba1-positive areas were always more intense when compared with that of TMEM119 in these brains, suggesting the possibility that there exist a number of Iba1-positive TMEM119- negative DAM-like microglia not only in NHD and AD brains but also in NC brains. For instance, we estimated that approximately one-third of Iba1+ microglia express TMEM119 in NHD brains. However, it is possible that an apparent inconsistency between microglial TMEM119 and Iba1 immunolabeling areas is attributable to a difference in the density of antigenic epitopes, the avidity of antibodies, or both. Furthermore, our results indicated that it is difficult to identify DAM solely by the loss of TMEM119 immunoreactivity on microglia in human brains.
TMEM119 is originally identified as an osteoblast induction factor (OBIF), composed of a type I transmembrane protein predominantly expressed in mouse osteoblasts with the location chiefly in the plasma membrane (18). OBIF-deficient mice exhibit severe bone hypoplasia accompanied by decreased expression of osteoblast marker genes (19). OBIF, induced by parathyroid hormone, interacts with several key regulators in the BMP2 signaling pathway, such as Smad1/5 and Runx2, to induce osteoblast differentiation (20). OBIF elevates the expression of ATF4, a transcription factor that plays a central role in osteoblast differentiation (21). However, at present, precise biological functions and endogenous ligands of TMEM119 and OBIF in human microglia and osteoblasts remain largely unknown.
In conclusion, TMEM119 is expressed on homeostatic microglia in NHD, AD, and NC brains. TMEM119-immunolabeled areas are not significantly different among the study population. TMEM119 expression on microglia might play a key role in steady-state brain maintenance. TMEM119 immunohistochemistry serves as a useful tool for investigating the biology and pathology of human microglia. However, it is difficult to identify DAM solely by the loss of TMEM119 immunoreactivity in these brains.
Acknowledgements
The authors thank Drs. Kenji Jinnai, Nobutaka Arai, Kiyotaka Nakamagoe, Nobutaka Motohashi, and Saburo Yagishita for providing us brain samples. This work was supported by the grants from the Research on Intractable Diseases, entitled "Clinicopathological and genetic studies of Nasu-Hakola disease" (H21-Nanchi- Ippan-201; H22-Nanchi-Ippan-136), the Ministry of Health, Labour and Welfare of Japan, and the grants from the JSPS KAKENHI (C25430054 and 16K07043) and the Dementia Drug Resource Development Center (DRC) project (S1511016), the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
References
- 1. Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol. 2011; 11:775-787. [DOI] [PubMed] [Google Scholar]
- 2. Michell-Robinson MA, Touil H, Healy LM, Owen DR, Durafourt BA, Bar-Or A, Antel JP, Moore CS. Roles of microglia in brain development, tissue maintenance and repair. Brain. 2015; 138:1138-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dubbelaar ML, Kracht L, Eggen BJL, Boddeke EWGM. The kaleidoscope of microglial phenotypes. Front Immunol. 2018; 9:1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haage V, Semtner M, Vidal RO, Hernandez DP, Pong WW, Chen Z, Hambardzumyan D, Magrini V, Ly A, Walker J, Mardis E, Mertins P, Sauer S, Kettenmann H, Gutmann DH. Comprehensive gene expression meta-analysis identifies signature genes that distinguish microglia from peripheral monocytes/macrophages in health and glioma. Acta Neuropathol Commun. 2019; 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010; 330:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Satoh J, Kino Y, Asahina N, Takitani M, Miyoshi J, Ishida T, Saito Y. TMEM119 marks a subset of microglia in the human brain. Neuropathology. 2016; 36:39-49. [DOI] [PubMed] [Google Scholar]
- 7. Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005; 308:1314-1318. [DOI] [PubMed] [Google Scholar]
- 8. Mosher KI, Wyss-Coray T. Microglial dysfunction in brain aging and Alzheimer's disease. Biochem Pharmacol. 2014; 88:594-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deczkowska A, Keren-Shaul H, Weiner A, Colonna M, Schwartz M, Amit I. Disease-associated microglia: A universal immune sensor of neurodegeneration. Cell. 2018; 173:1073-1081. [DOI] [PubMed] [Google Scholar]
- 10. Keren-Shaul H, Spinrad A, Weiner A, Matcovitch- Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, Amit I. A unique microglia type associated with restricting development of Alzheimer's disease. Cell. 2017; 169:1276-1290. [DOI] [PubMed] [Google Scholar]
- 11. Krasemann S, Madore C, Cialic R, et al. The TREM2- APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017; 47:566-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klünemann HH, Ridha BH, Magy L, Wherrett JR, Hemelsoet DM, Keen RW, De Bleecker JL, Rossor MN, Marienhagen J, Klein HE, Peltonen L, Paloneva J. The genetic causes of basal ganglia calcification, dementia, and bone cysts: DAP12 and TREM2. Neurology. 2005; 64:1502-1507. [DOI] [PubMed] [Google Scholar]
- 13. Bianchin MM, Capella HM, Chaves DL, Steindel M, Grisard EC, Ganev GG, da Silva Júnior JP, Neto Evaldo S, Poffo MA, Walz R, Carlotti Júnior CG, Sakamoto AC. Nasu-Hakola disease (polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy - PLOSL): a dementia associated with bone cystic lesions. From clinical to genetic and molecular aspects. Cell Mol Neurobiol. 2004; 24:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Satoh J, Tabunoki H, Ishida T, Yagishita S, Jinnai K, Futamura N, Kobayashi M, Toyoshima I, Yoshioka T, Enomoto K, Arai N, Arima K. Immunohistochemical characterization of microglia in Nasu-Hakola disease brains. Neuropathology. 2011; 31:363-375. [DOI] [PubMed] [Google Scholar]
- 15. Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991; 41:479-486. [DOI] [PubMed] [Google Scholar]
- 16. Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006; 112:389-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, Weissman IL, Chang EF, Li G, Grant GA, Hayden Gephart MG, Barres BA. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016; 113:E1738-E1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanamoto T, Mizuhashi K, Terada K, Minami T, Yoshikawa H, Furukawa T. Isolation and characterization of a novel plasma membrane protein, osteoblast induction factor (obif), associated with osteoblast differentiation. BMC Dev Biol. 2009; 9:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizuhashi K, Kanamoto T, Ito M, Moriishi T, Muranishi Y, Omori Y, Terada K, Komori T, Furukawa T. OBIF, an osteoblast induction factor, plays an essential role in bone formation in association with osteoblastogenesis. Dev Growth Differ. 2012; 54:474-480. [DOI] [PubMed] [Google Scholar]
- 20. Hisa I, Inoue Y, Hendy GN, Canaff L, Kitazawa R, Kitazawa S, Komori T, Sugimoto T, Seino S, Kaji H. Parathyroid hormone-responsive Smad3-related factor, Tmem119, promotes osteoblast differentiation and interacts with the bone morphogenetic protein-Runx2 pathway. J Biol Chem. 2011; 286:9787-9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanaka K, Kaji H, Yamaguchi T, Kanazawa I, Canaff L, Hendy GN, Sugimoto T. Involvement of the osteoinductive factors, Tmem119 and BMP-2, and the ER stress response PERK-eIF2α-ATF4 pathway in the commitment of myoblastic into osteoblastic cells. Calcif Tissue Int. 2014; 94:454-464. [DOI] [PubMed] [Google Scholar]


