SUMMARY
All viruses balance interactions between cellular machinery co-opted to support replication and host factors deployed to halt the infection. We use gene correlation analysis to perform an unbiased screen for host factors involved in influenza A virus (FLUAV) infection. Our screen identifies the cellular factor epidermal growth factor receptor pathway substrate 8 (EPS8) as the highest confidence pro-viral candidate. Knockout and overexpression of EPS8 confirm its importance in enhancing FLUAV infection and titers. Loss of EPS8 does not affect virion attachment, uptake, or fusion. Rather, our data show that EPS8 specifically functions during virion uncoating. EPS8 physically associates with incoming virion components, and subsequent nuclear import of released ribonucleoprotein complexes is significantly delayed in the absence of EPS8. Our study identifies EPS8 as a host factor important for uncoating, a crucial step of FLUAV infection during which the interface between the virus and host is still being discovered.
In Brief
Gene correlation analysis identifies host factors with functional impacts on influenza A virus replication. The top pro-viral factor, EPS8, enhances viral gene expression and titers. Larson et al. identify the step during influenza A virus entry when EPS8 functions, establishing EPS8 as a co-factor for virion uncoating.
Graphical Abstract

INTRODUCTION
Attachment and entry into a host cell is the first bottleneck virions encounter during infection. Virion entry requires efficient use of the host cell environment while evading cellular immune responses. Influenza A virus (FLUAV; Orthomyxoviridae: Alphainfluenzavirus), like all viruses, largely depends upon existing cellular machinery to successfully complete these initial stages of infection.
During the first step of infection, attachment, FLUAV hemagglutinin (HA) binds to the target cell via sialic acid linkages on host glycoproteins (Dou et al., 2018). Virions are internalized via receptor-mediated endocytosis and less frequently through an alternative macropinocytosis pathway (Matlin et al., 1981; de Vries et al., 2011). Once within endosomes, virions are trafficked toward the nucleus using the cytoskeletal components actin, dynein, and microtubules (Lakadamyali et al., 2003). The endosome matures and acidifies during cellular trafficking, and the virion interior is also acidified through the viral ion channel M2 (Pinto et al., 1992). The low pH in the endosome causes conformational changes in HA that drive fusion of the viral and endosomal lipid membranes, whereas the low pH within the virion causes the viral matrix protein M1 to dissociate from the inner membrane of the viral envelope (Bukrinskaya et al., 1982; Maeda and Ohnishi, 1980; Martin and Helenius, 1991; Zhirnov, 1990). Fusion of the two membranes releases a capsid-like viral core consisting of viral ribonucleoproteins (vRNPs) enclosed in an M1 shell-like structure into the cytoplasm. This complex engages the cellular aggresome and other host proteins to complete uncoating, and the released vRNPs are imported into the nucleus by cellular karyopherins (Banerjee et al., 2014; Melen et al., 2003; Miyake et al., 2019; O’Neill et al., 1995; Wang et al., 1997). Once in the nucleus, a pioneering round of transcription occurs on the incoming vRNPs that initiates replication and secondary rounds of transcription of the viral genome.
High-throughput screening approaches have expanded our knowledge of specific cellular cofactors involved in FLUAV infection, with many of these methods identifying host factors involved in viral entry. Gene disruption screens identified host factors involved in sialic acid metabolism used for attachment (Carette et al., 2009; Han et al., 2018). Vacuolar ATPases involved in endosomal acidification and other host factors facilitating fusion and uncoating were identified through small interfering RNA (siRNA) knockdown, proteomic, and overexpression screens (Banerjee et al., 2014; König et al., 2010; Lee et al., 2017; Mar et al., 2018; Yángüez et al., 2018). These studies also revealed previously unknown steps of FLUAV particle entry, such as the role of the aggresome in viral uncoating and the use of transportin 1 to debundle incoming RNPs prior to nuclear import (Banerjee et al., 2014; Miyake et al., 2019). Despite these discoveries, the mechanistic details of steps occurring after fusion remain poorly understood.
Here, we conducted a screen using gene correlation analysis to identify host factors involved in FLUAV infection. Gene correlation analysis exploits naturally occurring variations in gene expression across multiple cell lines without the need to exogenously manipulate the cellular environment. Variations in cellular gene expression were used to identify factors affecting a phenotype of interest, in this case susceptibility to FLUAV infection. We identified epidermal growth factor receptor (EGFR) pathway substrate 8 (EPS8) as a pro-viral cellular cofactor during the early stages of infection. We confirmed that EPS8 enhances FLUAV gene expression and replication, whereas knockout of EPS8 reduced susceptibility to infection. Stepwise dissection of the viral entry process revealed that EPS8 specifically facilitates uncoating of the viral core. Thus, we identified EPS8 as an important component of the FLUAV uncoating process, a necessary step for successful viral genome transcription and replication.
RESULTS
Gene Correlation Analysis Identifies Putative Enhancers and Suppressors of FLUAV Replication
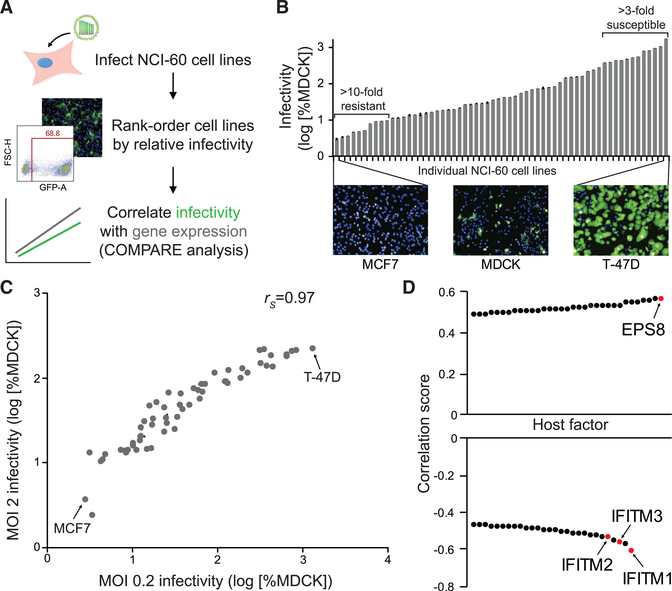
To overcome limitations of previous screening methodologies, we sought to identify both enhancers and suppressors of FLUAV replication in an unbiased manner. We used gene correlation analysis, which relied on inherent differences in gene expression among different cell lines and consequently did not require external manipulation of the cellular environment. The National Cancer Institute-60 (NCI-60) panel consists of 59 distinct cell lines with well-characterized transcriptomic profiles (Shankavaram et al., 2007; Weinstein and Pommier, 2003). The diversity of cell types and the depth of transcriptomic data permit high-confidence genome-wide correlations between cellular gene expression and infection susceptibility (Kondratowicz et al., 2013; Lenaerts et al., 2012; Schowalter et al., 2012). We therefore inoculated the NCI-60 panel of cell lines with a single-cycle variant of A/WSN/1933 (H1N1, WSN) encoding GFP (WSN-GFP) (Figure 1A). Using WSN-GFP, we specifically focused on host factors involved in early stages of infection up to and including viral gene expression and translation.
Figure 1. Gene Correlation Analysis Identifies Putative Enhancers and Suppressors of FLUAV Replication.
(A) Experimental workflow for NCI-60 screen. NCI-60 cell lines were inoculated with FLUAV encoding GFP, infections were visualized by fluorescence microscopy and quantified by flow cytometry at 24 hpi, and data were normalized to control MDCK cells inoculated in parallel.
(B) Infectivity at an MOI of 0.2 was determined relative to MDCK cells (mean of n = 3 ± SD). Images of highly resistant (MCF7) and hypersensitive (T-47D) infected cell lines are shown compared with the control MDCK cells. Data are representative of two biological replicates.
(C) Pairwise comparison of replicate NCI-60 screens performed at an MOI of 0.2 or 2 (mean of n = 3 ± SD; rs, Spearman’s correlation coefficient).
(D) COMPARE analysis of MOI 0.2 infectivity data identified top hits for putative pro-viral and anti-viral factors.
See also Figure S1.
The permissiveness of each cell line to WSN-GFP was determined and rank-ordered relative to Madin-Darby canine kidney (MDCK) cells, which are frequently used for the propagation of FLUAV (Figures 1A and 1B; Table S1). We detected a broad range of susceptibility after infecting cells at an MOI of 0.2. Relative to MDCK cells, 10 cell lines were highly refractory to WSN-GFP (at least a 10-fold decrease in infection rate), and 12 cell lines were highly permissive (at least a 3-fold increase in infection rate). An association between susceptibility, cell type, tumor type, or tissue of origin was not obvious. MCF7 breast tumor cells were the most refractory cell line, with a normalized infection rate of only about 3%, whereas T-47D cells, another breast tumor cell line, were the most susceptible, with an infection rate of approximately 1,300%. These data were highly reproducible with a strong correlation between results from two independent replicate screens (Figure S1A). To ensure that the assay captured the full dynamic range of susceptibility, especially for the highly resistant cell lines, the screen was repeated at an MOI of 2 (Figure S1B). Similar infectivity trends were detected at both MOIs, although the upper limit of the assay was reached for multiple cell lines at the higher MOI, at which effectively all cells were infected (Figures 1C and S1C). The number of infected cells increased at the higher MOI for most of the resistant cell lines, indicating that these cell lines are not completely refractory to FLUAV infection (Figure S1D).
The broad distribution of infectivity across the NCI-60 cell panel suggested that cell-intrinsic differences affected susceptibility to FLUAV infection. To identify cellular factors affecting FLUAV susceptibility, we calculated linear pairwise correlation coefficients between host gene expression within the NCI-60 panel of cell lines and susceptibility to infection using the COMPARE algorithm (Paull et al., 1989). We identified top hits for putative enhancers or suppressors of FLUAV infection on the basis of their strong correlation scores (Figure 1D; Table S2). Host genes identified as putative enhancing factors exhibited expression patterns that paralleled susceptibility to infection, yielding a positive correlation score. Conversely, expression of host genes identified as putative suppressive factors was inversely related to susceptibility, resulting in a negative correlation score. Notably, some of our strongest hits for suppressors of FLUAV infection were the interferon-inducible transmembrane proteins (IFITMs). IFITM1, IFITM2, and IFITM3 were previously characterized as potent inhibitors of FLUAV infection, providing confidence in our approach (Figure 1D; Brass et al., 2009). Most other candidate genes, including EPS8, were not previously associated with FLUAV susceptibility, revealing that gene correlation analysis can identify new host factors that regulate FLUAV infection.
EPS8 Enhances FLUAV Gene Expression and Titers
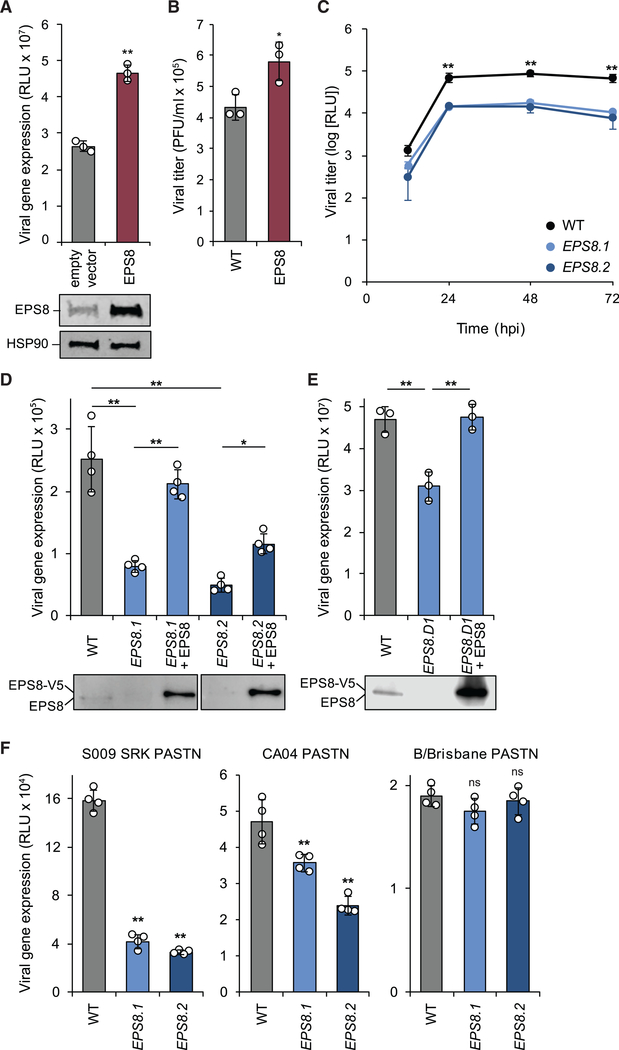
The putative enhancer with the strongest correlation score was EPS8, an adaptor protein involved in signaling via the EGFR and other pathways as well as modulating of actin dynamics (Figure 1D; Di Fiore and Scita, 2002; Hertzog et al., 2010). To validate the results of the screen and confirm a pro-viral function for EPS8, we assessed the effect of EPS8 on viral gene expression and replication. EPS8 was transiently overexpressed in HEK293T cells and infected with a replication-competent reporter version of WSN (WSN PA-Swap-2A-NanoLuc [PASTN]) to quantitatively measure viral gene expression (Tran et al., 2013). EPS8 overexpression increased viral gene expression during infection nearly 2-fold relative to the empty vector control (Figure 2A). Endogenous and overexpressed EPS8 levels were confirmed by immunoblot. We then assayed viral titers when EPS8 was stably overexpressed in human lung epithelial A549 cells. Viral titers 24 h post-infection (hpi) were increased by more than 15-fold in stable EPS8-overexpressing cells relative to wild-type (WT) cells (Figure 2B). Thus, overexpression of EPS8 enhances infection and replication in two different human cell lines, confirming the pro-viral correlation identified in the screen.
Figure 2. EPS8 Enhances FLUAV Gene Expression and Titers.
(A) 293T cells transiently overexpressing EPS8 were infected with WSN PASTN, and viral gene expression was assayed (mean of n = 4 ± SD). EPS8 expression was confirmed by immunoblot.
(B) A549 cells stably overexpressing EPS8 were infected with WSN (MOI 0.01), and viral titer was assayed at 24 hpi (mean of n = 3 ± SD).
(C) EPS8-edited A549 cells were infected with WSN PASTN, virus was harvested at the indicated times, and titers were assessed using luciferase activity (mean of n = 3 ± SD).
(D and E) EPS8-edited and complemented A549 (D) or 293 (E) cells were infected with WSN PASTN to assay viral gene expression (mean of n = 4 ± SD). EPS8 expression was confirmed by immunoblot.
(F) Viral gene expression was assayed in EPS8-edited cells infected with S009 SRK PASTN, CA04 PASTN, or B/Brisbane PASTN (mean of n = 4 ± SD).
*p < 0.05 and **p < 0.01; ns, not significant. (A) and (B) were analyzed using Student’s two-tailed t test, unequal variance. Multiple comparisons were made in (C)–(F) using a one-way ANOVA with post hoc Tukey honestly significant difference (HSD) test compared with WT A549 cells. All data are representative of three biological replicates. See also Figures S2–S4.
We next used CRISPR-Cas9 to generate clonal EPS8-knockout A549 cells. Sanger sequencing confirmed genotypic changes predicted to result in knockout of EPS8 in two independent clonal lines (EPS8.1 and EPS8.2) (Figures S2A and S2B). Immunoblotting for endogenous EPS8 revealed a dramatic reduction in EPS8 protein levels but not a complete loss in our edited clones (Figure S2C). EPS8.1 retained about 25% of the amount of EPS8 observed in the parental cells, whereas EPS8.2 levels were nearly undetectable. Editing occurred adjacent to the splice donor in exon 2 of EPS8, raising the possibility that alternative splice donors may be exploited to support the low levels of EPS8 protein expression detected (Figures S2B and S2D). These cell clones were used to further examine the importance of EPS8 during FLUAV infection.
Viral replication and gene expression were assayed in the EPS8-edited cells. Both EPS8.1 and EPS8.2 cell lines had defects in multicycle replication and viral gene expression assays. Viral titers were reduced by about 10-fold in both EPS8-edited lines compared with parental cells (Figure 2C). Viral gene expression was reduced 4- to 5-fold in A549 cells with edited EPS8 relative to WT cells (Figure 2D). The decrease in viral gene expression was more pronounced in EPS8.2, the cell line with the lower level of EPS8 expressed. Stable complementation with EPS8 rescued viral gene expression in both edited lines (Figure 2D), suggesting that the defects in gene expression were specifically due to decreases in EPS8 levels. To obtain a true knockout phenotype, EPS8 was edited in 293 cells (Figure S3). EPS8-knockout 293 cells (EPS8.D1) exhibited a significant decrease in viral gene expression, which was restored by transient complementation (Figure 2E). EPS8 editing or knockout thus decreases viral gene expression in two different cell lines.
Given that both cell types exhibited similar phenotypes, we continued our investigation using the edited A549 cell lines, as these cells are of lung origin and more closely represent natural target cells during influenza virus infection. We assessed whether EPS8 affected infection with reporter variants of other influenza virus isolates. Cells were inoculated with a reporter virus encoding an avian-background RNP from A/green-winged teal/OH/175/1983 in a WSN backbone (S009 SRK PASTN; H2N1) or a reporter version of A/California/04/2009 (CA04 PASTN; H1N1). We also infected EPS8-edited cells with the influenza B virus (FLUBV) B/Brisbane/60/2008 (B/Brisbane PASTN) (Figure 2F). Consistent with the results obtained using WSN, editing of EPS8 reduced viral gene expression for the influenza A strains S009 SRK PASTN and CA04 PASTN. Interestingly, EPS8 editing did not affect B/Brisbane PASTN gene expression (Figure 2F). We explored infection specificity further by assessing the relative infection rates of A549 cells overexpressing EPS8 in response to challenge by diverse viruses (Figure S4). EPS8 expression levels did not alter infection rates of Marburg virus (MARV) or Junín virus (JUNV). In contrast, EPS8 overexpression caused decreased Ebola virus (EBOV), Venezuelan equine encephalitis virus (VEEV), and Rift Valley fever virus (RVFV) infection rates. Hence, altering EPS8 expression does not generically affect viral replication. Together, these data confirm that EPS8 acts as a pro-viral host factor during FLUAV infection and exhibits specific effects on cell infectivity depending on the virus.
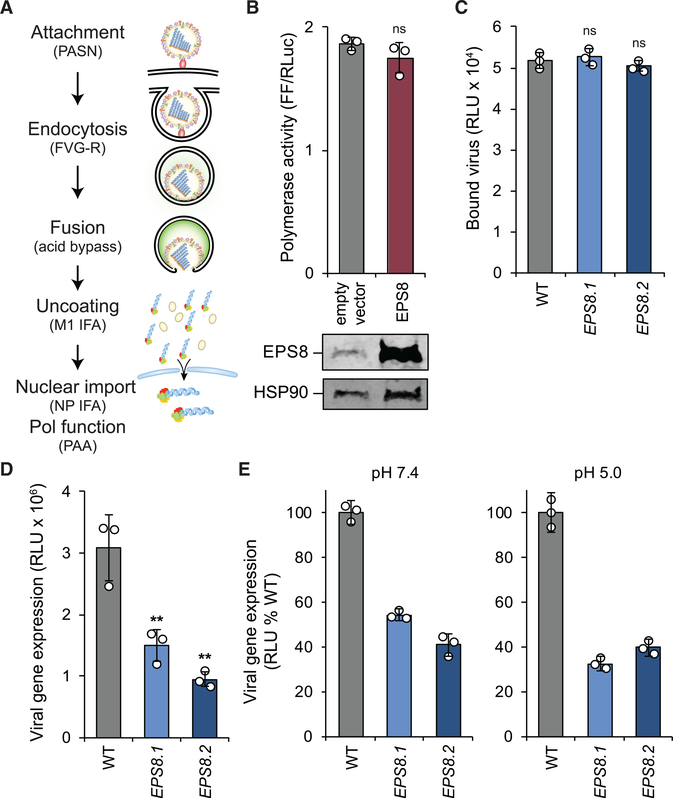
EPS8 Functions Post-fusion but before Viral Gene Expression during FLUAV Infection
The structure of the NCI-60 screen and our data indicated EPS8 functions in early stages of FLUAV replication. We therefore conducted a series of experiments to determine where in the viral replication cycle EPS8 functioned to enhance infection (Figure 3A). We first assessed whether EPS8 affects infection through a mechanism that directly affects viral polymerase activity. Polymerase activity was reconstituted in the absence of infection by expressing the heterotrimeric viral polymerase subunits PA, PB1, and PB2, nucleoprotein (NP), and a vRNA-like reporter encoding firefly luciferase. Polymerase activity was not statistically different in the presence or absence of exogenous EPS8 (Figure 3B). Immunoblotting confirmed high levels of exogenously expressed EPS8. Similarly, polymerase activity was indistinguishable when assays were performed in EPS8-knockout or complemented 293 cells (Figure S5A). These findings establish that EPS8-mediated enhancement of viral gene expression is not due to direct impacts on the viral polymerase but rather an upstream step in the early stages of infection.
Figure 3. EPS8 Functions Post-fusion but before Viral Gene Expression during FLUAV Infection.
(A) Early stages in the FLUAV replication cycle were systematically probed with the indicated reagents or assays detailed in the text. PASN, bioluminescent virions; FVG-R, recombinant FLUAV expressing VSIV-G; IFA, immunofluorescence assay; PAA, polymerase activity assay.
(B) Polymerase activity assays were performed in 293T cells expressing RNP components with or without exogenous EPS8. Firefly luciferase (FF) values were normalized to Renilla luciferase (RLuc) values within each sample. EPS8 expression was confirmed by immunoblot.
(C) Virion attachment was assayed in EPS8-edited cells incubated with bioluminescent PASN.
(D) WT or EPS8-edited cells were inoculated with FVG-R and viral gene expression was measured 8 hpi.
(E) Acid bypass assays were performed on virions attached to WT or EPS8-edited cells. Cells were transiently treated with buffers at physiological pH 7.4 to initiate canonical viral entry or acidic pH 5.0 to cause fusion at the cell surface. Viral gene expression was measured 8 h after treatment.
For all, data are mean of n = 3 ± SD. **p < 0.01; ns, not significant. (B) was analyzed using Student’s two-tailed t test, unequal variance. Multiple comparisons were made in (C)–(E) using a one-way ANOVA with post hoc Tukey HSD test compared with WT A549 cells. All data are representative of three biological replicates. See also Figure S5.
We probed each successive step that occurs early in the infectious cycle, beginning with viral attachment. WT or edited A549 cells were incubated with bioluminescent virions (PASN) that package nanoluciferase into the viral particle (Tran et al., 2015). Cells were incubated at 4°C to enable binding but prevent internalization of virions, and luciferase activity was assayed from the bound virions. No statistical difference was found between the amount of virus bound to WT and both EPS8-edited cell lines, indicating that EPS8 is not necessary for FLUAV attachment to cells (Figure 3C). To ascertain if EPS8 affects HA-mediated entry or the fusion process, we infected cells with FLUAV encoding a different entry protein, FVG-R, a recombinant virus expressing vesicular stomatitis Indiana virus glycoprotein (VSIV-G) instead of HA (Hao et al., 2008). Viral gene expression decreased in EPS8-edited cells infected with FVG-R compared with WT cells (Figure 3D). This observed decrease in viral gene expression was similar to the decrease demonstrated during infection with bona fide FLUAV (Figures 2D and 2E) and suggests that EPS8 does not specifically target HA-mediated entry.
Following attachment and entry, FLUAV traffics in an endosome that undergoes acidification, which results in fusion of the endosomal and viral membranes. The function of EPS8 during endosomal acidification and fusion was tested using an acid bypass assay. Acid bypass replaces the canonical entry route with fusion of viral and plasma membranes at the cell surface, depositing vRNPs into the cytoplasm where subsequent steps of infection then proceed as usual (Banerjee et al., 2014; Matlin et al., 1981). As in the attachment assay, virions bound to the surface of WT or edited cells at 4°C to synchronize infection. Cells were shifted to 37°C and transiently held at acidic conditions (pH 5.0) to initiate fusion at the cell surface or held at physiological conditions (pH 7.4) permitting canonical entry to proceed as a control. EPS8 editing resulted in a decrease in viral gene expression when infections were initiated at physiological pH (Figure 3E), consistent with prior data showing defects in gene expression during unsynchronized infections (Figures 2D and 2E). Bypassing canonical entry by treating cells with acidic conditions did not restore viral gene expression in the edited cells (Figure 3E), indicating that EPS8 does not function during endosomal acidification. Control experiments showed that the low-pH treatment used in our bypass assay ablates infectivity of cell-free virions (Figure S5B), indicating that viral gene expression observed in the bypass assays results from fusion at the plasma membrane and not endocytic uptake of residual virus that failed to fuse. Although indirect measures, neither acid bypass nor VSIV-G-mediated entry restored infectivity. Both of these entry pathways are distinct from canonical HA-mediated entry, suggesting EPS8 is unlikely to function at the discrete step of fusion. Together, these data establish that the effects of EPS8 during FLUAV infection are independent of virion attachment, endosomal entry, and HA-mediated fusion.
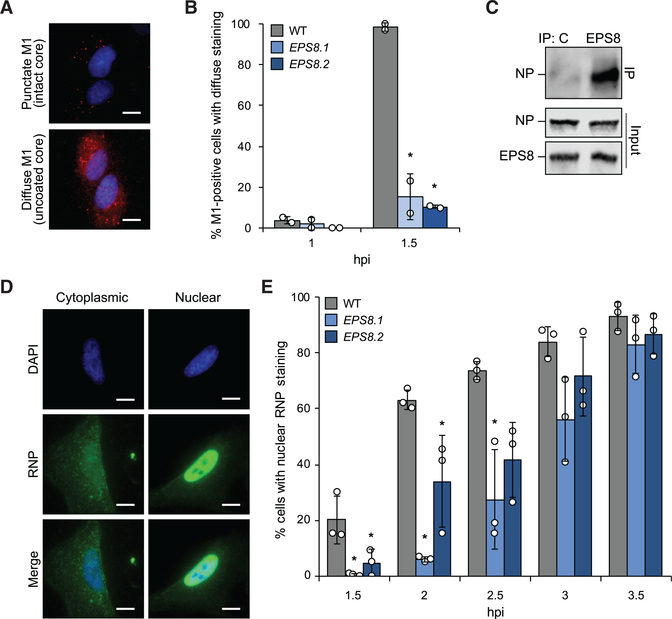
EPS8 Is Crucial for Viral Uncoating
Our line of experimentation indicated EPS8 functions at a step following release of the viral core into the cytoplasm but before viral gene expression. Therefore, we considered whether EPS8 facilitates viral uncoating. This process can be quantified by visualizing the redistribution of punctate matrix protein (M1) staining of intact particles to diffuse staining of M1 released throughout the cytosol (Figures 4A and S5A; Banerjee et al., 2013). WT and EPS8-edited cells were synchronously infected, and M1 localization was quantified at various times post-inoculation. As expected, most M1 staining was punctate in WT cells early in infection and then became diffuse at 1.5 hpi (Figure 4B). By contrast, uncoating was greatly delayed in both cell lines in which EPS8 was edited. Diffuse M1 staining was detected in only 10%–15% of EPS8-edited cells at 1.5 hpi compared with successful uncoating in almost all WT cells at the same time point. Immediately following fusion, viral cores are disassembled by the aggresome on the endosomal surface (Banerjee et al., 2014). Co-precipitations were used to probe how EPS8 might function during this period. Synchronized infections were initiated on EPS8-edited cells stably complemented with WT EPS8. NP specifically co-precipitated with EPS8 (Figure 4C). These data suggest EPS8 physically interacts with incoming viral cores, possibly through interactions with NP, the viral polymerase, M1, or bridged by cellular uncoating partners. Our results implicate EPS8 as an important host factor during viral uncoating.
Figure 4. EPS8 Is Crucial for Viral Uncoating.
(A) A549 cells synchronously infected with WSN were stained for M1 (red) and the nucleus (blue). Representative images show punctate M1 consistent with intact viral cores and diffuse M1 staining that occurs following viral uncoating.
(B) Quantification of diffuse staining in M1-positive cells (mean of n = 2 ± SD).
(C) EPS8-edited A549 cells complemented with EPS8 were infected and lysates subjected to immunoprecipitation. Co-precipitating NP and total NP and EPS8 expression were confirmed by immunoblot.
(D) WT A549 cells infected with WSN were stained for viral RNPs (green). Representative images show cytoplasmic RNP staining or nuclear RNP staining determined by colocalization with the nucleus (blue).
(E) Quantification of the number of cells with nuclear RNP staining at each time point (mean of n = 3 ± SD).
*p < 0.05, one-way ANOVA with post hoc Tukey HSD test compared with WT A549 cells. Scale bar, 20 μm. All data are representative of three biological replicates. See also Figure S6.
Uncoating releases vRNPs into the cytosol, where they are subsequently imported into the nucleus prior to viral gene expression. The defects in uncoating we detected in EPS8-edited cells predict that these cells should also exhibit delayed nuclear import. To test this possibility, we again used synchronized infections and immunofluorescence to examine the subcellular localization and kinetics of vRNP nuclear import over time. Staining for NP, the major protein component of vRNPs, revealed characteristic cytoplasmic localization of incoming vRNPs early during infection followed by distinct nuclear localization (Figure 4D). Discrete cytoplasmic and nuclear localizations of vRNPs were also detected in EPS8-edited cells (Figure S5B). Nuclearlocalized vRNPs were detected in WT cells as early as 1.5 hpi, and the number of cells with nuclear vRNP staining increased over time, consistent with the timing of viral uncoating reported above (Figure 4E). Cells lacking WT levels of EPS8, however, exhibited significantly delayed kinetics of nuclear import. Compared with WT cells, import rates in EPS8-edited cells were delayed by 1 h. This trend continued until 3.5 hpi when import in edited cells finally matched that of WT cells (Figure 4D). Although import was delayed in edited cells, it followed a similar trajectory to WT cells once initiated, suggesting that vRNP import was not directly altered by changes to EPS8 expression. Experiments were repeated in the presence of cycloheximide to test whether the nuclear localization signal captured incoming vRNPs or required de novo synthesis of NP. Similar localization of vRNPs into the nucleus was detected when infection progressed in the presence of cycloheximide, even if signal intensity was reduced (Figure S6C). Furthermore, as before, EPS8-edited cells exhibited delayed kinetics for NP nuclear localization. Thus, defects in uncoating (Figure 4B) result in delayed nuclear import (Figure 4E) and ultimately a reduction in viral gene expression (Figure 2D), reinforcing the conclusion that EPS8 is a key component of the cellular machinery used for viral uncoating.
DISCUSSION
Through gene correlation analysis, we conducted an unbiased genome-wide screen to identify host factors that have a functional impact on early stages of FLUAV replication. Our highest confidence pro-viral candidate was EPS8, a cytoplasmic protein involved in EGFR signaling and regulation of actin dynamics. We showed that EPS8 expression enhanced viral gene expression and titers, whereas loss of WT EPS8 caused defects in gene expression and viral replication. Stepwise investigation of the early stages of infection revealed that EPS8 functions independent of virion attachment, endosomal acidification, or HA-dependent fusion. Rather, EPS8 specifically functioned during the uncoating of the incoming viral cores. Defects in viral uncoating slowed the kinetics of vRNP nuclear import in EPS8-edited cells, corresponding with the overall delay in viral gene expression and replication in these cells. These data establish EPS8 as a cofactor important for viral uncoating during FLUAV infection.
The host factors used during FLUAV uncoating are not yet fully understood. Uncoating begins in the maturing endosome, where the drop in pH opens the M2 ion channel in the viral membrane (Pinto et al., 1992). The influx of potassium ions and protons into the virion interior initiates conformational changes that relax interactions between the matrix protein M1 and vRNPs, making the core competent for uncoating and disassembly of the RNP bundle (Stauffer et al., 2014). Following fusion of the viral and host membranes, the core requires further processing to fully disassemble. Unanchored ubiquitin chains packaged within the virion help activate the cellular aggresome on the late endosomal surface where mechanical forces have been proposed to accelerate uncoating and release of vRNPs into the cytosol, followed by debundling of vRNPs by cellular transportin 1 (Banerjee et al., 2014; Miyake et al., 2019). Our data now implicate EPS8 as another host factor important during these later stages of uncoating.
EGFR was previously implicated in FLUAV entry during virion internalization, and another EPS protein, EPS15, has been shown to play a role in regulating EGFR levels and endosome maturation during FLUAV uncoating (Eierhoff et al., 2010; Gschweitl et al., 2016). Our data, however, indicate that the role of EPS8 during FLUAV entry is divorced from its role in EGFR signaling. Unlike EPS15, loss of EPS8 did not alter levels of EGFR on A549 cells (Figure S7A). EPS8 was important for viral entry in both EGFR-positive A549 cells and EGFR-negative 293 cells (Figure S7A; Zhang et al., 2015). Finally, complementation of EPS8-edited cells with EPS8 lacking the EGFR binding domain (EPS8ΔEGFR) (Castagnino et al., 1995) rescued viral gene expression, possibly even better than WT EPS8 (Figure S7B). These independent lines of experimentation suggest EPS8 function during viral uncoating is independent of EGFR signaling. EPS8 is also involved in modulating actin dynamics (Hertzog et al., 2010). Actin has been implicated in the movement of virion-containing endosomes immediately after virion internalization and also plays a role in the discrete steps post-fusion but before uncoating is completed (Banerjee et al., 2014; Lakadamyali et al., 2003). A role for actin during post-fusion uncoating is the same step where our data revealed EPS8 functions, raising the possibility that the ability of EPS8 to engage and modulate actin dynamics is important for uncoating.
Although cells lacking EPS8 have decreased FLUAV gene expression, that was not the case during FLUBV infection. FLUAV and FLUBV are structurally similar, and it is tempting to generalize that the replication cycle is largely the same for the two viruses. Like FLUAV, FLUBV uses receptor-mediated endocytosis for entry (Shaw and Palese, 2013). Acidification of the FLUBV virion interior is facilitated by viral membrane protein and proton channel BM2, a FLUAV M2 homolog (Mould et al., 2003). FLUBV undergoes uncoating after fusion of viral and endosomal membranes, but many of the details of FLUBV uptake and uncoating are still unknown. Interestingly, cellular immune responses to FLUBV infection differ from those to FLUAV infection (Jiang et al., 2016; Mäkeläet al., 2015). Therefore, host processes involved in other steps of FLUBV infection may possibly also differ, as suggested by the discordant importance of EPS8 for FLUAV and FLUBV.
We also considered the possibility that other viruses using receptor-mediated endocytosis or similar internalization pathways could be affected by EPS8. A panel of RNA viruses using diverse cellular receptors and entry mechanisms was used to infect A549 cells overexpressing EPS8 (Figure S4). No obvious trends or associations with viral families or entry pathways were noted. Nonetheless, these results indicate that EPS8 enhancement of infection is specific to certain viruses, and the multifunctional nature of EPS8 may impart an anti-viral function for other viruses. In summary, our gene correlation analysis identified both pro- and anti-viral host factors with a functional impact on early stages of FLUAV replication without requiring artificial manipulation of the cellular environment. Through interrogation of early steps of FLUAV infection, we established EPS8 as a previously uncharacterized cofactor facilitating FLUAV uncoating.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andrew Mehle (amehle@wisc.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Authenticated stocks of 293T, A549, and MDCK cells were purchased from the American Type Culture Collection (ATCC). Parental and edited 293 cells were obtained from Synthego. MDCK-HA cells were a gift from P. Palese (Marsh et al., 2007). These cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS).The NCI-60 cell lines are a panel of 59 human breast, central nervous system, colon, lung, melanoma, ovarian, renal, and prostate cancer cell lines (Weinstein, 2006). The NCI-60 panel was obtained from the US National Cancer Institute’s Developmental Therapeutics Program (NCI DTP), Fort Detrick, Frederick, MD, USA. All NCI-60 panel cell lines were grown in Roswell Park Memorial Institute-1640 (RPMI-1640) medium supplemented with 10% heat-inactivated FBS. All cells were grown at 37°C with 5% CO2 and were regularly tested and verified free of mycoplasma contamination using MycoAlert (Lonza).
Viruses
Influenza A virus (FLUAV) strain H1N1 A/WSN/33 (WSN) was propagated in MDCK cells. The recombinant influenza A reporter viruses WSN PASTN (Tran et al., 2013), A/California/04/2009 PASTN (H1N1, CA04 PASTN) (Karlsson et al., 2015), WSN PASN (Tran et al., 2015), WSN with the polymerase from A/green-winged teal/ OH/175/1983 (H2N1) encoding PB2 S590/R591/K627 (S009 SRK PASTN), B/Brisbane/60/2008 (B/Brisbane) PASTN, and FVG-R (Hao et al., 2008) were rescued using the influenza virus reverse genetics system and prepared as previously described. WSN PASN was further purified by centrifugation through a 20% sucrose cushion to remove contaminating luciferase present in the media (Tran et al., 2015). WSN-GFP was amplified and titered on HAMDCK cells (Marsh et al., 2007).
Multicycle replication infections were performed by inoculating A549 cells at a multiplicity of infection (MOI) of 0.01 using virus diluted in virus growth medium (VGM) (DMEM supplemented with penicillin/streptomycin, 25 mM HEPES, 0.3% BSA) with 0.25 μg/ml TPCK-trypsin. Supernatants were collected at indicated times and titered by plaque assay on MDCK cells (Matrosovich et al., 2006) or by a Nano-Glo viral titer assay by inoculating MDCK cells with WSN PASTN and measuring luciferase activity (Karlsson et al., 2018; Tran et al., 2013).
Viral gene expression was measured by infecting cells with PASTN viruses. Virus was diluted in VGM with 0.25–0.5 μg/ml TPCK-trypsin for A549 cells or Opti-MEM I medium supplemented with 2% FBS for 293T and 293 cells. Viral gene expression was measured 8 hpi using a Nano-Glo luciferase assay kit (Promega).
Viral attachment was quantified by inoculating A549 cells with PASN. Purified virus was diluted in VGM with 0.25 μg/ml TPCK-trypsin, applied to cells for 45 mins at 4°C, and removed. Cells were washed with cold VGM and bound virions were detected by performing a Nano-Glo assay.
FVG-R infections were performed by inoculating A549 cells with virus diluted in Opti-MEM I medium supplemented with 0.2% FBS. Viral gene expression was measured 8 hpi using a Renilla luciferase assay system (Promega).
Infections with JUNV (Romero), EBOV, and MARV (Ci67) and infections with RVFV (ZH501) and VEEV (IC-SH3) were conducted under Biosafety Laboratory 4 and 3 conditions, respectively. Cells in 96-well format (30,000 cells per well) were infected at the indicated MOIs. After 1 hour, the inocula were removed, cells were washed with PBS, and replenished with fresh growth medium. VEEV and RVFV-infected plates were fixed in formalin 20 hours post-inoculation. All other infected plates were fixed 48 hours post-inoculation. Antigen staining and high-content quantitative image-based analysis were performed as previously described (Radoshitzky et al., 2010, 2016).
METHOD DETAILS
NCI-60 screen and COMPARE analysis
NCI-60 cell lines were seeded by groups of cell origin at 3 × 104 cells per well in 96-well plates and grown overnight. Cells were infected with WSN-GFP at MOIs 0.2 and 2. At 3 hours post-inoculation, the cells were washed with RPMI-1640 medium and fresh growth medium was added. WSN-GFP expression was measured by fluorescence microscopy 24 hours post-inoculation. Cells were fixed with 4% paraformaldehyde, and nuclei were stained with Hoechst 33342. WSN-GFP virus expression was detected by the Operetta-High Content Imaging System (PerkinElmer Inc.), and the percentage of GFP-positive cells were analyzed by Harmony4.1 software (PerkinElmer Inc.). WSN-GFP expression was evaluated by the flow cytometry (BD Biosciences, LSRFORTESSA) 24 hours post-inoculation. All infections were performed in triplicate, and two biological replicates performed for each MOI condition. Both approaches yielded similar results, and infectivity for each cell line was rank-ordered relative to MDCK cells. The relative infectivity of each cell line was log2-transformed and used as input for the COMPARE algorithm (Paull et al., 1989).
Knockout and stable expression of EPS8
The EPS8 locus was edited in A549 cells by lentiviral expression of CRISPR/Cas9 components. Vesicular stomatitis Indiana virus (VSIV) glycoprotein G-pseudotyped lentivirus was generated by transfecting 293T cells with the plasmids psPAX2, pMD2.G, and pLentiCRISPR (Addgene 52961; Sanjana et al., 2014) modified to encode a single-guide RNA (sgRNA) targeting EPS8 (5′-TCAAC TTACTTCATCTGAGA-3′, Figure S2). A549 cells were transduced with this virus, placed under puromycin selection (0.5 μg/ml), and single cells were cloned. Pooled 293 cells edited at the EPS8 locus were created by Synthego by transfecting cells with Cas9 RNPs containing an sgRNA targeting exon 5 (5′-GCACTTGACTACCTTTGTCC-3′) (Figure S3). 293 cells were single cell cloned. Edited alleles in both cell types were identified by PCR amplification of the locus, Sanger sequencing of the products, and inference of CRISPR edits (ICE) analysis (Hsiau et al., 2019) (Figures S2A, S2B, S3A, and S2B). Knockouts predicted by ICE analysis were assessed by immunoblot. Stable expression of EPS8 in cells was achieved by lentivirus gene delivery. The gene delivery vector pLX304-EPS8 was purchased from DNASU (HsCD00420355) and encodes the 822 amino acid splice variant (NCBI XP_024304650). pLX304-EPS8ΔEGFR was created by modifying pENTR223-EPS8 (DNASU HsCD00505776; (Seiler et al., 2014)) and subsequent Gateway recombination into pLX304 (Addgene 25890). Virus was produced by transfecting 293T cells with plasmids pLX304-EPS8 or pLX304-EPS8ΔEGFR, psPAX2, and pMD2.G. Wild-type and EPS8-edited A549 cells were transduced with these viruses and selected with blasticidin to obtain cells stably expressing EPS8 constructs.
Polymerase activity assay
293T or 293 cells were transfected with plasmids encoding WSN PA, PB1, PB2, and NP, a vNA-luciferase reporter, a Renilla luciferase control reporter, and EPS8 or an empty vector using TransIT-2020 (Mirus). Firefly luciferase and Renilla luciferase activity were assayed 24 hours post-transfection for 293T cells and 48 hours post-transfection for 293 cells. Firefly luciferase (FF) was normalized to Renilla luciferase (RLuc) within each sample. Expression of EPS8 was determined by immunoblot of cell lysates.
Acid bypass assays
Acid bypass with WSN PASTN was performed as described (Matlin et al., 1981; Mondal et al., 2017). Wild-type and EPS8-edited A549 cells were inoculated at an MOI of 0.1 with virus diluted in VGM with 0.25 μg/ml TPCK-trypsin for 1 hour at 4°C. The inoculum was removed and cells were washed with cold Dulbecco’s phosphate-buffered saline (DPBS). Inoculated cells were then either treated with 20 mM HEPES, pH 7.4 in 154 mM NaCl or 50 mM citrate, pH 5.0 in 154 mM NaCl for 45 s at 37°C. The inoculum and treatment buffer were removed and cells were washed with room temperature DPBS. Pre-warmed DMEM supplemented with 10% heat-inactivated FBS was added to the cells, infection progressed at 37°C for 8 hours, and viral gene expression was measured by a Nano-Glo assay.
Immunofluorescence assays
Wild-type and EPS8-edited A549 cells were grown on coverslips and inoculated with WSN at an MOI of 5 in VGM with 0.25 μg/ml of TPCK-trypsin for 1 hour at 4°C. Warm VGM was added to the cells and infection progressed for the indicated length of time at 37°C. Infection was also done in the presence of 1 mM cycloheximide at an MOI of 25 for detection of viral RNPs. Cells were fixed with 4% paraformaldehyde in DPBS for 20 minutes at room temperature, permeabilized with 0.1% Triton-X in 0.1 M glycine for 5 minutes at room temperature, and blocked in 3% BSA in DPBS overnight at 4°C. Cells were incubated sequentially with primary and secondary antibodies diluted in 3% BSA in DPBS: α-M1 (19 μg/ml) and chicken α-mouse AlexaFluor 594 (2 μg/ml); or α-RNP (1:1000) and donkey α-goat AlexaFluor 488 (2 μg/ml). Coverslips were mounted using mounting medium with 4’,6-diamidino-2-phenylindole (DAPI) stain (Vector Laboratories, H-1200) and imaged using 20X and 40X objectives on an EVOS FL Auto (ThermoFisher). For M1 staining, a minimum of 100 M1-positive cells at 1.5 hpi were counted across 10 random fields of view for each condition in 2 separate biological replicates. Similar quantification was performed at 1 hpi, although fewer M1-positive cells were present for all cell types. RNP localization was quantified by assessing a minimum of 100 cells across 10 random fields of view for each time point in each cell type across 3 separate biological replicates. Images were batch processed using ImageJ for quantification (Schneider et al., 2012). Representative images for cytoplasmic and nuclear RNP staining were batch-processed separately to show staining distribution.
EPS8 co-immunoprecipitations
Interactions between EPS8 and incoming RNPs was investigated in EPS8.1 A549 cells stably complemented with EPS8-V5. Cells were inoculated with WSN at an MOI of 25 diluted in cold VGM. Infections were synchronized by inoculating cells at 4°C for 1 hour. Warm VGM was added to the cells and infection progressed for 2.5 hours at 37°C. Cells were washed with cold PBS and lysed in co-IP buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors. Lysates were clarified and subjected to immunoprecipitation with 1 μg anti-V5 antibody or control rabbit IgG. Immune complexes were captured with protein A agarose resin, washed extensively with co-IP buffer, eluted, and analyzed by anti-RNP immunoblot to probe for NP.
QUANTIFICATION AND STATISTICAL ANALYSIS
Each assay was performed in technical triplicate or quadruplicate and represents at least three independent biological replicates with the exception of the immunofluorescence assays which represent at least two biological replicates. Mean and standard deviation were calculated, and statistical significance was tested using a two-tailed Student’s t test with unequal variance for pairwise comparison or a one-way ANOVA with a Tukey’s honestly significant difference (HSD) post hoc analysis for multiple comparisons.
DATA AND CODE AVAILABILITY
The source data for Figure 1 in the paper are available in Table S1.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-EPS8 | BD Biosciences | Cat# 610144; RRID: AB_397545 |
| Mouse anti-M1 HB-64 | Yewdell et al., 1981/ATCC | M2–1C6–4R3 (HB-64) |
| Goat polyclonal anti-RNP | BEI | NR-3133 |
| Mouse monoclonal anti-tubulin | Sigma | Cat# T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-HSP90α/β | Santa Cruz Biotechnology | Cat# sc-7947; RRID: AB_2121235 |
| Rabbit polyclonal anti-V5 | Bethyl Laboratories | Cat#A190–120A; RRID: AB_67586 |
| Mouse monoclonal anti-EGFR clone 2A2H10 | ProteinTech | 66455–1-Ig |
| Chicken polyclonal anti-mouse AlexaFluor 594 | Invitrogen | Cat # A-21201; RRID: AB_141630 |
| Donkey polyclonal anti-goat AlexaFluor 488 | Invitrogen | Cat# A-11055; RRID: AB_2534102 |
| Protein A agarose | Sigma | P7786 |
| Bacterial and Virus Strains | ||
| Influenza A virus: A/WSN/33 (H1N1; WSN) | rescued for this project | N/A |
| Influenza A virus: A/California/04/2009 (H1N1; CA04) | rescued for this project | N/A |
| Influenza B virus: B/Brisbane/60/2008 (B/Brisbane) | rescued for this project | N/A |
| Junín virus, Romero | USAMRIID virus stock | 23079 |
| Ebola virus | IRF-Frederick virus stock | IRF0259 |
| Marburg virus, Ci67 | USAMRIID virus stock | 18204 |
| Rift Valley fever virus, ZH501 | USAMRIID virus stock | 18205 |
| Venezuelan equine encephalitis virus, IC-SH3 | USAMRIID virus stock | 17539 |
| WSN-GFP | Marsh et al., 2007 | N/A |
| WSN-PASTN | Tran et al., 2013 | N/A |
| CA04 PASTN | Karlsson et al., 2015 | N/A |
| WSN-PASTN with viral polymerase from A/green-winged teal/ OH/175/1983 (H2N1) encoding PB2 S590/R591/K627 (S009 SRK PASTN) | This paper | N/A |
| B/Brisbane PASTN | This paper | N/A |
| PASN | Tran et al., 2015 | N/A |
| FVG-R | Hao et al., 2008 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TransIT-2020 | Mirus | MIR 5400 |
| ECL Prime Western Blotting Detection Reagent | GE Healthcare | GERPN2236 |
| Critical Commercial Assays | ||
| Nano-Glo luciferase assay kit | Promega | N1120 |
| Renilla luciferase assay system | Promega | E2810 |
| MycoAlert | Lonza | LT07–218 |
| Experimental Models: Cell Lines | ||
| Human: NCI-60 panel of cell lines | NCI DTP (https://dtp.cancer.gov) | NCI Anti-Cancer Cell Line Panel |
| Human: 293T | ATCC | CRL-3216 |
| Human: A549, male | ATCC | CCL-185 |
| Human: A549 EPS8 knockouts | this study | N/A |
| Canine: MDCK, female | ATCC | CCL-34 |
| Canine: MDCK-HA | Marsh et al., 2007 | N/A |
| Human: 293 | Synthego | N/A |
| Human: 293 EPS8 knockouts | Synthego | N/A |
| Recombinant DNA | ||
| Plasmid: pLX304-EPS8 | DNASU | HsCD00420355 |
| Plasmid: pLX304-EPS8 ΔEGFR | This paper | N/A |
| Plasmid: pENTR223-EPS8 | DNASU | HsCD00505776 |
| Plasmid: pLentiCRISPR | Sanjana et al., 2014 | Addgene 52961 |
| Plasmid: p3X-1T | Tran et al., 2013 | N/A |
| Plasmid: pCAGGS-NP | Neumann et al., 2005 | N/A |
| Plasmid: pHH21-vNA-Luc | Regan et al., 2006 | N/A |
| Plasmid: pRL-SV40 | Promega | E2231 |
| Plasmid: psPAX2 | D. Trono | Addgene 12260 |
| Plasmid: pMD2.G | D. Trono | Addgene 12259 |
| Software and Algorithms | ||
| COMPARE | Paull et al., 1989 | https://dtp.cancer.gov/databases_tools/compare.htm |
| ICE | Hsiau et al., 2019 | https://ice.synthego.com/#/ |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
Highlights.
Gene correlation analysis identifies host factors for influenza A virus replication
EPS8 is a pro-viral factor for influenza A virus replication
Cells lacking EPS8 have delayed virion uncoating and RNP import
EPS8 physically associates with vRNPs during uncoating
ACKNOWLEDGMENTS
We thank Drs. P. Palese, C. Brooke, N. Sherer, and Y. Kawaoka for reagents. We thank L. Bollinger for critically editing the manuscript. This work was funded by grant T32AI078985 to G.P.L., grant T32GM07215 to V.T., and grant R01AI125271 from the National Institutes of Health and a Shaw Scientist Award to A.M. A.M. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. This work was also supported in part through Battelle Memorial Institute’s prime contract with the National Institute of Allergy and Infectious Diseases (NIAID) under contract HHSN272200700016I (Y.C., S.Y., and J.H.K.). The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Departments of the Army, Defense, and Health and Human Services, or of the institutions and companies affiliated with the authors.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.064.
REFERENCES
- Banerjee I, Yamauchi Y, Helenius A, and Horvath P (2013). High-content analysis of sequential events during the early phase of influenza A virus infection. PLoS ONE 8, e68450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee I, Miyake Y, Nobs SP, Schneider C, Horvath P, Kopf M, Matthias P, Helenius A, and Yamauchi Y (2014). Influenza A virus uses the aggresome processing machinery for host cell entry. Science 346, 473–477. [DOI] [PubMed] [Google Scholar]
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, et al. (2009). The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinskaya AG, Vorkunova NK, Kornilayeva GV, Narmanbetova RA, and Vorkunova GK (1982). Influenza virus uncoating in infected cells and effect of rimantadine. J. Gen. Virol 60, 49–59. [DOI] [PubMed] [Google Scholar]
- Carette JEE, Guimaraes CPP, Varadarajan M, Park ASS, Wuethrich I, Godarova A, Kotecki M, Cochran BHH, Spooner E, Ploegh HLL, et al. (2009). Haploid genetic screens in human cells identify host factors used by pathogens. Science 326, 1231–1235. [DOI] [PubMed] [Google Scholar]
- Castagnino P, Biesova Z, Wong WT, Fazioli F, Gill GN, and Di Fiore PP (1995). Direct binding of eps8 to the juxtamembrane domain of EGFR is phosphotyrosine- and SH2-independent. Oncogene 10, 723–729. [PubMed] [Google Scholar]
- de Vries E, Tscherne DM, Wienholts MJ, Cobos-Jiménez V, Scholte F, García-Sastreı A, Rottier PJM, and de Haan CAM (2011). Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 7, e1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore PP, and Scita G (2002). Eps8 in the midst of GTPases. Int. J. Biochem. Cell Biol 34, 1178–1183. [DOI] [PubMed] [Google Scholar]
- Dou D, Revol R, Östbye H, Wang H, and Daniels R (2018). Influenza A virus cell entry, replication, virion assembly and movement. Front. Immunol 9, 1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eierhoff T, Hrincius ER, Rescher U, Ludwig S, and Ehrhardt C (2010). The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 6, e1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschweitl M, Ulbricht A, Barnes CA, Enchev RI, Stoffel-Studer I, Meyer-Schaller N, Huotari J, Yamauchi Y, Greber UF, Helenius A, and Peter M (2016). A SPOPL/Cullin-3 ubiquitin ligase complex regulates endocytic trafficking by targeting EPS15 at endosomes. eLife 5, e13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Perez JT, Chen C, Li Y, Benitez A, Kandasamy M, Lee Y, Andrade J, TenOever B, and Manicassamy B (2018). Genome-wide CRISPR/ Cas9 screen identifies novel host factors essential for influenza virus replication. Cell Rep. 23, 1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, and Kawaoka Y (2008). Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454, 890–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog M, Milanesi F, Hazelwood L, Disanza A, Liu H, Malabarba MG, Pasqualato S, Maiolica A, Confalonieri S, Clainche C, Le, et al. (2010). Molecular basis for the dual function of Eps8 on actin dynamics: bundling and capping. PLoS Biol. 8, e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau T, Conant D, Maures T, Waite K, Yang J, Kelso R, Holden K, Enzmann BL, and Stoner R (2019). Inference of CRISPR edits from Sanger trace data. bioRxiv. 10.1101/251082. [DOI] [PubMed] [Google Scholar]
- Jiang J, Li J, Fan W, Zheng W, Yu M, Chen C, Sun L, Bi Y, Ding C, Gao GF, et al. (2016). Robust Lys63-linked ubiquitination of RIG-I promotes cytokine eruption in early influenza B virus infection. J. Virol 90, 6263–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EA, Meliopoulos VA, Savage C, Livingston B, Mehle A, and Schultz-Cherry S (2015). Visualizing real-time influenza virus infection, transmission and protection in ferrets. Nat. Commun 6, 6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EA, Meliopoulos VA, Tran V, Savage C, Livingston B, Schultz-Cherry S, and Mehle A (2018). Measuring influenza virus infection using bioluminescent reporter viruses for in vivo imaging and in vitro replication assays. Methods Mol. Biol 1836, 431–459. [DOI] [PubMed] [Google Scholar]
- Kondratowicz AS, Hunt CL, Davey RA, Cherry S, and Maury WJ (2013). AMP-activated protein kinase is required for the macropinocytic internalization of ebolavirus. J. Virol 87, 746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, et al. (2010). Human host factors required for influenza virus replication. Nature 463, 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP, and Zhuang X (2003). Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U S A 100, 9280–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim J, Son K, d’Alexandry d’Orengiani AP, and Min JY (2017). Acid phosphatase 2 (ACP2) is required for membrane fusion during influenza virus entry. Sci. Rep. 7, 43893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts L, van Dam W, Persoons L, and Naesens L (2012). Interaction between mouse adenovirus type 1 and cell surface heparan sulfate proteoglycans. PLoS ONE 7, e31454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, and Ohnishi S (1980). Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett. 122, 283–287. [DOI] [PubMed] [Google Scholar]
- Mäkelä SM, Österlund P, Westenius V, Latvala S, Diamond MS, Gale M, and Julkunen I (2015). RIG-I signaling is essential for influenza B virus-induced rapid interferon gene expression. J. Virol 89, 12014–12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar KB, Rinkenberger NR, Boys IN, Eitson JL, McDougal MB, Richardson RB, and Schoggins JW (2018). LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat. Commun 9, 3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh GA, Hatami R, and Palese P (2007). Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 81, 9727–9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, and Helenius A (1991). Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67, 117–130. [DOI] [PubMed] [Google Scholar]
- Matlin KS, Reggio H, Helenius A, and Simons K (1981). Infectious entry pathway of influenza virus in a canine kidney cell line. J. Cell Biol 91, 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Matrosovich T, Garten W, and Klenk HD (2006). New low-viscosity overlay medium for viral plaque assays. Virol. J. 3, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K, Fagerlund R, Franke J, Kohler M, Kinnunen L, and Julkunen I (2003). Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem 278, 28193–28200. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Keusch JJ, Decamps L, Ho-Xuan H, Iketani S, Gut H, Kutay U, Helenius A, and Yamauchi Y (2019). Influenza virus uses transportin 1 for vRNP debundling during cell entry. Nat. Microbiol. 4, 578–586. [DOI] [PubMed] [Google Scholar]
- Mondal A, Dawson AR, Potts GK, Freiberger EC, Baker SF, Moser LA, Bernard KA, Coon JJ, and Mehle A (2017). Influenza virus recruits host protein kinase C to control assembly and activity of its replication machinery. eLife 6, e26910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould JA, Paterson RG, Takeda M, Ohigashi Y, Venkataraman P, Lamb RA, and Pinto LH (2003). Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell 5, 175–184. [DOI] [PubMed] [Google Scholar]
- Neumann G, Fujii K, Kino Y, and Kawaoka Y (2005). An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proc. Natl. Acad. Sci. U S A 102, 16825–16829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill RE, Jaskunas R, Blobel G, Palese P, and Moroianu J (1995). Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J. Biol. Chem 270, 22701–22704. [DOI] [PubMed] [Google Scholar]
- Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, Plowman J, and Boyd MR (1989). Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst 81, 1088–1092. [DOI] [PubMed] [Google Scholar]
- Pinto LH, Holsinger LJ, and Lamb RA (1992). Influenza virus M2 protein has ion channel activity. Cell 69, 517–528. [DOI] [PubMed] [Google Scholar]
- Radoshitzky SR, Dong L, Chi X, Clester JC, Retterer C, Spurgers K, Kuhn JH, Sandwick S, Ruthel G, Kota K, et al. (2010). Infectious Lassa virus, but not filoviruses, is restricted by BST-2/tetherin. J. Virol 84, 10569–10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshitzky SR, Pegoraro G, Chī XO, D Ng L, Chiang CY, Jozwick L, Clester JC, Cooper CL, Courier D, Langan DP, et al. (2016). siRNA screen identifies trafficking host factors that modulate alphavirus infection. PLoS Pathog. 12, e1005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan JF, Liang Y, and Parslow TG (2006). Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J. Virol 80, 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Reinhold WC, and Buck CB (2012). Entry tropism of BK and Merkel cell polyomaviruses in cell culture. PLoS ONE 7, e42181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler CY, Park JG, Sharma A, Hunter P, Surapaneni P, Sedillo C, Field J, Algar R, Price A, Steel J, et al. (2014). DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 42, D1253–D1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, Reimers MA, Scherf U, Kahn A, Dolginow D, et al. (2007). Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol. Cancer Ther. 6, 820–832. [DOI] [PubMed] [Google Scholar]
- Shaw ML, and Palese P (2013). Orthomyxoviruses In Fields Virology, Knipe DM and Howley PM, eds. (Lippincott Williams & Wilkins; ), pp. 1151–1185. [Google Scholar]
- Stauffer S, Feng Y, Nebioglu F, Heilig R, Picotti P, and Helenius A (2014). Stepwise priming by acidic pH and a high K+ concentration is required for efficient uncoating of influenza A virus cores after penetration. J. Virol 88, 13029–13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Moser LA, Poole DS, and Mehle A (2013). Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. J. Virol 87, 13321–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Poole DS, Jeffery JJ, Sheahan TP, Creech D, Yevtodiyenko A, Peat AJ, Francis KP, You S, and Mehle A (2015). Multi-modal imaging with a toolbox of influenza A reporter viruses. Viruses 33, 5319–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Palese P, and O’Neill RE (1997). The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol 71, 1850–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN (2006). Spotlight on molecular profiling: “integromic” analysis of the NCI-60 cancer cell lines. Mol. Cancer Ther. 5, 2601–2605. [DOI] [PubMed] [Google Scholar]
- Weinstein JN, and Pommier Y (2003). Transcriptomic analysis of the NCI-60 cancer cell lines. C. R. Biol 326, 909–920. [DOI] [PubMed] [Google Scholar]
- Yángüez E, Hunziker A, Dobay MP, Yildiz S, Schading S, Elshina E, Karakus U, Gehrig P, Grossmann J, Dijkman R, et al. (2018). Phosphoproteomic-based kinase profiling early in influenza virus infection identifies GRK2 as antiviral drug target. Nat. Commun 9, 3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang S, Yin L, Yang Y, Guan Y, Wang W, Xu H, and Tao N (2015). Quantification of epidermal growth factor receptor expression level and binding kinetics on cell surfaces by surface plasmon resonance imaging. Anal. Chem 87, 9960–9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhirnov OP (1990). Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology 176, 274–279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data for Figure 1 in the paper are available in Table S1.