Abstract
In type 1 diabetes (T1D), proinsulin is a major autoantigen and the insulin B:9–23 peptide contains epitopes for CD4 T cells in both mice and humans. This peptide requires C-terminal mutations for uniform binding in the proper position within the mouse IAg7 or human DQ8 MHCII peptide grooves and for strong CD4 T cell stimulation. Here we present structures showing how these mutations control CD4 T cell receptor (TCR) binding to these MHCII-peptide complexes. Our data reveal striking similarities between mouse and human CD4 TCRs in their interactions with these ligands. We also show how fusions between fragments of B:9–23 and of proinsulin C-peptide create chimeric peptides with activities as strong or stronger than the mutated insulin peptides. We propose transpeptidation in the lysosome as a mechanism that could accomplish these fusions in vivo, similar to the creation fused peptide epitopes for MHCI presentation shown to occur by transpeptidation in the proteasome. Were this mechanism unique to the pancreas and absent in the thymus, it could provide an explanation for how diabetogenic T cells escape negative selection during development but find their modified target antigens in the pancreas to cause T1D.
ONE SENTENCE SUMMARY
Structures show how similar modifications of an insulin peptide create strong CD4 T cell receptor agonists in type-1 diabetes in mice and humans.
INTRODUCTION
Insulin is a major target in type-1 diabetes (T1D) in humans and rodents (1). During the past several decades, many CD4 T cell clones have been isolated from NOD mice responsive to an epitope(s) in the B:9–23 insulin peptide. There has been disagreement about how these T cells target this peptide. Particularly controversial has been the position or “register” this peptide takes in the groove of the NOD IAg7 major histocompatibility complex (MHC) class II molecule (MHCII) for presentation to these T cells. Some have suggested registers 1 and 2 (R1 and R2) that put B:12–20 and B:13–21, respectively, in the p1 to p9 positions of the IAg7 binding groove because these registers provide IAg7 compatible anchor amino acids at p1, p4, p6 and p9 (2–5). However, our previous data argue strongly that nearly all diabetogenic NOD CD4 T cells recognize B:9–23 in register 3 (R3), which places B:14–22 (ALYLVCGER) in the p1 to p9 position (6–9). This register had not been previously considered because the peptide’s basic B:22R at p9 clashes with the IAg7 p9 pocket, which strongly prefers an acidic amino acid.
Our data showed that a substitution of E for R at p9 dramatically improved binding of the peptide in R3 converting the weakly stimulatory natural peptide into a very strong agonist for a subset of NOD B:9–23 specific T cells, generally referred to as Type A(2, 3). We also determined that for a second group of T cells, Type B (2, 3), an additional substitution at E to G at p8 (B:21), was needed to create the strong agonist for these T cells. Since p8 is usually a surface exposed amino acid in MHCII bound peptides, we postulated that the side chain of the p8E somehow interfered with the correct docking of the Type B T cell TCRs on the R3 IAg7-peptide complex. We have also studied human T cells specific for the B:9–23 peptide presented by human HLA-DQ8 (DQ8) (9–11). Like IAg7, the DQ8 beta chain has a polymorphism at β57 that creates a preference for an acidic amino acid at p9. The substitution of R to E at B:22 in the peptide also greatly improved the presentation of the insulin peptide by DQ8 to these human T cells, establishing R3 as the relevant register. Most recently, we have published high-resolution crystal structures of these modified peptides bound to IAg7 and DQ8 (9), confirming the R3 position and activity of the peptides.
Here we show the structures of a mouse Type A, a mouse Type B and a human Type A-like TCR, bound to their optimal versions of the MHCII-R3 insulin peptide ligands. The structures confirmed the R3 recognition of the T cells and showed that the specificity differences among mouse Type A and Type B T cells lies in how they deal with the amino acid at p8 (B:21) of the insulin peptide. Despite differences among the mouse and human TCRs in the sequences of their Vα and Vβ domains and their orientations on their ligands, there were some striking common features to the complexes pointing out the similarities in human and mouse both in how these ligands are formed and in how TCRs engage them. The structures also show how the peptide modifications were essential to the formation of the complexes, suggesting a role for modification of the peptide in vivo to initiate the CD4 T cell response in T1D. We show that the recently demonstrated activity of T cell chimeric epitopes formed by peptide fusion (12–19) could account for the required modifications. We predicted that internal proteolytic deletions of the appropriate portions of proinsulin between the B-chain and C-peptide could generate chimeric peptides similar to our modified ones. We show that synthetic versions of the predicted fused peptides act as super-agonists for a variety of mouse and human insulin reactive T cells.
RESULTS
Mouse and human CD4 T cells recognize B:9–23 bound similarly to IAg7 and DQ8, but with different docking modes.
Mouse and human CD4 T cells reactive to epitopes involving the B:9–23 insulin peptide have been reported and characterized in numerous publications (2, 3, 6, 7, 9, 11, 20–22). A subset of these are listed in Table 1, along with the sequences of their TCR Vα and Vβ CDR1, CDR2 and CDR3 loops (This table is an updated version of that presented in a previous publication (9)). For the structural studies presented here, we used three of these T cells: a NOD mouse Type A T cell, I.29 (6, 7); a NOD mouse Type B T cell, 8F10 (3, 9) and a human Type A-like T cell, T1D3, isolated from a T1D patient (9, 11). As described in the Materials and Methods, we used T cell hybridomas or TCR transduced T cell avatars for functional studies. For structural studies we prepared soluble versions of the TCRs, fusing the V-regions of the TCRs to the extra cellular domains of human Cα and Cβ, which were expressed separately in E. coli inclusion bodies, solubilized, mixed and refolded to prepare functional the soluble functional TCRs (23, 24).
Table 1 –
Properties of the TCRs of the mouse and human B:9–23 specific CD4 T cells
| Vα (TRAV)a | CDR1b | CDR2b | CDR3c | Jαa | Vβ(TRBV)a | CDR1b | CDR2b | CDR3c | Jβa | |
|---|---|---|---|---|---|---|---|---|---|---|
| I.29 | 15 (10) | DTASSY | IRSNVD | AASPSNSGGSNYKLT | 53 | 2 (1) | NSQYPW | LRSPGD | TCSAGLGYEQY | 2–7 |
| PCR1–10 | 13.1 (5D-4) | DSASNY | IRSNME | AASKTGGNNKLT | 56 | 11 (16) | ISGHSA | FRNQAP | ASSLDGGQGLEQY | 2–7 |
| 12–4.1 | 13.1 (5D-4) | DSASNY | IRSNME | AASGANSGGSNYKLT | 53 | 2 (1) | NSQYPW | LRSPGD | TCSPGLGNEQY | 2–7 |
| AS150 | 10.8 (13–1) | STTLNS | RLFYNP | AISSGSWQLI | 22 | 2 (1) | NSQYPW | LRSPGD | TCSADQNSYNSPLY | 1–6 |
| 4F7 | 13.1 (5D-4) | DSASNY | IRSNME | AGTGNYKYV | 33 | 2 (1) | NSQYPW | LRSPGD | TCSADQNQAPL | 1–5 |
| 8F10 | 13.1 (5D-4) | DSASNY | IRSNME | AASRRGSGGSNYKLT | 53 | 8.2 (13–2) | TNNHNN | SYGAGS | ASGGLGGDEQY | 2–7 |
| 8–1.1 | 13.1 (5D-4) | DSASNY | IRSNME | AASKTGGNNKLT | 56 | 12 (15) | VSGHND | FRSKSL | ASSLGWGDEQY | 2–7 |
| 12–4.4 | 13.1 (5D-4) | DSASNY | IRSNME | AASASGGSNTKLT | 53 | 12 (15) | VSGHND | FRSKSL | ASSPGQGTTLY | 1–3 |
| AS91 | 13.2 (5) | DSASNY | IRSNME | SRGNNNRIF | 31 | 1 (5) | HLGHNA | YNLKQL | ASSQLGGLDTQY | 2–5 |
| T1D-3 | 3 (17) | TSINN | IRSNER | ATDAGYNQGGKLI | 23 | 5.1 (5–1) | ISGHRS | YFSETQ | ASSAGNTIY | 1–3 |
| T1D-4 | 8 (13–1) | DSASNY | IRSNVG | AASKASNTGKLI | 3 | 8.3 (12–3) | ILGHNT | YRNRAP | ASLKATDTQY | 2–3 |
| T1D-10 | 2.2 (12–3) | NSAFQY | YTYSSG | ASSRGGGNTGELF | 26 | 7.1 (4–1) | HMGHRA | SYEKLS | ASSRGGGNTGELF | 2–2 |
The Vα, Jα, Vβ and Jβ are named using the classic nomenclature with the newer IMGT nomenclature for Vα and Vβ in parentheses.
CDR1 and CDR2 sequences are the 6 amino acids at the tips of these loops. Red amino acids are predicted to interact similarly with peptide or MHC.
CDR3 sequences are amino acids between the conserved C at the end of the V element and the conserved F in the FGXG motif of the J element.
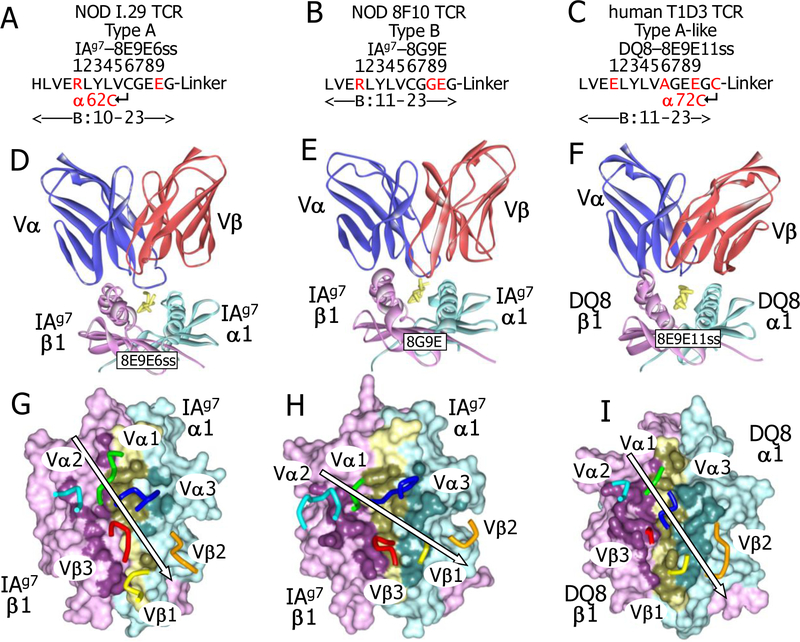
Also, as previously described (6, 7, 9), we prepared soluble versions of IAg7 and DQ8 bound to an epitope of insulin B:9–23, which was modified optimally for each of the T cells shown as schematically in Fig. 1A. Briefly, for all three ligands, the peptide was covalently attached to the N-terminus of the MHCII beta chain via a flexible linker (25) and carried substitutions at p1 and p9 to provide optimal anchor amino acids for either IAg7 (p1A>R and p9R>E) or DQ8 (p1A>E and p9R>E) in R3. The p1 mutations were used in constructs for soluble peptide/MHC complexes assure uniformity and stability of the covalent complexes, but they were not used for soluble peptides used in in vitro stimulation assays below.
Fig. 1. Mouse and human TCR engagement of IAg7 and DQ8 with a Register 3 bound insulin peptide.
. (A) Schematic representations of optimal versions of the insulin B:10–23 peptide for the I.29 (left, 8E9E6ss with IAg7), 8F10 (middle, 8G9E with IAg7) and T1D3 (right, DQ8 with DQ8) are shown. The core positions (p1-p9) of the peptide in the MHCII binding groove are numbered. Engineered disulfides are shown. Mutations from the natural sequences are in red. (B) Ribbons representation of the three TCRs docked on their optimal peptide/MHCII ligands are shown for I.29 (left), 8F10 (middle) and T1D3 (right) viewed from the C-terminal end of the peptide. Colors are: TCR Vα (blue), TCR Vβ (red), MHC α1 (cyan), MHC β1 (magenta) and peptide (yellow). (C) The footprints of the three TCRs on the solvent accessible surfaces of their MHCII-peptide ligands are shown from above: I.29, IAg7-8E9E6ss (left), 8F10, IAg7-8G9E (middle) and T1D3, DQ8–8E9E11ss (right). The surfaces are colored MHCIIα, cyan; MHCIIβ, magenta; peptide, yellow. The surface of atoms involved in TCR contact (≤4.5 Å from a TCR atom) are colored darker shades of the same colors. Also shown are tube representation the tips of the TCR CDR loops: αCDR1, green; αCDR2, cyan; αCDR3, blue; βCDR1, yellow; βCDR2, orange; βCDR3, red. For each complex, the arrow represents the angle of engagement of the TCR CDRs, placing αCDR2, βCDR3 and βCDR1 on one side and αCDR1, αCDR3, and βCDR1 on the other.
Additionally, for I.29, α62N in IAg7 was changed to C introducing a disulfide bond to the natural p6C in the peptide when the peptide bound to IAg7 in R3 (6). For 8F10, we changed B:21E to G at p8 to remove the p8E side chain from the surface, which is inhibitory for this and other Type B T cells (7, 9). For the human T1D3 T cell, we introduced C at the first amino acid of the linker (p11) and at DQ8 α72, while changing the natural B:19C at p6 to A in order to form an unambiguous disulfide bond between the C-terminus of the peptide and DQ8 alpha chain helix. These mutations were previously determined to create optimal stimulating ligands for the three CD4 T cells (6, 7, 9, 11). We refer to these three peptides below as 8E9E6ss, 8G9E and 8E9E11ss, respectively (Fig. 1A).
Our first attempts to co-crystalize the NOD mouse TCRs bound their IAg7 ligands produced crystals that contained only the free TCR. The crystal structures of these two free TCRs were solved (Materials and Methods and Table S1). They had crystallized with packing arrangements that in each crystal blocked the site for IAg7-peptide binding to the TCR. Examination of structures revealed several amino acids that appeared to be important this packing (Fig. S1). Since these amino acids were not on the TCR face predicted to interact with IAg7-peptide complex, we introduced mutations (Vβ 56K to A for I.29 and Cβ 202R to A for 8F10) at these positions to disrupt this crystal packing and re-expressed the proteins. In subsequent crystallization trials using these two of the modified mouse TCRs and the human T1D3 TCR, we obtained co-crystals with their optimal peptide ligands and were able to solve the structures of the ternary complexes (Table S1).
As seen in the scores of structures of other TCRs bound to MHC/peptide ligands (26) in our crystals of the TCRs bound to their MHCII/peptide complexes in the now familiar diagonal orientation, but with differences in the pitches and angles of docking (Fig. 1B,C). Views of the complexes from the C-terminal ends of the peptides (Fig. 1B) show that the I.29 TCR was pitched toward the IAg7 β chain helix of the IAg7-8E9E6ss complex, while the 8F10 and T1D3 TCRs sat more flatly on their IAg7-8G9E and DQ8–8E9E11ss ligands. Views from above (Fig. 1C) show that the CDRs of the I.29 and T1D3 TCRs were docked at a shallow angle of only about 30° to the peptide backbone, a feature that is at one extreme of the range seen with other TCR-MHCII complexes, while the 8F10 TCR sits on the IAg7-8G9E complex at an angle of about 60°, more typical of TCR-MHCII complexes (26).
The combination of these differences in pitch and angle led to different footprints among the TCRs on their ligands as measured by the number of atom-to-atom contacts and the buried surface area (BSA) at the interface (Fig. 1C, Table 2, Spreadsheet S1). The tilt and twist of the I.29 TCR created a footprint on IAg7 dominated by TCR interactions with the MHCII β1 helix at the expense of the α1 helix. The contacts usually seen between the Vβ CDR1 and CDR2 loops with the α1 helix of MHCII were absent, while the CDR3 of Vα now providing nearly all of the contact with this helix. Meanwhile, there was extensive I.29 TCR contact along the entire length of the IAg7 β1 helix, mediated by the Vα CDR1 and CDR2 as well as the CDR3s of Vα and Vβ. In contrast, the 8F10 and T1D3 TCR footprints were much more evenly spread in a conventional way over the MHC helices, with their Vβ CDR1 and CDR2 loops making multiple contacts with the MHC α1 helix and their Vα CDR1 and 2 loops doing the same with the IAg7 β1 helix. Thus, overall, these structures show that the MHCII/insulin specific TCRs bind their ligands in a variety of ways and confirm that the ligands of these three TCRs are indeed target insulin peptide bound to MHCII in R3.
Table 2 –
I.29, 8F10 and T1D3 TCR contacts with their ligands
| CDR Loop | CDR AA | # of Atom to Atom Contacts toa | CDR AA | # of Atom to Atom Contacts toa | CDR AA | # of Atom to Atom Contacts toa | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α | a | α | ||||||||||
| 1 | 26D | - | 30N | 26T | - | |||||||
| 27T | - | - | 27S | - | ||||||||
| 28A | - | 28I | - | |||||||||
| 30S | - | 29N | - | |||||||||
| BSA | 104.4(12.2) | BSA | 14.7(1.6) | BSA | 165.8(15.4) | |||||||
| 2 | 48D | 48L | - | |||||||||
| 50R | - | 50R | - | 50R | - | |||||||
| 51S | - | 51S | - | |||||||||
| 52N | - | 52N | - | 52N | - | |||||||
| 56K | - | |||||||||||
| BSA | 135.1(15.8) | BSA | 152.5(16.6) | BSA | 173.3(16.1) | |||||||
| 3 | 93R | - | ||||||||||
| 94S | - | 94R | - | 92A | - | |||||||
| 95N | - | 95G | - | 93G | 8 | |||||||
| 96S | 2 | 96S | 3 | 94Y | 25 | |||||||
| 97G | 13 | 97G | 5 | 95N | - | |||||||
| 98G | 5 | 98G | 7 | 96Q | 13 | |||||||
| 100N | - | 99S | 4 | |||||||||
| 101Y | - | |||||||||||
| BSA | 330.8(38.6) | BSA | 358.1(38.9) | BSA | 299.6(27.9) | |||||||
| 3 | 95A | - | 94L | 13 | 95A | - | ||||||
| 97L | 3 | 95G | - | 96G | - | |||||||
| 98G | - | 96G | - | 97N | - | |||||||
| 99Y | - | 97D | - | 98T | - | |||||||
| 100Y | - | |||||||||||
| BSA | 175.2(20.4) | BSA | 228.1(24.8) | BSA | 116.9(10.9) | |||||||
| 2 | 50F | 15 | ||||||||||
| 51Rc | - | 48Y | 22 | 51S | 4 | |||||||
| 52S | 1 | 55R | 22 | |||||||||
| 56N | 13 | |||||||||||
| BSA | 29.6(3.4) | BSA | 68.1(7.4) | BSA | 189.6(17.6) | |||||||
| 1 | 26N | 7 | 28G | - | ||||||||
| 29Q | - | 28N | 11 | 30R | 7 | |||||||
| 29N | 8 | 31S | - | |||||||||
| BSA | 82.3(9.6) | BSA | 99.0(10.8) | BSA | 130.5(12.1) | |||||||
| 23 | 81 | 109 | ||||||||||
| 857.3(100) | 920.4(100) | 1075.7(100) | ||||||||||
Atom to atom distances ≤ 4.5Å.
BSA= Å2 of Buried Surface Area (% of total BSA).
Red amino acids interact with p8 of the peptide
The p8 amino acid determines Type A vs. Type B recognition of the MHCII insulin complexes.
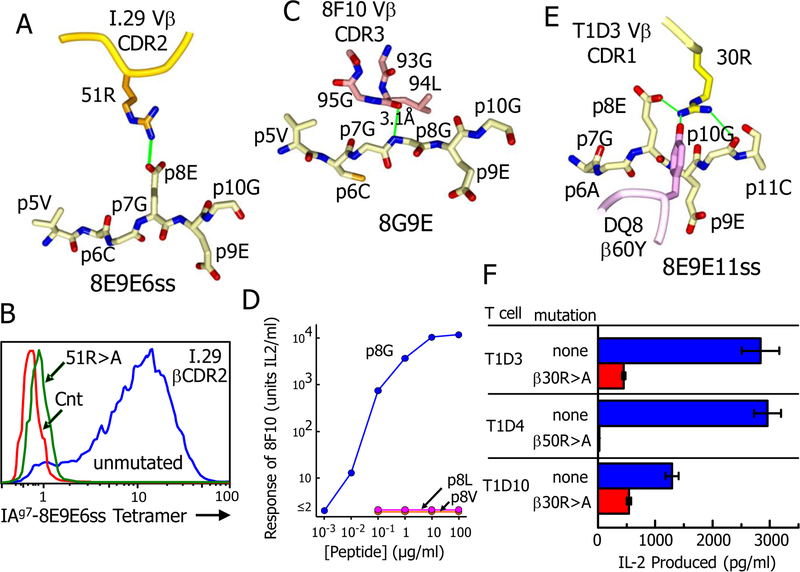
In the solved structures, the various interactions of the TCRs with the C-terminal ends of their optimal R3 bound peptides gave a structural explanation for the phenomena associated with the specificities of NOD mouse Type A and Type B CD4 T cells. Type A T cells are distinguished by the requirement for the natural p8E in the peptide for strong reactivity (7, 9). For the I.29 TCR, interaction with this amino acid was the only TCR contact at this end of the peptide. The unusual angle and tilt of engagement of the I.29 TCR with IAg7-8E9E6ss brought its Vβ2 CDR2 loop into a position such that β51R formed a salt bridge with the carboxylate of the p8E side chain (Fig. 2A, Table 2 and Spreadsheet S1). Our previous work had shown the importance of this contact in that removal of the p8E side chain by its mutation to G, reduced the peptide stimulatory potency by about 30-fold and drastically reduces the ability of the I.29 TCR to bind to the IAg7-8E9E6ss complex (9). Reciprocally, shown here (Fig. 2B) mutation of the TCR β51R to A eliminated the ability of the IAg7-8E9E6ss tetramer to bind to the I.29 T cell. Mutations to alanine of many of the other I.29 TCR amino acids making the most contact with the ligand in Table 2 also eliminated binding of the fluorescent IAg7-p8E9Ess tetramer to the I.29 T cell (Fig. S2). Many other B:9–23 reactive Type A T cells also use Vβ2 (Table 1), but, as yet, no Type B T cells have been reported to use this Vβ element. This suggests that the unusual rotation of the TCR and the selection of Vβ elements that contain an R in this position of CDR2 may be common feature of Type A T cells.
Fig. 2. TCR interactions with the peptide p8 amino acid determine Type A vs. Type B recognition.
(A) The salt bridge between β51R of the I.29 TCR βCDR2 loop and the p8E of the 8E9E6ss peptide is shown. (B) Binding of a soluble fluorescent tetramer version of the IAg7-8E9E6ss complex to three versions of the 5KC TCR─ T cell is shown: the untransduced 5KC control (red), 5KC transduced with the unmutated I.29 TCR (blue) and 5KC transduced with the I.29 TCR bearing an R>A mutation at the CDR2 β51 amino acid. The transductants were pre-screened for similar surface TCR expression. Addition data from this experiment are shown in Supplemental Fig. 2. (C) Extensive interaction of the 8F10 βCDR3 loop with the backbone of the 8G9E peptide in the region of p7G and p8G, approaching to within 3.1Å at the p8G backbone nitrogen. (D) Disruption of the 8F10 T cell response to the 8G9E peptide by mutation of the p8G to L or V. Representative of three separate experiments. (E) Extensive interaction is shown between β30R of the 8F10 TCR βCDR1 loop and the 8E9E11ss peptide from p8E to p10G, that includes a salt bridge to p8E as well as an H-bond to Y60 of the DQ8 β chain helix. (F) Inhibitory effect of mutating the β30R of the T1D3 TCR on the response of the T cell to soluble 8E9E peptide presented by M12.C3DQ8 APCs. The responses T1D4 and T1D10 to the peptide were also inhibited by mutation of β50R or β30R, respectively. Results are the average responses and SEM seen in three separate experiments.
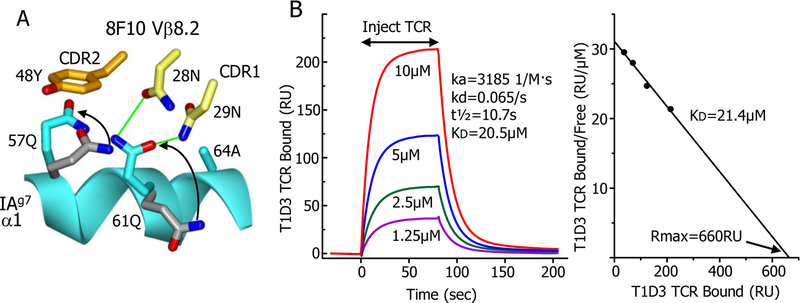
Like other Type B T cells (7), the response of 8F10 is inhibited by the natural p8E of the peptide, but the response improves dramatically when the p8E side chain is removed by mutation to G (7, 9). This finding is explained by the structure, in which the 8F10 Vβ8.2 CDR3 loop was positioned over the C-terminal end of the peptide, in close contact with the peptide backbone at p6C, p7G and p8G. The proximity of the CDR3 loop to the peptide leaves no room for a surface exposed sidechain at p8 (Fig. 2C). Consequently, leaving the natural p8E at this position (9) or replacing it with L or V (Fig. 2D) eliminated the ability of the peptide to stimulate the 8F10 T cell. The 8F10 TCR contains the well-studied Vβ8.2 element, which could explain its docking angle. Previously, β48Y in the CDR2 loop of Vβ8.2 has been shown repeatedly in TCR structures to favor a docking spot on the MHCII α chain helix between α57Q and α61Q (24, 27). In the 8F10 structure β48Y occupies this spot as well (Fig. 3A). Establishing this interaction requires the TCR to take the conventional docking angle seen the 8F10 complex. Another property of the 8F10 TCR is its second order binding kinetics to the IAg7-8G9E ligand, consistent with a fast on/fast off initial phase followed by a conformational change that leads to a slower off-rate (9). Examination of the IAg7-8G9E structure before (9) and after 8F10 TCR binding shows large rotational changes in the α57Q and α61Q side chains, aligning these amino acids for interaction with Vβ48Y and the 8F10 CDR1 loop, respectively (Fig. 3A). These changes in the ligand to accommodate the TCR could explain the biphasic kinetics and the docking angle.
Fig. 3. Other features of the 8F10 and T1D3 TCRs.
(A) A ribbon representation of a portion of the IAg7 α1 helix (cyan) is shown with the rotamer changes (arrows) in α57Q and α61Q side chains from their positions before (carbons - grey) and after (carbons - cyan) engagement by the 8F10 TCR. The subsequent interaction of these amino acids with the TCR Vβ8.2 CDR2 48Y (carbon – orange) and CDR1 28N and 29N (carbon – yellow) side chains are also shown with potential H-bonds in green. (B) Left panel - Surface plasmon resonance (SPR) data (RU, resonance units) for binding of various concentrations of the soluble T1D3 TCR to IAg7–8E9E11ss immobilized via a biotin tag in a BIAcore streptavidin flow cell. A flow cell containing immobilized IAg7–8G9E was used to correct the data for the fluid phase SPR signal. Standard BIAcore Biaeval software was used to fit the data to a first order a kinetic model and to calculate the kinetic association (kd, liters/mole▪sec) and dissociation (ka, 1/sec) rates. The overall dissociation constant (KD, μM) was calculated as the kd/ka. Right panel – Scatchard plot of the equilibrium SPR signal obtained with each concentration of the soluble T1D3 TCR tested. The KD was calculated as the negative slope of a straight line fit to the data. Rmax (predicted RU at infinite TCR concentration) is labeled. Results shown from a single experiment. Similar results were obtained in a second experiment.
The T1D3 T cell and the other human T cells in Table 1 also have a Type A-like phenotype. They respond strongly to DQ8–8E9E11ss and their responses are dramatically reduced by changing the peptide p8E to A (11). The T1D3 TCR binds to DQ8–8E9E11ss with an affinity typical of CD4 Tcells (Fig. 3B), but has a docking angle like the I.29 TCR (Fig. 1C). However, the T1D3 TCR does not have an R in its βCDR2 (Table 1). Its dependence on p8E is due to β30R in its CDR1, which, because of the conventional flat docking of the T1D3 TCR, was bought within range of p8E to form a salt bridge (Fig. 2E). Mutation of this R to A reduces the response to DQ8–8E9E11ss about 10-fold (Fig. 2F). Another human DQ8 plus insulin specific T cell, T1D10, bears hVβ7.1, which has a CDR1 R similarly positioned to that in T1D3 (Table 1). Mutation of this R to an A results in a modest reduction in T1D10’s response to DQ8 presented 8E9E11ss peptide (Fig. 2F) so engagement of DQ8 and the insulin peptide may be similar between these two human TCRs. Finally, the human T1D4 T cell uses the hVβ8.3 element, which has an R in its CDR2 at β50 similarly positioned to the β51R of mouse Vβ2 (Table 1). Mutation of this R to A eliminated the T1D4 response to DQ8 presented 8E9E11ss peptide predicting that this TCR would use a docking angle on DQ8 similar to that of the I.29 TCR on IAg7 (Fig. 2F).
These data confirm that the NOD and human T cells in Table 1 recognize the insulin peptide bound in R3, not R2 and R1 as previously suggested (2, 3). Consequently, the differences among these T cells in the recognition of the insulin peptide is much subtler than previous proposed and is confined to how the TCR Vβ domain is oriented and how it interacts with the amino acid at p8 in the R3 bound peptide. The natural E at this position can be either helpful or harmful depending on the particular TCR Vβ, thus defining the Type A vs. Type B phenotype.
Other shared features of TCR recognition of the insulin-MHCII complexes
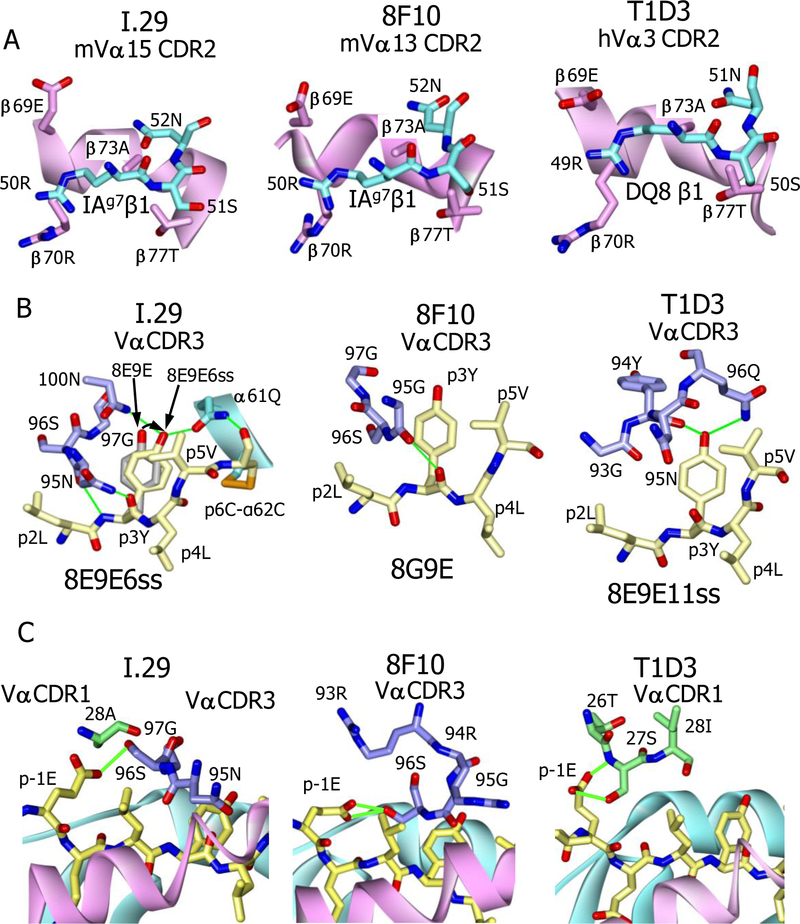
Despite the differences in orientation, footprint and specificity of the TCRs on their MHCII-peptide ligands, the complexes have several features in common that can explain some of the previous observations about the response T cells to insulin in T1D. For example, in the mouse, both the Type A and Type B CD4 T cell responses to the B:9–23 insulin peptide are dominated by CD4 T cells, such as 8F10, whose TCRs bear members of the Vα13 (TRAV5) family or in some cases, such as I.29, the related Vα15 (TRAV10) (Table 1). In the I.29 and 8F10 complex structures the Vα CDR1 and CDR2 regions sit in similar positions on the IAg7 β1 helix. Particularly striking is the fact that the CDR2 loops of these Vα’s are nearly identical in sequence (Table 1) and bind very similarly to the IAg7 β1 helix at a site highly conserved among MHCII β chains of most mammalian species (Fig. 4A). Although the Vα3 of the human T1D3 TCR is not particularly related to mouse Vα13 or Vα15, its CDR2 is very similar (Table 1) and it sits on the same conserved site of the DQ8 β chain (Fig. 4A). It is also worth noting that the human Vα8 of the T1D4 TCR is highly homologous to mouse Vα13, and its CDR2 is identical (Table 1). It seems likely that it would also have a similar docking of its αCDR2 on the DQ8 β chain.
Fig. 4. Conserved features of the three TCR complexes.
. (A) Similar interactions of the TCR Vα CDR2 loops (cyan) of the I.29 (left), 8F10 (middle) and T1D3 (right) with conserved amino acids (β69E to β77T, magenta) of the IAg7 or DQ8 β chain alpha helix. View is from within the peptide binding groove looking out to the β chain alpha helix. (B) Importance of the p3Y (B:16, yellow) for interaction with the Vα CDR3’s of the I.29 (left), 8F10 (middle) and T1D (right) TCRs. H-bonds are green. Also shown is how the introduced disulfide from p6 in the 8E9E6ss peptide to IAg7 α62 changes the position p3Y, realigning its OH (curved arrow) to make H-bonds both to the I.29 TCR Vα 100N and to α61Q of the IAg7 α chain. (C) The I.29 (left), 8F10 (middle) and T1D3 (right) TCRs all interact with the side chain of p-1E (B:13) of the peptide. Potential H-bonds or salt bridges are shown as green lines.
These conserved interactions between alpha helix of the MHCII β chain and the TCR Vα CDR2 loop could explain the heavy selection for certain Vα elements in the response this peptide. The interaction may create a flexible pivot point for initial TCR binding to the MHCII β chain via the Vα domain, while allowing the different Vβ domains to take different pitches and angles before settling to complete the interaction with the peptide and MHCII α chain. This would be a version of the two step TCR binding mechanism previously proposed (28).
Another feature common to the three complexes is the placement of insulin B:16Y in the center of the TCR footprints (Fig. 2C). This amino acid is very prominently exposed on the MHCII-peptide surfaces at p3 when the peptide is bound in R3 (11). Nakayama et al. (10) established the importance of this amino acid in mouse T1D, demonstrating that replacing it with an A in NOD mice prevents the development of diabetes. Mutation of this Y prevents TCR recognition by both the mouse and human CD4 T cells reactive to B:9–23. (10, 11). In agreement with these results, in our structures all three TCRs strongly interact with this amino acid in the complexes with multiple van der Waals and H-bond interactions. (Fig. 4B, Spreadsheet S1). Particularly noteworthy is the I.29 TCR complex with IAg7-8E9E6ss. Our previous work showed that the disulfide introduced between p6C to IAg7 α62C caused a small shift in the position of the p3Y side chain in the 8E9E6ss structure while improving considerably recognition of the complex by the I.29 TCR (7, 9). The I.29 complex structure suggests an explanation for this improved response (Fig. 3D). This shift perfectly aligns the p3Y OH to create bidentate H-bonds with the I.29 Vα 100N and IAg7 α61Q.
Finally, the focus of the TCRs on p3Y shifts their footprints toward the N-terminal part of the peptide. Consequently, all three TCRs now make a strong contact with the peptide E at p-1 (Fig. 4C). The p-1 amino acid is only occasionally contacted by TCRs interacting with MHCII ligands in the dozens of published structures of these complexes.
Could there be natural modifications of the B:9–23 peptide that create similar agonists?
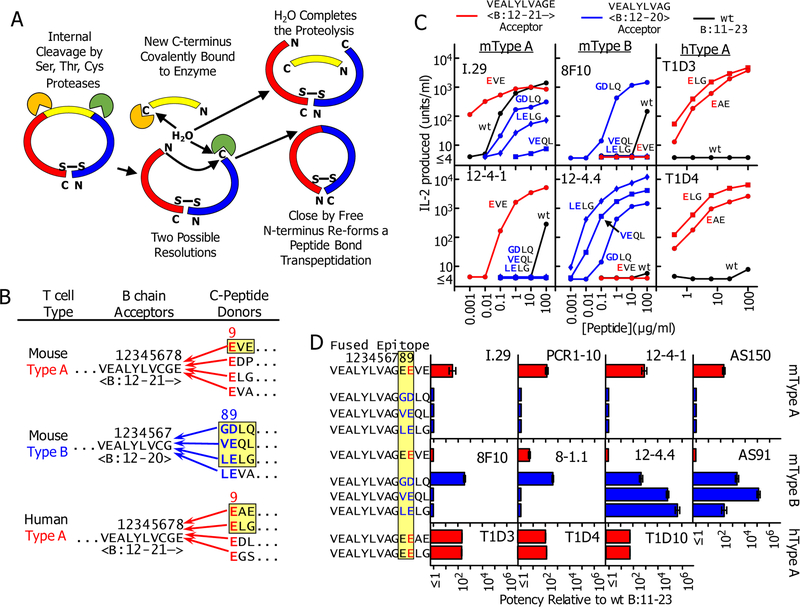
The data we present here reinforce our conclusions in previous studies that the B:9–23 peptide is recognized by NOD and human CD4 T cells bound to IAg7 or DQ8 in R3, and that mutations at the C-terminal end of the peptide are required to create an effective epitope (6–9, 11, 29). We have suggested that a C-terminal post-translational modification in the peptide may be required in vivo for induction of T1D and have proposed that these modifications could be accomplished by the process of transpeptidation (9, 16). Transpeptidation is a post-translational mechanism in which proteases can fuse two peptides to form a new chimeric peptide (reviewed in (14, 15)). As shown schematically in Fig. 5A, during proteolysis with serine, threonine or cysteine proteases, a transient covalent bond is formed between the enzyme and the newly generated carboxylate at the cleavage site. This bond can be resolved by water to complete the proteolysis or by the N-terminus of a nearby peptide, thus reforming a peptide bond and producing a new, chimeric, peptide. This reaction has long been studied in vitro and is known to play a role in the natural processing of proteins in vivo in micro-organisms, plants and animals. It can be particularly efficient when the donor and acceptor for the fusion are part of the same protein (15).
Fig. 5 -. How transpeptidation mediated internal deletions in proinsulin could form super-agonists similar to the mutant insulin peptides.
. (A) Schematic representation of how transpeptidation could cause internal deletions in proinsulin. See text for description. (B) Some potential transpeptidation protease mediated deletions in mouse or human proinsulin that could generate super-agonist fused epitopes with properties of 8E9E and 8G9E mutant peptides. (C) Peptides were synthesized joining the highlighted donor C-peptide sequences in panel (B) to the appropriate B chain acceptors shown. The natural B:19C at p6 was mutated to A to prevent peptide dimerization in vitro. The mouse peptides were titrated along with the wt insulin B:12–23 peptide using IAg7 presenting cells to stimulate IL2 production from the type A T cells (I.29 and 12–4.1) and two mouse type B T cells (8F10 and 12–4.4). The human peptides were tested similarly with two human T cells (T1D3 and T1D4) using DQ8 antigen presenting cells. The data are the average with SEM of the normalized results of three separate experiments (D) All of the mouse and human T cells in Table 1 (expect 7E6) were tested three times with the synthetic fused peptides as in (C). The data are presented as the relative stimulatory potency of the peptides with the SEM compared to the B11:23 control peptide calculated as described in the Material and Methods.
In recent years, transpeptidation by proteasomal theonine proteases to create neo-peptides presented by MHCI has been well-documented (12, 13, 18, 19). We have proposed that the lysosome should also be an ideal site for creation of chimeric neo-epitopes feeding into the peptide loading pathway for MHCII (9, 16). The high concentration of many different cysteine-cathespsins and serine proteases in the confined space of the lysosome creates an ideal milieu for transpeptidation like that found in the proteasome. We and others have suggested that neo-antigens formed in this way could create the epitopes driving T1D and other autoimmune diseases (9, 14–17). Since transpeptidation is enhanced by the proximity of donor and acceptor peptides, we have considered whether transpeptidation mediated deletions within proinsulin that fuse a B chain derived acceptor with a C-peptide derived donor could create super agonist versions of the B:9–23 similar to our mutated peptides.
Examples of potentially functional chimeric peptides, in mouse and in human, that could be generated by internal transpeptidation in proinsulin are shown in Fig. 5B. For the mouse Type A and human Type A-like T cells listed in Table 1, the acceptor would be an insulin B-chain fragment cleaved at B:21E to remove B:22R and donors would be peptides from C-peptide with an N-terminal E to replace the p9R. There are many possibilities within mouse and human C-peptide, a few of which are shown. For mouse Type B T cells, the acceptor would be formed by a cleavage of the B-chain at B:20G to remove both B:21E and B:22R. C-peptide donors would be peptides that have an E or D at the second position for p9 and some other amino acid other than E at the N-terminus for p8. The possibilities are G, V or L, thus replacing B:21–22 ER with GD, VE or LE. To test the predicted potencies of these chimeric peptides, we synthesized six of them (highlighted Fig. 5B) and compared their stimulating activity to that of the wt B:11–23 peptide with 11 of the 12 T cells listed in Table 1. Fig. 5C shows sample titrations of the peptides with two mouse Type A, two mouse Type B and two human Type A-like T cells, showing representative of the patterns of stimulation we obtained. Titrations with all 11 T cells were repeated three times and the average increases (±SEM) in potencies over the unmutated peptide are shown in Fig. 5D, determined as described in the Materials and Methods.
As predicted the B:12–21 acceptor peptide fused to the N-terminal fragment of mouse C-peptide, EVE, was every bit as effective with the NOD Type A T cells as the B:22R to E mutant peptide used in our previous studies (9), increasing potency on average about 100-fold. Also, as expected this fused peptide was virtually inactive with all of the NOD Type B T cells. Likewise, for the human Type A-like T cells fusing either the EAE or ELG fragments of human C-peptide to B:12–21, improved the potency of the peptide with the human Type A-like T cells about 100-fold.
For all of the NOD Type B T cells, fusion of B:12–20 to the mouse C-peptide fragment, GDLQ, increased its potency 100- to 1000-fold. The results with the VEQL and LELG were very different. 8F10 failed to respond to these peptides consistent with the structure of the 8F10 TCR bound to the 8G9E peptide bound to IAg7 (Fig. 2C) and the 8F10 stimulation data in Fig. 2D. Similarly, 8–1.1 failed to respond to these peptides. On the other hand, the 12–4.4 and AS91 T cells both responded to these peptides in several cases much better than they did to the GDLQ fused peptide or in our previous experiments with the 8G9E mutated peptide. For 12–4.4 the potency of the LELG fused peptide increased >500,000 fold and for AS91 the the potency of the VEQL fused peptide increased >100,000 fold (Fig. 5D). The GDLQ, VELQ, and LELG stimulated all of the Type A NOD T cells worse than the unmutated control peptide.
Together these results point out how the appropriate modifications of the C-terminus of the insulin B chain peptide can lead to enormous increases in the stimulating activity of these fused peptides for diabetogenic T cells.
DISCUSSION
For decades the NOD mouse and certain rat strains have been used as models for human T1D [reviewed in (30)]. While some have questioned whether these animal models are relevant to the human disease, the similarities in the genetic risk and in the CD4 autoimmune T cell response between humans and rodents are striking. One problem in using the rodent CD4 T cell responses in T1D to understand the corresponding human CD4 T cell response has been a lack of consensus on the molecular nature of the diabetogenic MHCII-peptide complexes driving the disease in these species. Nowhere has this problem been more apparent than in defining the CD4 T cell response to insulin, a major autoantigen in both humans and rodents. In NOD mice the insulin B chain peptide B:9–23 was reported decades ago to contain epitopes recognized by a variety of diabetogenic CD4 T cells (31), but how this peptide binds to IAg7 to form the pathogenic complex has remained a contentious issue. Different studies have proposed different binding positions or registers even for the same T cells (2, 3, 6, 7, 9).
Our studies presented here and previously resolve this confusion. We show that for the T cells listed in Table I, which include those previously reported to recognize R1 and R2 presented epitopes, the functional register is, in fact, R3, that places ALYLVCGER (B:14–22) in the core p1 to p9 positions in the IAg7 or DQ8 peptide binding groove. The extremely poor binding of this peptide in this register is due to the incompatibility of the B:22R with the p9 pocket of IAg7 and DQ8. Binding can be improved about 100-fold by changing this amino acid to the E which is optimal for peptide binding to either MHCII allele (6, 7, 32). Disulfides engineered between the peptide and the IAg7 or DQ8 α chain helix, which can form only when the peptide is bound in R3, show that this mutation is sufficient for forcing R3 binding (6, 7, 9, 11). Furthermore, we have shown that additionally mutating B:14A to R for IAg7 or to E for DQ8, while not required for optimal binding and T cell recognition, creates an optimal R3 p1 anchor for these MHCII alleles and does not interfere with peptide recognition by the set of T cells shown in Table 1 (6, 7, 9, 11). However, if the functional presenting register for these T cells were in fact R1 or R2, these B:14 mutated amino acids would lie on the surface at p3 (R1) or p2(R2) where they would be expected to interfere with TCR recognition.
Our structures presented here not only confirm our previous conclusions concerning R3 as the functional register for the insulin epitopes, but also show how mouse and human TCRs interact with these R3 presented epitopes. We conclude that for the human T cells and mouse Type A T cells in Table 1, the natural B:21E at p8 is an important part of the ligand interacting via a salt bridge with an R in the TCR. On the other hand, for mouse Type B T cells this E is very inhibitory and recognition is greatly improved by removal of its side chain by changing it to G or, for some T cells, a V or L. Moreover, even though the TCR Vα and Vβ segments and CDR3 sequences are quite different amongst the three T cells studied here, there are striking similarities in their complexes with their ligands. These include virtually identical interactions between the VαCDR2 loops with a highly conserved site on the MHCII β chain alpha helix and a focus by the TCR on the surface exposed B:16Y at p3, previously shown to be important for T1D development in the NOD mouse. These findings clear up much of the uncertainty in the literature about how this insulin epitope is bound and recognized by CD4 T cells.
However, our findings raised the question that, if these C-terminal modifications of this peptide are needed to create strong R3 MHCII binding and T cell stimulation, what is the nature of the real peptide driving the disease in vivo? We have suggested that there may be a role for post-translational modifications of this region of the peptide in T1D development (8, 9, 16). There are now multiple examples for how post-translational modifications can convert weak T cell autoantigens into strong or “heteroclitic” epitopes [reviewed in (8, 33)]. Specific examples are the conversion of arginine to citrulline in epitopes driving rheumatoid arthritis (34), conversion of glutamine to glutamic acid in celiac disease (35) and modifications to a myelin basic protein peptide in a mouse model of multiple sclerosis (36). As with our results with the B:9–23 peptide, in these three examples, the improving modification can be in either to an anchor or TCR contact residue.
Based on the recent findings that transpeptidation in the proteasome creates chimeric peptides for loading into MHCI (14, 15, 18, 19), we have proposed peptide fusion by transpeptidation as a means of creating the required modifications to the B:9–23 peptide (9, 16). Here we have synthesized several versions of proposed transpeptidation mediated deletions within mouse and human proinsulin, creating peptides every bit as active, and in some cases dramatically more active, than our original mutated peptides. A limitation to our hypothesis is that we have not yet identified the presence of these proinsulin chimeric peptides in vivo in the pancreas, a formidable task on which we continued to work. However, the requirement for fusion of a B chain acceptor to a C-peptide donor to complete these CD4 epitopes could explain why T1D patients commonly do not show hypersensitivity responses to daily injections of fully processed insulin, since mature insulin lacks the attached C-peptide, which might be required to complete the fusion.
A similar story has emerged for another NOD T1D antigen involving the WE14 peptide of chromogranin A (ChgA) (37). We have shown that the WE14 peptide binds very poorly to IAg7 because it fills only the p5 to p9 positions of the IAg7 binding groove. Its binding and recognition can be improved tremendously by adding the appropriate four amino acids to its N-terminus that fill the rest of the peptide binding groove. (7, 16). These amino acids also contribute to TCR binding at p2 and p3, while providing an optimal anchor amino acid at p4 (16). We suggested that this modification could be mimicked in vivo by transpeptidation and proposed a list of potential acceptors for a WE14 donor among various beta granule proteins (16). One of these, from proinsulin C-peptide would place TLAL at the p1 to p4 positions when fused to WE14. The Haskins also proposed this fusion and were able to find the fused peptide in lysates of a primary insulinoma tumor (17). In this same lysate, they also found the same C-peptide fragment fused to a peptide from islet amyloid polypeptide precursor completing the epitope for another diabetogenic CD4 T cell (17, 38). These remain the only CD4 functional chimeric epitopes reported to be found so far from an in vivo source.
An open question is still where these transpeptidation reactions might take place. We suggest that the relevant site is the lysosome, since it feeds into the MHCII loading pathway and has the right proteases and environment for the reaction. One possibility is that transpeptidation could be a by-product of crinophagy in beta cells, in which granule turnover is regulated by fusion with lysosomes, a process that is enhanced by beta cell stress (39). Alternately, lysosomes in antigen presenting cells, such as dendritic cells, macrophages or insulin-specific B cells, present either in the islet or in the pancreatic lymph node could be the site of the reaction.
Finally, the growing list of T cell autoantigen epitopes created by various mechanisms of peptide post-translational modification raises the question of whether these processes are unique to the periphery and absent in the thymic medullary epithelial cells responsible for T cell negative selection. If so, this could explain the escape of the pathogenic T cells from the thymus. Exploring this idea requires a much deeper an understanding of the antigen processing and MHC presentation pathways of these proteins in the thymus than is currently available.
MATERIALS AND METHODS
Study design
This study was designed to express soluble versions of the TCRs from human and mouse CD4 T cells and their ligands consisting of mouse IAg7 or human HLA-DQ8 MHCII alleles bound to various modified versions of the insulin B:9–23 peptide and then to solve the crystal structures of the TCR-ligand complexes. The structures were then used to analyze the nature of the TCR-ligand interfaces and relate this information to the controversies surrounding how proinsulin is a major CD4 T cell autoantigen in type-1 diabetes in both humans and mice. Based on these analyses we predicted that chimeric peptides formed by transpeptidation fusion of fragments of the B:9–23 peptide with those derived from proinsulin C-peptide would create super agonists for a diverse collection of human and mouse T cells. We synthesized the predicted epitopes and showed that their activity in stimulating these T cells were between 50–500,000 times better the natural insulin peptide.
T cell hybridomas and T cell avatars.
The origins, constructions and properties of the 12–4.1, 12–4.4, 8–1.1, PCR1–10, I.29, 8F10, AS150, and AS91 NOD mouse T cell hybridomas and TCR transduced avatars as well as the avatar versions of the human T1D3, T1D4 and T1D10 T cell clones used in these studies were described previously (2, 3, 6, 7, 9, 11, 20–22). Briefly, hybridomas were produced fusion to the TCR negative version of the mouse T cell tumor line, BW5147 (40). T cell avatars were produced by cloning sequence encoding the TCR V-domains fused to mouse Cα or Cβ into MSCV based vectors bearing an IRES followed by GFP or human nerve growth factor as a surrogate marker. Virus prepared from these constructions were used to transduce a mouse T cell hybridoma (5KC) that had previously been selected for the loss of its original functional TCR genes (41). The human avatars were transduced with human CD4 and the 12–4-1 T cell was transduced with a mutant version of human CD4 with high affinity for MHCII (42, 43) This system was also used to produce avatars expressing either the wildtype I.29 TCR or I.29 TCRs with single point mutations in its CDR loops. For testing the effects of these mutations the cells were selected for comparable levels of surface TCR. All cells were cultured in supplemented MEM with 10% FCS as previously described (40).
Antigen presentation assays and soluble peptides
For antigen presentation cells (APC) we used two versions of the M12.C3 B cell lymphoma, one expressing IAg7 (M12.C3.g7)(44) and the other expressing DQ8 (M12.C3-DQ8–8) (45). T cell hybridomas or TCR transduced avatars (105 cells) were mixed with 105 paraformaldehyde fixed (7) APCs and cultured overnight with various concentrations of peptide in a volume of 250μL. Secreted IL-2 was assayed with either a functional assay, following the growth and survival of the HT-2 IL-2 dependent cell line (40), or with an ELISA based assay for IL-2 (46). The relative potencies of the mutant and chimeric peptides relative to the wt insulin peptide were calculated from the shift in the IL-2 production vs. peptide titration curves along the peptide dose axis (40). Soluble peptides (>95% pure) were obtained from either CHI Scientific, Maynard MA or Schafer-N, Copenhagen, Denmark.
MHCII-peptide expression and purification
As previously described (7, 9), acid-base leucine zipper stabilized, soluble IAg7 and human HLA-DQ8 molecules with covalently attached peptides were produced in baculovirus infected insect cells and purified by immunoaffinity chromatography. For surface plasmon resonance or flow cytometry experiments, a biotinylation peptide tag attached to the C-terminus of the acidic half of the zipper was enzymatically biotinylated with BirA enzyme produced in our own laboratory, after purification of the molecule. For crystallization, the zippers and biotinylation tag were removed with papain.
TCR expression and purification
Sequence encoding V domains of wt and mutant mouse TCR I.29, 8F10; human TCR T1D3 were cloned separately into the pET30 bacterial expression system, in which the TCR V-domains were fused to sequence encoding the extracellular domains of human Cα or Cβ (23, 24). Mutated I.29 TCR (β56K to A) and 8F10 TCR (α169R to A) were prepared similarly. After IPTG induction of over-expression, the resultant proteins were solubilized from inclusion bodies in 8M urea buffer, mixed and refolded by gradual dialysis. The refolded TCR was further purified with a HiLoad Superdex 200 26/600 size chromatography column followed by a monoQ ion exchange chromatography. The mutated TCRs were tested for binding to their ligands to confirm mutations has no impact on TCR binding to their IAg7-peptide ligands.
Surface plasmon resonance measurements
Approximately 2000 RU of biotinylated DQ8–8E9E11ss or IAg7-8E9E were captured in the two separate flow cells of a BIAcore streptavidin BIAsensor chip. Various concentrations of the soluble refolded T1D3 TCR was injected and the association and dissociation kinetics in the recorded. The data for the DQ8–8E9E11ss flow cell was corrected for the fluid phase SPR signal using the data from the IAg7-8E9E flow cell as baseline. Kinetics was analyzed with BIAcore BIAEval 4 software.
Protein crystallization
Crystallization was performed by the hanging drop vapor diffusion method. WT TCR I.29 and 8F10 proteins were concentrated to 5mg/ml. I.29 crystals were grown from 20% MPD, 100mM sodium cacodylate at pH6.0, 50mM calcium acetate. 8F10 crystals were grown in 14% PEG 3350, 100mM sodium acetate at pH6.0. Both of I29 and 8F10 crystals were cryoprotected by well solution plus 25% glycerol. Examination of structures of free I.29 and 8F10 revealed that the crystal packing was likely to interfere with IAg7 binding. Therefore, a mutation was introduced into each TCR (β56K to A for I.29 and α169R to A for 8F10) predicted to inhibit this packing, while not affecting the TCR face predicted to interact with IAg7-peptide. The mutant TCRs were checked to be sure they had the same binding properties as the wild type ones. We were successful in crystallizing both mutant TCRs bound to their IAg7-peptide ligands. To obtain the I.29/IAg7-8E9E6ss complex crystals, IAg7-8E9E6ss and I.29 β56A mutant TCR were mixed at molar ratio of 1:1 at the concentrated of 10mg/ml. The crystals were grown in 12% PEG 20000, 100mM bicine pH9.0, 20% ethylene glycol. The I.29 complex crystals were cryoprotected by the well solution plus 15% ethylene glycol. Similarly, IAg7-8G9E/8F10 complex crystals were obtained by mixing IAg7-8G9E and 8F10 α169A mutant TCR at a 1:1 molar ratio at the concentration of 10 mg/ml, Crystals were grown in 11% PEG 3350, 100mM formate pH7.0 at room temperature. The 8F10 complex crystals were cryoprotected by the well solution plus 30% glycerol. Human T1D3/DQ8–8E9E11ss complex is crystalized in 10% PEG 8000, 100mM cacodylate pH6.2, 400mM NaCl at 4 °C, by mixing TCR T1D3 and DQ8–8E9E11ss with 1:1 molar ratio at the concentration of 12 mg/ml. The crystals were cryoprotected by well solution plus 30% glycerol.
Data collection, data processing and structural analysis
All diffraction data sets were collected at synchrotron beamline ID-24C at the Advanced Photon Source, Argonne National Laboratory using the Pilatus detector. The 8F10/IAg7-p8G9E complex data was processed by XDS software package at ID-24C beamline (47), all the rest collected data were processed with HKL2000 package (48), the structures were solved by molecular replacement method using Phaser (49) software and further refined by refmac5 (50) or Phenix (51), rebuilding of the structure was performed with Coot (52). NCONT in CCP4 (53) was used to analyze the atom-to-atom contacts between the TCRs and their ligands. Atoms within 4.5Å of each other were considered part of the interface. Contacts involving potential electron donors and acceptors (O or N) within 3.5Å were considered potential hydrogen bonds, or within 4.0Å, salt bridges. Other contacts were considered van der Waals contacts. Data collection and refinement statistics are shown in Supplemental Table S1. The contributions of the TCR CDR loops to the buried surface area (BSA) in the interface with their ligands were calculated with the CCP4 jsPISA program (54). Molecular superimpositions were performed with Swiss PDBViewer (55). Graphical representations were made with of Discovery Studio 3 (Accelrys).
Supplementary Material
Acknowledgments:
We thank Ella Kushnir and Christopher Brown for technical support as well as Kelly Bakke and Maria Millstein for administrative support. P.M and J.W.K acknowledge their previous long stranding support from the Howard Hughes Medical Institute.
Funding: This work was supported by NIH Grants 5T32-AI-074491 (Y.W.), ES-025797 and ES025885 (S.D.), AI-018785 (P.M. and J.W.K.), DK-032083 (T.S.), and P01AI-118688 (J.W.K.) as well as JDRF Grant 1-PNF-2015-126-A-R, University of Colorado CCTSI Grant KL2 TR001080, and a Boettcher Foundation Investigator award (S.D.). Additional financial support came from National Jewish Health (J.W.K and P.M), as well as the School of Medicine of University of Colorado at Denver and The Claire Friedlander Family Foundation (J.W.K). The synchrotron data were collected at Advanced Photon Source Argonne National Laboratory, Beamline 24 IDC, supported by NIH grant GM103403 and NIH-ORIP HEI Grant S10 RR029205.
Footnotes
SUPPLEMENTARY MATERIAL
Fig. S1. Mutations to prevent I.29 and 8F10 TCR dimerization.
Fig. S2. Extension of the mutational analysis of the I.29 TCR shown in Fig. 2b in the main manuscript.
Table S1 Crystallography Statistics
Spreadsheet S1 – Atom-to-atom contacts in the TCR-MHCII-Peptide complexes
Competing Interests: All of the authors declare they have no competing interests.
Data and materials availability: All of the cell lines and expression plasmids used in this work are available on request by J.W.K, M.N. and S.D. The coordinate and structure factors files for the 5 structures presented here have been validated and deposited in the RCSB Protein Data Bank for release at publication. Accession numbers are 6DFQ, 6DFS, 6DFV, 6DFW and 6DFX.
References
- 1.Eisenbarth GS, Moriyama H, Robles DT, Liu E, Yu L, Babu S, Redondo MJ, Gottlieb P, Wegmann D, Rewers M. Insulin autoimmunity: prediction/precipitation/prevention type 1A diabetes. Autoimmun Rev. 2002;1:139–145. [DOI] [PubMed] [Google Scholar]
- 2.Levisetti MG, Suri A, Petzold SJ, Unanue ER. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J Immunol. 2007;178:6051–6057. [DOI] [PubMed] [Google Scholar]
- 3.Mohan JF, Petzold SJ, Unanue ER. Register shifting of an insulin peptide-MHC complex allows diabetogenic T cells to escape thymic deletion. J Exp Med. 2011;208:2375–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan X, Unanue ER. Unique features in the presentation of insulin epitopes in autoimmune diabetes: an update. Current opinion in immunology. 2017;46:30–37. [DOI] [PubMed] [Google Scholar]
- 5.Wan X, Zinselmeyer BH, Zakharov PN, Vomund AN, Taniguchi R, Santambrogio L, Anderson MS, Lichti CF, Unanue ER. Pancreatic islets communicate with lymphoid tissues via exocytosis of insulin peptides. Nature. 2018;560:107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc Natl Acad Sci U S A. 2010;107:10978–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford F, Stadinski B, Jin N, Michels A, Nakayama M, Pratt P, Marrack P, Eisenbarth G, Kappler JW. Specificity and detection of insulin-reactive CD4+ T cells in type 1 diabetes in the nonobese diabetic (NOD) mouse. Proc Natl Acad Sci U S A. 2011;108:16729–16734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marrack P, Kappler JW. Do MHCII-presented neoantigens drive type 1 diabetes and other autoimmune diseases? Cold Spring Harb Perspect Med. 2012;2:a007765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Sosinowski T, Novikov A, Crawford F, Neau DB, Yang J, Kwok WW, Marrack P, Kappler JW, Dai S. C-terminal modification of the insulin B:11–23 peptide creates superagonists in mouse and human type 1 diabetes. Proceedings of the National Academy of Sciences. 2018;115:162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliott JF, Eisenbarth GS. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435:220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Chow IT, Sosinowski T, Torres-Chinn N, Greenbaum CJ, James EA, Kappler JW, Davidson HW, Kwok WW. Autoreactive T cells specific for insulin B:11–23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc Natl Acad Sci U S A. 2014;111:14840–14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanada K, Yewdell JW, Yang JC. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature. 2004;427:252–256. [DOI] [PubMed] [Google Scholar]
- 13.Vigneron N, Stroobant V, Chapiro J, Ooms A, Degiovanni G, Morel S, van der Bruggen P, Boon T, Van den Eynde BJ. An antigenic peptide produced by peptide splicing in the proteasome. Science. 2004;304:587–590. [DOI] [PubMed] [Google Scholar]
- 14.Cresswell P. Cell biology. Cutting and pasting antigenic peptides. Science. 2004;304:525–527. [DOI] [PubMed] [Google Scholar]
- 15.Berkers CR, de Jong A, Ovaa H, Rodenko B. Transpeptidation and reverse proteolysis and their consequences for immunity. Int J Biochem Cell Biol. 2009;41:66–71. [DOI] [PubMed] [Google Scholar]
- 16.Jin N, Wang Y, Crawford F, White J, Marrack P, Dai S, Kappler JW. N-terminal additions to the WE14 peptide of chromogranin A create strong autoantigen agonists in type 1 diabetes. Proc Natl Acad Sci U S A. 2015;112:13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delong T, Wiles TA, Baker RL, Bradley B, Barbour G, Reisdorph R, Armstrong M, Powell RL, Reisdorph N, Kumar N, Elso CM, DeNicola M, Bottino R, Powers AC, Harlan DM, Kent SC, Mannering SI, Haskins K. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351:711–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liepe J, Marino F, Sidney J, Jeko A, Bunting DE, Sette A, Kloetzel PM, Stumpf MP, Heck AJ, Mishto M. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science. 2016;354:354–358. [DOI] [PubMed] [Google Scholar]
- 19.Faridi P, Li C, Ramarathinam SH, Vivian JP, Illing PT, Mifsud NA, Ayala R, Song J, Gearing LJ, Hertzog PJ, Ternette N, Rossjohn J, Croft NP, Purcell AW. A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci Immunol. 2018;3. [DOI] [PubMed] [Google Scholar]
- 20.Daniel D, Gill RG, Schloot N, Wegmann D. Epitope specificity, cytokine production profile and diabetogenic activity of insulin-specific T cell clones isolated from NOD mice. Eur J Immunol. 1995;25:1056–1062. [DOI] [PubMed] [Google Scholar]
- 21.Simone E, Daniel D, Schloot N, Gottlieb P, Babu S, Kawasaki E, Wegmann D, Eisenbarth GS. T cell receptor restriction of diabetogenic autoimmune NOD T cells. Proc Natl Acad Sci U S A. 1997;94:2518–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama M, Castoe T, Sosinowski T, He X, Johnson K, Haskins K, Vignali DA, Gapin L, Pollock D, Eisenbarth GS. Germline TRAV5D-4 T-cell receptor sequence targets a primary insulin peptide of NOD mice. Diabetes. 2012;61:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tynan FE, Borg NA, Miles JJ, Beddoe T, El-Hassen D, Silins SL, van Zuylen WJ, Purcell AW, Kjer-Nielsen L, McCluskey J, Burrows SR, Rossjohn J. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J Biol Chem. 2005;280:23900–23909. [DOI] [PubMed] [Google Scholar]
- 24.Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozono H, White J, Clements J, Marrack P, Kappler J. Production of soluble MHC class II proteins with covalently bound single peptides. Nature. 1994;369:151–154. [DOI] [PubMed] [Google Scholar]
- 26.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. [DOI] [PubMed] [Google Scholar]
- 27.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. [DOI] [PubMed] [Google Scholar]
- 28.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–556. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama M, McDaniel K, Fitzgerald-Miller L, Kiekhaefer C, Snell-Bergeon JK, Davidson HW, Rewers M, Yu L, Gottlieb P, Kappler JW, Michels A. Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects. Proc Natl Acad Sci U S A. 2015;112:4429–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullen Y. Development of the Nonobese Diabetic Mouse and Contribution of Animal Models for Understanding Type 1 Diabetes. Pancreas. 2017;46:455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wegmann DR, Gill RG, Norbury-Glaser M, Schloot N, Daniel D. Analysis of the spontaneous T cell response to insulin in NOD mice. J Autoimmun. 1994;7:833–843. [DOI] [PubMed] [Google Scholar]
- 32.Suri A, Walters JJ, Gross ML, Unanue ER. Natural peptides selected by diabetogenic DQ8 and murine I-A(g7) molecules show common sequence specificity. J Clin Invest. 2005;115:2268–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyle HA, Mamula MJ. Autoantigenesis: the evolution of protein modifications in autoimmune disease. Current opinion in immunology. 2012;24:112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegner N, Lundberg K, Kinloch A, Fisher B, Malmstrom V, Feldmann M, Venables PJ. Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunological reviews. 2010;233:34–54. [DOI] [PubMed] [Google Scholar]
- 35.Sollid LM, Jabri B. Celiac disease and transglutaminase 2: a model for posttranslational modification of antigens and HLA association in the pathogenesis of autoimmune disorders. Current opinion in immunology. 2011;23:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. [DOI] [PubMed] [Google Scholar]
- 37.Stadinski BD, Delong T, Reisdorph N, Reisdorph R, Powell RL, Armstrong M, Piganelli JD, Barbour G, Bradley B, Crawford F, Marrack P, Mahata SK, Kappler JW, Haskins K. Chromogranin A is an autoantigen in type 1 diabetes. Nat Immunol. 2010;11:225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiles TA, Delong T, Baker RL, Bradley B, Barbour G, Powell RL, Reisdorph N, Haskins K. An insulin-IAPP hybrid peptide is an endogenous antigen for CD4 T cells in the non-obese diabetic mouse. J Autoimmun. 2017;78:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandberg M, Borg LA. Intracellular degradation of insulin and crinophagy are maintained by nitric oxide and cyclo-oxygenase 2 activity in isolated pancreatic islets. Biol Cell. 2006;98:307–315. [DOI] [PubMed] [Google Scholar]
- 40.White J, Kappler J, Marrack P. Production and characterization of T cell hybridomas. Methods Mol Biol. 2000;134:185–193. [DOI] [PubMed] [Google Scholar]
- 41.White J, Pullen A, Choi K, Marrack P, Kappler JW. Antigen recognition properties of mutant V beta 3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. J Exp Med. 1993;177:119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang XX, Li Y, Yin Y, Mo M, Wang Q, Gao W, Wang L, Mariuzza RA. Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex. Proc Natl Acad Sci U S A. 2011;108:15960–15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams T, Krovi HS, Landry LG, Crawford F, Jin N, Hohenstein A, DeNicola ME, Michels AW, Davidson HW, Kent SC, Gapin L, Kappler JW, Nakayama M. Development of T cell lines sensitive to antigen stimulation. J Immunol Methods. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abiru N, Maniatis AK, Yu L, Miao D, Moriyama H, Wegmann D, Eisenbarth GS. Peptide and major histocompatibility complex-specific breaking of humoral tolerance to native insulin with the B9–23 peptide in diabetes-prone and normal mice. Diabetes. 2001;50:1274–1281. [DOI] [PubMed] [Google Scholar]
- 45.Michels AW, Landry LG, McDaniel KA, Yu L, Campbell-Thompson M, Kwok WW, Jones KL, Gottlieb PA, Kappler JW, Tang Q, Roep BO, Atkinson MA, Mathews CE, Nakayama M. Islet-derived CD4 T-cells targeting proinsulin in human autoimmune diabetes. Diabetes. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allicotti G, Borras E, Pinilla C. A time-resolved fluorescence immunoassay (DELFIA) increases the sensitivity of antigen-driven cytokine detection. J Immunoassay Immunochem. 2003;24:345–358. [DOI] [PubMed] [Google Scholar]
- 47.Xds Kabsch W. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods enzymol. 1997;276:307–326. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krissinel E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 2015;43:W314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.