Abstract
Cancer is one of the leading causes of death worldwide, despite large efforts to improve understanding of cancer biology and develop treatments. Efforts to improve cancer treatment are limited by the complexity of the local milieu in which cancer cells exist. The tumor microenvironment (TME) consists of a diverse population of tumor cells and stromal cells with immune constituents, microvasculature, extracellular matrix (ECM), and gradients of oxygen, nutrients, and growth factors. The TME is not recapitulated in traditional models used in cancer investigation, limiting the translation of preliminary findings to clinical practice. Advances in three-dimensional (3D) cell culture, tissue engineering, and microfluidics have led to development of “cancer-on-a-chip” platforms that expand the ability to model the TME in vitro and allow for high through-put analysis. Here we discuss advances in the development of cancer-on-chip platforms, implications for drug development, challenges to leveraging this technology for improved cancer treatment, and future integration with artificial intelligence for improved predictive drug screening models.
Keywords: Cancer models, organ-on-a-chip, microfluidics, chemotherapy, artificial intelligence
1. Introduction
Cancer is a leading cause of death worldwide, with an estimated 17.0 million new cancer cases and 9.5 million cancer-related deaths reported by the International Agency for Research on Cancer in 2018 [1]. By 2040, global incidence will grow to 27.5 million new cancer cases, with cancer-related mortality at an estimated 16.3 million [1]. Cancer is the second leading cause of death in the United States, with approximately 1.74 million new cancer cases and 600,000 cancer-related deaths in 2018 (American Cancer Society) [1, 2] Despite extensive efforts by many research groups to improve the understanding of cancer biology, identify novel therapeutics, and push these advances into clinical practice, cancer remains a global burden to human healthcare and a prominent cause of death.
Malignant tumors often undergo the process of metastasis, disseminating cancer cells to distant locations in the body [3]. During metastasis, cancer cells leave the primary tumor site and travel to other regions of the body via the blood or lymphatic system, forming new tumors in other organs or tissues [3]. Metastatic tumors drastically increase patient mortality and decrease the efficacy of clinical treatments [4]. While patients diagnosed with localized tumors can often be successfully treated with surgery and/or radiation, with relatively high survival rates, a diagnosis of metastatic cancer often designates a terminal illness, with a five year survival rate less than twenty percent for half of all cancer sites[5]. The development of cancer metastases often necessitates the use of chemotherapeutic drugs, which enter the body’s circulation and travel, withinflicting cytotoxicity on tumor cells and therefore impacting tumor growth and, ideally, curing the patient [6].
The staggeringly high mortality and morbidity rates associated with cancer highlight the need for more efficacious therapies. Drug discovery and development for cancer treatment has been slow in its clinical translation due to a high attrition rate during drug development [7]. Despite robust research efforts, only 5.1% of anti-cancer drugs that enter phase I clinical trials receive Food and Drug Administration (FDA) approval [8]. One reason for this low translation rate is the poor ability of disease and drug screening models to predict patient outcomes [7]. The improved ability to more accurately and rapidly identify drug candidates and eliminate ineffective drugs as potential candidates would drastically improve the drug development process and accelerate the rate of clinical translation of [7, 9].
A central challenge in translating research advances from pre-clinical models to patient therapies and treatments is the immense complexity of the tumor microenvironment (TME). The TME is a complex niche created by each tumor and influenced by a tumor’s interactions [10, 11]. It is comprised of the non-cancerous cells within a tumor that support tumor cell growth. The TME consists of a heterogeneous population of stromal and immune cells, microvasculature, Eextracellular matrix (ECM), and the proteins produced and secreted by tumor cells. The TME is characterized by its specific mechanical properties and complex gradients of oxygen, nutrients, and growth factors. These components influence tumor biology and play a role in invasion, metastasis, and treatment outcomes[12]. A comprehensive review of TME components’ respective roles in these processes and cancer treatment can be found in previously published review papers [10, 13]. Current preclinical models for anti-cancer drug screening fall into two categories, in vitro and in vivo models. The simplest, and most commonly used, in vitro model is the traditional two-dimensional (2D) culture of immortalized cell lines [14]. 2D culture is relatively inexpensive and allows for high-throughput analysis and is therefore commonly used in studies aiming to elucidate cancer biology or identify novel chemotherapeutic agents. However, despite their advantages, these models subject cancer cells to artificially 2D growth conditions and lack key components of the TME that influence cancer biology and drug response, including the stroma, ECM, tumor mechanical properties, and intertumoral gradients [14, 15]. As a result, the predictive value of these models is limited. Another traditional pre-clinical tool for cancer studies is in vivo animal models. Animal models for cancer mimic the tumor microenvironment to a greater fidelity than simplified 2D cell culture [16-18]. These models are often used to study an individual patient’s tumor ex vivo via patient-derived xenograft (PDX) models. However, despite their advantages, these models have several limitations. While these models provide a three-dimensional (3D) environment, a comparison between two vastly different species cannot be made with high accuracy. As a result, there is often a disparity between the outcomes of cancer related drug studies in animal models and patient trials. In addition, animal studies are often cost prohibitive. Thus, an alternative model is needed to provide efficient and effective drug screening to guide the initial selection of chemotherapeutic agents for cancer patients. This need drives the development of 3D models, which seek to integrate the advantages of in vivo and in vitro techniques for improved cancer studies and drug development.
Recently, much effort has been put into the development of organ-on-a-chip platforms that recapitulate both the biology and physiology of in vivo human tumors [7, 19-21]. The majority of these platforms are designed to mimic crucial functions of organs and tissues, enabling the investigation of pharmacokinetics and pharmacodynamics [19]. These approaches employ the use of multichannel, 3D microfluidic chips to simulate the mechanics, activity and physiological response of organs. Cancer-on-a-chip models are at the frontier of nanomedicine and offer promising utility as sophisticated microsystems to elucidate the mechanisms of cancer biology and improve anti-cancer drug development. As new organ-on-a-chip platforms are developed, becoming increasingly higher throughput, large data sets are generated, bringing forth new challenges and opportunities. The development of these high-throughput organ-on-a-chip platforms gives rise to the application of deep learning-based analysis processes for high-throughput drug screening, making way for a new avenue of cutting-edge research[22]. In this review, we will highlight seminal cancer-on-a-chip papers and new advances in the field, incorporating discussion on key features of the TME and the challenges associating with recapitulating its features. We will discuss recent applications of cancer-on-a-chip models for pathomorphological and drug development studies, with special emphasis on the integration of and challenges associated with combining these platforms with machine learning and data processing technologies. Future perspectives on how cancer-on-a-chip and machine learning algorithms can synergize to improve anti-cancer drug development will be considered.
2. Anti-Cancer Drug Development and the Need for Improved Predictive Models
The anti-cancer drug develpment process starts with the identification of effective compounds via pre-clinical models. These compounds are then evaluated further in sequential human clinical trials to assess safety, dosing, and efficacy, comparing the drug in question to the current standard of care. Unfortunately, the majority of compounds identified as effective in pre-clinical models are found to benot safe or efficacious in these later studies [8, 23]. The low predictive value of pre-clinical models increases the cost and resources expended during the drug development process. Such clinical trial failures point to the need for improved pre-clinical models that mimic the TME with high fidelity [1, 18, 23].
In addition to the challenges associated with current pre-clinical models, progress in anti-cancer drug development is hindered by the complexity of cancer biology. Cancer biology is highly heterogenous and complex on several levels: among tumor subtypes, individual patients, and separate tumors within one patient. Selection of chemotherapeutic agents is currently dominated by evidence generated from randomized clinical trials (RCTs), in which patients are assigned to treatment groups by chance. Given the ongoing development of many new chemotherapeutics and the necessity to monitor the stability of previously developed agents, it is impossible to perform RCTs to investigate all FDA approved drugs for each tumor type, let alone trials to investigate all drug combinations. An individual patient’s cancer biology is unique and tumors of the same histologic subtype often show vastly different responses to therapies. Thus, initiatives for precision, or personalized, medicine have emerged [24]. This emerging field is based on the principle that the genetic and molecular information of an individual patient can be used to deploy more effective, less toxic, and patient-specific treatments [24]. Furthermore, in order to personalize the chemotherapy regimens given to a patient and optimize the chance of an effective response, there exists the need to develop robust models to perform drug screening for both general cancer subtypes and for an individual patient’s tumors.
3. Comparison of Pre-Clinical Cancer Models
3.1. Two-Dimensional Monoculture
In traditional 2D cultures, cells are grown as an adherent monolayer in a culture dish, attached to the plastic dish surface [25, 26]. Assays derived from 2D monolayers are easy to use, low cost, and high throughput. However, despite their advantages, these assays are quite limited in their predictive value. One such limitation is the inability of 2D cell cultures to mimic the native structure of tissues and tumors [14, 26]. The 2D culture environment does not recapitulate the cell-cell and cell-environment interactions present in native tumor [14, 26]. These interactions are fundamental to cell proliferation, cell differentiation, gene and protein expression, stimuli response, drug metabolism, and other cellular functions [14, 27]. Another limitation of 2D culture models is that cells in an adherent monolayer have infinite, homogenous access to key nutrients, including oxygen and metabolites [14, 26]. In vivo, cancer cells have more variable access to nutrients and oxygen due to natural tumor architecture [27]. Because of these disadvantages, there exists the need to find alternate models which better mimic the native cancer microenvironment.
3.2. Transwell Model
Transwell assays are used to study the invasion and migrations of cancer cells [28]. These assays use a cell culture insert made of a porous, polymeric membrane that allows for migration through the pores [29, 30]. Transwell assay applications include migration, invasion, and transendothelial migration [29, 30]. Migration assays, the simplest Transwell-based assay, seed cancer cells on top of the polymeric membrane insert and measure the ability of the cells to translocate though the membrane’s pores [28-30]. Invasion assays are more complex in that they add a layer of ECM on top of the porous membrane and characterize cancer cell migration through the ECM [28-30]. Transwell assays are used as both a tool for drug screening and a model for studying cancer cell migration, invasion, extravasation, and matrix remodeling. The Transwell-based assay serves as a straightforward in vitro technique for studying a tumor’s ability to metastasize to a secondary site. However, despite their advantages, Transwell assays study the motility of individual cells and, as a result, is not an optimal tumor model.
3.3. 3D Culture Models
3D culture models use various matrices or scaffolds for cancer cells to grow and ECM, thus mimicking important components of the TME. Scaffolds support cell attachment, growth, and morphogenesis [29]. These scaffolds are typically made from natural or synthetic materials, such as gelatin, collagen, alginate, hyaluronic acid, polyethylene glycol, or polylactide, polyatcide-co-glycolide and various other polymers [7]. These scaffold-based approaches are ideal in that they have similar mechanical and physical properties to the native ECM and TME [7]. More recently, alternative approaches to scaffolds have been developed, including the creation of cell spheroids via bioprinting [7]. These structures have improved perfusion due to their vascularization but are limited by technical challenges and their capacity to recapitulate complex tissue types [21]. Various other methods have been developed, e.g. the use of non-adhesive polyethylene glycol di-methacrylate hydrogel microwells to produce cancer cell multicellular aggregates [31], hanging drop cultures, and spinner cultures. While these methods can be improved upon, these models are an important foundation for more novel platforms, such as cancer-on-a-chip, as they model tumor-tumor cell interactions, native ECM, and may be designed to recapitulate the biophysical properties of native tumor.
3.4. Animal Models
Preclinical animal models are a necessary component in the process of anti-cancer drug development and discovery [32]. These in vivo models capture physiological complexity with higher fidelity than 2D monoculture techniques [7]. While these models have vastly improved our understanding of cancer, they are also limited in their capacity. One such shortcoming of animal models is their limited translatability to humans. The inability of animal models to fully recapitulate human cancer physiology is evidenced by the failure in clinical trials of drugs identified in preclinical results [7]. To improve the replicative value of in vivo animal studies, PDX tumor models have been established. PDX models are created by implanting cancer cells or tissues derived from patient tumors into immunodeficient mice [33]. PDX models are used extensively in cancer research, asthey simulate human tumor biology in vivo. While these models better replicate human tumor biology, the use of immunocompromised animals impedes analysis of the immune system’s response to a tumor [17, 34]. An additional challenge with PDX models is the establishment rate. Previous research reports a successful formation rate of implanted tumors as being 39.2% [35]. As animal models are expensive, highly regulated, and limited by a low initiation rate, there are constraints to the number of studies that can be done, preventing the PDX model from being a high-throughput assay. In addition, the procedure of creating PDX models takes months to establish, making PDXs logistically difficult for use in making timely clinical decisions [36]. Despite their advantages, animal models do not practically allow for the high-throughput assessment of multiple combinations of chemotherapeutics, highlighting further the need for high-throughput platforms to be used in precision medicine to identify anti-cancer drugs on a patient-specific basis. A thorough discussion of pre-clinical models and their advantages and disadvantages can be found in previously published reviews [14, 26, 28, 37].
4. Cancer-on-a-Chip Platform
4.1. Organ-on-a-Chip Structure and Function
In recent years, organ-on-a-chip platforms have significantly advanced for several applications, such as preclinical drug screening and disease modeling. Organs-on-a-chip are microdevices with miniaturized tissues to model human organ physiology in vitro [38]. Organ-on-a-chip devices incorporate microfluidics with 3D tissues to recapitulate native organ complexity and cues, for example electrical signals, fluid flow, and biochemical cues. Organ-on-a-chip has many advantages, including improved recapitulation of native microenvironment, simplicity, decreased cost, and reproducibility. The microfluidic chip components are traditionally made of polydimethylsiloxane (PDMS), which has ideal properties, such as transparency, low toxicity to cells, and high permeability to O2 and CO2 gases [39]. Cell are cultured in small chambers within these miniature chips, either in 2D monolayers or 3D suspensions, to emulate organ tissues. Membranes may be integrated into the microfluidic chips, creating multiple channels and separating the cells[40-47]. The microfluidic components of organ-on-a-chip platforms recapitulate in vivo conditions, such as flow, pressure, and nutrient levels. Organ-on-a-chips are also able to expose the cells or tissues to ontrolled laminar flow of fluids, improving the accuracy of biomarker-identification and drug-screening [48]. Examples of organ-on-a-chip platforms include thrombosis-on-a-chip [49], alveolus-on-a-chip [50], lung-on-a-chip [45], gut-on-a-chip [45]. More details of organ-on-a-chip development and applications can be found in previously published reviews [38, 51, 52].
4.2. Organ-on-a-Chip Advantages
Organ-on-a-chip platforms are miniaturized, reducing the required sample sizes and materials consumed during in vitro testing [53]. As a result, organ-on-a-chip testing is less costly than alternative preclinical models, such as animals. In addition, organs-on-a-chip perhaps offer an ethical advantage and alternative to animal models. Due to the miniaturized nature of these platforms, it has become possible to use materials from a single animal to runs hundreds of tests, instead of running a single test on hundreds of animals. The small size and low cost of organs-on-a-chip allow for accelerated research and testing, as many samples can be run on one device. Another advantage of organs-on-a-chip is the ability to recapitulate native microenvironments, modelling mechanical stresses, nutrient diffusion, and fluid flow, for example. These advantages have sparked interest in combining organ-on-a-chip platforms with other 3D models, such as organoid cultures. While 3D organoids may recapitulate a singular organ, organ-on-chip platforms mimic how these organs interact with their in vitro environment. Further discussion on the combination of 3D organoid cultures with organs-on-a-chip can be found in recent reviews [53, 54].
4.3. Cancer-on-a-Chip Platforms
Cancer-on-a-chip platforms were developed by using cancer and tumor-derived cells and matrix materials inside of previously developed cancer-on-a-chip platforms. The microfluidic devices developed for organ-on-a-chip platforms have shown to be promising for applications in large scale, high-throughput anti-cancer drug screening, study of metastatic cancer processes, and screening of drugs against a patient’s individual tumor (personalized medicine) [7, 13, 21, 55]. In the subsequent sections, we will focus our discussion on cancer-on-a-chip models that recapitulate components of the TME. A discussion of cancer-on-a-chip to evaluate nanomedicine [19] and for personalized medicine applications [56] can be found in recently published reviews.
5. Applications of Cancer-on-a-Chip Technologies
The development of cancer-on-a-chip systems has greatly expanded the ability of in vitro models to recapitulate the TME. With the recent development of dynamic culture systems and the advent of organ-on-a-chip platforms, which offer spatial and temporally controlled microenvironments, the improved cancer modelling has become an attractive prospect for studying both cancer biology and treatment options [57]. Although this field is still in its infantile stage, its progress is expected to grow exponentially. Cancer-on-a-chip models offer many advantages. First, they allow researchers to mimic elements of the TME in isolation or in concert, including the addition of cancer cells to stromal cells, immune constituents, vasculature, and oxygen, nutrient, or growth factor gradients. Cancer-on-a-chip systems also allow for non-invasive real time monitoring of crucial cellular parameters and recapitulate the complex cellular and extracellular microenvironment of tumors. These abilities allow for the investigation into the role microenvironmental features play in the progressing stages of cancer metastasis [24]
Cancer-on-a-chip models have been used in to evaluate aspects of cancer biology in several different malignancies, examining local tumor invasion, metastasis and angiogenesis, as well as serving as models for immunotherapy research and drug screening [4, 7, 29]. These studies examined several types of cancer as model systems, including some of the most common and deadly cancers, e.g. breast [36, 58, 59], lung [20, 60] and pancreatic cancer [15]. It is very important to work on these frontiers, but, nevertheless, other cancer models should be developed both to address special characteristics and the subtypes of cancers, as well as the comprehensive spectrum of cancers, including those of the head and neck.
5.1. Cancer-on-a-Chip Systems Model Tumor Morphology and Drug Response with High Fidelity
Cancer-on-a-chip systems that recapitulate in vivo cancers and tumors are fundamental to improved strategies for anti-cancer drug selection. In 2014, Vidi et al. developed a cancer-on-a-chip model, mimicking cancer mammary ducts [61]. The breast-on-a-chip device was comprised of a breast luminal epithelium monolayer on a semicircular acrylic support. Tumor cells grown in these channels were different morphologically than the same cells cultured on a traditional flat surface. Additionally, tumor nodules cultured in these channels displayed a different anti-cancer drug sensitivity compared to their flat and monoculture counterparts, providing new insight for the design and testing of cancer therapies.
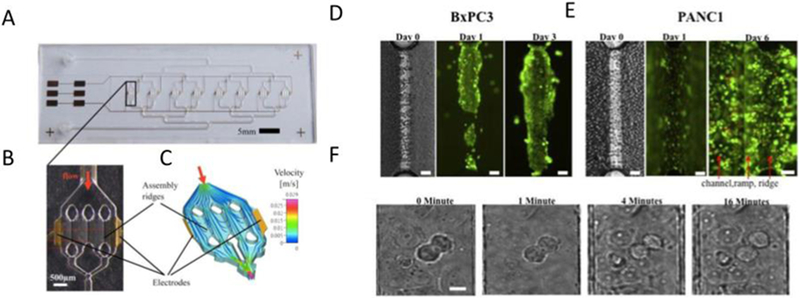
An important component of developing a new platform is the comparison of its results to the existing standard. In 2017, Beer et al. compared their pancreatic ductal adenocarcinoma-on-a-chip model to other in vitro and to in vivo PDX models (Figure 1) [15]. Their HepaChip consisted of eight chambers, each containing three 1 mm × 60 μm cell culture regions, which were coated with collagen. These cell culture regions were irradiated by UV light, creating acid groups to which collagen was bound. Electrodes were integrated on the wall of each chamber, creating dielectrophoretic forces. pancreatic Ductal adenocarcinoma (PDAC) human cell lines were cultured inside of the polymer chambers. The combination of microfluidics and dielecrophoresis assembled in vitro micro-organs. The experimental results show morphological and growth characteristics more like that of spheroid cultures than the 2D culture. Compared to traditional 2D pre-clinical platforms, the HepaChip model is better at capturing cell-cell and cell-ECM interactions, and thus it is more biomimetic, representing a more predictive model with potential to be useful in the development of personalized pancreatic cancer treatment.
Figure 1:
Photographs and simulation of the HepaChip. (A) Image of the chip, with 8 culture chambers, fluid inlet and outlet and gold electrodes. (B) Enlarge view of single chamber, with 2 electrodes and 3 assembly ridges. (C) Simulation of the flow and cell trajectory inside of the culture chamber. (D) Live/dead staining of BxPC3, growing on the assembly ridge. (E) Live/dead staining of PANC1, spread on well channel walls and bottom. (F) Mitosis of MxPC3, observed after 16h culture. Reproduced under the terms of the Creative Commons License. [15] 2017 Nature.
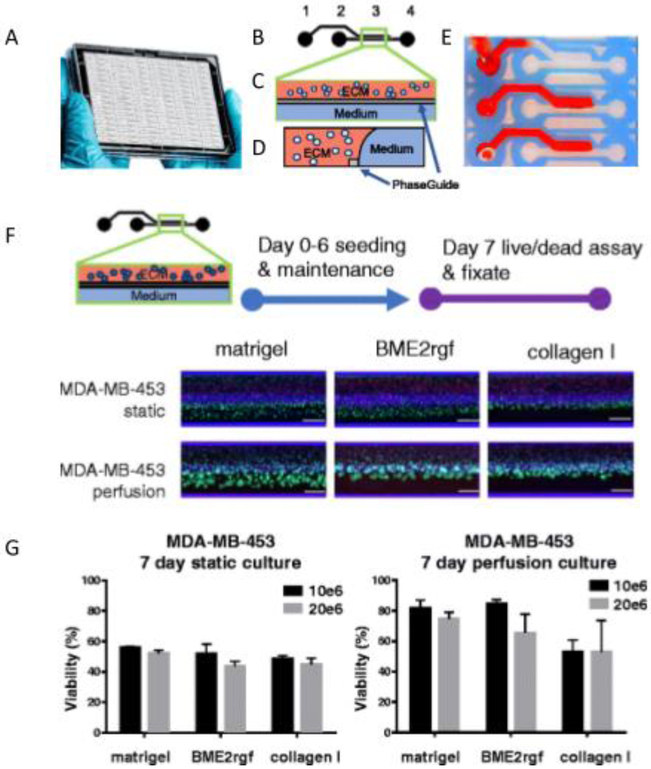
Another more recent study by Lanz et al. utilized breast cancer cell lines for developing a high throughput breast-cancer-on-a-chip to study the response of triple negative breast cancer cell lines (MDA-MB-453, MDA-MB-231, AND HC1937) to anti-cancer therapeutic drugs (paxlitaxel, olaparib, and cisplastin) [36]. Several conditions were evaluated, including cell seeding density, ECM composition, biomechanical conditions, and the response to therapy, as compared to those seen in 2D cultures [36]. Differences in drug response were observed different ECM materials )Matrigel vs BME2rgf vs collagen 1). This microfluidic platform allowed for the simultaneous culture of 96 perfused microtissues (approximately 10 cells per data point). Additional advantages include the use of small quantities of material and the ability to perform drug screening using patient-derived samples. This strategy is a vast improvement from previous 3D culture techniques, as it allows for constant perfusion of the culture medium. While the tested system did not entirely capture in vivo complexities, this strategy presents a more high throughput and efficient system for testing and raises the possibility for use in developing personalized medicine by determining appropriate drug sensitivity and predicting individual patient response in a real-time fashion.
5.2. Cancer-on-Chip Systems Model Mechanical Properties of the Tumor Microenvironment
In addition to modelling tumor morphology and drug response, cancer-on-a-chip platforms also offer improved modelling of the TME’s mechanical properties, such as its stiffness, which plays a role in cancer progression [62]. In 2017, Hassell et al. studied non-small-cell lung cancer (NSCLC) in an organ-on-a-chip device (Figure 2) [60]. This NSCLC-on-a-chip model recapitulated organ microenvironment-specific growth, tumor dormancy, and response to tyrosine kinase inhibitor (TKI), a therapy used in vivo in human patients. This new platform revealed a newly observed mechanical sensitivity of NSCLCs. TKI therapeutic response was discovered to be sensitive to the physical cues of breathing motions, with mechanical breathing motions perhaps suppressing NSCLC response to TKI therapy. This new finding elucidates understanding of NSCLC and can help to explain therapy resistance in patients with lung tissue that remains aerated and mobile, as mechanical strain leads to the downregulation of epidermal growth factor receptor (EGFR), which is partly responsible for decreased response to therapy in persistent tumors [60]. The understanding of this previously unstudied mechanism has implications in future mitigation of drug resistance and development of efficacious therapies.
Figure 2:
Human Orthotopic Lung Cancer-on-a-Chip Model. (A) Schematic of a cross-section through the designed microfluidic chip. (B) Micrograph of a cross-section of the two central channels of the alveolus chip taken via fluorescence micrograph. (C) Immunofluorescence micrograph of a cluster of GFP labelled NSCLC cells, implanted in the airway chip. (D) Quantification of NSCLC densities after implantation in the chip. (E) Growth pattern of GFP labelled lung cancer cells within the epithelial monolayer. (F) Lung cancer cell growth dynamics. Reproduce with permission. [60] 2017 Elsevier.
These findings further validate the need for the development of dynamic systems and platforms, which take into consideration the components of the TME [63]. Future work need not only consider traditional properties, such as cell type and ECM, but also dynamic properties, such as mechanical stiffness, to mimic in vivo events and develop more reliable predictive models.
5.3. Cancer-on-Chip Systems Model Tumor Immune Microenvironment
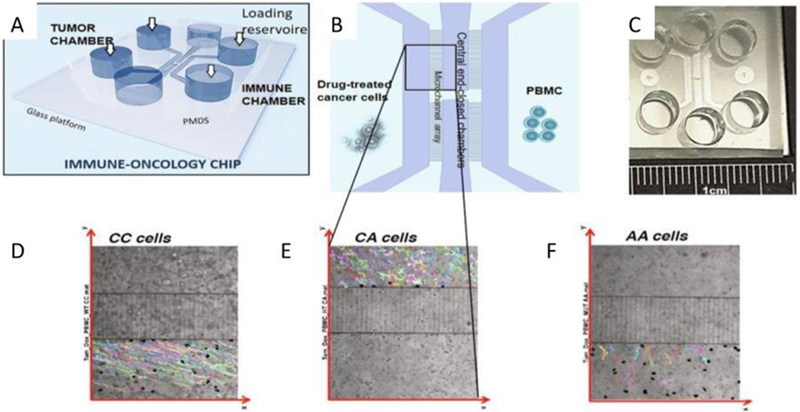
Immune system elements play a large role in the TME, with crosstalk between the immune and cellular TME components [64]. Interactions within the TME contribute to cancer initiation (carcinogenesis), progression and metastasis. After a tumor escapes immune recognition, , the TME affects immune cell behavior and the two play a synergistic role in tumor progression [65]. Organ-on-a-chip platforms can be useful in the development of immuno-oncology models. Such a model was shown by Biselli et al. in their work on mononuclear and cancer cells (Figure 3) [66]. The presented cancer-on-chip model studied the crosstalk between immune cells, leukocytes and human breast cancer cells treated with chemotherapeutics. The study compared leukocytes with and without the FRP1 gene and concluded that leukocytes lacking the FPR1 gene do not recognize the chemotherapy-treated cells, while leukocytes with expression of FPR1 perform random walks, drifting towards the tumor cells and establishing persistent interactions with them [66]. These findings demonstrate the capacity and necessity to develop immune-oncology methods using the organ-on-a-chip platform, as immune response is a key component of the in vivo environment (evidenced by the cell-cell interactions of immune and tumor cells) [66, 67]. A detailed discussion on crosstalk between the TME and immune system can be found in a recent review paper [64].
Figure 3:
(A) General schematic of the immune-oncology chip, whose design features six reservoirs for cells loading and culture medium replacement and four compartments for cell culture. (B) Detailed view of the four chambers. (C) Picture of the whole device. (D-F) Trajectories of FPR1 CC cells, FPR1 CA cells, and FPR1 AA cells (respectively). Reproduced under the terms of the Creative Commons License. [66] 2017 Nature.
5.4. Cancer-on-Chip Systems Model Tumor Angiogenesis, Microvasculature, and Lymphatics
Angiogenesis plays a critical role in both the growth and metastatic spread of tumor cells [68, 69]. The TME can regulate angiogenesis, via ECM molecules and growth factors within the TME [70]. Increased vascularity has been observed, even in the bone marrow, in patients with hematological malignancies [68]. Despite this observation, the role of angiogenesis in regulating hematological malignancies is not well understood [68]. In 2016, Zheng et al. used a 3D microfluidic angiogenesis-on-a-chip to study the unique morphogenic signatures of angiogenesis induced by leukemic cells, with or without bone marrow stromal cell co-culture [68]. The microfluidic device was fabricated from PDMS by soft lithography, forming three parallel microchannels, separated by trapezoidal posts. The central channel was filled with collagen 1 and the side channels with endothelial and leukemia derived cells. The role of leukemic cells on angiogenic induction and vessel formation was observed. This model has not yet been achieved via existing 2D culture techniques. Angiogenesis in the bone marrow is a highly dynamic process and is critically dependent on both cell-cell and cell-matrix interactions, which are abundant in the native bone marrow microenvironment, presenting the critical need for functional angiogenic assays.
The lymphatic system serves as a method through which many cancers can be disseminated. New microfluidic flow systems make it possible to study the role of lymphatic capillary microenvironment in the lymphatic invasion of mammary adenocarcinoma cells [69]. This platform allows for the quantification of cell transmigration and its dynamics, revealing that both luminal and transluminal flow are important in increasing tumor transmigration, as opposed to the previous belief that this behavior was a result of luminal flow alone. This study provides new insights on flow-mediated regulation of lymphatic tumor migration and presents a new tool for exploration of cancer therapy, allowing for medium to high throughput studies.
5.5. Cancer-on-Chip Systems Model Cancer Invasion and Metastasis
Cancer metastases contribute to over 90% of cancer-related mortalities [71]. Thus, the use of experimental models to effectively represent the metastatic microenvironment is warranted. Metastasis-on-a-chip platforms allow for the study of important aspects of the metastasis process, such as physiochemical factors from the tumor stroma and heterocellular interactions, which influence cell migration, as well as physicochemical gradients, which lead to tumor cell motility and invasion [21]. An early study to visualize metastatic progression in 2007 by Yates et al. developed a system to visualize the interactions between tumor cells and target organs to where they metastasize [72]. This study investigated hepatic cells with prostate and breast carcinoma cells, examining tumor cell invasion and expansion. They found that tumor cells were unable to grow without a supporting hepatic microtissue, due to the absence of paracrine functionality or liver structure support. The developed system served as a crucial model for examining tumor-host interactions during the processes of metastasis and invasion, circumventing the limitations of previous models.
More recently, microfluidic metastasis-on-a-chip models have been designed to more accurately study cancer progression [59]. These studies investigate the various stages of cancer metastasis, lymphangiogenesis [73] and angiogenesis, intravasation [74, 75], arrest, organ-specific extravasation [76-79], and the formation of micro-metastases. Additionally, studies looked at invasion rate as a method for studying cancer [80]. In 2018, Hao et al. developed a bone-on-a-chip model the aid in the study of breast cancer metastasis to bone tissue [58]. This new bone-on-chip design is miniaturized, increasing experimental throughput, and facilitates easy and frequent observations. Fundamental markers of breast cancer colonization of the bone were observed and confirmed with in vivo collected data.
Another organ-on-a-chip study constructed a multi-organ microfluidic chip platform to investigate lung cancer metastasis [20]. This multi-organ-on-a-chip system consisted of 4 organs, one upstream lung and three downstream organs, with three parallel microchannels formed by PDMS. Bronchial epithelial, lung cancer, microvascular endothelia, and fibroblast cells were grown in the lung organ, and astrocytes, osteocytes, and hepatocytes were grown in the three downstream organs, mimicking the metastasis of lung cancer to the brain, bone, and liver. Damage to the astrocytes, osteocytes, and hepatocytes validated metastasis in the organs-on-a-chip system.
Migration and invasion studies in cancer-on-a-chip models improved upon the use of more traditional assays, such as transwell cultures and scratch-wound assays. Recent work from Toh et al. utilized a microfluidic cancer-on-chip cell migration model which resolved different aspects of cell intravasation (the invasion of cancer cells into a blood or lymphatic vessel) in a biologically relevant 3D microenvironment [81]. The described platform incorporates a 3D microenvironment, which plays a critical role in the invasive properties of cancer cells, with a microfluidic system, creating an appealing model for the testing of anti-migration and anti-invasion cancer drugs, which can be multiplexed to allow for high throughput assays. The development of new tumor models will be crucial in improving management of cancers and the prognosis of cancer patients and, as a result, may help ultimately in the reduction of healthcare costs [82].
5.6. Cancer-on-Chip Systems for Use in Chemotaxis Studies
The cell types and factors in the TME influence the occurrence of cancer migration modes [83]. Microfluidic organ-on-a-chip systems have been used to study the chemotaxis, or migration due to chemical gradients, of cancer cells [84]. Aung et al. developed a cancer-on-chip platform with cancer spheroids encapsulated with gelatin methacryloyl (GelMA) hydrogel, surrounded by an endothelial cell barrier [84]. For this study, they harnessed the chemoattracted-induced motility of human umbilical vein endothelial cells (HUVEC) and cancer spheroids to control organization within the microfluidic device. Cancer cell migration was observed in relation to the presence and location of the chemotactic source. Although this study uses an endothelial cell-cancer cell coculture, this approach provides a framework for establishing platforms with the same level of complexity as physiological tumors.These models bridge the gap between in vitro cell culture and in vivo animal experiments and serve as a promising platform for studying tumor behaviors in the vascular system [85]. More complex, hybrid cancer-on-a-chip models incorporate 3D tumor tissue models, such as spheroids, resulting in more advanced and high-performance models [86] [87]. In the future, these models will have elucidated mechanisms for cancer invasion, such as chemotaxis, and present the possibility of developing migration inhibitory drugs [84].
5.7. Cancer on a Chip Platform for Cancer Treatment and Drug Development
Improved 3D culture models have been developed with the prospect of accelerating the selection of therapies by improving the ability to predict anti-cancer drug responses [36]. One avenue of new cancer-on-a-chip studies involves the use of 3D microfluidic devices for improved anti-cancer drug screening and selection. Examples of these new strategies include a 3D high-throughput perfused microfluidic platform for testing new breast cancer therapies, developed by Lanz et al. (Figure 4) [36], and microfluidic platform for studying biomolecular characteristics of pancreatic ductal adenocarcinoma cells, developed by Beer et al. [15]. These strategies employ microfluidic-based devices, which show promise to be used for personalized pharmacological testing. Microfluidic systems can be employed for the fabrication of drug delivery systems having precisely controlled size and shape, rigidity as well as drug-loading. In addition, microfluidic systems can be efficiently used for the evaluation of drug-releasing preparations [88]. Because organ-on-a-chip platforms can recapitulate human physiology and pathophysiology, they can be effectively helpful in translating new therapeutics to the clinic including advanced nanomedicine [19].
Figure 4:
Microtiter cancer-on-a-chip plate for anti-cancer breast cancer drug testing. (A) Photo of the OrganoPlate platform. (B-D) Closeup, top, and side view of an individual channel, respectively. (E) Photo demonstrating the filling of an ECM channel. (F) Epifluorescence microscopy images showing morphology and viability of MDA-MB-453 in Matrigel®, BME2rgf and collagen I, under both static and perfusion conditions. (G) Quantification of the effect of ECM composition, seeding density, and perfusion or static conditions on cell viability. Reproduced under the terms of the Creative Commons License. [36] 2017 BioMed Central.
Hassell et al. finds that microenvironmental cues elicited by cells, as well as mechanical cues, significantly influence non-small-cell lung cancer growth in vitro [60]. More importantly, their data demonstrates the ability of orthotopic cancer chip models to mimic growth patterns observed in vivo in patients and is consistent with that of human clinical trials, a feature which had not previously been recapitulated in vitro [60]. Other strategies to mimic the TME include the use of multicellular aggregates (“microtumors”) of subtype-specific breast cancer cells, by Singh et al. [31], human-on-chip microvascular assay for visualization of tumor cell extravasation dynamics, by Chen et al. [76], as well as development of an array of gut-on-a-chips for drug development [89]. The ultimate value in these new developments lies in their potential to allow for high-throughput drug screening of chemotherapeutics on ex vivo models of individual patients’ tumors.
6. High Throughput Cancer-on-Chip Studies for Large Data Generation
Cancer-on-a-chip models are advantageous for preclinical drug screening, as they can be designed to allow for high-throughput analysis of anti-tumor drug response and other biological parameters [19, 90]. In order to generate the large quantities of data needed to appropriately predict drug efficacy and potential side effects, as is seen in clinical trials, high-throughput systems need to be developed. With the use of cancer-on-a-chip platforms, this type of large data creation and large scale analysis is possible.
6.1. Large Data Generating Cancer-on-Chip Studies
In order to perform high-throughput studies, a large number of devices must be fabricated with high fidelity, reproducibility and homogeneity or a device must have the capacity to run many tests on a single chip. In 2017, Chen et al. published a protocol extension describing the fabrication of a microfluidic device for modelling early metastasis in Nature Protocols [76]. Their device was made from PDMS and featured three hydrogel regions, separated by channels for media. Microposts marked the device between each region. The central region was filled with a fibrin gel and human umbilical vein endothelial cell (HUVEC) suspension and the two peripheral regions were filled with a fibrin gel and human lung fibroblast suspension. Their device served as a model for microcirculation, representing transendothelial migration and early metastasis. They reported the capacity to fabricate and seed up to 36 devices at a time without impacting cell viability. Coupled with rapid quantification, their large number of devices per experiment is expected to allow for high parametric and throughput study, generating a large quantity of data.
To address the challenges associated with current in vitro and in vivo preclinical models, such as the need for large numbers of cell or animal materials, researchers have developed microdevices with up to thousands of microwells, allowing for high-throughput testing. In 2014, Zhang et al. developed a microfluidic device with 4000 ultraminiaturized wells for high-throughput monitoring of chemotactic migration and invasion [91]. Their multi-well invasion (MI) chip was fabricated from PDMS using photolithography. The MI-chip was comprised of four compartments, each containing ten, ten by ten microwell arrays, equaling 4000 microwells. They fabricated both round (200 μm diameter) and square (200 × 200 μm) wells, with a depth of 160 μm. The MI-chips were used to perform 3D cell invasion assays with breast cancer cell lines, to validate the chip’s capacity to be used as a model for studying metastatic breast cancer. The MI-chip used a small cell sample size (less than 1000 cells), allowing it to be used in the future with limited cell sources, such as primary tumor cell samples. The chip can be used in the future to run many tests at one time or run tests on rare samples, making it a useful future tool for clinicians to evaluate the behavior of cancer cells and anti-cancer drug regimens.
Another method for generating large data sets via organ-on-a-chip is the fabrication of multiple models on a single device. To our knowledge, there is not currently published literature joining multiple cancer-on-a-chip platforms. However, groups have multiplexed organ-on-a-chip devices together for other applications. Work in the organ-on-a-chip field reports the use of an array of gut-on-a-chip devices joined together, comprising a total of 357 gut tubes [89]. The OrganoPlate (shown in Figure 5) is a 384-well plate platform housing 40 networks of microfluidic channels. Each OrganoPlate housed 40 epithelial gut tubes, which were tested against drug compounds at different concentrations to study the effect on epithelial barrier integrity. The study generated over 20,000 data-points, making it the largest reported organ-on-chip data set thus far. This study’s high-throughput nature shows the promise of organ-on-a-chip platforms for use as new, efficient, and reliable pre-clinical models, with applications in anti-cancer drug testing [89, 92].
Figure 5:
(A) Photograph of the bottom of an OrganoPlate, showing 40 microfluidic channel networks and the top of a 384 well plate device. (B) Zoomed in photograph of a single microfluidic channel network, with three channels joining in the center. (C, D) Horizontal and vertical cross section. (E) 3D sketch of the chip, comprised of a tubule, an extracellular matrix gel, and a perfusion lane. Reproduced under the terms of the Creative Commons License. [89] 2017 Nature.
6.2. Large Data Management and Extraction of Information
With high-throughput platforms, such as organ-on-a-chip, generating unprecedented quantities of data, there lies the need for appropriate data management and analysis systems. While cancer-on-a-chip platforms have been rapidly developing, machine learning algorithms to manage this data have been developing in parallel. New data management strategies should incorporate four core pillars. The first pillar in data management is the hardware implementation of microchips, with appropriate sensors and microsystems to measure the desired parameters. The second pillar of large data management is data collections, transmission, and storage. The third pillar required is advanced machine learning algorithms to extract information from the available huge data sets. Finally, the fourth pillar concerns the interpretation of obtained data and its applications in the discovery of new theories.
Cancer-on-a-chip platform sensors continuously measure attributes of all cells within the device. The data flow can be of extremely high volume; for example, if there are 50,000 cells under investigation, each with 20 measured factors per minute, then one would receive approximately two gigabytes of data each day (variable by the coding method of the collected data). With several parallel platforms and prolonged data collection (potentially months of collection), the size of the collected data may be exceedingly large.
After data collection, the next large stage is the big data processing. For this purpose, we rely on highly sophisticated machine learning (ML) algorithms, which can be divided into two categories: supervised or unsupervised learning [93]. These algorithms take into consideration the number of cells (N), number of parameters or attributes (M) and the number of samples (T). In this example, we can define health status as H and assign binary values of 0 (dead cell) and 1 (fully healthy cell). After collecting big size data, we can build some model (dynamic or otherwise) for the relationship between cellular attributes and health. The data is typically divided into two parts, with one part (the learning portion) used for modeling and the second used for validation; the first part should be large enough in size to capture important mapping relations between the attributes and the output status H. Other alternative ML algorithms to be used include recurrent type networks with deep learning structure. There are many ML algorithms with different concepts and criteria. A crucial component to any model, however, is keeping the complexity at enough to obtain a good generalized model but avoid creating unnecessary complexity, which may give high errors in the validation and testing phase.
There are other alternative approaches to supervised learning which are not discussed in this review. More information regarding ML algorithms can be found in other sources [93]. Another challenge of big data processing is the interpretation of the ML algorithm results. Advanced artificial intelligence (AI) algorithms may be used to help human experts interpret ML outcomes. The discussed methodologies will aid in the processing and interpretation of data collected from future high-throughput, large data generating studies. With real-time data collection and processing, in the future, researchers will be able to perform trials on millions of therapeutic agents against a patient’s specific tumor. Microfluidics, sensors, computing facilities, smart algorithms, and intelligent micro-automated systems can be joined as the basis of advanced systems for next-generation anti-cancer drug design and development [7, 94].
7. Challenges and Future Work
Recent studies involving cancer-on-a-chip technologies have made great strides in better recapitulating the TME and in vivo cancer microenvironment. However, there is still work to be done identifying the fundamental TME elements needed to better mimic cancer tissue, for both the study of cancer biology and improved predictive ability of pre-clinical models for anti-cancer drug development. The development of personalized cancer-on-a-chip platforms for patients’ primary tumor tissues will be a large step forward in harnessing the capabilities of cancer-on-a-chip platforms. This advancement will result in precision medicine and personalized oncology. Incorporation of TME elements, such as oxygen concentration and cytokine concentration gradients, will increase the complexity of cancer-on-a-chip models, improving the predictive power of these platforms. Once developed, there will likely be many challenges in the adoption of these technologies: i.e.) standardization and validation against current models.
With the adoption of organ-on-a-chip technologies, future oncology treatment is expected to be vastly different than today’s regimens As we move towards personalized oncology models, one predicted outcome is the use of patient derived cells and extracted ECMs in chip technologies, capturing the biochemical, biophysical and mechanical cues of the in vivo human cancer microenvironment. Another advantage of cancer-on-a-chip technologies is the capability for high throughput, personalized screening of anti-cancer drug treatment and therapies. The capabilities of these platforms may also be expanded and used for more innovative cancer detection [51], e.g. on-chip blood tests to replace bone biopsies for multiple myeloma [81].
8. Conclusions
Once cancer-on-a-chip technologies are fully realized, the current regimented practice of choosing specific chemotherapeutics based solely on tumor type will seem imprecise and inaccurate. Personalized cancer chemotherapy will eventually be adopted and the use of cancer-on-a-chip models will become the clinical standard, allowing for more precise and individual function-based selection of chemotherapeutics. After the predictive power of these models are demonstrated, on-a-chip tests will serve a central role in the development and approval of new cancer therapeutics, replacing current pre-clinical models. Adoption of these new technologies will both accelerate and decrease the costs of the drug development process and increase the precision of cancer therapy, benefiting patients, physicians and care-providers, as well as pharmaceutical and insurance companies.
Table 1:
Summary of Cancer-on-Chip Studies
| No. | Cancer type | Method | Result | Authors | Year | Ref. |
|---|---|---|---|---|---|---|
| 1. | Breast cancer | Microfluidic 3D in vitro model for breast cancer metastasis to bone | 3D in vitro data on extravasation and micrometastasis generation of breast cancer cells within bone microenvironment | Bersini et al. | 2014 | (51) |
| 2. | Breast cancer | 3D bone-on-chip for bone metastasis study of breast cancer cells | Unique hallmarks of breast cancer bone in colonization observed, previously only seen in vivo | Hao et al. | 2018 | (50) |
| 3. | Breast cancer | Disease-on-a-chip model in which cancer grows within phenotypically normal breast luminal epithelium on semicircular acrylic supports | Mimicry of tumor environment provides a framework for the design and test of anti-cancer therapies | Vidi et al. | 2014 | (54) |
| 4. | Breast cancer | 3D high-throughput microfluidic platform for screening of three triple negative breast cancer lines against several anti-cancer drugs | High-throughput organ on a chip platform to select therapies in personalized medicine | Lanz et al. | 2017 | (12) |
| 5. | Cancer immune interactions | Organ-on-chip tool to evaluate cancer-immune cell interactions | Quantitative confirmation of the essential role of FPR1 in cancer chemotherapy response | Biselli et al. | 2017 | (60) |
| 6. | Lung cancer | Multi-organ microfluidic chip mimicking in vivo microenvironment of lung cancer metastasis | Multi-organ system provides useful tool to investigate cell-cell interactions in metastasis | Xu et al. | 2016 | (53) |
| 7. | Lung | Organ-on-chip model recapitulates orthotopic lung cancer growth and therapeutic response | Discovery of mechanical stimuli dependent TKI therapy resistance | Hassell et al. | 2017 | (52) |
| 8. | Pancreatic ductal adenocarcinoma | Microfluidic 3D platform for culturing pancreatic ductal adenocarcinoma cells | Growth characteristics closer to those of cells grown as spheroids than as classical 2 dimensional (2D) in vitro cultures. | Beer et al. | 2017 | (13) |
Acknowledgements
The authors acknowledge funding from the National Institutes of Health (AR057837, AR066193, EB021857-01A1, HL137193-01, EB024403-01-01, CA214411-01A1, EB023052-01A1, HL140618-01, GM126831, HL140951, AR073135, and AR069564), Department of Defense: BiofabUSA Quick Start Project, and American Heart Association Transformational Project Award (18TPA34230036).
References
- 1.Society AC, Global Cancer Facts & Figures 4th Edition. American Cancer Society: Atlanta, 2018. [Google Scholar]
- 2.Siegel RL; Miller KD; Jemal A, CA: a cancer journal for clinicians 2018, 68 (1), 7–30. DOI 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Institute NC, In NCI Dictionary of Cancer Terms, National Institute of Health. [Google Scholar]
- 4.Ma Y-HV; Middleton K; You L; Sun Y, Microsystems &Amp; Nanoengineering 2018, 4, 17104 DOI 10.1038/micronano.2017.104. [DOI] [Google Scholar]
- 5.Steeg PS, Nature Reviews Cancer 2016, 16, 201 DOI 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shewach DS; Kuchta RD, Chemical reviews 2009, 109 (7), 2859–2861. DOI 10.1021/cr900208x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hachey SJ; Hughes CCW, Lab on a Chip 2018, 18 (19), 2893–2912. DOI 10.1039/C8LC00330K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smietana K; Siatkowski M; Moller M, Nature reviews. Drug discovery 2016, 15 (6), 379–80. DOI 10.1038/nrd.2016.85. [DOI] [PubMed] [Google Scholar]
- 9.Kola I; Landis J, Nature reviews. Drug discovery 2004, 3 (8), 711–5. DOI 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 10.Whiteside TL, Oncogene 2008, 27 (45), 5904–5912. DOI 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M; Zhao J; Zhang L; Wei F; Lian Y; Wu Y; Gong Z; Zhang S; Zhou J; Cao K; Li X; Xiong W; Li G; Zeng Z; Guo C, J Cancer 2017, 8 (5), 761–773. DOI 10.7150/jca.17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swartz MA; Iida N; Roberts EW; Sangaletti S; Wong MH; Yull FE; Coussens LM; DeClerck YA, Cancer research 2012, 72 (10), 2473–80. DOI 10.1158/0008-5472.Can-12-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z; Samanipour R; Kim K, Organ-on-a-Chip Platforms for Drug Screening and Tissue Engineering In Biomedical Engineering: Frontier Research and Converging Technologies, Jo H; Jun H-W; Shin J; Lee S, Eds. Springer International Publishing: Cham, 2016; pp 209–233. [Google Scholar]
- 14.Kapałczyńska M; Kolenda T; Przybyła W; Zajączkowska M; Teresiak A; Filas V; Ibbs M; Bliźniak R; Łuczewski Ł; Lamperska K, Archives of medical science : AMS 2018, 14 (4), 910–919. DOI 10.5114/aoms.2016.63743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beer M; Kuppalu N; Stefanini M; Becker H; Schulz I; Manoli S; Schuette J; Schmees C; Casazza A; Stelzle M; Arcangeli A, Scientific Reports 2017, 7 (1), 1325 DOI 10.1038/s41598-017-01256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakrabarti R; Kang Y, Methods in molecular biology (Clifton, N.J.) 2015, 1267, 367–80. DOI 10.1007/978-1-4939-2297-0_18. [DOI] [PubMed] [Google Scholar]
- 17.Cekanova M; Rathore K, Drug design, development and therapy 2014, 8, 1911–21. DOI 10.2147/dddt.S49584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Day C-P; Merlino G; Van Dyke T, Cell 2015, 163 (1), 39–53. DOI 10.1016/j.cell.2015.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang YS; Zhang YN; Zhang W, Drug discovery today 2017, 22 (9), 1392–1399. DOI 10.1016/j.drudis.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Z; Li E; Guo Z; Yu R; Hao H; Xu Y; Sun Z; Li X; Lyu J; Wang Q, ACS applied materials & interfaces 2016, 8 (39), 25840–25847. DOI 10.1021/acsami.6b08746. [DOI] [PubMed] [Google Scholar]
- 21.Portillo-Lara R; Annabi N, Lab Chip 2016, 16 (21), 4063–4081. DOI 10.1039/c6lc00718j. [DOI] [PubMed] [Google Scholar]
- 22.Urban G; Bache KM; Phan D; Sobrino A; Shmakov AK; Hachey SJ; Hughes C; Baldi P, IEEE/ACM Transactions on Computational Biology and Bioinformatics 2018, 1–1. DOI 10.1109/TCBB.2018.2841396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhandapani M; Goldman A, Journal of molecular biomarkers & diagnosis 2017, 8 (5), 356 DOI 10.4172/2155-9929.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray JW; Mills GB, Cancer discovery 2015, 5 (11), 1130–1132. DOI 10.1158/2159-8290.CD-15-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breslin S; O'Driscoll L, Drug discovery today 2013, 18 (5-6), 240–9. DOI 10.1016/j.drudis.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Duval K; Grover H; Han L-H; Mou Y; Pegoraro AF; Fredberg J; Chen Z, Physiology (Bethesda, Md.) 2017, 32 (4), 266–277. DOI 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 27.Pampaloni F; Reynaud EG; Stelzer EH, Nature reviews. Molecular cell biology 2007, 8 (10), 839–45. DOI 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 28.Katt ME; Placone AL; Wong AD; Xu ZS; Searson PC, Frontiers in bioengineering and biotechnology 2016, 4, 12–12. DOI 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai H-F; Trubelja A; Shen AQ; Bao G, 2017, 14 (131), 20170137. DOI doi: 10.1098/rsif.2017.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Justus CR; Leffler N; Ruiz-Echevarria M; Yang LV, Journal of visualized experiments : JoVE 2014, (88), 51046 DOI 10.3791/51046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh M; Mukundan S; Jaramillo M; Oesterreich S; Sant S, Cancer research 2016, 76 (13), 3732–43. DOI 10.1158/0008-5472.Can-15-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruggeri BA; Camp F; Miknyoczki S, Biochemical pharmacology 2014, 87 (1), 150–61. DOI 10.1016/j.bcp.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 33.Lai Y; Wei X; Lin S; Qin L; Cheng L; Li P, Journal of hematology & oncology 2017, 10 (1), 106–106. DOI 10.1186/s13045-017-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carvalho MR; Lima D; Reis RL; Correlo VM; Oliveira JM, Trends in biotechnology 2015, 33 (11), 667–678. DOI 10.1016/j.tibtech.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Moro M; Bertolini G; Caserini R; Borzi C; Boeri M; Fabbri A; Leone G; Gasparini P; Galeone C; Pelosi G; Roz L; Sozzi G; Pastorino U, Scientific Reports 2017, 7 (1), 6689 DOI 10.1038/s41598-017-06912-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanz HL; Saleh A; Kramer B; Cairns J; Ng CP; Yu J; Trietsch SJ; Hankemeier T; Joore J; Vulto P; Weinshilboum R; Wang L, BMC cancer 2017, 17 (1), 709–709. DOI 10.1186/s12885-017-3709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimlin LC; Casagrande G; Virador VM, Molecular carcinogenesis 2013, 52 (3), 167–82. DOI 10.1002/mc.21844. [DOI] [PubMed] [Google Scholar]
- 38.Ronaldson-Bouchard K; Vunjak-Novakovic G, Cell Stem Cell 2018, 22 (3), 310–324. DOI 10.1016/j.stem.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitesides GM, Nature 2006, 442, 368 DOI 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 40.Achyuta AK; Conway AJ; Crouse RB; Bannister EC; Lee RN; Katnik CP; Behensky AA; Cuevas J; Sundaram SS, Lab Chip 2013, 13 (4), 542–53. DOI 10.1039/c2lc41033h. [DOI] [PubMed] [Google Scholar]
- 41.Brown JA; Pensabene V; Markov DA; Allwardt V; Neely MD; Shi M; Britt CM; Hoilett OS; Yang Q; Brewer BM; Samson PC; McCawley LJ; May JM; Webb DJ; Li D; Bowman AB; Reiserer RS; Wikswo JP, Biomicrofluidics 2015, 9 (5), 054124 DOI 10.1063/1.4934713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musah S; Mammoto A; Ferrante TC; Jeanty SSF; Hirano-Kobayashi M; Mammoto T; Roberts K; Chung S; Novak R; Ingram M; Fatanat-Didar T; Koshy S; Weaver JC; Church GM; Ingber DE, Nature Biomedical Engineering 2017, 1, 0069 DOI 10.1038/s41551-017-0069 https://www.nature.com/articles/s41551-017-0069#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang KJ; Mehr AP; Hamilton GA; McPartlin LA; Chung S; Suh KY; Ingber DE, Integrative biology : quantitative biosciences from nano to macro 2013, 5 (9), 1119–29. DOI 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 44.Kim HJ; Ingber DE, Integrative biology : quantitative biosciences from nano to macro 2013, 5 (9), 1130–40. DOI 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 45.Kim HJ; Huh D; Hamilton G; Ingber DE, Lab Chip 2012, 12 (12), 2165–74. DOI 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 46.Huh D; Matthews BD; Mammoto A; Montoya-Zavala M; Hsin HY; Ingber DE, Science (New York, N.Y.) 2010, 328 (5986), 1662–8. DOI 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Booth R; Kim H, Lab Chip 2012, 12 (10), 1784–92. DOI 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- 48.Zheng F; Fu F; Cheng Y; Wang C; Zhao Y; Gu Z, Small (Weinheim an der Bergstrasse, Germany) 2016, 12 (17), 2253–82. DOI 10.1002/smll.201503208. [DOI] [PubMed] [Google Scholar]
- 49.Zhang YS; Davoudi F; Walch P; Manbachi A; Luo X; Dell'Erba V; Miri AK; Albadawi H; Arneri A; Li X; Wang X; Dokmeci MR; Khademhosseini A; Oklu R, Lab on a chip 2016, 16 (21), 4097–4105. DOI 10.1039/c6lc00380j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jain A; Barrile R; van der Meer AD; Mammoto A; Mammoto T; De Ceunynck K; Aisiku O; Otieno MA; Louden CS; Hamilton GA; Flaumenhaft R; Ingber DE, Clinical pharmacology and therapeutics 2018, 103 (2), 332–340. DOI 10.1002/cpt.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaushik G; Leijten J; Khademhosseini A, Stem Cells 2016, 35, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B; Korolj A; Lai BFL; Radisic M, Nature Reviews Materials 2018, 3 (8), 257–278. DOI 10.1038/s41578-018-0034-7. [DOI] [Google Scholar]
- 53.Fetah K; Tebon P; Goudie MJ; Eichenbaum J; Ren L; Barros N; Nasiri R; Ahadian S; Ashammakhi N; Dokmeci MR; Khademhosseini A, Progress in Biomedical Engineering 2019, 1 (1), 012001 DOI 10.1088/2516-1091/ab23df. [DOI] [Google Scholar]
- 54.Takebe T; Zhang B; Radisic M, Cell Stem Cell 2017, 21 (3), 297–300. DOI 10.1016/j.stem.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 55.Sleeboom JJF; Eslami Amirabadi H; Nair P; Sahlgren CM; den Toonder JMJ, Disease Models & Mechanisms 2018, 11 (3). DOI 10.1242/dmm.033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sontheimer-Phelps A; Hassell BA; Ingber DE, Nature Reviews Cancer 2019, 19 (2), 65–81. DOI 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- 57.Elmusrati M; Ashammakhi N, 2018, 29 (7), 1682–1683. DOI 10.1097/scs.0000000000004703. [DOI] [PubMed] [Google Scholar]
- 58.Hao S; Ha L; Cheng G; Wan Y; Xia Y; Sosnoski DM; Mastro AM; Zheng SY, Small (Weinheim an der Bergstrasse, Germany) 2018, 14 (12), e1702787 DOI 10.1002/smll.201702787. [DOI] [PubMed] [Google Scholar]
- 59.Bersini S; Jeon JS; Dubini G; Arrigoni C; Chung S; Charest JL; Moretti M; Kamm RD, Biomaterials 2014, 35 (8), 2454–61. DOI 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hassell BA; Goyal G; Lee E; Sontheimer-Phelps A; Levy O; Chen CS; Ingber DE, Cell reports 2017, 21 (2), 508–516. DOI 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 61.Vidi PA; Maleki T; Ochoa M; Wang L; Clark SM; Leary JF; Lelievre SA, Lab Chip 2014, 14 (1), 172–7. DOI 10.1039/c3lc50819f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emon B; Bauer J; Jain Y; Jung B; Saif T, Comput Struct Biotechnol J 2018, 16, 279–287. DOI 10.1016/j.csbj.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahn J; Sei YJ; Jeon NL; Kim Y, Bioengineering (Basel, Switzerland) 2017, 4 (3), 64 DOI 10.3390/bioengineering4030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parente P; Parcesepe P; Covelli C; Olivieri N; Remo A; Pancione M; Latiano TP; Graziano P; Maiello E; Giordano G, Gastroenterology research and practice 2018, 2018, 7530619 DOI 10.1155/2018/7530619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Upadhyay S; Sharma N; Gupta KB; Dhiman M, Journal of cellular biochemistry 2018, 119 (7), 5028–5042. DOI 10.1002/jcb.26663. [DOI] [PubMed] [Google Scholar]
- 66.Biselli E; Agliari E; Barra A; Bertani FR; Gerardino A; De Ninno A; Mencattini A; Di Giuseppe D; Mattei F; Schiavoni G; Lucarini V; Vacchelli E; Kroemer G; Di Natale C; Martinelli E; Businaro L, Sci Rep 2017, 7 (1), 12737 DOI 10.1038/s41598-017-13070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Agliari E; Biselli E; De Ninno A; Schiavoni G; Gabriele L; Gerardino A; Mattei F; Barra A; Businaro L, Scientific Reports 2014, 4, 6639 DOI 10.1038/srep0663910.1038/srep06639https://www.nature.com/articles/srep06639#supplementary-informationhttps://www.nature.com/articles/srep06639#supplementary-information . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng Y; Sun Y; Yu X; Shao Y; Zhang P; Dai G; Fu J, Advanced healthcare materials 2016, 5 (9), 1014–24. DOI 10.1002/adhm.201501007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pisano M; Triacca V; Barbee KA; Swartz MA, Integrative biology : quantitative biosciences from nano to macro 2015, 7 (5), 525–33. DOI 10.1039/c5ib00085h. [DOI] [PubMed] [Google Scholar]
- 70.Mittal K; Ebos J; Rini B, Seminars in oncology 2014, 41 (2), 235–51. DOI 10.1053/j.seminoncol.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 71.Chaffer CL; Weinberg RA, Science (New York, N.Y.) 2011, 331 (6024), 1559–64. DOI 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 72.Yates C; Shepard CR; Papworth G; Dash A; Beer Stolz D; Tannenbaum S; Griffith L; Wells A, Advances in cancer research 2007, 97, 225–46. DOI 10.1016/s0065-230x(06)97010-9. [DOI] [PubMed] [Google Scholar]
- 73.Kim S; Chung M; Jeon NL, Biomaterials 2016, 78, 115–28. DOI 10.1016/j.biomaterials.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 74.Lee H; Park W; Ryu H; Jeon NL, Biomicrofluidics 2014, 8 (5), 054102 DOI 10.1063/1.4894595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zervantonakis IK; Hughes-Alford SK; Charest JL; Condeelis JS; Gertler FB; Kamm RD, Proceedings of the National Academy of Sciences of the United States of America 2012, 109 (34), 13515–13520. DOI 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen MB; Whisler JA; Frose J; Yu C; Shin Y; Kamm RD, Nature protocols 2017, 12 (5), 865–880. DOI 10.1038/nprot.2017.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeon JS; Bersini S; Gilardi M; Dubini G; Charest JL; Moretti M; Kamm RD, 2015, 112 (1), 214–219. DOI 10.1073/pnas.1417115112 %J Proceedings of the National Academy of Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Q; Liu T; Qin J, Lab Chip 2012, 12 (16), 2837–42. DOI 10.1039/c2lc00030j. [DOI] [PubMed] [Google Scholar]
- 79.Jeon JS; Zervantonakis IK; Chung S; Kamm RD; Charest JL, PLOS One 2013, 8 (2), e56910 DOI 10.1371/journal.pone.0056910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lei KF; Tseng HP; Lee CY; Tsang NM, Sci Rep 2016, 6, 25557 DOI 10.1038/srep25557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toh Y-C; Raja A; Yu H; van Noort D, Bioengineering (Basel, Switzerland) 2018, 5 (2), 29 DOI 10.3390/bioengineering5020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caballero D; Kaushik S; Correlo VM; Oliveira JM; Reis RL; Kundu SC, Biomaterials 2017, 149, 98–115. DOI 10.1016/j.biomaterials.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 83.Roussos ET; Condeelis JS; Patsialou A, Nat Rev Cancer 2011, 11 (8), 573–587. DOI 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aung A; Theprungsirikul J; Lim HL; Varghese S, Lab Chip 2016, 16 (10), 1886–98. DOI 10.1039/c6lc00184j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang R; Zheng W; Liu W; Zhang W; Long Y; Jiang X, Sci Rep 2015, 5, 17768 DOI 10.1038/srep17768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sart S; Tomasi RFX; Amselem G; Baroud CN, Nature Communications 2017, 8 (1), 469 DOI 10.1038/s41467-017-00475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi Y; Hyun E; Seo J; Blundell C; Kim HC; Lee E; Lee SH; Moon A; Moon WK; Huh D, Lab Chip 2015, 15 (16), 3350–7. DOI 10.1039/c5lc00514k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang H; Zhu Y; Shen Y, Small (Weinheim an der Bergstrasse, Germany) 2018, 14 (28), e1800360 DOI 10.1002/smll.201800360. [DOI] [PubMed] [Google Scholar]
- 89.Trietsch SJ; Naumovska E; Kurek D; Setyawati MC; Vormann MK; Wilschut KJ; Lanz HL; Nicolas A; Ng CP; Joore J; Kustermann S; Roth A; Hankemeier T; Moisan A; Vulto P, Nature Communications 2017, 8 (1), 262 DOI 10.1038/s41467-017-00259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kashaninejad N; Nikmaneshi MR; Moghadas H; Kiyoumarsi Oskouei A; Rismanian M; Barisam M; Saidi MS; Firoozabadi B, Micromachines (Basel) 2016, 7 (8), 130 DOI 10.3390/mi7080130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y; Zhou L; Qin L, Journal of the American Chemical Society 2014, 136 (43), 15257–15262. DOI 10.1021/ja5072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ashammakhi N; Elmusrati M, 2018, 273847 DOI 10.1101/273847 %J bioRxiv. [DOI] [Google Scholar]
- 93.Alpaydin E, Introduction to Machine Learning. The MIT Press: Cambridge, Massachusetts, 2016. [Google Scholar]
- 94.Kongadzem E Machine Learning Application: Organs-On-a-Chip in Parallel. Faculty of Technology and Innovations, University of Vassa, Vassa, Finland, 2018. [Google Scholar]