Abstract
Wearable, multisensor, consumer devices that estimate sleep are now commonplace, but the algorithms used by these devices to score sleep are not open source, and the raw sensor data is rarely accessible for external use. As a result, these devices are limited in their usefulness for clinical and research applications, despite holding much promise. We used a mobile application of our own creation to collect raw acceleration data and heart rate from the Apple Watch worn by participants undergoing polysomnography, as well as during the ambulatory period preceding in lab testing. Using this data, we compared the contributions of multiple features (motion, local standard deviation in heart rate, and “clock proxy”) to performance across several classifiers. Best performance was achieved using neural nets, though the differences across classifiers were generally small. For sleep-wake classification, our method scored 90% of epochs correctly, with 59.6% of true wake epochs (specificity) and 93% of true sleep epochs (sensitivity) scored correctly. Accuracy for differentiating wake, NREM sleep, and REM sleep was approximately 72% when all features were used. We generalized our results by testing the models trained on Apple Watch data using data from the Multi-ethnic Study of Atherosclerosis (MESA), and found that we were able to predict sleep with performance comparable to testing on our own dataset. This study demonstrates, for the first time, the ability to analyze raw acceleration and heart rate data from a ubiquitous wearable device with accepted, disclosed mathematical methods to improve accuracy of sleep and sleep stage prediction.
Keywords: sleep tracking, ambulatory sleep monitoring, machine learning, validation, mathematical modeling of sleep
Statement of Significance.
Use of consumer sleep trackers is widespread, but because the type of data returned from the devices is often proprietary (e.g. “Fitbit steps”) and the algorithms are typically trade secret, most are not used by the clinical and research communities. We wrote our own code to directly access the accelerometer on the Apple Watch. We then recorded raw acceleration, along with heart rate data as measured via photoplethysmography in the Apple Watch, during the night while subjects underwent the gold standard for sleep tracking, polysomnography. We compared the output of multiple classification algorithms to ground truth polysomnography to determine best performance. This sets the stage for greater transparency in the use of wearables to assess sleep on a large scale.
Introduction
An estimated 50–70 million individuals in the United States are impacted by sleep that is inadequate in duration or quality [1]. The negative effects of sleep loss are even more profound when the poor sleep quality or shortened sleep duration takes place on a chronic, daily basis, rather than as a singular disturbance. The gold standard for sleep measurement is the polysomnogram (PSG), which requires a sleep lab, sleep technician, and monitoring of multiple physiological parameters [2]. As such, polysomnography is generally restricted to the assessment of sleep for only one or two nights. Longitudinal, ambulatory sleep measurement can benefit a number of populations, including patients with suspected sleep disorders, workers in occupations where any impairment in alertness is high risk (i.e. transportation workers), and healthy individuals who desire improved sleep for maximal cognitive and physical performance and optimal health.
The current method accepted by the medical and scientific community for objective, longitudinal sleep measurement in the ambulatory setting is actigraphy [3, 4]. Actigraphy refers to the use of FDA-approved, wrist-worn accelerometry devices that measure movement to estimate sleep. A large body of peer-reviewed evidence has assessed performance of actigraphy against PSG. However, actigraphy has significant inadequacies that limit its use: actigraphs are expensive compared to the consumer sleep trackers which are already owned by millions of individuals, actigraphs record only movement, and they struggle to correctly classify wake events during the attempted sleep period [5–8].
Logistically, actigraphs typically require in-person set-up and data recovery (given the lack of Bluetooth or cloud capability of most platforms), and at least two contacts with a trained individual on the sleep medicine or research team are required. In addition, seamless integration is lacking between actigraphy software and the electronic health record or other platforms to manage health and wellness.
Consumer marketed wearables are a tempting solution to the problem of ambulatory sleep tracking given ease of use, widespread availability, measurement of multiple biological signals, low cost, and opportunity for integration with other health technology products. However, the minimal validation of consumer sleep trackers and their associated outputs against PSG has precluded use in clinical, research, and occupational settings [9–13].
Even when devices are validated against PSG once, both device firmware and associated software are frequently updated by the manufacturer. As algorithms that determine sleep metrics are rarely disclosed, such updates could make previous validation studies irrelevant. These barriers to validation and the lack of transparency surrounding the associated software’s sleep scoring methods have historically reduced enthusiasm for consumer marketed wearable use in medicine and research. Overcoming these barriers is of great interest, as a growing body of evidence has begun to reveal the potential clinical and research utility of commercially available products [14, 15].
The current generation of consumer marketed wearable devices that claim to measure sleep use multisensor data acquisition, typically microelectromechanical systems (MEMS) accelerometers and photoplethysmography (PPG) [16, 17]. MEMS accelerometers are the ubiquitous sensors used in mobile and wearable devices to measure motion and are widely validated for the assessment of physical activity and energy expenditure. Over the past decade, the technology underlying MEMS accelerometers has rapidly advanced and allowed for increased memory and battery capacity, wide acceleration range, minute size, and low cost. Importantly, raw acceleration signal can be extracted from MEMS accelerometers prior to processing by manufacturer algorithms [18]. PPG is an optical technique that measures blood volume changes which has been validated to accurately measure heart rate in multiple contexts [15, 19]. The utility of consumer-available PPG is underscored by recent FDA clearance of a mobile application that analyzes PPG signal acquired by the Apple Watch for over-the-counter use to evaluate for irregular heart rhythms [15, 20].
On top of the rapid progress in sensor development, technological advances have expanded our ability to analyze the vast amount of data they collect. Machine learning techniques and other advanced computational methods that make use of the current capabilities of computing power, memory, and storage to classify novel input data are well-suited for the prediction of sleep metrics from massive amounts of sensor acquired signals. Therefore, the weighted sum algorithms [21–25] that have formed the cornerstone of existing actigraphy software programs are likely to be outperformed by newer techniques.
Lastly, for over 40 years, mathematical models have described the biological properties of sleep-wake control. Specifically, sleep is governed by the well-described two-process model comprised of the circadian oscillator and homeostatic sleep drive [26, 27]. Homeostatic drive accumulates with prolonged wakefulness and is opposed by the mounting circadian alerting signal such that a consolidated period of wakefulness is maintained during the daytime [28, 29]. At night, conversely, the central circadian clock maintains a low alerting signal to promote consolidation of the nocturnal sleep period [28, 29]. Additionally, an ultradian cycle of alternating non-rapid eye movement (NREM) and rapid eye movement (REM) sleep stages is superimposed on the two-process model. These interactions have been studied at length and are built into more recent mathematical models of human sleep [30, 31].
Given the ability to numerically simulate such models of the circadian clock, one can consider predicted circadian phase over the course of the night as an additional input to a sleep/wake classification algorithm. With a sufficiently long window of recorded activity patterns, a circadian input can be estimated and provided as a feature alongside the traditionally incorporated measurements of motion and heart rate used in algorithms applied to wearable data.
Therefore, the primary goal of this study was to collect raw acceleration and heart rate data from the MEMS accelerometer and PPG housed within the Apple Watch and use modern classification methods to distinguish sleep from wake and determine sleep stages as compared to gold-standard PSG. The secondary goal was to assess how the incorporation of a “clock proxy” term that represents the changing circadian propensity for sleep over the night influenced performance across all classifiers. Finally, to generalize our algorithms beyond the Apple Watch accelerometer and PPG, models trained on our dataset were tested on an independently collected dataset from the Multi-ethnic Study of Atherosclerosis (MESA) cohort, which consists of motion data from actigraphy-derived activity counts and heart rate via pulse oximetry from co-recorded PSG.
Methods
Study protocol
Procedure.
After approval by the University of Michigan Institutional Review Board, 39 subjects were recruited for participation in the study. Written informed consent was obtained and an exclusion criteria questionnaire was used to ensure participants did not have a known diagnosis of the following: restless legs syndrome, sleep-related breathing disorders, insomnia, parasomnias, central disorders of hypersomnolence, cardiovascular disease (congenital heart disease, congestive heart failure, coronary artery disease, myocardial infarction, cardiac arrhythmias), peripheral vascular disease, vision impairment not correctable by glasses or contacts, or other disorders expected to result in significant neurological or psychiatric impairment. Individuals who participated in night shift work or transmeridian travel greater than two time zones within the month prior to enrollment were excluded. Significant excessive daytime sleepiness was ruled out by use of the Epworth Sleepiness Scale (ESS) to ensure participants did not score >10 (indicative of excessive daytime sleepiness) [32].
After enrollment, participants were provided with an Apple Watch (Apple Inc.) which was applied to the wrist and a mobile application developed by OW that contained a digital sleep diary and psychomotor vigilance test (results from which are not discussed here). On the final night of the 7- to 14-day ambulatory recording period, the patients presented to the University of Michigan Sleep and Chronophysiology Laboratory and underwent attended PSG. During the entirety of the PSG recording, subjects continued to wear the Apple Watch, and data was transmitted in real-time to servers housed at the University of Michigan. OW’s code for accessing the accelerometer and heart rate data in the Apple Watch is online at https://github.com/ojwalch/sleep_accel.
Subjects that demonstrated PSG findings suggestive of REM sleep behavior disorder (loss of normal REM atonia in the submentalis electromyogram lead combined with motor behaviors and vocalizations directly observed by the registered polysomnographic technologist [RPSGT] during stage REM sleep; 1 subject) or obstructive sleep apnea (apnea-hypopnea index of at least five per hour of sleep based on the respiratory event scoring described below; 3 subjects) were removed from analysis. The PSG records of excluded subjects were reviewed by a board-certified sleep medicine physician after RPSGT scoring. Four additional subjects were removed from the subject pool due to incomplete data. In cases where the battery on the Apple Watch failed before the sleep opportunity ended, the data was cropped to include only those time points for which valid data existed.
Ambulatory recording.
Apple Watch (Series 2 and 3, Apple Inc) devices were worn continuously during the 7- to 14-day ambulatory recording period with the exception of a nightly interruption to charge the device. The 7- to 14-day ambulatory recording period allowed for estimation of each subject’s daily activity patterns, which were used to generate predictions of circadian phase used as the “clock proxy” feature.
Laboratory PSG and Apple Watch recording.
Subjects underwent an 8-hour sleep opportunity monitored with PSG with lights out at the time of habitual bedtime. PSG was conducted in accordance with the technical specifications of the American Academy of Sleep Medicine (AASM) [2] with the exception of the oronasal thermistor and nasal pressure transducer. Bilateral frontal, central and occipital electroencephalogram (EEG) recorded with use of the International 10–20 system of electrode placement, bilateral electrooculogram (EOG) recorded from the supraorbital and infraorbital ridges, chin electromyogram (EMG), thoracic and abdominal respiratory inductance plethysmography (RIP) belts, snore microphone, pulse oximetry, and electrocardiogram (ECG) with use of two leads were recorded.
Electrophysiological signals for the first eight subjects were recorded on a Vitaport 3 (TEMEC Instruments B.V., The Netherlands) data acquisition system, while all others were recorded on a Grael HD-PSG/EEG Diagnostic Amplifier System using Compumedics Profusion SLEEP4 Online Acquisition and Analysis Software (Compumedics, USA Inc., Charlotte, NC). All data were digitized at 256 Hz and stored off-line for visual staging and scoring using standard AASM scoring criteria [2]. Given absent oronasal thermistor and nasal pressure transducer, respiratory inductance plethysmography sum (RIPsum) and dual thoracoabdominal RIP belts were used as alternative apnea and hypopnea sensors, respectively. Hypopnea rule 1A was used.
Concurrent to the monitoring of sleep via PSG, raw acceleration and heart rate were recorded from the Apple Watch and transmitted to a secure server. The Apple Watch uses a triaxial MEMS accelerometer that measures acceleration in the x, y, and z directions, in units of g (9.8 m/s2). Heart rate is measured by the Apple Watch with PPG on the dorsal aspect of the wrist. Raw acceleration signal and heart rate data are obtained from the device by creating a “Workout Session” and using functions built-in to the iOS WatchKit and HealthKit frameworks.
Analysis
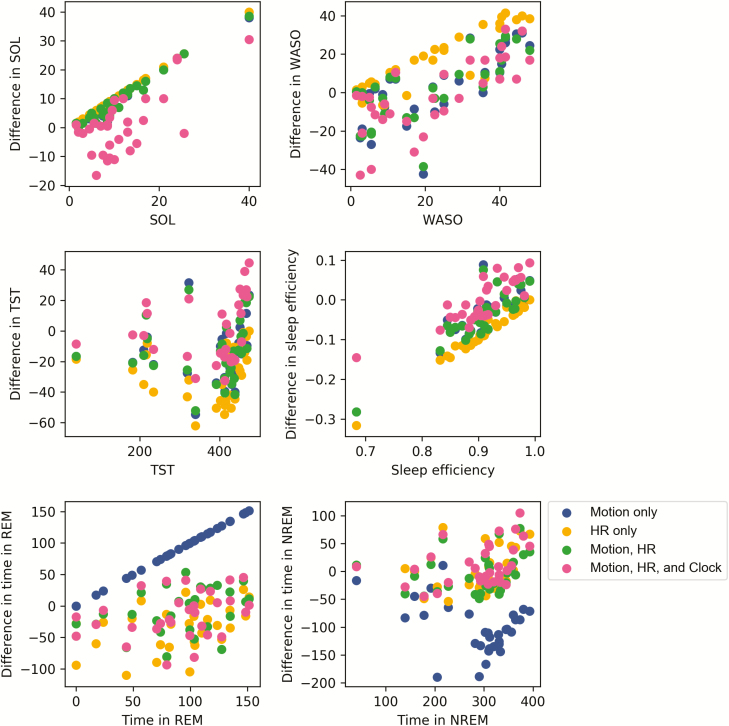
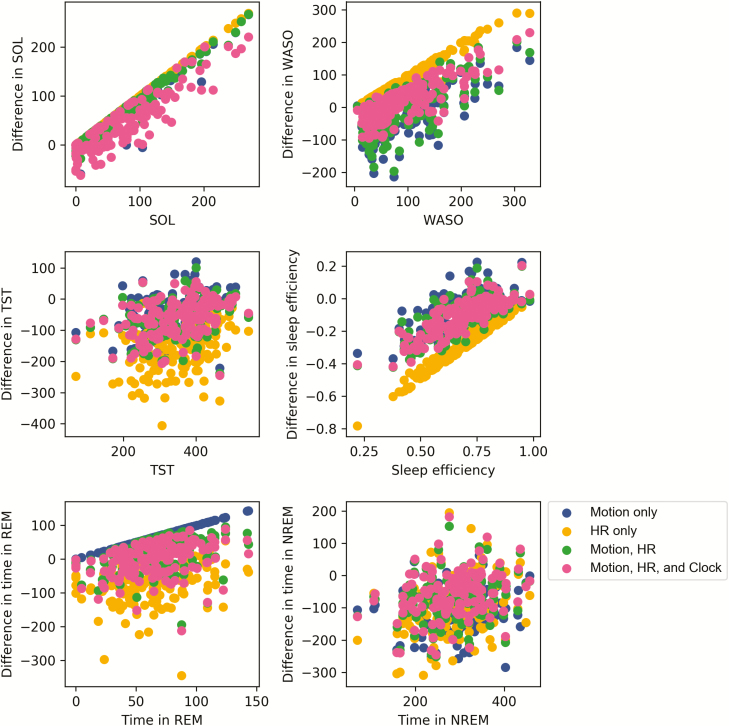
Summary PSG parameters (time in bed [TIB], total sleep time [TST], sleep onset latency [SOL], wake after sleep onset [WASO], sleep efficiency [SE], REM sleep minutes, and NREM sleep minutes) were assessed with descriptive methods. Bland-Altman plots were created to visualize agreement and heteroscedasticity (Figure 5) [33]. Epoch-by-epoch classifier output comparison to PSG is detailed below.
Figure 5.
Bland-Altman plots for TST (minutes), SOL (minutes), WASO (minutes), SE (fraction), stage REM sleep (minutes), and NREM sleep (minutes) as predicted by the neural net classifier. The differences in classifier-produced values versus PSG values are plotted on the y-axis and the corresponding ground truth PSG values are plotted on the x-axis. Sleep metrics were computed using the same fixed thresholds for wake and REM for all subjects.
Feature and algorithm selection.
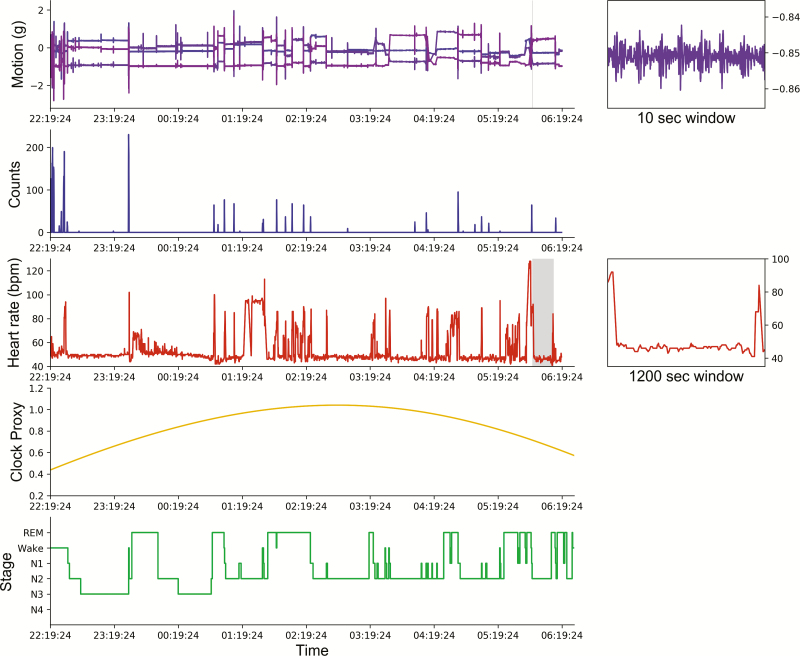
Three types of features were considered as inputs to the classification algorithms tested: motion (activity counts, converted from raw acceleration in m/s2 using the method outlined in [34]), heart rate, and a “clock proxy” term representing simulated input to sleep from the circadian clock. Every sample classified by the algorithms corresponds to a 30-second epoch scored during PSG. When classifying each 30-second epoch, features are cropped to a local window of 10 minutes around the scored epoch. Sample data for one subject’s PSG and Apple Watch recordings are shown in Figure 1.
Figure 1.
Sample data from one subject’s night of sleep. From top: Motion, raw acceleration from the Apple Watch microelectromechanical system accelerometer (in x, y, and z directions); Counts, acceleration processed as activity counts using code from [34]; Heart rate, heart rate from Apple Watch photoplethysmography; “Clock proxy,” predicted from ambulatory recording with Apple Watch; Stages, hypnogram from scored polysomnography. Insets on the right show zoomed-in version of the data on the left, with selected windows marked with gray overlays.
Motion feature.
Acceleration was returned from the Apple Watch as three vectors representing acceleration in the x, y, and z directions, and a fourth, representing the timestamp of the measurement in seconds since January 1, 1970 (UNIX or epoch time). The acceleration in each direction was returned in units of g. In general, data were sampled at approximately 50 Hz, with two exceptions: (1) motion was sampled at 20 Hz for the first two subjects due to battery life concerns and (2) occasionally, short windows of time with missing data would occur, likely due to server-side issues during the real-time sleep night data collection. Less than 3% of the total recording time was affected in this way, and interpolation was used to estimate counts during missing time points. Data from the first two subjects were included only after it was verified that doing so did not meaningfully change the results.
To make trained classifiers backwards compatible with historical data collection methods, we converted our raw acceleration data to activity counts using MATLAB code available online and validated in the work of te Lindert and colleagues. The final activity count feature was arrived at by convolving the window with a Gaussian (σ = 50 seconds).
Heart rate feature.
Heart rate was measured by PPG from the Apple Watch and returned in beats per minute sampled every several seconds. This signal was interpolated to have a value for every 1 second, smoothed and filtered to amplify periods of high change by convolving with a difference of Gaussians filter (σ 1 = 120 seconds, σ 2 = 600 seconds). Each individual was normalized by dividing by the 90th percentile in the absolute difference between each heart rate measurement and the mean heart rate over the sleep period. The standard deviation in the window around the scored epoch was used as the representative feature for heart rate. While this represents variation in heart rate, it is distinct from ECG-based definitions of heart rate variability.
“Clock proxy” and time-based feature.
By “clock proxy,” we refer to a feature meant to approximate the changing drive of the circadian clock to sleep over the course of the night. The clock-proxy feature was determined by two separate ways. The first way was to use a fixed cosine wave, shifted relative to the time of recording start, which rose and fell over the course of the night. This way of computing the clock proxy term is attractive because it only requires the time of recording as an input.
In an effort to incorporate longitudinal, personalized information about the subjects’ circadian clocks, such as their phase at the time of sleep onset, we also computed the clock proxy feature using well-validated mathematical model of the circadian clock [35]. Most models of the human circadian clock require light input in order to predict circadian phase. The Apple Watch does not currently allow developer access to a light sensor; however, it does allow access to steps data via HealthKit. To arrive at the clock proxy feature, steps data imported from the Apple Watch was used in place of light data, with the rationale that walking or running typically takes place in a lit environment. The imported steps data was used to infer a “typical” daily pattern of rest and activity, specific to each subject, and converted to estimated light using a simple steps-to-light function; specifically, if steps were above a threshold, the “light” was assumed to be one of three levels depending on the time of day: 50 lux between 10:00 pm and 07:00 am, 500 lux during the evening between 04:00 pm and 10:00 pm, 500 lux in the morning between 07:00 am and 10:00 am, and 1000 lux between 10:00 am and 4:00 pm. The normalized output from the model of the circadian clock could then be used as the estimated “clock proxy” feature.
The full circadian clock model predictions were used for the results presented in the main body of the manuscript. Differences between the full circadian clock model feature, the cosine feature, and a feature which is just time since recording onset (as employed in [36]) are described in the Supplementary Materials.
Algorithm training and selection.
Logistic regression, k-nearest neighbors, a random forest classifier, and a neural net (multilayer perceptron, MLP) were used as candidate models in our comparison of different classification algorithms. Pre-built tools from scikit-learn (version 0.20.3) [37] for Python (Python Software Foundation. Python Language Reference, version 3.7. Available at http://www.python.org) were used for each implementation. All code used to perform the analysis and generate the figures in this paper is available at https://github.com/ojwalch/sleep_classifiers. The hyperparameters searched for each classifier are provided in Supplementary Table S1.
Validation against PSG
Initially, all training and testing was done within the Apple Watch dataset. Classification of sleep stage (either sleep/wake or wake/NREM/REM) by each of the models considered was compared to PSG in an epoch-by-epoch analysis. Epochs were aligned with Apple Watch recordings using seconds since January 1, 1970 (UNIX) timestamps.
Models were trained and tested using both Monte Carlo cross-validation and leave-one-out cross-validation. For Monte Carlo cross-validation with sleep/wake classification, the dataset was randomly split 50 times into a training set (approximately 70% of the subjects) and a testing set (approx. 30%), and for wake/NREM/REM classification, the dataset was randomly split 20 times at the same training and testing proportions. In the leave-one-out cross-validation, a single subject was held out for testing, and the model was trained on the remaining subjects. No samples in the training set were ever used in the corresponding testing set, nor were samples from a single subject ever simultaneously used in both the training and testing sets. Parameters were tuned for each training dataset to minimize the risk of overfitting.
Using Monte Carlo cross-validation, the classification ability of each algorithm across all feature sets considered was summarized using receiver operating characteristic (ROC) curves and precision-recall curves. ROC curves are created by varying a threshold parameter and plotting the true positive and false-positive rates at all thresholds against each other [38]. An ROC curve presents all possible true and false-positive rates for the model, rather than a single true/false positive rate pair in isolation. Doing so allows flexibility in model creation: the choice of threshold can be driven by the relative importance of achieving highly accurate detection of sleep epochs versus highly accurate detection of wake epochs. Higher area under the ROC curve (AUC) suggests that the model is better able to distinguish classes.
Due to class imbalance between sleep and wake, precision-recall curves were also plotted with wake as the positive class. In this case, the recall (on the x-axis) is the fraction of wake epochs scored correctly, and the precision (on the y-axis) shows the fraction of all epochs labeled wake that were truly wake [39]. In this way, one can see how often the classifier labels epochs as the less frequent class erroneously across all thresholds.
Each ROC and precision-recall curve for sleep/wake classification using the Apple Watch dataset represents the average performance across all 50 training and testing sets, with new subdivisions of the data generated at each iteration. Likewise, each ROC curve for wake/NREM/REM classification represents the average performance across all 20 training and testing sets. To visualize performance of wake/NREM/REM classification, one versus rest plots were also created and included in the supplement, also with 20 training/testing splits.
Leave-one-out cross-validation was used to understand subject variability in classifier performance. From the results of training on all subjects but one and testing on the remaining subject, histograms of specificity, sensitivity, and accuracy across subjects were constructed.
Use of the MESA dataset
The National Sleep Research Resource (NSRR) provides access to the data from MESA, a multicenter longitudinal investigation of factors associated with the development of subclinical cardiovascular disease and the progression to clinical cardiovascular disease [40–42]. A diverse sample of 6814 black, white, Hispanic, and Chinese-American men and women were recruited for participation in 2000–2002. From 2010 to 2013, 2237 participants also were enrolled in a Sleep Exam (MESA Sleep) which included full overnight unattended PSG and 7-day wrist-worn actigraphy. For the purpose of this study, a subset of the data (188 subjects; chosen for computational feasibility) with co-recorded actigraphy and PSG data was extracted and processed for use as an independent testing set. Given the different data collection methodology, the motion and local standard deviation in heart rate features corresponded to direct activity counts from actigraphy and heart rate during PSG, respectively. Heart rate was derived from pulse oximetry, which uses PPG, increasing the comparability of the Apple Watch (training) and MESA (testing) set. The “clock proxy” feature was derived from the ambulatory actigraphy recording for each MESA participant.
Results
Demographic and summary PSG data
Summary sleep variables are provided for the 31 subjects (21 female) in Table 1. The average age of participants was 29.4 years (σ = 8.52 years).
Table 1.
Age and summary sleep statistics from the Apple Watch (PPG, MEMS)-PSG training set
| Parameter | Mean (SD) | Range |
|---|---|---|
| Age (years) | 29.42 (8.52) | 19.0–55.0 |
| TST (minutes) | 427.87 (38.87) | 318.5–474.0 |
| TIB (minutes) | 472.56 (27.03) | 373.0–490.0 |
| SOL (minutes) | 14.97 (10.1) | 2.0–44.0 |
| WASO (minutes) | 28.73 (22.8) | 2.0–92.0 |
| SE (%) | 90.48 (5.54) | 77.0–97.9 |
| Time in REM (minutes) | 107.15 (31.22) | 44.14–194.32 |
| Time in NREM (minutes) | 320.77 (39.11) | 227.47–393.3 |
REM, rapid eye movement sleep; NREM, non-rapid eye movement sleep.
Algorithm comparisons
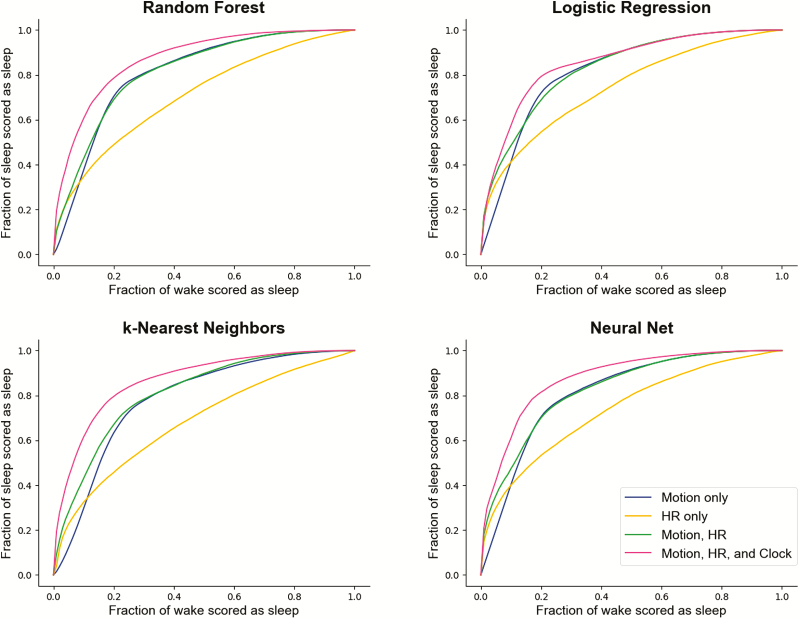
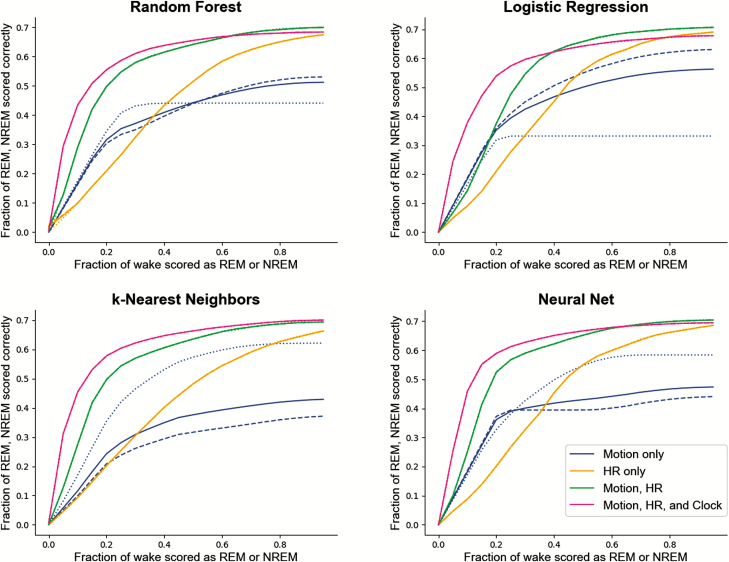
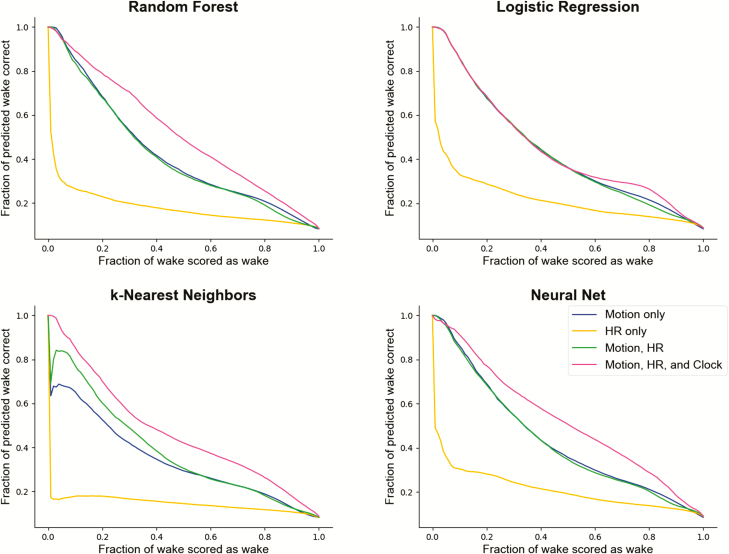
Across every algorithm surveyed, performance was best when all available features—motion, heart rate, and clock proxy—were used as inputs to the classifier. ROC curves summarizing the performance of each classifier for sleep/wake and sleep stage classification are shown in Figures 2 and 4. Precision-recall plots for wake classification in the sleep-wake classifier are shown in Figure 3. Bland-Altman plots to visualize the differences between classifier and PSG values (y-axis) versus PSG values (x-axis) were constructed for TST, SOL, WASO, SE, stage REM sleep, and NREM sleep (Figure 5). This plot was generated using fixed thresholds for wake and REM sleep . While a difference choice of fixed thresholds, or choosing a different threshold for each person using an additional hold-out set after training, would change this plot, it can still be used to identify inter-individual differences and show how the motion-only classifier struggles with distinguishing REM and NREM.
Figure 2.
ROC curves across multiple classifiers and features for differentiating sleep and wake. The x-axis represents the fraction of true wake epochs incorrectly classified as sleep and the y-axis represents the fraction of true sleep epochs correctly classified as sleep. ROC curves are generated by applying the full range of possible thresholds to the class probabilities assigned to each epoch by the classifiers.
Figure 4.
ROC curves across multiple classifiers and features for classifying wake/NREM/REM. Each point on the x-axis represents the fraction of wake epochs classified incorrectly, with any wake epoch classified either as NREM or REM sleep counting as a false positive. The y-axis summarizes REM and NREM accuracy rates. To choose a threshold for distinguishing REM and NREM sleep, a binary search was performed to find the value that minimized the difference between REM accuracy and NREM accuracy. In the case of the motion-only feature set, the dashed and dotted lines correspond to the REM and NREM accuracies (respectively), and the solid line is their average. For all others, the solid line represents the average of the REM and NREM accuracies, which could be made nearly identical through the choice of the appropriate threshold. NREM, non-rapid eye movement; REM, rapid eye movement.
Figure 3.
Precision-recall curves across multiple classifiers and features for differentiating sleep and wake. The x-axis represents the fraction of true wake epochs correctly classified as wake and the y-axis represents the fraction of all epochs labeled as wake by the classifier that were correct.
Performance metrics for sleep/wake classification across all classifiers surveyed are summarized in Tables 2–5. The fraction of true sleep epochs scored correctly (also referred to as sensitivity in the sleep literature, when sleep is treated as the positive class), the fraction of true wake epochs scored correctly (specificity), accuracy, AUC, and Cohen’s kappa values were determined every time the model was tested (on a reserved portion of the data not used for training), and averaged across trials. Similar performance metrics for wake/NREM/REM classification are in Table 6.
Table 2.
Sleep/wake differentiation performance by logistic regression across different feature inputs in the Apple Watch (PPG, MEMS) dataset
| Accuracy | Wake correct (specificity) | Sleep correct (sensitivity) | κ | AUC | |
|---|---|---|---|---|---|
| Motion | 0.794 | 0.725 | 0.8 | 0.277 | 0.819 |
| 0.871 | 0.549 | 0.9 | 0.343 | ||
| 0.892 | 0.476 | 0.93 | 0.361 | ||
| 0.905 | 0.415 | 0.95 | 0.367 | ||
| HR | 0.776 | 0.512 | 0.8 | 0.174 | 0.743 |
| 0.852 | 0.326 | 0.9 | 0.187 | ||
| 0.875 | 0.266 | 0.93 | 0.191 | ||
| 0.889 | 0.215 | 0.95 | 0.182 | ||
| Motion, HR | 0.792 | 0.707 | 0.8 | 0.269 | 0.83 |
| 0.87 | 0.546 | 0.9 | 0.341 | ||
| 0.892 | 0.475 | 0.93 | 0.36 | ||
| 0.905 | 0.417 | 0.95 | 0.369 | ||
| Motion, HR, and Clock Proxy | 0.8 | 0.798 | 0.8 | 0.31 | 0.854 |
| 0.871 | 0.556 | 0.9 | 0.347 | ||
| 0.892 | 0.471 | 0.93 | 0.358 | ||
| 0.905 | 0.411 | 0.95 | 0.364 |
Fraction of wake correct, fraction of sleep correct, accuracy, κ, and AUC for sleep-wake predictions of logistic regression with use of motion, HR, clock proxy, or combination of features. HR, heart rate.
Table 3.
Sleep/wake differentiation performance by k-nearest neighbors across different feature inputs in the Apple Watch (PPG, MEMS) dataset
| Accuracy | Wake correct (specificity) | Sleep correct (sensitivity) | κ | AUC | |
|---|---|---|---|---|---|
| Motion | 0.789 | 0.672 | 0.8 | 0.255 | 0.803 |
| 0.866 | 0.483 | 0.9 | 0.3 | ||
| 0.887 | 0.405 | 0.93 | 0.307 | ||
| 0.9 | 0.345 | 0.95 | 0.307 | ||
| HR | 0.768 | 0.406 | 0.8 | 0.117 | 0.682 |
| 0.845 | 0.237 | 0.9 | 0.117 | ||
| 0.868 | 0.172 | 0.93 | 0.103 | ||
| 0.882 | 0.12 | 0.95 | 0.082 | ||
| Motion, HR | 0.79 | 0.678 | 0.8 | 0.255 | 0.81 |
| 0.867 | 0.496 | 0.9 | 0.308 | ||
| 0.889 | 0.431 | 0.93 | 0.327 | ||
| 0.903 | 0.38 | 0.95 | 0.338 | ||
| Motion, HR, and Clock Proxy | 0.8 | 0.797 | 0.8 | 0.309 | 0.868 |
| 0.877 | 0.627 | 0.9 | 0.391 | ||
| 0.897 | 0.535 | 0.93 | 0.404 | ||
| 0.909 | 0.458 | 0.95 | 0.402 |
Fraction of wake correct, fraction of sleep correct, κ, and AUC for sleep-wake predictions of k-nearest neighbor classifier with use of motion, HR, clock proxy, or combination of features. HR, heart rate.
Table 4.
Sleep/wake differentiation performance by random forest classifier across different feature inputs in the Apple Watch (PPG, MEMS) dataset
| Accuracy | Wake correct (specificity) | Sleep correct (sensitivity) | κ | AUC | |
|---|---|---|---|---|---|
| Motion | 0.793 | 0.713 | 0.8 | 0.27 | 0.81 |
| 0.869 | 0.53 | 0.9 | 0.329 | ||
| 0.891 | 0.457 | 0.93 | 0.346 | ||
| 0.904 | 0.399 | 0.95 | 0.352 | ||
| HR | 0.771 | 0.454 | 0.8 | 0.142 | 0.708 |
| 0.849 | 0.282 | 0.9 | 0.152 | ||
| 0.872 | 0.221 | 0.93 | 0.149 | ||
| 0.886 | 0.174 | 0.95 | 0.14 | ||
| Motion, HR | 0.792 | 0.707 | 0.8 | 0.267 | 0.816 |
| 0.869 | 0.519 | 0.9 | 0.322 | ||
| 0.89 | 0.448 | 0.93 | 0.339 | ||
| 0.904 | 0.394 | 0.95 | 0.349 | ||
| Motion, HR, and Clock Proxy | 0.799 | 0.789 | 0.8 | 0.303 | 0.871 |
| 0.879 | 0.653 | 0.9 | 0.405 | ||
| 0.901 | 0.579 | 0.93 | 0.433 | ||
| 0.914 | 0.513 | 0.95 | 0.444 |
Fraction of wake correct, fraction of sleep correct, accuracy, κ, and AUC for sleep-wake predictions of random forest classifier with use of motion, HR, clock proxy, or combination of features. HR, heart rate.
Table 5.
Sleep/wake differentiation performance by neural net across different feature inputs in the Apple Watch (PPG, MEMS) dataset
| Accuracy | Wake correct (specificity) | Sleep correct (sensitivity) | κ | AUC | |
|---|---|---|---|---|---|
| Motion | 0.793 | 0.714 | 0.8 | 0.276 | 0.815 |
| 0.87 | 0.542 | 0.9 | 0.342 | ||
| 0.891 | 0.467 | 0.93 | 0.358 | ||
| 0.904 | 0.408 | 0.95 | 0.364 | ||
| HR | 0.775 | 0.506 | 0.8 | 0.174 | 0.737 |
| 0.851 | 0.323 | 0.9 | 0.187 | ||
| 0.874 | 0.263 | 0.93 | 0.19 | ||
| 0.887 | 0.208 | 0.95 | 0.177 | ||
| Motion, HR | 0.792 | 0.707 | 0.8 | 0.272 | 0.828 |
| 0.868 | 0.528 | 0.9 | 0.333 | ||
| 0.89 | 0.461 | 0.93 | 0.353 | ||
| 0.904 | 0.408 | 0.95 | 0.364 | ||
| Motion, HR, and Clock Proxy | 0.801 | 0.816 | 0.8 | 0.322 | 0.878 |
| 0.881 | 0.675 | 0.9 | 0.424 | ||
| 0.901 | 0.596 | 0.93 | 0.449 | ||
| 0.913 | 0.523 | 0.95 | 0.455 |
Fraction of wake correct, fraction of sleep correct, accuracy, κ, and AUC for sleep-wake predictions of neural net classifier with use of motion, HR, clock proxy, or combination of features. HR, heart rate.
Table 6.
Sleep stage classification accuracy across different features and classifiers in the Apple Watch (PPG, MEMS) dataset
| Wake correct | NREM correct | REM correct | Best accuracy | κ | ||
|---|---|---|---|---|---|---|
| Logistic regression | Motion | 0.6 | 0.506 | 0.332 | 0.71 | 0.085 |
| HR | 0.6 | 0.452 | 0.453 | 0.698 | 0.033 | |
| Motion, HR | 0.6 | 0.625 | 0.625 | 0.701 | 0.161 | |
| Motion, HR, Clock | 0.6 | 0.623 | 0.623 | 0.699 | 0.13 | |
| k-Nearest neighbors | Motion | 0.6 | 0.294 | 0.532 | 0.698 | 0.072 |
| HR | 0.6 | 0.402 | 0.402 | 0.671 | 0.108 | |
| Motion, HR | 0.6 | 0.607 | 0.605 | 0.711 | 0.227 | |
| Motion, HR, Clock | 0.6 | 0.648 | 0.647 | 0.721 | 0.243 | |
| Random forest | Motion | 0.6 | 0.397 | 0.441 | 0.702 | 0.075 |
| HR | 0.6 | 0.434 | 0.434 | 0.676 | 0.165 | |
| Motion, HR | 0.6 | 0.615 | 0.615 | 0.695 | 0.293 | |
| Motion, HR, Clock | 0.6 | 0.638 | 0.638 | 0.686 | 0.302 | |
| Neural net | Motion | 0.6 | 0.394 | 0.498 | 0.713 | 0.084 |
| HR | 0.6 | 0.454 | 0.454 | 0.698 | 0.04 | |
| Motion, HR | 0.6 | 0.622 | 0.622 | 0.723 | 0.256 | |
| Motion, HR, Clock | 0.6 | 0.651 | 0.65 | 0.723 | 0.277 |
Performance metrics for wake/NREM/REM classification across multiple classifiers with use of motion, HR, clock proxy, or combination of features. NREM and REM Correct refer to the fraction of NREM and REM sleep epochs scored correctly when a threshold is chosen so they are as close as possible, while maintaining the fraction of correctly scored wake epochs at 0.6. Best accuracy refers to the highest accuracy found during the threshold search, and κ is the Cohen’s kappa for that accuracy. HR, heart rate.
Sleep/wake classification
In the case of binary sleep/wake classification, local heart rate standard deviation by itself (without motion) was consistently the lowest performing feature set for the classifiers, scoring roughly 24%–33% of wake epochs correctly (specificity) when the fraction of sleep epochs scored correctly (sensitivity) was fixed at 90% across classifiers. The motion-only feature set identified 48%–55% of wake epochs correctly when the fraction of correct sleep epochs was fixed at 90%.
Combining motion and heart yielded few improvements to sleep/wake classification over motion-only for binary sleep/wake classification (adding only roughly 3% to the fraction of wake scored correctly in k-nearest neighbors at the 95% threshold for the fraction of sleep epochs scored correctly). The inclusion of the clock proxy improved the fraction of wake epochs scored correctly by about 14% (when the fraction of sleep epochs scored correctly was fixed at 90%) when added to motion and heart rate in both the random forest and neural net classifiers.
AUC is greatest when all three features are considered and a neural net is used as the classifier (AUC = 0.878). The differences between the types of classifiers, however, are much less pronounced than those between choices of feature sets. For instance, the AUC of the logistic regression classifier for all features is 0.854, roughly 3% lower than the AUC of the neural net classifier trained on all features, while the difference between AUC for the heart rate-only versus motion-only logistic regression classifiers is approximately 10%.
Wake/NREM/REM classification
Two different approaches were employed for the analysis of the wake/NREM/REM classifier performance: traditional ROC curves, and one versus rest ROC curves.
Typically, ROC curves are generated for binary classification problems. In cases where there is more than one class, as in wake/NREM/REM classification, the definition of “true positive” on the y-axis is ambiguous; therefore, one versus rest ROC curves for each class were also used; that is, wake versus not wake, REM versus not REM, and NREM versus not NREM. This reduces the classification problem to a binary one. These plots are shown in Supplementary Figures S1S2S3.
Additional ROC curves are found in Figure 4 and summarize the performance in all three classes by replacing “true positive” with the accuracy where REM and NREM performance is (approximately) equal. These multi-class staging ROC curves were generated by applying two thresholds to the probabilities returned from the classifier. The first was applied to achieve a desired wake false positive rate; i.e. the fraction of wake epochs scored incorrectly, either as REM or NREM sleep.
For those epochs not scored as wake under that threshold, a second threshold was chosen for the REM and NREM class probabilities that made their respective accuracies (i.e. fraction of each class classified correctly) as close to equal as possible. It is important to note that choosing these thresholds requires knowledge of ground truth classifications; thus, these plots should be taken only as an exploration of model properties when ground truth is known.
This process was repeated for a spread of desired wake false-positive rates ranging from 0 to 1 in steps of 0.05 in order to achieve full coverage along the x-axis of Figure 4. In every case but for classifiers trained on the motion-only feature set, it was possible to choose a threshold that made the REM and NREM accuracies essentially equal (the dotted and dashed lines in Figure 4 show the NREM and REM accuracies, with the solid line showing their average).
Choosing thresholds that make the fractions of NREM and REM sleep classified correctly approximately equal does not generally yield the highest accuracy. This occurs because more time is spent in NREM sleep than in REM sleep in a typical night, and as such, the fraction of NREM sleep classified correctly is proportionally more important to accuracy than the fraction of REM sleep classified correctly. Table 6 includes the highest accuracy values found during the threshold search, along with their corresponding κ values.
Motion by itself is the weakest predictor of NREM and REM. The average of the REM and NREM accuracies for motion (solid blue lines in Figure 4) is lower than other feature sets, with either the fraction of REM sleep scored correctly (dashed line) or NREM sleep scored correctly (dotted line) being extremely low. Decreasing the threshold for one class does not fix this, as the accuracy of the other falls rapidly in response.
Heart rate, while only improving performance minimally over motion alone in sleep/wake classification, plays a much more significant role in wake/NREM/REM classification (Figure 4). With the inclusion of heart rate, it was possible to change thresholds without experiencing dramatic changes in the NREM and REM accuracies, as occurred with the motion-only feature set. Heart rate furthermore improved the NREM/REM accuracy (found by choosing the threshold that makes them approximately equal) by 15%–25% across classifiers when included as a feature on top of motion.
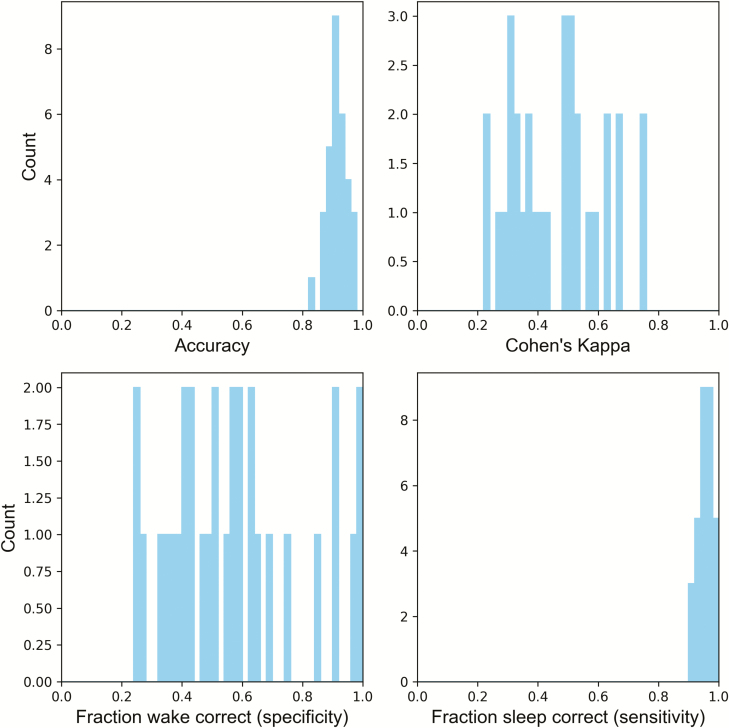
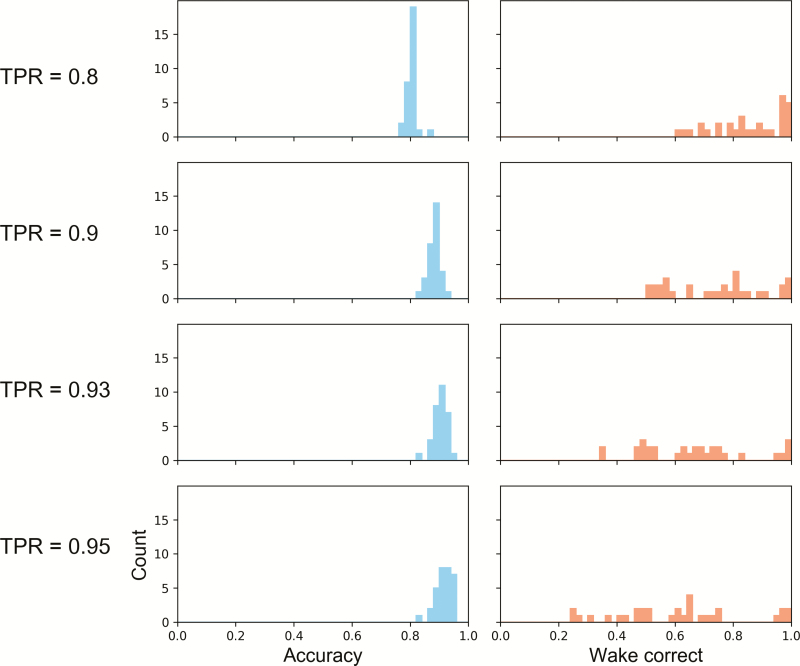
Individual performance
Variability in performance between subjects is visualized by the histograms in Figures 6 and 7. In these histograms, one subject is omitted while the rest are used to train a neural net classifier. In Figure 6, the same fixed threshold that the wake probability must exceed for an epoch to be counted as wake is used for all subjects. In Figure 7, the threshold is chosen so that the fraction of sleep epochs scored as sleep meets the “true positive rate” values specified for each row.
Figure 6.
Histograms of performance when training on all subjects but one, and testing on the omitted subject using the neural net classifier. For all tested subjects’ sleep nights, an epoch was counted as wake if its probability exceeded 0.3 .
Figure 7.
Histograms of performance when training on all subjects and testing on one subject who was omitted from training. The two performance measures plotted are accuracy and specificity. Here, specificity refers to the fraction of wake epochs scored correctly as wake. Each row corresponds to a fixed “true positive rate” (TPR), referring to the fraction of sleep epochs scored correctly as sleep. For each row, the threshold separating sleep and wake was chosen to match the fixed true positive rate. As the required true positive rate increases, the likelihood of a wake epoch being scored as sleep increases; hence, there is a skew towards lower values in the specificity histograms as TPR increases.
Algorithm testing in MESA dataset
Models for each classifier were trained using all subjects from the Apple Watch dataset, saved as files, and used to test unseen data from the MESA subcohort with co-recorded actigraphy and PSG. Summary sleep variables are summarized from the 188 subjects (90 female) of the MESA testing set in Table 7. The average age of participants was 68.78 years (σ = 8.81).
Table 7.
Age and summary sleep statistics from the MESA (pulse oximetry, actigraphy)-PSG testing set
| Parameter | Mean (SD) | Range |
|---|---|---|
| Age (years) | 68.82 (8.81) | 56.0–89.0 |
| TST (minutes) | 356.33 (87.56) | 70.0–591.0 |
| TIB (minutes) | 470.54 (85.18) | 199.0–770.0 |
| WASO (minutes) | 91.6 (59.73) | 4.5–303.5 |
| SE (%) | 75.68 (13.08) | 27.24–98.38 |
| AHI | 17.57 (16.2) | 0.0–78.7 |
AHI, apnea-hypopnea index.
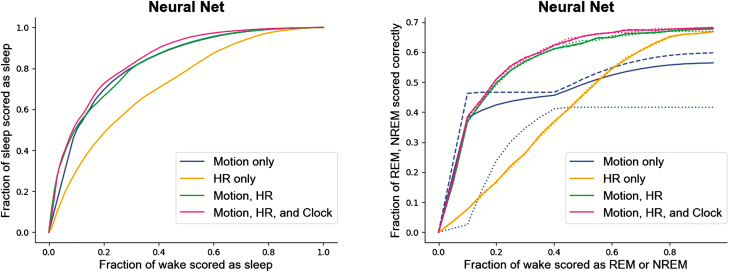
In Figure 8, ROC curves are shown comparing performance of the neural net model against PSG in the MESA subcohort with different feature sets, for both sleep/wake (A), and wake/NREM/REM (B) classification. As in the Apple Watch dataset, including more features improves the ability of the model to differentiate sleep from wake, with heart rate-only yielding the weakest performance for sleep/wake classification.
Figure 8.
MESA dataset (n = 188) validation of neural net models trained on the Apple Watch-PSG dataset. Left) Sleep/wake differentiation performance in MESA dataset Right) Wake/NREM/REM classifier performance in MESA dataset. The dashed lines represent REM accuracy for the motion-only feature set; the dotted lines represent NREM accuracy for the motion-only feature set. For all, solid lines represent the average of the NREM and REM accuracies, chosen to be as close to equal as possible via threshold selection. NREM, non-rapid eye movement; REM, rapid eye movement.
The neural net sleep/wake classifier, trained using all features on the entirety of the Apple Watch data set and tested on the MESA subcohort, scored 60% of wake epochs correctly, 90% of sleep epochs correctly, and demonstrated a Cohen’s Kappa (κ) of 0.525 and an area under the ROC curve of 0.845. The wake/NREM/REM neural net classifier achieved a best accuracy of 69%, and a corresponding κ of 0.4. Performance metrics for testing the neural net classifier with the MESA cohort are provided in Tables 8 and 9, and Bland-Altman plots for sleep metrics using a neural net model trained on the Apple Watch-PSG dataset and tested on the MESA dataset are presented in Figure 9.
Table 8.
Sleep/wake differentiation performance by the neural net classifier across different feature inputs in the MESA dataset
| Accuracy | Wake correct (specificity) | Sleep correct (sensitivity) | κ | AUC | |
|---|---|---|---|---|---|
| Motion | 0.768 | 0.702 | 0.8 | 0.486 | 0.822 |
| 0.785 | 0.543 | 0.9 | 0.473 | ||
| 0.783 | 0.472 | 0.93 | 0.447 | ||
| 0.777 | 0.413 | 0.95 | 0.416 | ||
| HR | 0.699 | 0.487 | 0.8 | 0.294 | 0.718 |
| 0.726 | 0.359 | 0.9 | 0.292 | ||
| 0.729 | 0.304 | 0.93 | 0.273 | ||
| 0.729 | 0.262 | 0.95 | 0.254 | ||
| Motion, HR | 0.767 | 0.697 | 0.8 | 0.482 | 0.827 |
| 0.786 | 0.546 | 0.9 | 0.476 | ||
| 0.785 | 0.477 | 0.93 | 0.452 | ||
| 0.78 | 0.42 | 0.95 | 0.423 | ||
| Motion, HR, and Clock Proxy | 0.774 | 0.72 | 0.8 | 0.501 | 0.845 |
| 0.803 | 0.599 | 0.9 | 0.525 | ||
| 0.805 | 0.542 | 0.93 | 0.514 | ||
| 0.803 | 0.493 | 0.95 | 0.495 |
Fraction of wake correct, fraction of sleep correct, accuracy, κ, and AUC for sleep-wake predictions of neural net classifier with use of motion, HR, clock proxy, or combination of features. HR, heart rate.
Table 9.
Sleep stage classification accuracy across different features by the neural net classifier in the MESA dataset
| Wake correct | NREM correct | REM correct | Best accuracy | κ | |
|---|---|---|---|---|---|
| Motion | 0.6 | 0.466 | 0.411 | 0.668 | 0.352 |
| HR | 0.6 | 0.37 | 0.364 | 0.624 | 0.243 |
| Motion, HR | 0.6 | 0.611 | 0.609 | 0.667 | 0.372 |
| Motion, HR, Clock | 0.6 | 0.622 | 0.625 | 0.686 | 0.403 |
Figure 9.
Quantifying sleep metrics performance in the MESA dataset (n = 188) using a model trained on the Apple Watch-PSG dataset. Bland-Altman plots for TST (minutes), SOL (minutes), WASO (minutes), SE (fraction), stage REM sleep (minutes), and NREM sleep (minutes) as classified by a neural net classifier. The differences in classifier values versus PSG values are plotted on the y-axis (actual - predicted) and the corresponding ground truth PSG values are plotted on the x-axis. Sleep metrics were computed using the same fixed thresholds for wake and REM for all subjects.
Discussion
This study demonstrates, for the first time, the ability of a widely used consumer wearable device to estimate sleep stages using investigator, as opposed to manufacturer, developed algorithms and the generalizability of these algorithms to data collected by traditional methods. Compared to PSG, our neural net model applied to Apple Watch-derived heart rate, motion, and a computed circadian estimate demonstrated sleep/wake differentiation with 93% of true sleep epochs scored correctly and 60% of true wake epochs scored correctly and REM-NREM sleep stage differentiation accuracy of 72%. Although various consumer marketed wearable devices that employ MEMS accelerometers and PPG have been compared to PSG, these validation studies are dependent entirely on preprocessed outputs from the manufacturers’ proprietary algorithms. Our study is novel in our use of raw motion and heart rate data, readily accessible from the Apple Watch, to develop and optimize sleep stage estimation algorithms and in doing so disclose our methodology. Additionally, we incorporate a clock proxy term as a feature based on the a priori knowledge of sleep-wake regulation. Finally, we show the generalizability of our algorithms by testing them on a dataset (MESA) collected via entirely different means in a population with significantly different demographics.
Comparison to consumer wearable devices
Our algorithm differentiated sleep from wake with an accuracy of 90% and a specificity (true wake epochs scored correctly) of 60% (when at the 93% sensitivity threshold). Therefore, our results are similar to previously reported performance of actigraphy and in line with past work validating consumer wearable devices that estimate sleep with proprietary algorithms (see [43] for a comprehensive review).
When specifically comparing our findings to the performance of current generation, off-the-shelf consumer wearable devices that use PPG and accelerometry, reported sensitivity and specificity are similar. The FitBit Charge HR, FitBit Charge 2, Jawbone UP3, and FitBit Alta HR have all been validated in epoch-by-epoch analyses against PSG in adolescents and adults [12, 44–46]. The sensitivity (fraction of true sleep epochs scored as sleep) reported in these investigations ranged from 95% to 97% and specificity (fraction of true wake epochs scored as wake) was reported at 39%–62%.
For sleep stage prediction, the Fitbit Charge 2 was able to achieve 81% accuracy for stage N1+N2, 49% accuracy for N3, and 74% accuracy for stage REM [44]. Cook and colleagues evaluated the accuracy of the Jawbone UP3 in a group with suspected disorders of central hypersomnolence and found accuracy of 56% for N1 + N2, 82% for N3, and 72% for REM [45]. The same group evaluated the FitBit Alta HR in the same patient population and found accuracy of 73% for N1 + N2, 89% for N3, and 89% for stage REM [46]. It is important to note is that the N1 + N2, N3, and REM estimation performance values noted above are simply agreement between device output and PSG without taking into account the potential for this agreement to occur by chance as would be reflected by the kappa statistic.
Our classifier is able to achieve an accuracy of 72% for the three-stage classifier, with balanced class accuracies (where a threshold is chosen so the REM accuracy equals the NREM accuracy) occurring at roughly 65% accuracy for each class. Our classifiers generally perform worse than the consumer wearable devices described above. One reason for this could be that additional processing, beyond real-time individual epoch classification, could be employed in these algorithms to improve results; e.g. choosing thresholds to match appropriate percentages of time spent in each stage of sleep. Such an approach would likely improve sleep/wake classification in the general population, while worsening it populations of atypical sleepers. Further, given that these validation studies are limited by manufacturer preprocessed data and undisclosed algorithms, the reported performance from a single study remain relevant only to that specific device, firmware, and software iteration and cannot be replicated for vigor or generalized to similar wearables. Finally, because the comparator studies are purely validation of the proprietary, undisclosed algorithm output, we are unable to draw conclusions regarding the candidate causes of the discrepancies in performance.
To our knowledge, there are only two published studies that, similar to our work, extracted raw signal from MEMS accelerometer and PPG sensors to develop, optimize, and validate sleep-wake scoring algorithms.
Fonseca and colleagues [16] trained ECG heart rate variability based sleep-wake scoring and sleep staging algorithms on the SIESTA dataset and validated the performance of these algorithms, applied to wrist-worn PPG and accelerometer signal, against PSG on an independent testing set. The final selected algorithm was found to yield a sensitivity to wake of 58%, and accuracy of 92% and a Cohen’s kappa (κ) of 0.55. For three classes (wake, NREM and REM), the classifier achieved a κ of 0.46 and accuracy of 73% while the four class (wake, N1+N2, N3, and REM) classifier, demonstrated a κ of 0.42 and accuracy of 59%.
Beattie and colleagues trained and validated algorithms with use of raw motion and PPG signal (from the FitBit device) co-recorded with at home PSG [47]. In the epoch-by-epoch analysis, the fraction of wake epochs correctly identified as wake (specificity) was 69% and the fraction of sleep epochs correctly identified as sleep (sensitivity) was 95%. Cohen’s kappa of the four-class classifier was 0.52.
Validation within our PPG-MEMS accelerometer (Apple Watch) dataset demonstrated sleep-wake accuracy (90%), specificity (fraction of true wake epochs scored correctly, 60%) and sensitivity (fraction of true sleep epochs scored correctly, 93%) that was similar to Fonseca and colleagues and approaching that of Beattie and colleagues; however, our κ was somewhat worse at 0.455. Importantly, in the more difficult problem of three-class sleep stage classification (wake, NREM, and REM) although our accuracy of 72% was similar to that of [16] and [47] our best κ of 0.3 was markedly worse than the κ values reported in both studies. It is possible that this could be due to the sampling rate of the Apple Watch heart rate via PPG (every 8–10 seconds), due to differences in the collection of heart rate and motion collected via the Apple Watch versus other wearables, or our own choice of parameters in the classification. Our hope is that in making all data and code open source, other groups can use the same data we have to improve upon our results. Interestingly, in our independent validation of our algorithm on the MESA dataset, κ for sleep-wake and three-class sleep staging were 0.525 and 0.4 respectively; the potential explanation of the improved performance on the MESA dataset is explored later in the discussion.
Implications of classifier and feature selection
We surveyed four different classifiers in this work: logistic regression, k-nearest neighbors, random forest, and neural nets. While the classifier methods differ in their ability to distinguish wake from sleep and differentiate sleep stages, these differences are not particularly pronounced. However, feature inclusion significantly impacts performance. As an example, the AUC for all classifiers is significantly increased when heart rate and motion are taken together, versus heart rate alone. Moreover, the inclusion of a feature that exploits the known circadian control of sleep using longitudinal data measurably improves performance: fixing the percentage of sleep epochs scored correctly at 90%, the percentage of wake epochs scored correctly in the MESA dataset increases 5% when the estimated circadian phase (“clock proxy”) is included as a feature.
Use of the clock proxy described above is a step towards integrating machine learning predictions with a priori knowledge of the physiology of human sleep. We calculated the circadian input in two ways, as a fixed cosine wave, shifted relative to the time of recording start and with a well-validated mathematical model of the circadian clock [35] that takes into account the longitudinal activity to compute estimated circadian phase. We chose to use the second model as our final approach in computing the clock proxy to include more personalized information about the individual’s circadian state (for instance, if they are trying to fall asleep at too early a phase). The inherent way individuals typically use wearable devices, wearing the device daily for extended durations, provides the opportunity to include long term ambulatory data as an input to sleep-wake estimation. The disadvantage of this computation is that it did require reliance on Apple’s proprietary steps calculation function, which reduces transparency.
Apart from our use of the clock proxy to assist with sleep-wake scoring many possible extensions of this idea exist. The sleep homeostat is not included in our predictions, nor are known feedback mechanisms between REM, NREM, and wake-promoting parts of the brain. To further integrate derived physical models and statistical predictions, the exchange of information would need to be bi-directional. For example, consider the sleep homeostat, which decreases during sleep and increases during wake. If the classifier is highly confident that the subject is awake, and thus, that the homeostat is increasing rather than decreasing, this information could be used to affect classification of epochs later in the night. A generalization of combining mathematical modeling with statistical methods of classifying sleep could be to incorporate model predictions that change in response to information from the classifier; e.g. using differential equations with a Kalman filter. These methods may be considered in future work to monopolize on the known biological properties of sleep as inputs for improved algorithm performance.
Generalizability
In addition to our goal of estimating sleep from consumer-available sensors in a transparent manner with validation against PSG, we wanted to ensure our work was generalizable and device agnostic. Therefore, we used a method described by te Lindert and Van Someren to convert MEMS accelerometer signal into the activity counts used by traditional actigraphy. The conversion provided by te Lindert and Van Someren has vast implications for sleep medicine and research. Firstly, as they note, backward compatibility confers the ability to pool different cohorts and analyze the objective, longitudinal sleep data with the same algorithms, regardless of the device from which the data was derived. Further, given the rapid expansion in computing power and data storage, the ability to convert between raw MEMS accelerometer signal and activity counts provides a greater wealth of wrist-worn motion data collected alongside ground truth PSG. Investigators may, therefore, use previously collected actigraphy and PSG data from well-established cohorts such as those contained within the NSRR, to develop and test algorithms that can be applied to the current generation of wearable devices.
We used their code to ensure that our algorithms, developed from Apple Watch data and PSG in a population of 31 healthy individuals, could be tested for performance on a much larger, more diverse population. Indeed, despite the differences in motion and heart rate data acquisition in the MESA cohort (traditional actigraphy and pulse oximetry), our algorithms demonstrated excellent sleep/wake prediction compared to PSG. One particularly intriguing finding is that the best kappa values were obtained when our algorithm was validated on the MESA dataset. Performance on the unseen MESA testing set actually exceeded our best kappa during validation within the Apple Watch dataset. One reason for this could be fundamental differences between the data acquisition methods—i.e. MEMS accelerometer versus actigraphy and wrist-worn PPG versus finger worn, medical-grade PPG (pulse oximetry). It could also be that differences in the sleep-wake characteristics of the two subject populations change the predictive ability of the classifier. In our Apple Watch dataset, WASO was only sometimes accompanied by significant movement; in the MESA cohort, we have qualitatively observed that wake after recording onset was often associated with significant motion (e.g. the subject was standing up and moving around), making wake easier to classify.
The availability of algorithms that span both raw acceleration and activity counts, obtained through different sensors, will standardize ambulatory sleep tracking for both research and clinical practice. This methodology, and the data sharing required to support its use, allows for continued utilization of established resources while promoting innovation.
Limitations
Despite the strengths, this study is not without limitations. Our training dataset was comprised of relatively young, healthy individuals free of sleep disorders. Because local heart rate standard deviation was used as an input to the model, the ability of the model to estimate sleep is likely predicated on the presence of a functioning autonomic nervous system and performance could be reduced in the setting of cardiovascular disease as well as sleep-disordered breathing, insomnia, and periodic limb movements of sleep [48–56]. Additionally, to extend this work to clinical populations, further algorithm validation in other disorders must take place. For example, a condition that affects motion during sleep, such as REM behavior disorder, could significantly impact our results. Further, the incorporation of the clock proxy may require a normal functioning circadian timing system that interacts as expected with the sleep homeostat, which may be altered in certain sleep disorders.
The individuals in our training set demonstrated high SE which could lead to falsely low specificity (i.e. if few wake epochs exist on PSG, the sleep/wake classifier has a reduced opportunity to correctly designate wake epochs and is more susceptible to noise). Our concerns about this limitation are mitigated given the preserved performance when we test models trained on our Apple Watch data with data from the MESA cohort, which was comprised of PSG records with lower sleep efficiencies.
In our training set, “ground truth” PSG labeling was based on the staging of a single registered polysomnographic technologist (RPSGT), which is not infallible. We did not require subjects to wear the watch on their non-dominant wrist, which could be a confounding factor; although other investigators have not found this to be the case [47]. Additionally, user errors with the app and problems with the server collecting data resulted in loss of data for four subjects in the study. Improved user interface and stability in the open-source application developed for this project will be needed before the app can be deployed at a broader scale.
As demonstrated by the Bland-Altman plots of sleep metrics, the ability of the classifier to accurately quantify sleep varies across different values of SOL, WASO, TST, and SE. Although our classifier had improved specificity compared to actigraphy and most current generation multisensor wearables, the problem of greater inaccuracy with larger amounts of wake during the attempted sleep period persists. The broad distribution of how well the classifier performs could limit the utility in practical use cases. Future work should continue to focus on improvement of algorithm specificity for wake; further, different populations may require different algorithms to most accurately measure sleep with wearable devices.
Conclusion
Algorithms that estimate sleep from actigraphy have existed for decades. The initial algorithms used thresholds to decide sleep and wake applied to motion count data from actigraphs that had been processed with understandable, disclosed methods. State-of-the-art classifiers are no longer so transparent. Each method comes with large numbers of tunable parameters, with the meaning of each specific to the classifier in use. Easily summarizing a classifier as an expression or table in a paper, as was done in the past, is no longer feasible.
In addition to the growing complexity of classification algorithms, we now have many more sources of data than the limited set available from actigraph devices. Although each device returns data processed in a slightly different way, the rapid growth of wearable sensor capabilities provides access to new streams of data for use in classification.
Other medical fields have demonstrated the ability of new technology to produce FDA cleared, over-the-counter adjunct evaluation tools; for example, home pregnancy tests, glucometers, and more recently, the Apple Watch irregular heart rate detection capability. Because sleep health is marked by the convergence of behavior and biology, sleep medicine is an obvious beneficiary of instruments that lie on the interface of consumer technology and medicine. However, the field of sleep medicine has remained somewhat resistant to the use of consumer marketed sensors given the lack of transparency in data acquisition and analysis, and the lack of a feasible, efficient method to validate the vast number of devices and associated software [13].
The adoption of affordable, ubiquitous sensors holds significant potential for growing our understanding of sleep and increasing the reach of sleep medicine. Achieving this potential requires wearable manufacturers to allow access to raw sensor data, an intact infrastructure for data sharing of resources with overlapping wearable sensor and scored PSG data, open-source code and disclosed algorithms such as those presented here. This work sets the stage to harness commercial devices for sleep research at large scales.
Supplementary material
Supplementary material is available at SLEEP online.
Figure S1. One vs Rest ROC curves for the REM vs not REM classification problem.
Figure S2. One vs Rest ROC curves for the NREM vs not NREM classification problem.
Figure S3. One vs Rest ROC curves for the wake vs not wake classification problem.
Figure S4. Comparing adding model-generated circadian drive (pink), cosine (purple), and time since recording start (gray), to motion and heart rate (green). This plot was generated by repeating Monte Carlo cross validation ten times in the manner described in the main text. Here, “clock” refers to the circadian model, and “time” refers to time since recording start.
Funding
This work was supported by the Exercise & Sport Science Initiative University of Michigan; Mobile sleep and circadian rhythm assessment for enhanced athletic performance (U056400) M-Cubed; Analyzing light, human sleep and circadian rhythms through smartphones (U049702), and NSF DMS 1714094.
Acknowledgments
Thanks to Mallory Newsted, Jennifer Zollars, and the University of Michigan Sleep and Circadian Research Laboratory for their assistance.
Conflict of interest statement. O.W. has given talks at Unilever events and received honorariums/travel expenses. She is the CEO of Arcascope LLC, a company that makes circadian rhythms software. D.F. is the CSO of Arcascope and has equity in the company. Arcascope did not sponsor this research. C.G. receives royalties from UpToDate.
Data repository
The data described in this manuscript is available on PhysioNet.
References
- 1. Institute of Medicine (US) Committee on Sleep M, Research. The national academies collection: reports funded by National Institutes of Health. In: Colten HR, Altevogt BM, eds. Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem. Washington (DC): National Academies Press (US).National Academy of Sciences; 2006. doi: 10.17226/11617 [DOI] [PubMed] [Google Scholar]
- 2. Berry RB, et al. ; for the American Academy of Sleep Medicine.The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications. Version 2.3.0 ed. Darien, IL: American Academy of Sleep Medicine; 2016. [Google Scholar]
- 3. Ancoli-Israel S, et al. The role of actigraphy in the study of sleep and circadian rhythms. Sleep. 2003;26(3):342–392. [DOI] [PubMed] [Google Scholar]
- 4. Smith MT, et al. Use of actigraphy for the evaluation of sleep disorders and circadian rhythm sleep-wake disorders: an American academy of sleep medicine clinical practice guideline. J Clin Sleep Med. 2018;14(7):1231–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marino M, et al. Measuring sleep: accuracy, sensitivity, and specificity of wrist actigraphy compared to polysomnography. Sleep. 2013;36(11):1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Souza L, et al. Further validation of actigraphy for sleep studies. Sleep. 2003;26(1):81–85. [DOI] [PubMed] [Google Scholar]
- 7. Blood ML, et al. A comparison of sleep detection by wrist actigraphy, behavioral response, and polysomnography. Sleep. 1997;20(6):388–395. [PubMed] [Google Scholar]
- 8. Paquet J, et al. Wake detection capacity of actigraphy during sleep. Sleep. 2007;30(10):1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee J, et al. Consumer sleep tracking devices: a critical review. Stud Health Technol Inform. 2015;210:458–460. [PubMed] [Google Scholar]
- 10. Russo K, et al. Consumer sleep monitors: is there a baby in the bathwater? Nat Sci Sleep. 2015;7:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ko PR, et al. Consumer sleep technologies: a review of the landscape. J Clin Sleep Med. 2015;11(12):1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Zambotti M, et al. Measures of sleep and cardiac functioning during sleep using a multi-sensory commercially-available wristband in adolescents. Physiol Behav. 2016;158:143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khosla S, et al. ; American Academy of Sleep Medicine Board of Directors. Consumer sleep technology: an American academy of sleep medicine position statement. J Clin Sleep Med. 2018;14(5):877–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baron KG, et al. Feeling validated yet? A scoping review of the use of consumer-targeted wearable and mobile technology to measure and improve sleep. Sleep Med Rev. 2018;40:151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tison GH, et al. Passive detection of atrial fibrillation using a commercially available smartwatch. JAMA Cardiol. 2018;3(5):409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fonseca P, et al. Validation of photoplethysmography-based sleep staging compared with polysomnography in healthy middle-aged adults. Sleep. 2017;40(7). doi: 10.1093/sleep/zsx097. [DOI] [PubMed] [Google Scholar]
- 17. Goldstone A, et al. Actigraphy in the digital health revolution: still asleep? Sleep. 2018;41(9). doi: 10.1093/sleep/zsy120. [DOI] [PubMed] [Google Scholar]
- 18. Troiano RP, et al. Evolution of accelerometer methods for physical activity research. Br J Sports Med. 2014;48(13):1019–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spierer DK, et al. Validation of photoplethysmography as a method to detect heart rate during rest and exercise. J Med Eng Technol. 2015;39(5):264–271. [DOI] [PubMed] [Google Scholar]
- 20. FDA. Statement from FDA Commissioner Scott Gottlieb, M.D., and Center for Devices and Radiological Health Director Jeff Shuren, M.D., J.D., on agency efforts to work with tech industry to spur innovation in digital health https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/UCM620246.htm.
- 21. Cole RJ, et al. Automatic sleep/wake identification from wrist activity. Sleep. 1992;15(5):461–469. [DOI] [PubMed] [Google Scholar]
- 22. Sadeh A, et al. Activity-based sleep-wake identification: an empirical test of methodological issues. Sleep. 1994;17(3):201–207. [DOI] [PubMed] [Google Scholar]
- 23. Jean-Louis G, et al. Sleep estimation from wrist movement quantified by different actigraphic modalities. J Neurosci Methods. 2001;105(2):185–191. [DOI] [PubMed] [Google Scholar]
- 24. Kushida CA, et al. Comparison of actigraphic, polysomnographic, and subjective assessment of sleep parameters in sleep-disordered patients. Sleep Med. 2001;2(5):389–396. [DOI] [PubMed] [Google Scholar]
- 25. Kripke DF, et al. Wrist actigraphic scoring for sleep laboratory patients: algorithm development. J Sleep Res. 2010;19(4):612–619. [DOI] [PubMed] [Google Scholar]
- 26. Borbély AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1(3):195–204. [PubMed] [Google Scholar]
- 27. Daan S, et al. Timing of human sleep: recovery process gated by a circadian pacemaker. Am J Physiol. 1984;246(2 Pt 2):R161–R183. [DOI] [PubMed] [Google Scholar]
- 28. Dijk DJ, et al. Paradoxical timing of the circadian rhythm of sleep propensity serves to consolidate sleep and wakefulness in humans. Neurosci Lett. 1994;166(1):63–68. [DOI] [PubMed] [Google Scholar]
- 29. Czeisler CA, et al. Human sleep: its duration and organization depend on its circadian phase. Science. 1980;210(4475):1264–1267. [DOI] [PubMed] [Google Scholar]
- 30. Booth V, et al. Physiologically-based modeling of sleep-wake regulatory networks. Math Biosci. 2014;250:54–68. [DOI] [PubMed] [Google Scholar]
- 31. Phillips AJ, et al. A quantitative model of sleep-wake dynamics based on the physiology of the brainstem ascending arousal system. J Biol Rhythms. 2007;22(2):167–179. [DOI] [PubMed] [Google Scholar]
- 32. Johns MW. A new method for measuring daytime sleepiness: the Epworth sleepiness scale. Sleep. 1991;14(6):540–545. [DOI] [PubMed] [Google Scholar]
- 33. Bland J, et al. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):301–310. [PubMed] [Google Scholar]
- 34. te Lindert BH, et al. Sleep estimates using microelectromechanical systems (MEMS). Sleep. 2013;36(5):781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Forger DB, et al. A simpler model of the human circadian pacemaker. J Biol Rhythms. 1999;14(6):532–537. [DOI] [PubMed] [Google Scholar]
- 36. Fonseca P, et al. A comparison of probabilistic classifiers for sleep stage classification. Physiol Meas. 2018;39(5):055001. [DOI] [PubMed] [Google Scholar]
- 37. Pedregosa F, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 38. Hanley JA, et al. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. [DOI] [PubMed] [Google Scholar]
- 39. Saito T, et al. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015;10(3):e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bild DE, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. [DOI] [PubMed] [Google Scholar]
- 41. Dean DA II, et al. Scaling up scientific discovery in sleep medicine: the national sleep research resource. Sleep. 2016;39(5):1151–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang GQ, et al. The National Sleep Research Resource: towards a sleep data commons. J Am Med Inform Assoc. 2018;25(10):1351–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DE Zambotti M, et al. Wearable Sleep Technology in Clinical and Research Settings. Med Sci Sports Exerc. 2019;51(7):1538–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. de Zambotti M, et al. A validation study of Fitbit Charge 2™ compared with polysomnography in adults. Chronobiol Int. 2018;35(4):465–476. [DOI] [PubMed] [Google Scholar]
- 45. Cook JD, et al. Ability of the multisensory jawbone UP3 to quantify and classify sleep in patients with suspected central disorders of hypersomnolence: a comparison against polysomnography and actigraphy. J Clin Sleep Med. 2018;14(5):841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cook JD, et al. Ability of the Fitbit Alta HR to quantify and classify sleep in patients with suspected central disorders of hypersomnolence: a comparison against polysomnography. J Sleep Res. 2019;28(4):e12789. [DOI] [PubMed] [Google Scholar]
- 47. Beattie Z, et al. Estimation of sleep stages in a healthy adult population from optical plethysmography and accelerometer signals. Physiol Meas. 2017;38(11):1968–1979. [DOI] [PubMed] [Google Scholar]
- 48. de Zambotti M, et al. Nighttime cardiac sympathetic hyper-activation in young primary insomniacs. Clin Auton Res. 2013;23(1):49–56. [DOI] [PubMed] [Google Scholar]
- 49. Allena M, et al. Periodic limb movements both in non-REM and REM sleep: relationships between cerebral and autonomic activities. Clin Neurophysiol. 2009;120(7):1282–1290. [DOI] [PubMed] [Google Scholar]
- 50. Gottlieb DJ, et al. Restless legs syndrome and cardiovascular disease: a research roadmap. Sleep Med. 2017;31:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nannapaneni S, et al. Periodic limb movements during sleep and their effect on the cardiovascular system: is there a final answer? Sleep Med. 2014;15(4):379–384. [DOI] [PubMed] [Google Scholar]
- 52. Thomas RJ, et al. Differentiating obstructive from central and complex sleep apnea using an automated electrocardiogram-based method. Sleep. 2007;30(12):1756–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Narkiewicz K, et al. Cardiovascular variability characteristics in obstructive sleep apnea. Auton Neurosci. 2001;90(1-2):89–94. [DOI] [PubMed] [Google Scholar]
- 54. Lurie A. Hemodynamic and autonomic changes in adults with obstructive sleep apnea. Adv Cardiol. 2011;46:171–195. [DOI] [PubMed] [Google Scholar]
- 55. Bonnet MH, et al. Hyperarousal and insomnia: state of the science. Sleep Med Rev. 2010;14(1):9–15. [DOI] [PubMed] [Google Scholar]
- 56. Tobaldini E, et al. Heart rate variability in normal and pathological sleep. Front Physiol. 2013;4:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.