Abstract
Chronic periodontitis has a polymicrobial biofilm etiology and interactions between key oral bacterial species, such as Porphyromonas gingivalis and Treponema denticola contribute to disease progression. P. gingivalis and T. denticola are co-localized in subgingival plaque and have been previously shown to exhibit strong synergy in growth, biofilm formation and virulence in an animal model of disease. The motility of T. denticola, although not considered as a classic virulence factor, may be involved in synergistic biofilm development between P. gingivalis and T. denticola. We determined the role of T. denticola motility in polymicrobial biofilm development using an optimized transformation protocol to produce two T. denticola mutants targeting the motility machinery. These deletion mutants were non-motile and lacked the gene encoding the flagellar hook protein of the periplasmic flagella (ΔflgE) or a component of the stator motor that drives the flagella (ΔmotB). The specificity of these gene deletions was determined by whole genome sequencing. Quantitative proteomic analyses of mutant strains revealed that the specific inactivation of the motility-associated gene, motB, had effects beyond motility. There were 64 and 326 proteins that changed in abundance in the ΔflgE and ΔmotB mutants, respectively. In the ΔflgE mutant, motility-associated proteins showed the most significant change in abundance confirming the phenotype change for the mutant was related to motility. However, the inactivation of motB as well as stopping motility also upregulated cellular stress responses in the mutant indicating pleiotropic effects of the mutation. T. denticola wild-type and P. gingivalis displayed synergistic biofilm development with a 2-fold higher biomass of the dual-species biofilms than the sum of the monospecies biofilms. Inactivation of T. denticola flgE and motB reduced this synergy. A 5-fold reduction in dual-species biofilm biomass was found with the motility-specific ΔflgE mutant suggesting that T. denticola periplasmic flagella are essential in synergistic biofilm formation with P. gingivalis.
Keywords: polymicrobial biofilm, chronic periodontitis, periplasmic flagella, motility, quantitative proteomics
Introduction
Chronic periodontitis is a polymicrobial disease believed to be initiated by changes in the bacterial species composition of subgingival plaque biofilms, and subsequent dysregulation of the host immune response (Hajishengallis and Lamont, 2012). It is linked with the overgrowth of a small number of oral microbial species within subgingival plaque biofilms accreted to the surface of the tooth root (Wiebe and Putnins, 2000; Byrne et al., 2009). Treponema denticola and Porphyromonas gingivalis are pathobionts associated with chronic periodontitis due to their strong association with the clinical measurements of severe periodontal disease, such as periodontal pocket depth and bleeding on probing (Lamont and Jenkinson, 1998; Socransky et al., 1998; Holt and Ebersole, 2005; Dashper et al., 2011). The study of their interactions in polymicrobial biofilms is important to understand chronic disease initiation and progression.
T. denticola and P. gingivalis synergistically form biofilms in vitro (Yamada et al., 2005; Zhu et al., 2013). Intriguingly, T. denticola cells in monospecies biofilms lose their characteristic spiral morphology but retain it when grown in polymicrobial biofilms with P. gingivalis (Zhu et al., 2013). The spiral morphology of T. denticola is closely related to its periplasmic flagella (Ruby et al., 1997), which in turn are required for motility (Li et al., 1996). T. denticola motility therefore may play a role in synergistic biofilm formation with P. gingivalis. Unlike extracellularly flagellated bacteria, T. denticola remains motile in a highly viscous environment, likely due to the protection of the flagella in the periplasmic space, away from direct contact with the external environment (Klitorinos et al., 1993). The ability of T. denticola to move in highly viscous environments may be beneficial for its movement through polymicrobial biofilms. Together with the presence of a chemotaxis system that allows it to move in response to environmental stimuli, T. denticola may create pores in the biofilm matrix as they move through the biofilms (Houry et al., 2012), allowing better nutrient penetration and waste removal, thereby contributing to a larger biofilm biomass.
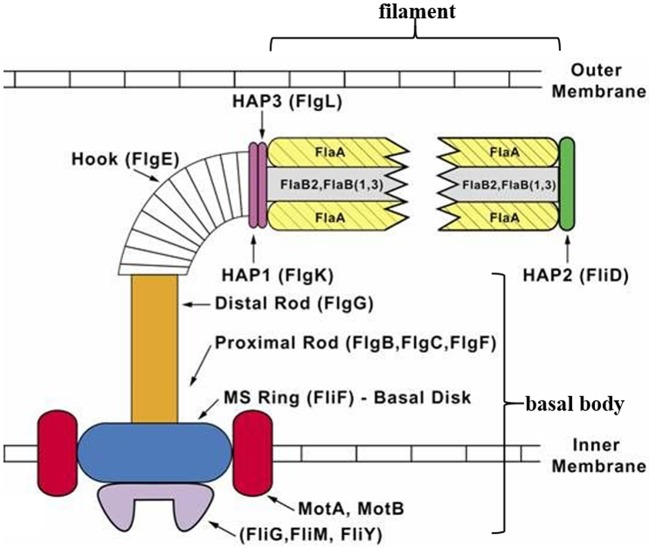
Similar to a typical bacterial flagellum, a spirochete periplasmic flagellum can be divided into three parts: a basal body, hook, and filament (Figure 1). The basal body is composed of a rod and a motor-switch complex embedded in the inner membrane. The motor is made up of two parts: the rotor and the stator. The rotor is made up of proteins FliF, FliG, FliM, and FliY (Limberger, 2004; Morimoto and Minamino, 2014). The stator, which is comprised of two integral membrane proteins (MotA and MotB), is an ion channel complex coupling transmembrane ion movement with flagellar rotation (Kojima and Blair, 2001; Chevance and Hughes, 2008; Kojima et al., 2009; Morimoto and Minamino, 2014). The flagellar hook structure consists of one major polypeptide, FlgE, and serves as a universal joint that transmits torque produced by the motor in the basal body to the flagellar filament, whose rotation results in specific movements of the cell body that enables the cell to move (Berg, 1976; Limberger, 2004). The flagellar filament of T. denticola is made up of three filament outer layer proteins (FlaA1-3) and three filament core proteins (FlaB1-3). The flagellar filaments originate at opposite ends of the same cell and overlap at mid-cell (Chan et al., 1993). Each T. denticola cell commonly possesses four periplasmic flagella, two anchored at each end of the cell that overlap in the middle of the cell (Izard et al., 2008).
Figure 1.
Diagrammatic representation of a T. denticola periplasmic flagellum. The periplasmic flagellum is made up of three parts: a basal body, hook, and filament. The flagellar hook (FlgE) connects and transmits torque produced by the motor [rotor (FliF, FliG, FliM, and FliY) and stator (MotA and MotB)] in the basal body, to the flagellar filament which then rotates. Diagram was adapted and modified from Limberger (2004).
T. denticola is a slow-growing, fastidious and highly fragile obligate anaerobe (Salvador et al., 1987; Wardle, 1997), making its cultivation and handling in the laboratory difficult. The characterization of the molecular detail of T. denticola virulence has been hampered by its low transformation efficiency, lack of shuttle plasmids and small number of selectable markers (Li and Kuramitsu, 1996; Li et al., 1996, 2015; Chi et al., 1999; Kuramitsu et al., 2005; Bian and Li, 2011; Godovikova et al., 2015). Despite two decades of work a relatively small number of T. denticola transformants have been reported in the literature and many laboratories have reported modifications of the original published protocol for successful transformation (Ishihara et al., 1998; Limberger et al., 1999; Chi et al., 2002; Lux et al., 2002; Abiko et al., 2014; Godovikova et al., 2015).
In this study, we generated two T. denticola motility mutants to investigate the role of T. denticola motility in synergistic biofilm formation with P. gingivalis. The flgE and motB genes were targeted as their inactivation in T. denticola or other bacteria resulted in mutants with impaired motility (Li et al., 1996; Houry et al., 2010; Sultan et al., 2015). The mutants generated were characterized for their morphology, motility, growth, binding with P. gingivalis and protein expression profiles. Finally, the ability of the mutants to form dual-species biofilms with P. gingivalis was determined.
Materials and Methods
Bacterial Strains and Culture Conditions
P. gingivalis W50 and T. denticola ATCC 33520 and ATCC 35405 were obtained from the culture collection of the Oral Health Cooperative Research Centre, The University of Melbourne. The bacteria were maintained in an anaerobic workstation (MK3; Don Whitley Scientific) at 37°C. Planktonic P. gingivalis cultures were routinely grown in Brain Heart Infusion (BHI) medium containing 37 g/L Brain Heart Infusion (Becton, Dickinson and Company), L-cysteine hydrochloride (0.5 mg/mL), and hemin (5 μg/mL). T. denticola was grown in Oral Bacterium Growth Medium (OBGM) (Veith et al., 2009). When needed, agarose (UltraPure™ Low Melting Point Agarose, Thermo Fisher Scientific Inc., USA) and antibiotics (described in detail below) were added to the medium. Growth of bacterial cultures was monitored by measuring absorbance at a wavelength of 650 nm (A650) and cells were harvested by centrifugation. Culture purity was routinely checked by Gram staining and colony morphology.
Construction of T. denticola Motility Mutants
T. denticola ATCC 33520 motility mutants were constructed where the open reading frame of flgE and motB were deleted. These two genes are found in the same operon driven by the fla promoter; recombination cassettes included the fla promoter downstream of the erythromycin resistance gene to enable transcription of the remainder of the operon. Briefly, recombination cassettes for the deletion of flgE (HMPREF9722_RS03025) and motB (HMPREF9722_RS03040) were constructed using the PCR-based splicing by overlap-extension (SOE) method (Horton et al., 1989). T. denticola genomic DNA was purified using the DNeasy® Blood and Tissue kit (QIAGEN Pty.) according to the manufacturer's instructions. Genomic DNA was resuspended in deionized water and subject to PCR using the primers shown in Supplementary Table 1. A schematic representation of the mutational approach is shown in Supplementary Figure 1. The upstream and downstream regions of the target gene as well as fla promoter were amplified by PCR from the chromosomal DNA of T. denticola 33520 whilst the ermAM gene was amplified from the shuttle vector pHS17 (Fletcher et al., 1995). The four amplicons were fused into a single fragment using the SOE method (Horton et al., 1989). The final fragment was cloned into a pGEM®-T Easy vector, yielding pHN-ΔflgE and pHN-ΔmotB. The fidelity of each recombination cassette was confirmed by DNA sequencing.
Each plasmid construct (10 μg) was linearized by digestion with NotI then electroporated into T. denticola ATCC 33520 cells as previously published (Li et al., 1996) with some modifications. Briefly, 2-days (exponential phase; A650 of 0.16–0.28) T. denticola ATCC 33520 cultures were decanted into centrifuge tubes in the anaerobic chamber. The cells were harvested by centrifugation (4,000 g, 10 min, 4°C) and the subsequent washing and resuspension steps were conducted anaerobically at 4°C. All resuspension steps were performed using 1 mL pipette tips which had ~1 cm cut off to reduce shear forces on cells. The cells were washed twice instead of three times to reduce handling and loss of cells. Sterile 10% (v/v) glycerol used for all wash and resuspension steps was pre-reduced in the anaerobic chamber for at least 16 h before use. Electroporation was carried out as previously described (Li et al., 1996), typically producing a time constant of 4.0–4.7 ms. T. denticola cells were then immediately suspended in 1.2 mL of pre-reduced OBGM in the anaerobic chamber and incubated overnight at 37°C. Transformants were selected on OBGM agar plates containing 0.8% (w/v) agarose supplemented with 40 μg/mL erythromycin. The OBGM agar medium was pre-equilibrated to 37°C before plating of the electroporated cells to avoid thermal shock. To screen for the presence of the appropriate homologous recombination event, the resulting transformants were grown in OBGM (2 mL) containing the appropriate antibiotics whereupon a small volume of culture (~25 μL) was subject to PCR with the appropriate oligonucleotide primers (Supplementary Table 1).
Genomic Sequencing
Genome sequencing was performed using an Ion Torrent Personal Genome Machine (PGM; Thermo Fisher Scientific,) according to the protocols of the manufacturer unless otherwise stated. Briefly, 1 μg of T. denticola genomic DNA was fragmented to ~400 bp using a Covaris M220 Focused-ultrasonicator™ (TrendBio, Australia). A 1 μL aliquot of sheared DNA was visualized using a LabChip GX Touch 24 Nucleic Acid Analyzer (PerkinElmer, USA) to ensure a peak fragment size of 400 bp. The DNA was end-repaired (Ion Xpress™ Plus Fragment Library Kit) and purified (Agencourt™ AMPure™ XP Kit, Beckman Coulter). Barcoded adaptors were ligated to the DNA and nick-repaired (Ion Xpress™ Plus Fragment Library Kit; Ion Xpress™ Barcode Adapters Kit) and then purified again (Agencourt™ AMPure™ XP Kit). The labeled library was then size-selected again using the Pippin Prep™ DNA Size Selection System (Sage Science), aiming for a target-peak size of ~480 bp collected over a specified range. Following sample purification (Agencourt™ AMPure™ XP Kit) the concentration of the unamplified library was determined using qPCR (Ion Library TaqMan® Quantitation Kit). All of the prepared libraries were at an adequate concentration which did not require further amplification. Barcoded libraries were pooled in equimolar amounts of 26 pM to ensure an equal representation of each barcoded library in the sequencing run. The library was then used to prepare enriched, template-positive Ion PGM™ Hi-Q™ Ion Sphere™ Particles (ISPs) using the Ion OneTouch™ 2 System (Ion PGM™ Hi-Q™ OT2 Kit). The recovered template positive ISPs were enriched using the Ion OneTouch™ ES Instrument and Ion OneTouch™ ES Supplies Kit, then loaded onto an Ion 318™ Chip v2 BC and sequenced using the Ion PGM™ Hi-Q™ Sequencing Kit and Ion PGM™ Instrument. The resulting sequencing reads were downloaded from the Torrent Server and analyzed using Geneious R8.1.9 (Biomatters Ltd, New Zealand) with comparison made to Treponema denticola ATCC 33520 NCBI Reference Sequences NZ_AGDS00000000.1 and NZ_KB445542.1, and the sequence of our laboratory T. denticola 33520 strain. In depth examination of predicted amino acid substitutions due to single nucleotide polymorphisms was performed using NCBI BLASTp followed by COBALT Constraint-based Multiple Alignment Tool (Papadopoulos and Agarwala, 2007).
Swimming Assay
To compare the motility of T. denticola mutants with that of the wild-type ATCC 33520, a swimming assay in semisolid OBGM agar [OBGM supplemented with 0.4% (w/v) Molecular Grade Agarose (Bioline) and 1% (w/v) BD Difco™ gelatin (Bacto Laboratories Pty. Ltd.)] was developed based on previously published motility assays (Lux et al., 2002; Bian et al., 2013). The OBGM agar plates were pre-reduced overnight in an anaerobe chamber. T. denticola cells grown to exponential growth phase in OBGM were harvested by centrifugation (4,000 g, 6 min, 25°C) and gently suspended in an appropriate volume of deionized water. The bacterial suspension (2 μL; 107 cells) was carefully injected into the semisolid OBGM agar using a pipettor, ensuring that all of the suspension was below the surface. The plates were dried for 20 min at room temperature before they were incubated anaerobically at 37°C for 10 days. Images of T. denticola turbid plaques were obtained using a Fujifilm LAS-3000 Imager.
Cryo-Electron Microscopy (Cryo-EM) and Scanning Electron Microscopy (SEM)
Unwashed T. denticola whole cells were prepared for cryo-EM and imaged at the Bio21 Advanced Microscopy Facility, The University of Melbourne, as previously described (Chen et al., 2011) with the following modification. The images were acquired in FEI Tecnai G2 F30 equipped with a FEI Ceta 4 × 4 k CMOS camera and operated at 200 or 300 kV. T. denticola and P. gingivalis biofilms were prepared and imaged using SEM on a Phillips XL30 Gold-emission scanning 203 electron microscope (Phillips, Eindhoven, The Netherlands) at a voltage of 2 kV as described previously (Mitchell et al., 2010).
Autoaggregation and Coaggregation Assays
Autoaggregation of T. denticola strains and P. gingivalis W50 as well as coaggregation between T. denticola strains and P. gingivalis were evaluated as previously described (Abiko et al., 2014) with some modifications. T. denticola and P. gingivalis cells were grown to exponential phase before they were harvested by centrifugation (4,000 g, 10 min, 25°C). Cells were washed twice with coaggregation buffer (20 mM phosphate buffer, pH 8.0, 1 mM CaCl2, 1 mM MgCl2, 150 mM NaCl) and suspended in the coaggregation buffer to an A650 of 0.5. For the autoaggregation assay, 1 mL of the cell suspension was placed in a cuvette and the height of the bacterial aggregates was monitored for 7 h. For the coaggregation assay, equal volumes of T. denticola and P. gingivalis cell suspensions were combined in a cuvette to give an A650 of 0.5. The height of the bacterial aggregates was monitored for 7 h by measurement with a ruler.
Quantitative Proteomic Analyses
Whole cell samples were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% (w/v) NuPAGE® Novex® Bis-Tris precast mini-gel. The gel was run at 150 V for ~10 min and the whole gel band from well to dye front was excised from the SimplyBlue™ SafeStain stained SDS-PAGE gel. In-gel digestion was performed after reduction with 10 mM DTT and alkylation with 50 mM iodoacetamide using sequencing-grade-modified trypsin (Promega) overnight at 37°C, as previously published (Mortz et al., 2001). The digestion was stopped by the addition of TFA to a final concentration of 0.1% (v/v). The fluid was transferred to a new tube for analysis by LC-MS/MS as previously described (Glew et al., 2017).
Raw MS files were analyzed by MaxQuant (Ver 1.5.3.30) (Cox et al., 2014) using label-free quantification (LFQ) searching against the whole T. denticola ATCC 35405 protein database containing a total of 2,786 protein sequences obtained from the Comprehensive Microbial Resource Website (cmr.jcvi.org). The default parameters were used for LFQ. Data analysis was performed in Excel, using the MaxQuant output file “proteinGroups.txt.” Any contaminating eukaryotic proteins, such as human keratins or serum proteins from the growth media were removed from the list of identified proteins. LFQ intensity values were used to produce ratios of paired samples and the geometric mean of these ratios was calculated. Statistical analysis was performed using paired, two-tailed Student's t-tests. The abundance of a protein was considered to have significantly changed between two samples if there was ≥1.5- or ≤0.67-fold change in the geometric mean and a p-value of <0.05.
Static Biofilm Assay
Static biofilm assays were carried out essentially as previously described (Dashper et al., 2014). Briefly, T. denticola and P. gingivalis cells were grown to exponential phase and diluted with fresh pre-reduced OBGM to A650 of 0.15 where necessary. For monospecies biofilms, 2 mL of each cell suspension was aliquoted into each well of a CELLSTAR® 12-well flat-well plate (Greiner, Sigma-Aldrich). For dual-species biofilms, equal volumes of T. denticola and P. gingivalis cell suspensions were aliquoted into each well of the 12-well flat-well plate and mixed. After inoculation, the plates were incubated at 37°C anaerobically for 1 h, sealed with a Greiner multiwell plate sealer (Sigma-Aldrich) and further incubated at 37°C anaerobically for 5 days. The attached biofilms were quantified by crystal violet staining. Following incubation, the medium in each well was decanted and each well was gently rinsed with 2.1 mL of deionized water. The plates were air-dried and the biofilms were stained with 0.1% (v/v) crystal violet for 30 min at RT, then rinsed twice with 2.5 mL of deionized water. The plates were air-dried and destained with 2.1 mL of 99% (v/v) ethanol and the absorbance of each well at 540 nm (A540) was measured using a Wallac VICTOR3™ 1420 Multilabel Counter (PerkinElmer, USA). A Kruskal-Wallis rank sum test was conducted with the Conover-Imam test for the statistical analysis of the biofilm data (Kruskal and Wallis, 1952; Conover and Iman, 1979).
Results
T. denticola Motility Mutants
T. denticola ATCC 33520 motility mutants were generated via electroporation as previously described, with some modifications (Capone et al., 2007). Significantly, T. denticola cells were washed twice instead of thrice under strictly anaerobic conditions to improve transformation efficiency. Positive ErmR clones resulting from double-crossover homologous recombination following electroporation with linearized pHN-ΔflgE and pHN-ΔmotB were verified by PCR using the primer pairs 5′flgE-F/3′flgE-R and 5′motB-F/3′motB-R, respectively. The modified transformation protocol used in this study resulted in consistently achieving 98–100% true positive transformants and eliminated the complication of spontaneous Erm resistant colonies commonly reported for earlier protocols (Bian et al., 2012). Validated deletant strains of T. denticola ΔflgE and ΔmotB were used for further analyses.
Genomic sequencing was performed to confirm the veracity of the mutants [NCBI Bioproject accession number PRJNA561478]. Both mutants had the appropriate target genes deleted from their genomes. Between the two mutants, a total of four different loci were affected by single nucleotide polymorphisms (SNPs), with one found in both mutants (Supplementary Table 2). None of the SNPs were found in the T. denticola 33520 laboratory strain. These SNPs resulted in amino acid substitutions in two motility related proteins and were predicted to have no effect on protein structure or function as the substituted amino acids were all found in nature in at least one other T. denticola strain, as determined using COBALT (data not shown). The only locus which was subject to a frame shift occurred in ΔflgE whereby the insertion of a single C in hypothetical protein HMPREF9722_RS03985 resulted in ΔflgE missing the final 5% of this protein. Interestingly 14% of all sequencing reads at this locus in the WT also had this identical C insertion suggesting that this mutation may possibly be a common mutation emerging in WT. This hypothetical protein was detected in ΔFlgE using a proteomics approach (see below).
Phenotypic Characterization of T. denticola Wild-Type, ΔflgE and ΔmotB
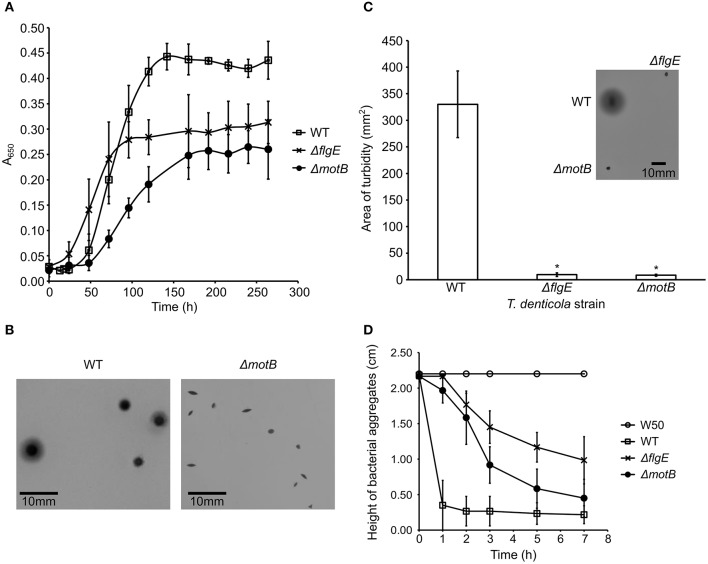
The planktonic growth of T. denticola ΔmotB and ΔflgE mutants was impaired compared with the wild-type. The mutants had a lower maximum cell density at stationary phase and a longer mean generation time than wild-type (Figure 2A). The mean generation times for wild-type, ΔflgE and ΔmotB were 14 ± 3, 22 ± 3, and 24 ± 5 h, respectively. When grown on agar plates, the colonies of ΔflgE (data not shown) and ΔmotB were small, dense, and pinpoint-shaped. The edges of the colonies were defined instead of diffusing outward like that of the wild-type (Figure 2B). Swimming assay analysis revealed that the mutants were non-motile as they were unable to swim outward from the point of inoculation and form a turbid plaque like those of the wild-type (Figure 2C). Autoaggregation assays were carried out to examine the ability of P. gingivalis W50, T. denticola wild-type and mutants to bind to themselves. P. gingivalis did not autoaggregate in this assay whereas T. denticola wild-type autoaggregated at the highest rate, followed by ΔmotB and finally ΔflgE (Figure 2D).
Figure 2.
Characterization of T. denticola wild-type, ΔflgE, ΔmotB. (A) Growth of T. denticola wild-type and mutants. T. denticola cultures at stationary phase were inoculated into fresh OBGM medium at t = 0. A650 of the cultures was measured at 6–24 h intervals for 264 h. The mean generation time was calculated based on the rate of change of A650 during the exponential growth phase of cultures (N = 3). The mean generation times were statistically analyzed using a one way ANOVA with both mutants ΔmotB and ΔflgE significantly different (p < 0.05) to WT. (B) Colony morphologies of T. denticola wild-type and ΔmotB. T. denticola cells were inoculated into OBGM agar [0.8% (w/v) agarose] and poured into petri dishes which were then incubated anaerobically for 2 weeks. The colonies formed by ΔflgE had a similar morphology to the colonies of ΔmotB. (C) Swimming assay of T. denticola wild-type and mutants. T. denticola cells harvested at exponential phase were spotted on semisolid OBGM agar [0.4% (w/v) agarose and 1% (w/v) gelatin] and incubated anaerobically for 10 days before the area of turbid plaque was measured. The data are presented as means plus standard deviations (N = 29) and were analyzed by Students' T-test. Values that were significantly different (p < 0.05) from the value for T. denticola wild-type are indicated by an asterisk (*). A representative image of the swimming assays of T. denticola wild-type, ΔflgE, ΔmotB after 10 days of anaerobic incubation is shown. (D) Autoaggregation of P. gingivalis W50, T. denticola wild-type and mutants. T. denticola and P. gingivalis cells harvested at exponential phase were washed twice with coaggregation buffer and adjusted to A650 of 0.5. The height of bacterial aggregates was monitored for 7 h. The data are presented as means and standard deviations (N = 3).
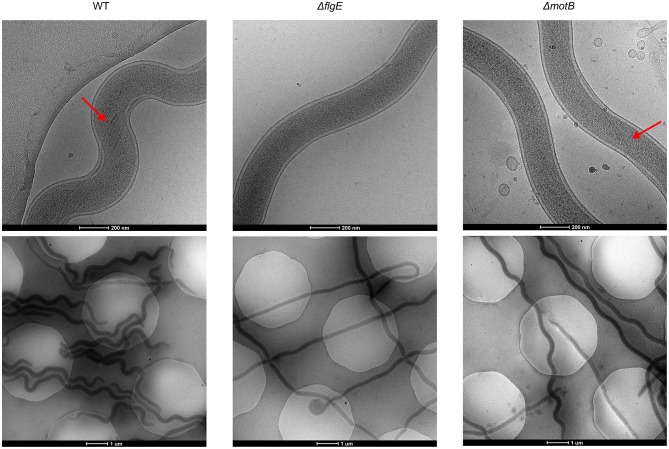
Cryo-Electron Microscopy
T. denticola wild-type and mutant cells in planktonic exponential growth phase were imaged with cryo-electron microscopy to examine periplasmic flagella and to determine cellular morphology (Figure 3). Four or five periplasmic flagella were distinguishable in T. denticola wild-type cells and no periplasmic flagella were observed in ΔflgE. Only two or three periplasmic flagella were distinguishable in ΔmotB and not all cells or cell segments of ΔmotB showed visible flagella. T. denticola wild-type cells adopted an irregular twisted morphology with both planar and helical regions. The ΔmotB cells appeared to have less spirality than the wild-type while many cells or cell segments of ΔflgE had limited or no spirality and appeared rod-shaped.
Figure 3.
Representative cryo-Electron Microscopy images of T. denticola wild-type, ΔflgE, and ΔmotB. T. denticola cells were grown to exponential phase and viewed directly under cryo-EM. Red arrows point to the periplasmic flagella.
Quantitative Proteomics of ΔflgE and ΔmotB
In order to identify proteins that changed in abundance in the motility mutants relative to the wild-type, whole cell lysates were examined using quantitative proteomics. The proteins FlgE (TDE2768) and MotB (TDE2765) were found in wild-type but not in the ΔflgE and ΔmotB mutants, respectively, further confirming the deletion of these genes in the mutants.
There were 64 and 326 proteins that changed in abundance in ΔflgE and ΔmotB, respectively, relative to the wild-type (Supplementary Tables 3–5). Proteins that significantly changed in abundance were sorted into functional categories on the basis of clusters of orthologous groups (COGs) (Tatusov et al., 1997, 2003). Of the 64 proteins in ΔflgE that changed in abundance, 42 significantly decreased and 22 significantly increased. COG category N (cell motility) had the highest number of proteins (19%) (Table 1) that changed in abundance in ΔflgE indicating the wider effects of this mutation on flagella and motility. Of the 326 proteins in ΔmotB that changed in abundance, 229 significantly decreased and 97 significantly increased. COGs related to metabolism contained the highest number of proteins (28%), followed by cellular processes and signaling (25%) and information storage and processing (10%) (Table 1).
Table 1.
Proteins significantly changed in abundance in T. denticola ΔflgE and ΔmotB relative to wild-type, grouped by COG category.
| COGa | Number of proteins | ||
|---|---|---|---|
| ΔflgE | ΔmotB | ||
| Information storage and processing | J | 3 | 18 |
| K | 1 | 5 | |
| L | 2 | 9 | |
| Cellular processes and signaling | D | 1 | 4 |
| M | 3 | 14 | |
| N | 12 | 14 | |
| O | 3 | 12 | |
| T | 1 | 23 | |
| U | 2 | 9 | |
| V | – | 4 | |
| Metabolism | C | 5 | 12 |
| E | 2 | 22 | |
| F | 3 | 10 | |
| G | 1 | 16 | |
| H | 1 | 7 | |
| I | 2 | 6 | |
| P | 3 | 17 | |
| Q | – | 2 | |
| Poorly or not characterized | R | – | 8 |
| S | 17 | 94 | |
| N/A | 2 | 20 | |
| Total | 64 | 326 | |
One-letter abbreviations for the functional COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; U, intracellular trafficking, secretion, and vesicular transport; O, post-translational modification, protein turnover, chaperones; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
Thirty-five proteins changed in abundance in both mutants with the largest number being in COG category N (Table 2). All of these proteins, including those expressed from the fla operon (TDE2764-TDE2768), FlaB (TDE1004), FlaA (TDE1408, TDE1409, TDE1712), and a flagellar hook-associated protein FlgL (TDE2353), significantly decreased in abundance in both mutants, except TDE0119 (FliS) which decreased in abundance in ΔmotB but increased in abundance in ΔflgE (Table 3).
Table 2.
Proteins significantly changed in abundance in both T. denticola ΔflgE and ΔmotB or ΔmotB only, grouped by COG category.
| COGa | Number of proteins | |||
|---|---|---|---|---|
| Changed in abundance in both mutants | Increased in abundance in ΔmotB only | Decreased in abundance in ΔmotB only | ||
| Information storage and processing | J | 2 | 12 | 4 |
| K | 1 | 1 | 3 | |
| L | 2 | 2 | 5 | |
| Cellular processes and signaling | D | – | 3 | 1 |
| M | – | 2 | 12 | |
| N | 10 | – | 5 | |
| O | 1 | 2 | 9 | |
| T | 1 | 5 | 17 | |
| U | 1 | 3 | 3 | |
| V | – | 1 | 3 | |
| Metabolism | C | 1 | 6 | 5 |
| E | 1 | 6 | 15 | |
| F | 2 | 4 | 4 | |
| G | – | 5 | 11 | |
| H | – | 3 | 4 | |
| I | – | – | 6 | |
| P | 3 | 1 | 14 | |
| Q | – | – | 2 | |
| Poorly or not characterized | R | – | 1 | 6 |
| S | 5 | 23 | 63 | |
| N/A | 5 | 6 | 13 | |
| Total | 35 | 86 | 205 | |
One-letter abbreviations for the functional COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; U, intracellular trafficking, secretion, and vesicular transport; O, post-translational modification, protein turnover, chaperones; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
Table 3.
Proteins significantly changed in abundance in both ΔmotB and ΔflgE mutants relative to wild-type (ratio ≥1.5 and ≤0.67, p < 0.05).
| Locus tag | Protein description | Wild-type abundancea | ΔmotB ratiob | ΔflgE ratiob | COGc |
|---|---|---|---|---|---|
| TDE0114 | Iron-dependent transcriptional regulator | 3.46E+07 | 0.59 | 0.55 | K |
| TDE0119 | Flagellar protein FliS | 1.49E+06 | 0.00 | 2.71 | N |
| TDE0383 | Hypothetical protein | 5.62E+06 | 0.00 | 0.00 | S |
| TDE0423 | Hypothetical protein | 1.85E+07 | 0.27 | 0.53 | – |
| TDE0463 | Purine nucleoside phosphorylase (DeoD) | 5.41E+07 | 0.55 | 0.62 | F |
| TDE0501 | Hypothetical protein | 9.24E+06 | 0.00 | 0.50 | – |
| TDE0689 | 5-Methylthioribose kinase | 1.51E+06 | 0.00 | 0.00 | S |
| TDE0758 | Iron compound ABC transporter, periplasmic iron compound-binding protein, putative | 6.66E+06 | 4.14 | 0.23 | P |
| TDE0781 | Ribosomal protein S8 (rpsH) | 4.62E+07 | 1.61 | 0.65 | J |
| TDE0984 | Oligopeptide/dipeptide ABC transporter, permease protein, putative | 5.71E+06 | 8.79 | 0.00 | P |
| TDE0985 | Oligopeptide/dipeptide ABC transporter, periplasmic peptide-binding protein, putative | 3.22E+08 | 5.20 | 0.33 | E |
| TDE0986 | Oligopeptide/dipeptide ABC transporter, ATP-binding protein | 2.92E+06 | 12.92 | 0.00 | P |
| TDE1004 | Flagellar filament core protein (FlaB) | 5.28E+08 | 0.24 | 0.01 | N |
| TDE1208 | DNA topoisomerase I (TopA) | 6.03E+07 | 1.93 | 1.79 | L |
| TDE1234 | Hypothetical protein | 3.64E+05 | 3.81 | 1.51 | – |
| TDE1318 | Hypothetical protein | 7.60E+05 | 0.00 | 0.00 | – |
| TDE1408 | Flagellar filament outer layer protein (FlaA) | 5.92E+08 | 0.29 | 0.04 | N |
| TDE1409 | Flagellar filament outer layer protein (FlaA) | 5.59E+08 | 0.28 | 0.04 | N |
| TDE1483 | Conserved hypothetical protein | 1.28E+08 | 1.52 | 0.30 | S |
| TDE1712 | Flagellar filament outer layer protein (FlaA) | 1.85E+09 | 0.18 | 0.02 | N |
| TDE1727 | Conserved hypothetical protein | 1.03E+08 | 0.54 | 0.46 | O |
| TDE1754 | Desulfoferrodoxin/neelaredoxin | 6.80E+07 | 2.71 | 0.54 | C |
| TDE1919 | Conserved domain protein | 1.64E+07 | 0.63 | 1.90 | S |
| TDE2043 | Signal recognition particle-docking protein FtsY (ftsY) | 5.55E+06 | 0.00 | 0.64 | U |
| TDE2085 | Amino acid kinase family protein | 7.21E+07 | 0.46 | 1.55 | F |
| TDE2087 | Translation initiation factor IF-1 (infA) | 4.30E+07 | 1.68 | 0.61 | J |
| TDE2302 | HD domain protein | 1.09E+07 | 0.00 | 0.64 | T |
| TDE2353 | Flagellar hook-associated protein (FlgL) | 1.83E+06 | 0.00 | 0.59 | N |
| TDE2611 | Conserved hypothetical protein | 2.62E+06 | 0.32 | 1.52 | S |
| TDE2721 | Helicase domain protein | 3.38E+06 | 2.70 | 0.67 | L |
| TDE2764 | Flagellar protein FliL | 6.46E+07 | 0.12 | 0.50 | N |
| TDE2765 | Flagellar motor rotation protein B (MotB) | 3.44E+07 | 0.00 | 0.35 | N |
| TDE2766 | Motility protein A (MotA) | 3.46E+07 | 0.42 | 0.44 | N |
| TDE2768 | Flagellar hook protein FlgE | 4.39E+07 | 0.33 | 0.00 | N |
| TDE2779 | Hypothetical protein | 5.95E+07 | 0.10 | 0.31 | – |
Shading indicates proteins predicted to be organized in an operon.
The abundance of each protein in the wild-type T. denticola wild-type was calculated from the average IBAQ intensity from three replicates.
Geometric mean of ratios, from three replicates, produced from the LFQ intensity of protein in mutant relative to that of protein in wild-type. Ratio of ≥1.5 indicates that the protein had increased in abundance in mutant relative to wild-type and ratio of ≤0.67 indicates that the protein had decreased in abundance in mutant relative to wild-type. Zero ratio indicates that the protein was identified in ATCC 33520 but not in the mutant.
One-letter abbreviations for the functional COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; U, intracellular trafficking, secretion, and vesicular transport; O, post-translational modification, protein turnover, chaperones; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
Of the proteins that increased in abundance in ΔmotB only, COG category J (translation, ribosomal structure and biogenesis) contained the highest number of proteins (Table 2). These proteins include several aminoacyl-tRNA synthetases, such as valyl-tRNA synthetase (TDE1364), tryptophanyl-tRNA synthetase (TDE1588), and phenylalanyl-tRNA synthetase (TDE1927), suggesting an increased usage of the corresponding amino acids valine, tryptophan and phenylalanine in ΔmotB. There was also an increase in the abundance of recombination protein A (RecA; TDE0872), DNA topoisomerase I (TopA; TDE1208), transcription termination factor Rho (Rho; TDE1503) and a rod-shaped determining protein MreB (TDE1349). The metabolic enzymes desulfoferrodoxin/neelaredoxin (TDE1754), FAD-dependent oxidoreductase (TDE2643), and Na+-translocating NADH-quinone reductase E subunit (NqrE; TDE0834) were also among the proteins that increased in abundance in ΔmotB. Two methyl-accepting chemotaxis proteins (MCPs; TDE1009 and TDE2549), two metallo-beta-lactamase family proteins (TDE1444 and TDE1541) and several ABC transporter proteins (TDE0758, TDE0983–TDE0987) also increased in abundance in ΔmotB.
Of the proteins that decreased in abundance in ΔmotB only, COG category T (signal transduction mechanisms) contained the highest number of proteins (Table 2). FleN (TDE2685), an MCP (TDE2783), two chemotaxis proteins (CheX and CheY) and two penicillin binding proteins (PBPs; TDE1314 and TDE1352) were among the proteins that decreased in abundance. T. denticola virulence factors including Msp (TDE0405), hemolysin (TDE1669), and the glycine reductase complex proteins GrdD (TDE0239) and GrdE2 (TDE2120) also decreased in abundance.
Coaggregation Assay of T. denticola Strains With P. gingivalis
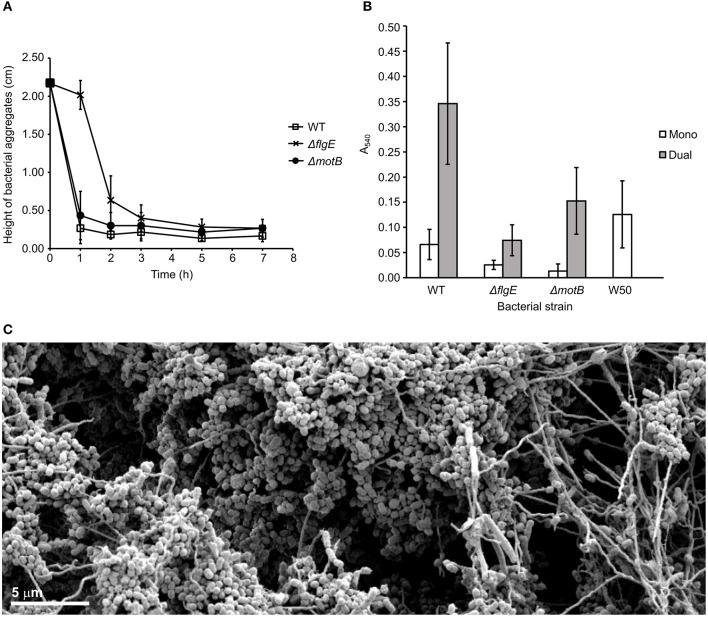
T. denticola ΔmotB coaggregated with P. gingivalis at the same rate as wild-type but ΔflgE coaggregated with P. gingivalis at a slower rate than the wild-type (Figure 4A).
Figure 4.
Coaggregation and biofilm formation of T. denticola wild-type, ΔflgE, ΔmotB with P. gingivalis W50. (A) Coaggregation of T. denticola wild-type and mutants with P. gingivalis W50. T. denticola ATCC 33520 and P. gingivalis cells at exponential growth phase were harvested, washed twice with coaggregation buffer and adjusted to A650 of 0.5 with the coaggregation buffer. Equal volumes of T. denticola and P. gingivalis cell suspensions were combined. The height of bacterial aggregates was monitored for 7 h. The data are presented as means plus standard deviations (N = 3). (B) Monospecies and dual-species static biofilms of T. denticola ATCC 33520 wild-type, ΔflgE, ΔmotB with P. gingivalis W50. T. denticola and P. gingivalis cells were grown to exponential phase, diluted to A650 of 0.15 and grown anaerobically for 5 days in a 12-well plate, either in a monospecies or dual-species culture. The resultant biofilm was stained with crystal violet and the total biomass determined spectrophotometrically. The data are presented as means and standard deviations (N = 23–27) and were analyzed using a Kruskal-Wallis with Conover-Imam test. All values were significantly different (p < 0.05) except between the following pairs: ΔmotB (mono); wild-type (mono) and ΔflgE (dual); as well as wild-type (dual). (C) SEM image of a biofilm containing T. denticola wild type ATCC 35405 (the long thin spirochaete) and P. gingivalis W50 (the grape-like coccobacillus). The image shows the motile T. denticola facilitating expansion of the biofilm microcolonies by forming bridge structures while carrying P. gingivalis.
Biofilm Assay of T. denticola Strains With P. gingivalis
A biofilm assay was used to determine the ability of T. denticola wild-type and motility mutants to form monospecies biofilms and dual-species biofilms with P. gingivalis W50. The T. denticola motility mutants ΔflgE and ΔmotB formed monospecies biofilms with significantly less biomass than those formed by the wild-type (Figure 4B). A synergy was observed between T. denticola wild-type and P. gingivalis in dual-species biofilm formation, as demonstrated by an ~2-fold higher biomass of the dual-species biofilms than the sum of the monospecies biofilms (Figure 4B). The motility of wild type T. denticola facilitated expansion of the biofilm through the formation of bridges and transport of P. gingivalis (Figure 4C; Zhu et al., 2013). T. denticola ΔflgE showed a considerably reduced ability to form dual-species biofilms with P. gingivalis compared with the wild-type, with a 5-fold reduction in biomass. Interestingly, ΔflgE dual-species biofilms were smaller than P. gingivalis monospecies biofilms (p < 0.05). ΔmotB formed biofilms with P. gingivalis that had a 2-fold lower biomass than those formed with the wild-type (Figure 4B).
Discussion
Although not generally referred to as classic virulence factors in most human pathogens, motility and chemotaxis are undoubtedly important in bacterial-host interactions and disease progression (Dashper et al., 2011). The combination of a chemotaxis system and motility enables efficient nutrient acquisition, avoidance of toxic substances and translocation to optimal colonization sites by bacteria. It is thus important in the survival and proliferation of bacteria, especially for nutritionally fastidious organisms, such as the oral treponemes. Approximately 5–6% of the genome of sequenced treponemes is dedicated to motility and chemotaxis (Seshadri et al., 2004). The motility of T. denticola is dependent on its periplasmic flagella which, unlike the exposed flagella of most motile bacteria, are located in the periplasmic space between the outer and cytoplasmic membranes of the cells (Chi et al., 1999). T. denticola ΔflgE and ΔmotB mutants lacking specific genes involved in the motility machinery were generated to investigate the importance of T. denticola periplasmic flagella and/or motility in synergistic biofilm formation with P. gingivalis.
Traditionally the roles of gene products in bacterial phenotypes and behaviors are determined by the specific inactivation or deletion of the gene under investigation. In this study we confirmed the specific nature of gene deletion by sequencing the genomes of the mutants. The genome of ΔMotB contained no predicted effects on protein structure other than the absence of an ORF encoding motB. The genome of ΔFlgE contained a deletion of flgE ORF and only one other change causing a truncation of the last 5% of hypothetical protein, HMPREF9722_RS03985. The phenotypic effects of this truncation remain unknown, nevertheless proteomics data revealed that this protein is present in ΔFlgE. However, quantitative proteomic analyses of the mutants revealed that the specific deletion of the motB gene from the T. denticola genome had wide ranging effects on the overall protein abundance profiles of the mutants.
Among the 35 proteins that changed in abundance in both mutants, 10 were related to flagella and motility. All of them were reduced in abundance in both mutants, except FliS which had increased in abundance in ΔflgE. Completion of the hook-basal body structure serves as a checkpoint for transcriptional regulation of flagellum synthesis (Hughes et al., 1993). Therefore, the deletion of motB and flgE may have negatively regulated the transcription of genes from within the fla operon where motB and flgE are located as well as those that are not located in the fla operon. These included fliS, flgL, flaA, and flaB genes. It is also possible that the regulation was mediated at the post-transcriptional level as was seen in the reduced abundance of FlaA and FlaB proteins in a Borrelia burgdorferi ΔflgE mutant (Sal et al., 2008). Moreover, the presence of MotB might also act as a checkpoint for the expression of chemotaxis genes as several chemotaxis-related proteins were differentially regulated in ΔmotB. Together, these results suggest that flagellum synthesis and the expression of chemotaxis and motility genes utilizes a finely regulated process where there is a series of transcriptional and/or post-transcriptional controls.
Both ΔmotB and ΔflgE were non-motile. Although T. denticola ATCC 33520 is commonly observed to have four periplasmic flagella (Izard et al., 2008), in this study up to five periplasmic flagella were occasionally observed (Figure 3). As expected, the ΔflgE mutant was deficient in periplasmic flagella as it lacks the flagellar hook protein, FlgE, necessary for flagella assembly (Li et al., 1996). Periplasmic flagella were observed in ΔmotB mutants, but at a lower number than the wild-type and not all cell segments of the mutants showed visible flagella (Figure 3). The reduction in periplasmic flagella number in ΔmotB is consistent with the quantitative proteomic results which showed a reduction in the abundance of the flagellar filament proteins, FlaA and FlaB, in this mutant relative to wild-type. Furthermore, FleN, a protein involved in the regulation of flagella number in Pseudomonas aeruginosa (Dasgupta et al., 2000), was also decreased in abundance in ΔmotB. A T. denticola non-motile mutant lacking FliG, a flagellar motor protein, had a markedly decreased number of flagellar filaments and the flagellar filaments were usually shorter in length than those of the wild-type (Slivienski-Gebhardt et al., 2004). In addition, there was a reduction in the FlaA and FlaB proteins in the ΔfliG mutant (Slivienski-Gebhardt et al., 2004). The similar phenotype of the ΔfliG mutant with ΔmotB suggested the importance of the flagellar motor in the complete assembly of T. denticola periplasmic flagella.
The cellular morphologies of ΔmotB and ΔflgE were different to that of the wild-type. The T. denticola periplasmic flagella contribute to the irregular twisted morphology of the bacterium, which is the predominant form adopted by the cells in planktonic exponential phase, with the other being the regular right-handed helical form (Ruby et al., 1997). T. denticola wild-type cells of both strains 35405 and 33520 with the outer membrane removed and T. denticola mutants lacking periplasmic flagella lost their irregular morphology and adopted a helical form (Ruby et al., 1997). In this study, an irregular morphology was observed in T. denticola wild-type cells. However, unlike the previous study, the ΔflgE mutant lacking periplasmic flagella adopted a rod shape instead of a helical shape (Figure 3). This result is more similar to that observed in B. burgdorferi where a ΔflaB mutant deficient in periplasmic flagella lost its flat-wave morphology and adopted a rod shape (Motaleb et al., 2000). The reason for the discrepancy in the observed morphology of ΔflgE mutant in this study with the previous study is unclear since the same strain of T. denticola harvested at the same growth phase was used and both mutants were deficient in periplasmic flagella. Differences in the growth media and the temperature of incubation between studies were evident but it is uncertain how these would affect the cell morphology. The most likely reason for these discrepancies may lie in the specific mutation of the genomes. In the previous study the T. denticola 33520 aflagellated mutant was a spontaneous occurring mutant that remained genetically uncharacterized. Furthermore, the T. denticola 35405 specific FlgE- mutant which was created by deletion of flgE from the chromosome by Erm cassette insertion did not involve reintroduction of the promoter sequence downstream of the Erm cassette in the genome as has been done in this study. In the present study we have used RT-PCR (results not shown) to show that transcription of the six operon genes (motA, motB, fliL, fliM, fliY, and fliP) downstream of deleted flgE remain unaffected by the introduction of ermAM. Furthermore, the phenotypic effect of the possible truncation of the C-terminus of hypothetical protein HMPREF9722_RS03985 in ΔFlgE in the present study remains unknown.
When compared to wild-type, ΔmotB appeared less spiral and more rod-like which could be caused by the observed reduction in the periplasmic flagella number. Alternatively, it might be a result of the change in abundance of proteins that are involved in the morphology of bacteria, as observed in the spirochete Leptospira (Slamti et al., 2011). These proteins include the penicillin binding proteins (PBPs) and rod shape-determining cytoskeleton protein MreB, which likely controls cell morphology through their interactions with the peptidoglycan layer (Divakaruni et al., 2007). The PBPs were reduced in abundance while MreB increased in abundance in the ΔmotB mutant, suggesting a switch in the mechanism of cell shape maintenance. Interestingly, the cellular localization of the MreB cytoskeleton in Bacillus subtilis requires a transmembrane ion motive force (IMF); disruption of the IMF resulted in a rapid delocalization and loss of helicity of the MreB cytoskeleton (Strahl and Hamoen, 2010). As the IMF is predicted to be disrupted in ΔmotB, it is possible that the cells increased the production of MreB in order to compensate for the delocalization of MreB. Furthermore, the altered cellular morphology could also be caused by a lack of flagellar rotation. In B. burgdorferi, flagellar rotation is required for the flat-wave morphology of the bacterium and the deletion of motB in B. burgdorferi caused part of the cells to be rod shaped (Sultan et al., 2015). Overall, these results suggest that the presence of properly assembled, functional periplasmic flagella is an important determinant of the characteristic spiral morphology of T. denticola.
The loss of motility impaired the growth of ΔmotB and ΔflgE. One of the reasons for this could be a lack of nutrient accessibility as a result of the loss of motility. In addition, the generation of the transmembrane IMF through the ion channel complex formed by MotA and MotB may be an important source of energy for T. denticola growth. The lack of MotB, especially the plug segment required for suppression of undesirable flow through the MotA/B ion channel complex (Hosking et al., 2006; Morimoto et al., 2010) may have resulted in transmembrane ion leakage that negatively affected energy and growth. There was a significant increase in the abundance of an oligopeptide/dipeptide ABC transporter made up of proteins TDE0983–TDE0987 in ΔmotB, possibly to compensate for a reduction in oligopeptides or dipeptides usually transported into the cell by IMF-driven systems. This transport system, which relies on ATP hydrolysis to drive the transport of substrates across the cell membrane, would be more inefficient than the IMF-driven system and would result in ΔmotB growing slower than the wild-type, as reflected in the growth studies.
In addition, the disruption of MotB increased the abundance of a number of proteins involved in the bacterial stress response, including desulfoferrodoxin/neelaredoxin (TDE1754), RecA (TDE0872), DNA topoisomerase I (TopA; TDE1208), and transcription termination factor Rho (TDE1503) (Brennan et al., 1987; Jovanovic et al., 2000; Italiani et al., 2002; Liu et al., 2011). Furthermore, two aminoacyl-tRNA synthetases, valyl-tRNA synthetase (TDE1364) and phenylalanyl-tRNA synthetase (TDE1927), involved in the editing of misacylated tRNAs (Ling et al., 2009) and proposed to be critical for bacterial stress responses and survival (Bullwinkle and Ibba, 2016) were also increased in abundance. The observed reduction in the abundance of the PBPs, which are essential for the synthesis of the peptidoglycan layer of the cell (Sauvage et al., 2008), suggested that ΔmotB were under peptidoglycan stress. The increased abundance of the metallo-beta-lactamase family proteins (TDE1444 and TDE1541) is likely to be a response to peptidoglycan stress as the cells may perceive the reduction in peptidoglycan synthesis as a result of PBPs inhibition and thus produce more metallo-beta-lactamases, the enzymes that catalyze the hydrolysis of beta-lactam antibiotics that inhibit PBPs (Palzkill, 2013). Overall, these results show the importance of the MotA/B proton channel complex in T. denticola.
The Na+-translocating NADH/quinone reductase E subunit (NqrE; TDE0834) increased in abundance in the ΔmotB mutant. NqrE is part of the Na+-translocating NADH/quinone oxidoreductase (Na+-NQR) respiratory complex found in prokaryotes (Barquera, 2014). Na+-NQR translocates sodium ions across the membrane, generating an electrochemical Na+ gradient which energizes important functions in the cell, including rotation of the Na+-dependent flagella for motility (Barquera, 2014). The membrane-embedded stators, Mot complexes, harness energy of either transmembrane H+ or Na+ ion gradients to power flagellar rotation. There are two distinct types of Mot stators with different ion specificities. In B. subtilis, MotA and MotB constitute the H+-coupled Mot while MotP and MotS constitute a Na+-coupled Mot (Ito et al., 2005; Terahara et al., 2008). In Vibrio, the force-generating unit of the Na+-driven flagellar motor is composed of four components: PomA, PomB, MotX, and MotY (Li et al., 2011). The use of Na+ ion gradients is often associated with elevated pH and sodium concentrations (Mulkidjanian et al., 2008).
The increase in abundance of TDE0834 (NqrE) in ΔmotB may suggest that the mutant has switched to the usage of Na+ as a coupling ion for flagellar motor rotation, instead of H+, as a result of IMF disruption. The switching to the use of Na+ instead of, or in addition to, H+ for flagellar motor rotation may be an adaptation of T. denticola to the increase in pH and salinity in an inflamed periodontal pocket during chronic periodontitis. It has been reported that the progression of periodontitis is associated with a rise in pH of the gingival sulcus (Zilm et al., 2010; Barros et al., 2016). Maintaining high levels of IMF would become increasingly difficult for T. denticola as the external concentration of H+ ions become lower. In addition, the concentration of Na+ ions in the periodontal pocket increases as a result of bleeding and tissue inflammation (Barros et al., 2016). Switching to the use of Na+ ions may thus be more advantageous for T. denticola in this relatively alkaline and sodium-rich environment. This hypothesis is supported by a study which shows that the MotA/B of Bacillus clausii, an alkaliphilic bacterium, is bifunctional with respect to ion-coupling capacity in that it couples motility to sodium at high pH (pH > 8.5) but uses protons at lower pH (pH < 8.5) (Terahara et al., 2008). B. clausii MotA/B increases its use of sodium as the pH becomes increasingly alkaline (Terahara et al., 2008) and presumably the IMF is getting smaller. The observed increase in the component of Na+-NQR in ΔmotB with the predicted disruption in IMF may thus suggest that the ion-coupling pattern of T. denticola MotA/B changes as the relative magnitudes of the transmembrane sodium and proton motive forces change.
In this study, T. denticola formed detectable monospecies biofilms. Although generally regarded as a poor biofilm former (Davey, 2006; Biyikoglu et al., 2012), P. gingivalis W50 was also able to form a biofilm (Figure 4B). Compared to the wild-type, both T. denticola motility mutants had reduced abilities to form a mono-species biofilm. Together with the autoaggregation results (Figure 2D), this indicated the importance of motility in the binding of T. denticola cells for monospecies biofilm formation under static conditions. However, it should be noted that the mutants were less spiral than the wild-type and the reduced autoaggregation and biofilm forming ability could be a result of the changed morphology of the mutants.
T. denticola and P. gingivalis W50 form dual-species biofilms synergistically. T. denticola periplasmic flagella were shown to be important for synergistic biofilm formation with P. gingivalis. In agreement with previous studies (Vesey and Kuramitsu, 2004; Yamada et al., 2005), the absence of periplasmic flagella in ΔflgE attenuated its biofilm forming ability with P. gingivalis in static biofilm assays. ΔflgE also displayed a reduced rate of coaggregation with P. gingivalis. Although the abundance of the surface proteins involved in the binding of T. denticola and P. gingivalis, including dentilisin and the major sheath protein (Hashimoto et al., 2003; Rosen et al., 2008), in ΔflgE was comparable to those of wild-type, there could be changes in other unidentified surface components that are involved in the binding of T. denticola and P. gingivalis. Interestingly, the dual-species biofilms of ΔflgE grown together with P. gingivalis were smaller than the mono-species biofilms of P. gingivalis, suggesting an inhibitory effect from the absence of periplasmic flagella to dual-species biofilm formation under static conditions. This appears to be further supported by the observation that ΔmotB contains a lower number of periplasmic flagella and reduced spirality compared with WT and produces a dual species biofilm intermediate in size compared with those of flagellated WT and the unflagellated ΔflgE. However, caution is required when drawing such conclusions about the effects of a reduced number of flagella in ΔmotB dual species biofilm since the absence of MotB also had extensive effects on proteins not directly involved in motility, including changes in expression of several major surface proteins which may also effect dual species biofilm production.
The comprehensive quantitative analysis of the proteomes of T. denticola mutants revealed the wide effects of motility gene inactivation on T. denticola proteomes; this technique may be used to help identify changes in protein expression that contribute to mutant phenotypes. T. denticola periplasmic flagella were shown to be important for synergistic biofilm formation with P. gingivalis.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Bioproject accession number PRJNA561478.
Author Contributions
HN conducted the majority of the benchwork as part of her Ph.D. under the supervision of NS, SD, and CB. SD, NS, CB, and ER devised the project. The mutant creation and confirmation were conducted by NS, CB, BH, and HN. Proteomic analyses of the mutants were carried out by PV. Y-YC carried out the electron microscopy imaging and analyses. SL and HN executed the biofilm analyses. HN, SD, CB, NS, and ER interpreted the results and wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Drs. Shuai Nie, Ching-Seng Ang, and Nicholas Williamson for the acquisition of Orbitrap LCMS/MS data and their technical support through the Mass Spectrometry and Proteomics Facility at Bio21 Institute, The University of Melbourne, Australia.
Footnotes
Funding. This research was funded by the National Health and Medical Research Council of Australia Grant ID 1083600 and the Australian Government Department of Industry, Innovation and Science Grant ID 20080108. HN was the recipient of an International Post-graduate Research Scholarship from the Australian Federal Government.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00432/full#supplementary-material
A schematic representation of the mutational approach (A) flgE mutant construction (B) motB mutant construction.
Primers used in this study.
Nucleotide polymorphisms of T. denticola 33520 mutants determined by genomic sequencing.
Proteins significantly changed in abundance in T. denticola ΔflgE mutant relative to wild-type (ratio ≥1.5 and ≤0.67, p < 0.05).
Proteins significantly changed in abundance in T. denticola ΔmotB mutant relative to wild-type (ratio ≥1.5 and ≤0.67, p < 0.05).
Proteins that were not detected in T. denticola ATCC 33520 but were detected in ΔmotB (p < 0.05).
References
- Abiko Y., Nagano K., Yoshida Y., Yoshimura F. (2014). Major membrane protein TDE2508 regulates adhesive potency in Treponema denticola. PLoS ONE 9:e89051. 10.1371/journal.pone.0089051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquera B. (2014). The sodium pumping NADH:quinone oxidoreductase (Na(+)-NQR), a unique redox-driven ion pump. J. Bioenerg. Biomembr. 46, 289–298. 10.1007/s10863-014-9565-9 [DOI] [PubMed] [Google Scholar]
- Barros S. P., Williams R., Offenbacher S., Morelli T. (2016). Gingival crevicular fluid as a source of biomarkers for periodontitis. Periodontol. (2000) 70, 53–64. 10.1111/prd.12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H. C. (1976). How spirochetes may swim. J. Theor. Biol. 56, 269–273. 10.1016/S0022-5193(76)80074-4 [DOI] [PubMed] [Google Scholar]
- Bian J., Fenno J. C., Li C. (2012). Development of a modified gentamicin resistance cassette for genetic manipulation of the oral spirochete Treponema denticola. Appl. Environ. Microbiol. 78, 2059–2062. 10.1128/AEM.07461-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J., Li C. (2011). Disruption of a type II endonuclease (TDE0911) enables Treponema denticola ATCC 35405 to accept an unmethylated shuttle vector. Appl. Environ. Microbiol. 77, 4573–4578. 10.1128/AEM.00417-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J., Liu X., Cheng Y. Q., Li C. (2013). Inactivation of cyclic Di-GMP binding protein TDE0214 affects the motility, biofilm formation, and virulence of Treponema denticola. J. Bacteriol. 195, 3897–3905. 10.1128/JB.00610-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biyikoglu B., Ricker A., Diaz P. I. (2012). Strain-specific colonization patterns and serum modulation of multi-species oral biofilm development. Anaerobe 18, 459–470. 10.1016/j.anaerobe.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Dombroski A. J., Platt T. (1987). Transcription termination factor rho is an RNA-DNA helicase. Cell 48, 945–952. 10.1016/0092-8674(87)90703-3 [DOI] [PubMed] [Google Scholar]
- Bullwinkle T. J., Ibba M. (2016). Translation quality control is critical for bacterial responses to amino acid stress. Proc. Natl. Acad. Sci. U.S.A. 113, 2252–2257. 10.1073/pnas.1525206113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S. J., Dashper S. G., Darby I. B., Adams G. G., Hoffmann B., Reynolds E. C. (2009). Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol. Immunol. 24, 469–77. 10.1111/j.1399-302X.2009.00544.x [DOI] [PubMed] [Google Scholar]
- Capone R. F., Ning Y., Pakulis N., Alhazzazi T., Fenno J. C. (2007). Characterization of Treponema denticola pyrF encoding orotidine-5′-monophosphate decarboxylase. FEMS Microbiol. Lett. 268, 261–267. 10.1111/j.1574-6968.2006.00589.x [DOI] [PubMed] [Google Scholar]
- Chan E. C., Siboo R., Keng T., Psarra N., Hurley R., Cheng S. L., et al. (1993). Treponema denticola (ex Brumpt 1925) sp. nov., nom. rev., and identification of new spirochete isolates from periodontal pockets. Int. J. Syst. Bacteriol. 43, 196–203. 10.1099/00207713-43-2-196 [DOI] [PubMed] [Google Scholar]
- Chen Y., Peng B., Yang Q., Glew M. D., Veith P. D., Cross K. J., et al. (2011). The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol. Microbiol. 79, 1380–1401. 10.1111/j.1365-2958.2010.07530.x [DOI] [PubMed] [Google Scholar]
- Chevance F. F., Hughes K. T. (2008). Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6, 455–465. 10.1038/nrmicro1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B., Chauhan S., Kuramitsu H. (1999). Development of a system for expressing heterologous genes in the oral spirochete Treponema denticola and its use in expression of the Treponema pallidum flaA gene. Infect. Immun. 67, 3653–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B., Limberger R. J., Kuramitsu H. K. (2002). Complementation of a Treponema denticola flgE mutant with a novel coumermycin A1-resistant T. denticola shuttle vector system. Infect. Immun. 70, 2233–2237. 10.1128/IAI.70.4.2233-2237.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover W. J., Iman R. L. (1979). On Multiple-Comparisons Procedures. Technical Report LA-7677-MS, Los Alamos Scientific Laboratory.
- Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., Mann M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta N., Arora S. K., Ramphal R. (2000). fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182, 357–364. 10.1128/JB.182.2.357-364.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashper S., O'brien-Simpson N., Liu S. W., Paolini R., Mitchell H., Walsh K., et al. (2014). Oxantel disrupts polymicrobial biofilm development of periodontal pathogens. Antimicrob. Agents Chemother. 58, 378–385. 10.1128/AAC.01375-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashper S. G., Seers C. A., Tan K. H., Reynolds E. C. (2011). Virulence factors of the oral spirochete Treponema denticola. J. Dent. Res. 90, 691–703. 10.1177/0022034510385242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. E. (2006). Techniques for the growth of Porphyromonas gingivalis biofilms. Periodontol. (2000) 42, 27–35. 10.1111/j.1600-0757.2006.00183.x [DOI] [PubMed] [Google Scholar]
- Divakaruni A. V., Baida C., White C. L., Gober J. W. (2007). The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 66, 174–188. 10.1111/j.1365-2958.2007.05910.x [DOI] [PubMed] [Google Scholar]
- Fletcher H. M., Schenkein H. A., Morgan R. M., Bailey K. A., Berry C. R., Macrina F. L. (1995). Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63, 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glew M. D., Veith P. D., Chen D., Gorasia D. G., Peng B., Reynolds E. C. (2017). PorV is an outer membrane shuttle protein for the Type IX secretion system. Sci. Rep. 7:8790. 10.1038/s41598-017-09412-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godovikova V., Goetting-Minesky M. P., Shin J. M., Kapila Y. L., Rickard A. H., Fenno J. C. (2015). A modified shuttle plasmid facilitates expression of a flavin mononucleotide-based fluorescent protein in Treponema denticola ATCC 35405. Appl. Environ. Microbiol. 81, 6496–6504. 10.1128/AEM.01541-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Lamont R. J. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27, 409–419. 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Ogawa S., Asai Y., Takai Y., Ogawa T. (2003). Binding of Porphyromonas gingivalis fimbriae to Treponema denticola dentilisin. FEMS Microbiol. Lett. 226, 267–271. 10.1016/S0378-1097(03)00615-3 [DOI] [PubMed] [Google Scholar]
- Holt S. C., Ebersole J. L. (2005). Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex', a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. (2000) 38, 72–122. 10.1111/j.1600-0757.2005.00113.x [DOI] [PubMed] [Google Scholar]
- Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. (1989). Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68. 10.1016/0378-1119(89)90359-4 [DOI] [PubMed] [Google Scholar]
- Hosking E. R., Vogt C., Bakker E. P., Manson M. D. (2006). The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364, 921–937. 10.1016/j.jmb.2006.09.035 [DOI] [PubMed] [Google Scholar]
- Houry A., Briandet R., Aymerich S., Gohar M. (2010). Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 156, 1009–1018. 10.1099/mic.0.034827-0 [DOI] [PubMed] [Google Scholar]
- Houry A., Gohar M., Deschamps J., Tischenko E., Aymerich S., Gruss A., et al. (2012). Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 109, 13088–13093. 10.1073/pnas.1200791109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Gillen K. L., Semon M. J., Karlinsey J. E. (1993). Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262, 1277–1280. 10.1126/science.8235660 [DOI] [PubMed] [Google Scholar]
- Ishihara K., Kuramitsu H. K., Miura T., Okuda K. (1998). Dentilisin activity affects the organization of the outer sheath of Treponema denticola. J. Bacteriol. 180, 3837–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiani V. C., Zuleta L. F., Marques M. V. (2002). The transcription termination factor Rho is required for oxidative stress survival in Caulobacter crescentus. Mol. Microbiol. 44, 181–194. 10.1046/j.1365-2958.2002.02865.x [DOI] [PubMed] [Google Scholar]
- Ito M., Terahara N., Fujinami S., Krulwich T. A. (2005). Properties of motility in Bacillus subtilis powered by the H+- coupled MotAB flagellar stator, Na+-coupled MotPS or hybrid stators MotAS or MotPB. J. Mol. Biol. 352, 396–408. 10.1016/j.jmb.2005.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard J., Hsieh C., Limberger R. J., Mannella C. A., Marko M. (2008). Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J. Struct. Biol. 163, 10–17. 10.1016/j.jsb.2008.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic T., Ascenso C., Hazlett K. R., Sikkink R., Krebs C., Litwiller R., et al. (2000). Neelaredoxin, an iron-binding protein from the syphilis spirochete, Treponema pallidum, is a superoxide reductase. J. Biol. Chem. 275, 28439–28448. 10.1074/jbc.M003314200 [DOI] [PubMed] [Google Scholar]
- Klitorinos A., Noble P., Siboo R., Chan E. C. (1993). Viscosity-dependent locomotion of oral spirochetes. Oral Microbiol. Immunol. 8, 242–244. 10.1111/j.1399-302X.1993.tb00567.x [DOI] [PubMed] [Google Scholar]
- Kojima S., Blair D. F. (2001). Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40, 13041–13050. 10.1021/bi011263o [DOI] [PubMed] [Google Scholar]
- Kojima S., Imada K., Sakuma M., Sudo Y., Kojima C., Minamino T., et al. (2009). Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 73, 710–718. 10.1111/j.1365-2958.2009.06802.x [DOI] [PubMed] [Google Scholar]
- Kruskal W. H., Wallis W. A. (1952). Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47, 583–621. 10.1080/01621459.1952.10483441 [DOI] [Google Scholar]
- Kuramitsu H. K., Chi B., Ikegami A. (2005). Genetic manipulation of Treponema denticola. Curr. Protoc. Microbiol. Chapter 12:Unit 12B.12. 10.1002/9780471729259.mc12b02s00 [DOI] [PubMed] [Google Scholar]
- Lamont R. J., Jenkinson H. F. (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62, 1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Kuramitsu H. K. (1996). Development of a gene transfer system in Treponema denticola by electroporation. Oral Microbiol. Immunol. 11, 161–165. 10.1111/j.1399-302X.1996.tb00352.x [DOI] [PubMed] [Google Scholar]
- Li H., Ruby J., Charon N., Kuramitsu H. (1996). Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J. Bacteriol. 178, 3664–3667. 10.1128/jb.178.12.3664-3667.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Kojima S., Homma M. (2011). Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells 16, 985–999. 10.1111/j.1365-2443.2011.01545.x [DOI] [PubMed] [Google Scholar]
- Li Y., Ruby J., Wu H. (2015). Kanamycin resistance cassette for genetic manipulation of Treponema denticola. Appl. Environ. Microbiol. 81, 4329–4338. 10.1128/AEM.00478-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limberger R. J. (2004). The periplasmic flagellum of spirochetes. J. Mol. Microbiol. Biotechnol. 7, 30–40. 10.1159/000077867 [DOI] [PubMed] [Google Scholar]
- Limberger R. J., Slivienski L. L., Izard J., Samsonoff W. A. (1999). Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J. Bacteriol. 181, 3743–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J., Reynolds N., Ibba M. (2009). Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63, 61–78. 10.1146/annurev.micro.091208.073210 [DOI] [PubMed] [Google Scholar]
- Liu I. F., Sutherland J. H., Cheng B., Tse-Dinh Y.-C. (2011). Topoisomerase I function during Escherichia coli response to antibiotics and stress enhances cell killing from stabilization of its cleavage complex. J. Antimicrob. Chemother. 66, 1518–1524. 10.1093/jac/dkr150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux R., Sim J. H., Tsai J. P., Shi W. (2002). Construction and characterization of a cheA mutant of Treponema denticola. J. Bacteriol. 184, 3130–3134. 10.1128/JB.184.11.3130-3134.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell H. L., Dashper S. G., Catmull D. V., Paolini R. A., Cleal S. M., Slakeski N., et al. (2010). Treponema denticola biofilm-induced expression of a bacteriophage, toxin-antitoxin systems and transposases. Microbiology 156, 774–788. 10.1099/mic.0.033654-0 [DOI] [PubMed] [Google Scholar]
- Morimoto Y. V., Che Y. S., Minamino T., Namba K. (2010). Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS Lett. 584, 1268–1272. 10.1016/j.febslet.2010.02.051 [DOI] [PubMed] [Google Scholar]
- Morimoto Y. V., Minamino T. (2014). Structure and function of the bi-directional bacterial flagellar motor. Biomolecules 4, 217–234. 10.3390/biom4010217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortz E., Krogh T. N., Vorum H., Gorg A. (2001). Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 1, 1359–1363. [DOI] [PubMed] [Google Scholar]
- Motaleb M. A., Corum L., Bono J. L., Elias A. F., Rosa P., Samuels D. S., et al. (2000). Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. U.S.A. 97, 10899–10904. 10.1073/pnas.200221797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian A. Y., Dibrov P., Galperin M. Y. (2008). The past and present of the sodium energetics: may the sodium-motive force be with you. Biochim. Biophys. Acta 1777, 985–992. 10.1016/j.bbabio.2008.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palzkill T. (2013). Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 1277, 91–104. 10.1111/j.1749-6632.2012.06796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos J. S., Agarwala R. (2007). COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. 10.1093/bioinformatics/btm076 [DOI] [PubMed] [Google Scholar]
- Rosen G., Genzler T., Sela M. N. (2008). Coaggregation of Treponema denticola with Porphyromonas gingivalis and Fusobacterium nucleatum is mediated by the major outer sheath protein of Treponema denticola. FEMS Microbiol. Lett. 289, 59–66. 10.1111/j.1574-6968.2008.01373.x [DOI] [PubMed] [Google Scholar]
- Ruby J. D., Li H., Kuramitsu H., Norris S. J., Goldstein S. F., Buttle K. F., et al. (1997). Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J. Bacteriol. 179, 1628–1635. 10.1128/jb.179.5.1628-1635.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sal M. S., Li C., Motalab M. A., Shibata S., Aizawa S., Charon N. W. (2008). Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J. Bacteriol. 190, 1912–1921. 10.1128/JB.01421-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador S. L., Syed S. A., Loesche W. J. (1987). Comparison of three dispersion procedures for quantitative recovery of cultivable species of subgingival spirochetes. J. Clin. Microbiol. 25, 2230–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258. 10.1111/j.1574-6976.2008.00105.x [DOI] [PubMed] [Google Scholar]
- Seshadri R., Myers G. S., Tettelin H., Eisen J. A., Heidelberg J. F., Dodson R. J., et al. (2004). Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. U.S.A. 101, 5646–5651. 10.1073/pnas.0307639101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamti L., De Pedro M. A., Guichet E., Picardeau M. (2011). Deciphering morphological determinants of the helix-shaped leptospira. J. Bacteriol. 193, 6266–6275. 10.1128/JB.05695-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slivienski-Gebhardt L. L., Izard J., Samsonoff W. A., Limberger R. J. (2004). Development of a novel chloramphenicol resistance expression plasmid used for genetic complementation of a fliG deletion mutant in Treponema denticola. Infect. Immun. 72, 5493–5497. 10.1128/IAI.72.9.5493-5497.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky S. S., Haffajee A. D., Cugini M. A., Smith C., Kent R. L., Jr. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144. 10.1111/j.1600-051X.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- Strahl H., Hamoen L. W. (2010). Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. U.S.A. 107, 12281–12286. 10.1073/pnas.1005485107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan S. Z., Sekar P., Zhao X., Manne A., Liu J., Wooten R. M., et al. (2015). Motor rotation is essential for the formation of the periplasmic flagellar ribbon, cellular morphology, and Borrelia burgdorferi persistence within Ixodes scapularis tick and murine hosts. Infect. Immun. 83, 1765–1777. 10.1128/IAI.03097-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., Koonin E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinform. 4:41. 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Koonin E. V., Lipman D. J. (1997). A genomic perspective on protein families. Science 278, 631–637. 10.1126/science.278.5338.631 [DOI] [PubMed] [Google Scholar]
- Terahara N., Krulwich T. A., Ito M. (2008). Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on motors Proc. Natl. Acad. Sci. U.S.A. 105, 14359–14364. 10.1073/pnas.0802106105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith P. D., Dashper S. G., O'brien-Simpson N. M., Paolini R. A., Orth R., Walsh K. A., et al. (2009). Major proteins and antigens of Treponema denticola. Biochim. Biophys. Acta 1794, 1421–1432. 10.1016/j.bbapap.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Vesey P. M., Kuramitsu H. K. (2004). Genetic analysis of Treponema denticola ATCC 35405 biofilm formation. Microbiology 150, 2401–2407. 10.1099/mic.0.26816-0 [DOI] [PubMed] [Google Scholar]
- Wardle H. M. (1997). The challenge of growing oral spirochaetes. J. Med. Microbiol. 46, 104–116. 10.1099/00222615-46-2-104 [DOI] [PubMed] [Google Scholar]
- Wiebe C. B., Putnins E. E. (2000). The periodontal disease classification system of the American Academy of Periodontology–an update. J. Can. Dent. Assoc. 66, 594–597. [PubMed] [Google Scholar]
- Yamada M., Ikegami A., Kuramitsu H. K. (2005). Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol. Lett. 250, 271–277. 10.1016/j.femsle.2005.07.019 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Dashper S. G., Chen Y. Y., Crawford S., Slakeski N., Reynolds E. C. (2013). Porphyromonas gingivalis and Treponema denticola synergistic polymicrobial biofilm development. PLoS ONE 8:e71727. 10.1371/journal.pone.0071727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilm P. S., Mira A., Bagley C. J., Rogers A. H. (2010). Effect of alkaline growth pH on the expression of cell envelope proteins in Fusobacterium nucleatum. Microbiology 156, 1783–1794. 10.1099/mic.0.035881-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A schematic representation of the mutational approach (A) flgE mutant construction (B) motB mutant construction.
Primers used in this study.
Nucleotide polymorphisms of T. denticola 33520 mutants determined by genomic sequencing.
Proteins significantly changed in abundance in T. denticola ΔflgE mutant relative to wild-type (ratio ≥1.5 and ≤0.67, p < 0.05).
Proteins significantly changed in abundance in T. denticola ΔmotB mutant relative to wild-type (ratio ≥1.5 and ≤0.67, p < 0.05).
Proteins that were not detected in T. denticola ATCC 33520 but were detected in ΔmotB (p < 0.05).
Data Availability Statement
The datasets generated for this study can be found in the NCBI Bioproject accession number PRJNA561478.