Abstract
A new study reveals that a novel serotonin sensor in the spore-forming gut bacteria, Turicibacter sanguinis, might be important for host lipid and steroid metabolism. These findings support the emerging concept that bidirectional signalling pathways can influence bacterial community structure and exert effects on host physiology.
The gut microbiota is located at the interface between the external world and the host environment. The composition of this complex community contributes to homeostasis and overall health, whereas alterations in the microbial ecosystem are increasingly associated with metabolic and autoimmune diseases, inflammation, and neurological disorders. Knowledge of the factors that regulate bacterial colonization in the gut has advanced in large part due to studies performed in the genus Bacteroides1, yet our understanding of the molecular mechanisms underlying the relationship between the host intestine and distinct microorganisms remains largely unknown. Now, a new study published in Nature Microbiology by Fung et al.2 presents findings in support of a role for gut-derived serotonin in mediating host–microorganism communication within the intestine.
Serotonin is a neurotransmitter involved in a wide range of functions in the central nervous system and periphery, including mood, social behaviour, sleep, cognition and gastrointestinal motility. The majority of serotonin in the body is produced by enterochromaffin cells present in the epithelial lining of the gastrointestinal tract. Upon release, serotonin diffuses from both the basal and apical surfaces of enterochromaffin cells, where it is well positioned to interact with the host mucosal surface and gut microorganisms. Previous studies have shown that gut bacteria promote enterochromaffin cell serotonin production via microbial metabolites such as short-chain fatty acids3,4. In addition, selective colonization of spore-forming bacteria in the intestine of germ-free mice alters gut motility and haemostasis, presumably through enhanced serotonin bioavailability4. These studies support a direct role for bacterial metabolism on mucosal serotonin levels and subsequent actions on host physiological responses.
To address the question of whether cross-talk between mucosal serotonin and the gut microbiome is bidirectional in nature and can lead to altered functional responses in bacteria, Fung and colleagues enhanced serotonin levels in mice2. Through oral supplementation of serotonin or genetic modifications to limit serotonin reuptake, they found that increased faecal serotonin levels were associated with alterations in the structure of the gut microbial community, specifically an increase in the relative abundance of spore-forming bacteria2. Bioinformatic approaches revealed that spore-forming bacteria of the genus Turicibacter express the protein CUW_0748, which has sequence and predicted homology to mammalian serotonin transporter (SERT), a membrane transporter responsible for reuptake and inactivation of serotonin in many organs, including the gut. Culture experiments showed that the bacterial species Turicibacter sanguinis was capable of serotonin uptake and, further, that this effect was inhibited by the selective serotonin reuptake inhibitor (SSRI) fluoxetine2. Heterologous expression of CUW_0748 in a host bacterium that does not natively express this protein revealed low levels of serotonin uptake, suggesting that additional machinery for serotonin import might exist in T. sanguinis or, alternatively, that the protein was not correctly expressed. Transcriptome profiling of T. sanguinis in response to serotonin alone or combined exposure with fluoxetine yielded robust changes in gene expression, further supporting a role for serotonin-mediated effects on bacterial responses2.
To determine whether bacterial function was similarly altered, the authors next examined colonization of T. sanguinis in the intestine. Interestingly, colonization of T. sanguinis was decreased in the small intestine and colon in response to treatment with serotonin and fluoxetine as compared to vehicle or serotonin alone, suggesting an inhibitory effect of fluoxetine on bacterial colonization in the presence of serotonin2. To further investigate these effects on host physiology, transcriptome profiling of the intestines of mice colonized with T. sanguinis was performed and revealed differential expression of genes involved in steroid and lipid metabolism2. These findings corresponded with a reduction in monoenoic triglycerides and the observation that fat cells appeared smaller with multiple lipid droplets, supporting the overarching concept that gut serotonin-sensing bacteria such as Turicibacter might contribute to host metabolic and homeostatic mechanisms (Fig. 1).
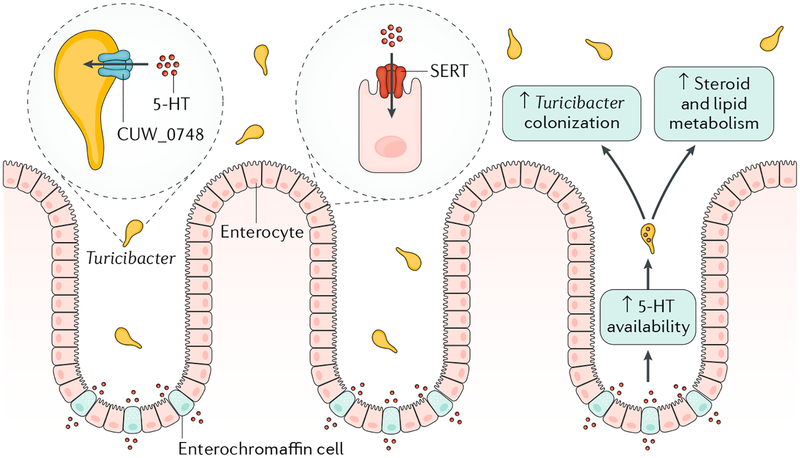
Fig. 1 |. Turicibacter alters host physiology via a novel serotonin sensor.
Serotonin (5-HT) released from host enterochromaffin cells interacts with Turicibacter in the lumen via a novel serotonin sensor, CUW_0748, which has homology to the serotonin transporter (SERT) expressed on host enterocytes. Under conditions of increased 5-HT availability in the mouse intestine, enhanced 5-HT detection by Turicibacter leads to increased colonization, alterations in host bacterial community structure and subsequent changes in steroid and lipid metabolism.
Collectively, these data could have important clinical implications for antidepressant use. A human twin study found a correlation between exposure to the SSRI class of anti-depressants and decreased abundance of faecal Turicibacter5. As altered abundance of Turicibacter has been shown in mouse models of depression and obesity6, it will be interesting to examine whether Turicibacter plays a part in host anti-depressive effects of SSRIs and/or their adverse effects (for example, weight gain through altered lipid homeostasis, metabolic syndrome and/or altered gastrointestinal motility). Given previous findings showing that colonization of spore-forming bacteria enhances whole-gut transit and colonic motility4, it remains possible that levels of Turicibacter can influence gastrointestinal motor patterns. Interestingly, decreased Turicibacter abundance has been shown in irritable bowel syndrome7, warranting future investigations to explore the contribution of individual species of Turicibacter to symptoms of disrupted motility commonly observed in disorders of altered mucosal serotonin bioavailability and/or transport.
This work also suggests that further investigation of host–microorganism interactions involving other species of spore-forming bacteria and gut-derived serotonin is required. The bacterial class Clostridia showed robust responses to exogenous serotonin delivery or genetic modifications of serotonin reuptake2, yet it is not known whether these effects are mediated by CUW_0748 or another serotonin-sensing molecule. Future research will yield alternative mechanisms responsible for bacterial responses to host serotonin and their subsequent effects on host physiology.
Along with Bacteroidetes, bacteria of the phylum Firmicutes comprise the majority of the intestinal microbiome. Of Firmicutes, the genus Turicibacter has been described as highly heritable in mice as well as humans, drawing from analyses performed on the faecal microbiome in monozygotic twins8. Although relatively little is known on the role of specific species of Turicibacter, increasing associations with inflammatory conditions, cancer and altered metabolism strengthen their potential contributions to the heritability of host health and disease8. Along these lines, an investigation in 2019 by Kemis et al. used quantitative trait locus analyses in mice to map overlapping gene loci for Turicibacter, plasma cholic acid and an ileal bile acid transporter9. They found that increased bile acid availability in the small intestine affected microbial metabolism of bile acids by Turicibacter, in turn reducing circulating levels of cholic acid. Importantly, these data illustrate the utility of systems genetic approaches to further uncover bidirectional signalling molecules mediating host–microorganism interactions with relevance to metabolic disease.
In conclusion, the present study by Fung and colleagues identifies a novel host–microorganism interaction mediated by intestinal bacteria evolved to respond to gut-derived serotonin. It also paves the way for future studies focusing on how specific bacteria of the gut microbial community sense and respond to gut-derived neurotransmitters and their subsequent effects on host physiology. If such studies are able to be reproduced in humans, these mechanisms have the potential to influence novel therapeutic interventions involving Turicibacter and/or related microbial approaches10.
Footnotes
Competing interests
The authors declare no competing interests
References
- 1.Wexler AG & Goodman AL An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol 2, 17026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fung TC et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat. Microbiol 10.1038/s41564-019-0540-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reigstad CS et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 29, 1395–1403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yano JM et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson MA et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat. Commun 9, 2655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung MJ et al. Chronic repression of mTOR complex 2 induces changes in the gut microbiota of diet-induced obese mice. Sci. Rep 6, 30887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang X et al. Fecal microbiota alterations associated with diarrhea-predominant irritable bowel syndrome. Front. Microbiol 9, 1600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodrich JK et al. Cross-species comparisons of host genetic associations with the microbiome. Science 352, 532–535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kemis JH et al. Genetic determinants of gut microbiota composition and bile acid profiles in mice. PLOS Genet. 15, e1008073 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margolis KG & Gershon MD Enteric Neuronal Regulation of Intestinal Inflammation. Trends Neurosci. 39, 614–624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]